Abstract
Genomic data have the potential to revolutionize the delineation of conservation units (CUs) by allowing the detection of adaptive genetic variation, which is otherwise difficult for rare, endangered species. In contrast to previous recommendations, we propose that the use of neutral versus adaptive markers should not be viewed as alternatives. Rather, neutral and adaptive markers provide different types of information that should be combined to make optimal management decisions. Genetic patterns at neutral markers reflect the interaction of gene flow and genetic drift that affects genome-wide variation within and among populations. This population genetic structure is what natural selection operates on to cause adaptive divergence. Here, we provide a new framework to integrate data on neutral and adaptive markers to protect biodiversity.
Conservation units in the genomics age
New tools for an old problem: delineating conservation units using genomic data
The rapid increase in the availability of genomic data (see Glossary) is quickly transforming how long-standing questions are addressed and answered in evolution [1–4], ecology [5], and now conservation [6–8]. Genomics has the potential to revolutionize understanding of adaptive differentiation and the delineation of CUs within species [7,9]. In particular, next-generation sequencing makes it easier to integrate information from neutral and adaptive loci to characterize CUs and adaptive differentiation within a population genetics framework. For the past two decades, microsatellite loci have been the most commonly used marker type for delineating CUs, but microsatellite analysis requires a relatively expensive and laborious marker development stage and only yields a few variable loci (typically 10–20). By contrast, next-generation sequencing makes it possible to simultaneously discover and genotype thousands of single nucleotide polymorphisms (SNPs). This, combined with the much lower per-locus cost of next-generation sequencing compared with microsatellite analysis, is quickly making next-generation sequencing the method of choice in population genetics.
What are CUs and why are they important?
Broadly speaking, CUs are population units identified within species that are used to help guide management and conservation efforts [9,10]. Identifying CUs is an essential first step in conservation so that managers and policy makers know the boundaries of the population units that they are trying to conserve. It is not possible to assess the status of a population or develop a management strategy to increase population growth rates without first knowing where a population begins and ends. In addition, some types of CU receive legal protection, as described below.
The two most commonly discussed conservation units are evolutionarily significant units (ESUs) and management units (MUs). Although there are many definitions of ESUs (Box 1), an ESU can generally be defined as a population or group of populations that warrant separate management or priority for conservation because of high genetic and ecological distinctiveness [9,11–14]. Identification and conservation of ESUs has been an important focus of conservation because maintenance of different ESUs will maximize evolutionary potential in the face of environmental change. Moreover, major intraspecific units, such as ESUs, are granted legal protection in many countries, including the USA (under the Endangered Species Act), Canada (Species at Risk Act), and Australia (Endangered Species Protection Act).
Box 1. Proposed definitions of ESUs.
Ryder (1986): populations that represent significant adaptive variation based on concordance between sets of data derived by different techniques [11].
Waples (1991): populations that are reproductively separate from other populations (e.g. as inferred from molecular markers) and that have distinct or different adaptations and that represent an important evolutionary legacy of a species [13].
Dizon et al. (1992): populations that are distinctive based on morphology, geographic distribution, population parameters, and genetic data [61].
Moritz (1994): populations that are reciprocally monophyletic for mtDNA and that show statistically significant divergence of allele frequencies at nuclear loci [12].
Avise (1994): sets of populations derived from consistently congruent gene phylogenies [62].
Vogler and DeSalle (1994): groups that are diagnosed by characters that cluster individuals or populations to the exclusion of other such clusters [63].
US Fish and Wildlife Service (USFWS) and National Marine Fisheries Service (NMFS) (1996): [US policy for recognition of discrete population segments (DPSs)]: (i) discreteness of the population segment in relation to the remainder of the species to which it belongs; and (ii) the significance of the population segment to the species to which it belongs [64]. This DPS policy is a further clarification of Waples’ [13] Pacific salmon ESU policy that applies to all species under the US Endangered Species Act (ESA).
Crandall et al. (2000): populations that lack: (i) ‘ecological exchangeability’ (i.e. they have different adaptations or selection pressures [e.g. life histories, morphology, quantitative trait locus (QTL) variation, habitat, predators, etc.] and different ecological roles within a community); and (ii) ‘genetic exchangeability’ (e.g. they have had no recent gene flow, and show concordance between phylogenetic and geographic discontinuities) [14].
Fraser and Bernatchez (2001): a lineage demonstrating highly restricted gene flow from other such lineages within the higher organizational level (lineage) of the species [10].
At a smaller scale, MUs are populations that are demographically independent [12]. That is, their population dynamics (growth rate) depend on local birth and death rates rather than on immigration. The identification of these units, similar to ‘stocks’ recognized in fisheries biology, is useful for short-term management goals, such as delineating hunting or fishing areas, setting local harvest quotas, and monitoring habitat and population status [15]. As such, MUs are typically smaller than ESUs and there may be many MUs within an ESU. Maintaining multiple MUs is crucial for ensuring the long-term persistence of species [16].
Why is it important to conserve adaptive differentiation among CUs?
In addition to delineating CUs, a long-standing but elusive goal has been to detect and conserve adaptive differences among CUs. There may be important adaptive differences among ESUs or MUs that go unaccounted for when making management decisions [17,18]. Recognizing adaptive differences among CUs is particularly important in two respects. First, understanding patterns of adaptive differentiation is crucial when prioritizing which populations to conserve and focus management efforts on [19]. For example, if resources were only available to conserve two populations, then all other things being equal, the two most adaptively divergent populations might warrant the highest conservation priority to maintain adaptive variation within the species. Second, understanding adaptive differentiation is also of the utmost importance when deciding which populations to use as sources for translocation, supplementation, and assisted migration efforts [20]. Supplementing a declining population with individuals from a source population adapted to a very different environment can lead to outbreeding depression [21]. Thus, knowledge of patterns of adaptive differentiation is essential for making wise conservation choices.
Aims of paper
A consideration of how to use genomic data to delineate CUs is extremely timely. There is an increasing number of threatened and exploited species for which CUs need to be defined at the same time that such genomic data are becoming cheaper and easier to obtain [22–24]. We have three goals in this paper. Our first is to outline several considerations when using genomic data to delineate CUs. Our second is to provide a new framework for using geno-mic data to delineate CUs and characterize adaptive differentiation among them and to discuss how to use this information to inform management decisions. Our last goal is to highlight important remaining questions in this field and suggest future research to address them. We do not discuss which sequencing methods and platforms are the most appropriate for collecting and analyzing genomic data, as this topic has been covered extensively elsewhere [7,22,24].
Important considerations when using genomic data to define CUs
When should genomics be used to define CUs?
Before using genomic data to define CUs, the first consideration is whether a population genomic approach should be used as opposed to a more standard approach such as a population genetic analysis using microsatellite loci. We argue that genomic data will usually be better than micro-satellite data for delineating CUs, as genomic data allow quantification of adaptive variation. Microsatellite data can be used to define ESUs and MUs, but are generally inadequate for characterizing adaptive patterns. Thus, genomic data should be used in cases where significant adaptive differentiation is expected among ESUs or MUs, because it is in these cases that genomics is most powerful.
Species that are most likely to have significant adaptive differences among CUs are those with: (i) significant environmental gradients across their ranges or stark environmental variation among habitat patches that could result in strong divergent selection [25,26]; (ii) large effective population sizes (Ne) in which selection overpowers genetic drift [27]; and (iii) low migration rates (m), such that gene flow does not constrain adaptive divergence [28–30]. Conversely, species that are least likely to have significant adaptive differences among CUs are those with: (i) little or no environmental variation across their ranges; (ii) small Ne (e.g. highly endangered species with only a few individuals remaining or species with historically small Ne; [31]); and (iii) high m. However, precaution should be exercised when judging that there is little environmental variation across the range of a species, as there may be ‘cryptic’ environmental variation that is not obvious at first, such as variation in disease prevalence or soil type [32,33]. Moreover, even in the absence of extensive local adaptation, genomic data will provide greater resolution than will microsatellites for delineating ESUs and MUs because of a huge increase in the number of loci. Thus, even in these cases, a genomic approach is arguably superior for delineating CUs.
Which loci should be used for delineating different types of CU?
Once it is decided to use a genomic approach to delineate CUs and genomic data have been collected, the next consideration is which classes of loci should be used for delineating different types of CU. Options include neutral loci, presumably adaptive loci [e.g. FST or linkage disequilibrium (LD) outliers [26,34,35]], specific genes of known function, or all loci. Importantly, in contrast to some previous authors (e.g. [36]), we believe that neutral and adaptive loci should both be used in a hierarchical approach to define CUs and characterize adaptive differences among CUs, rather than be considered as alternatives. In addition, when making a decision about which loci to use, it is important to consider which evolutionary processes affect variation at these different classes of marker. Genetic variation at neutral loci is shaped by mutation, recombination, genetic drift, and gene flow [29]. Variation at adaptive loci, by definition, is also influenced by selection. Thus, neutral versus adaptive loci may group populations differently, depending on spatial patterns of drift, gene flow, and selection (Box 2).
Box 2. Grouping populations using all, outlier, or non-outlier SNPs.
Grouping populations using high-FST outlier loci (presumably under divergent selection) might give significantly different results than groupings using non-outlier loci (presumably neutral), particularly when patterns of gene flow do not match patterns of adaptive divergence. For example, if divergent selection is strong despite high gene flow, high FST values are expected at loci under selection, but low FST at neutral loci. Thus, neutral loci may be a poor proxy for patterns of adaptive differentiation [17,65].
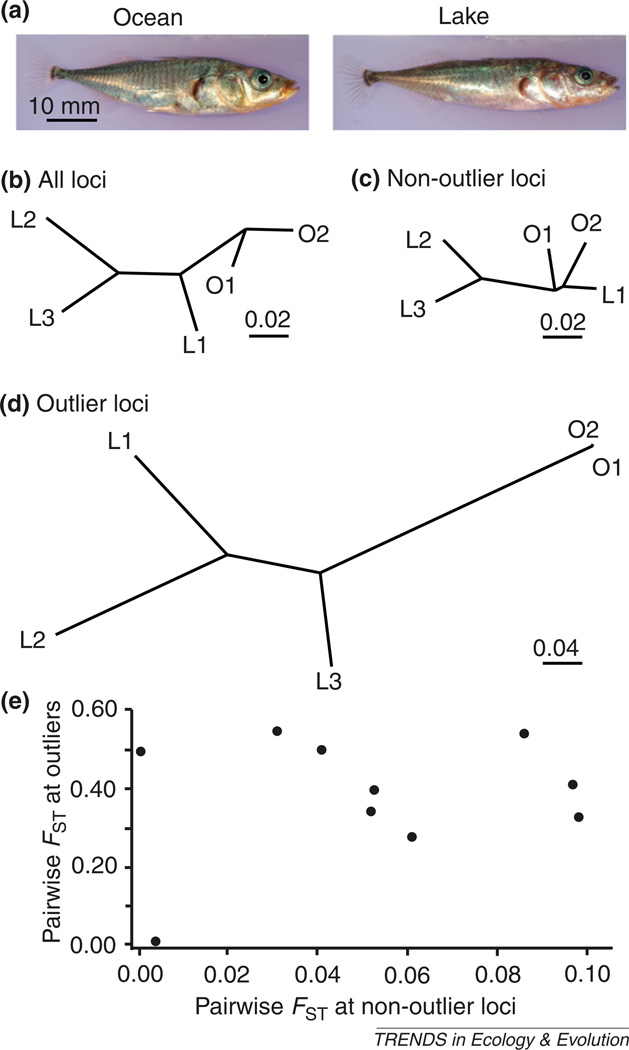
To illustrate this point, we tested whether population relationships differ based on all loci, outliers, or non-outliers using a threespine stickleback (Gasterosteus aculeatus) data set consisting of five populations (three freshwater and two ocean; Figure Ia), 20 fish per population, and 12 648 SNP loci (genotyped in at least ten individuals in each population [26]). Threespine sticklebacks have colonized and adapted to freshwater lakes multiple times [66]. Thus, we expected divergent selection at loci involved in adaptation to freshwater, making it an excellent case study for testing the effects of using outliers versus non-outliers on population groupings.
Our analysis involved three main steps. First, we tested for loci that were global FST outliers (top 5% of significance values) using goodness-of-fit G test statistics corrected for multiple comparisons [26,67,68]. We used G test statistics rather than a model-based approach such as BAYESCAN [51] because this latter method assumes an island model, which does not hold here. Second, we calculated pairwise FST [26] among all populations using all loci, just outliers, or just non-outliers. Lastly, we constructed neighbor-joining dendrograms to visualize population relationships based on these pairwise FST values [69].
The topology and branch lengths of the population dendrograms based on all loci (Figure Ib), non-outlier loci (Figure Ic), or outlier loci (Figure Id) differed significantly from each other. Moreover, Mantel tests indicate that there was no relationship between FST at outliers and FST at non-outliers (R2 = 0.04, P = 0.58; Figure Ie), indicating presumably neutral loci (non-outliers) do not predict adaptive differentiation.
This example shows that outlier loci can provide information about adaptive differentiation that is not apparent from neutral loci, but which is important for making conservation decisions. For example, if sticklebacks in O1 were declining and managers wanted to supplement this population, the non-outlier dendrogram suggests that L1 is the most similar and appropriate source. However, the outlier dendrogram reveals that O2 is most similar to O1 at presumably adaptive loci and, therefore, a better source.
Taking into consideration these differences in the evolutionary processes affecting neutral and adaptive loci is important for matching markers to the type of CU being delineated. For example, because MUs are demographically independent units with restricted gene flow, they should be delineated using neutral markers that are shaped by gene flow and Ne, not by selection. Finally, we argue that it is ill advised to characterize patterns of adaptation among CUs using single or a few loci of known function (i.e. candidate genes) because this only provides information about one or a few previously identified traits. Instead, a whole-genome approach can identify loci that are associated with the multitude of traits and loci that contribute to adaptation.
Which analyses should be used to identify CUs?
Another fundamental consideration is which analyses to use to delineate groups. Several options are available, including population dendrograms, such as unweighted pair group method with arithmetic averages (UPGMA [37]) and neighbor-joining (NJ [37]); multivariate statistical approaches such as principle component analysis (PCA [9]); Bayesian clustering algorithms, such as STRUCTURE [38] and GENELAND [39]; and newly developed landscape genomic approaches that test for correlations between genomic variation at neutral or adaptive loci and landscape structure [18,40–44]. In choosing an analysis, it is important to assess whether the class of marker being used (e.g. neutral or adaptive loci) meets the assumptions of the given analysis. For example, it may be inappropriate to use STRUCTURE to group populations using adaptive loci because this algorithm assumes Hardy–Weinberg proportions and linkage equilibrium; these assumptions are not appropriate for loci under selection. Thus, because different classes of marker should be used for delineating different types of CU, as discussed above, the most appropriate type of analysis will also depend not only on the biological context and the nature of the genomic data, but also on the type of CU being delineated.
New genomic framework for delineating CUs and quantifying adaptive differentiation
Premises of new genomic framework
Based on the above considerations, we developed a new framework for delineating CUs and quantifying adaptive differences among them (Box 3). Our framework is based on two premises. First, different classes of marker should be used for delineating ESUs versus MUs. Second, there may be important adaptive differences among ESUs and MUs that should be tested for and quantified using loci under divergent selection. Below, we describe each of these points in more detail.
Box 3. Workflow for delineating CUs and characterizing adaptive differentiation.
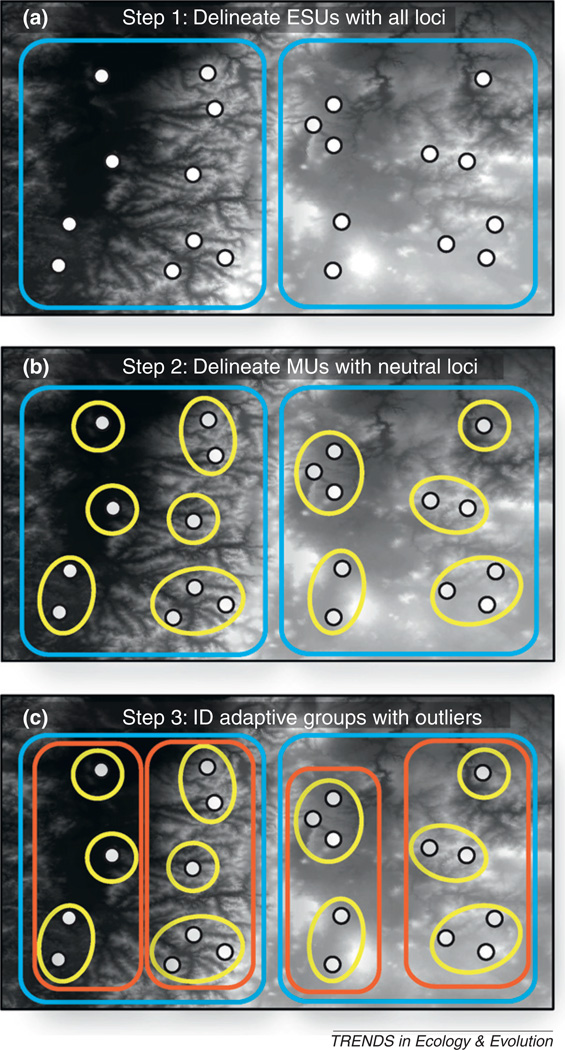
Figure I outlines a general workflow for delineating CUs and testing for adaptive differentiation among CUs using genomic data (e.g. SNP data).
Step 1
Delineate ESUs, the largest intraspecific CUs, using all loci, both FST outlier loci (presumably adaptive), and non-outlier loci (presumably neutral). We recommend using both neutral loci and loci under selection for defining ESUs because ESUs are shaped by both neutral (e.g. historical isolation) and adaptive (e.g. divergent selection) processes. In this example, two ESUs were identified, one on each side of a mountain chain.
Step 2
Define MUs using non-outlier loci, as MUs are defined as demographically independent populations with restricted gene flow among them. In this case, MUs contained from one to three sampling localities.
Step 3
Quantify adaptive differentiation among MUs using outlier loci, for example using the procedure described in Box 2 for threespine sticklebacks. Here, MUs cluster into a high and a low elevation group at outlier loci within each ESU, suggesting elevation is the major axis of adaptive differentiation.
The first premise of our new framework is that different classes of marker should be used to define different CUs. Specifically, we argue that all loci, both neutral and presumably adaptive loci (e.g. FST outlier loci), should be used to delineate ESUs. This is because ESUs are major, intra-specific units that have been historically isolated from each other and that likely have important adaptive differences among them. By contrast, MUs should be demarcated using only neutral loci because, as explained above, MUs are demographically independent units that are de-fined by restricted gene flow. Palsbøll et al. [15] provide guidelines on setting quantitative thresholds for designating populations as MUs. A thorough discussion of thresholds for ESUs is also needed, but is beyond the scope of this article.
The second idea behind our new framework is that, once ESUs and MUs have been delineated as described above, adaptive differentiation among them should be quantified using loci exhibiting signatures of divergent selection, such as elevated FST. The advantage of using outlier loci to characterize adaptive divergence, rather than a few genes of known function, is that the outlier approach should capture adaptation to multiple dimensions of environmental variation. In addition, we argue that SNP loci are more appropriate for quantifying adaptive differentiation than are gene expression approaches because divergence at SNPs reflects genetically based variation (which is required for adaptive divergence), whereas divergence in gene expression can also be caused by environmental differences among populations. A potential disadvantage of using outlier loci is that some outliers may be false positives (i.e. statistical artifacts not truly indicative of divergent selection; [45]), and other loci under weak selection may be missed as false negatives [46]. Nonetheless, the goal is not to identify all individual loci causing adaptation; given the large number of markers assayed with genomic techniques, averaging across outlier loci is still expected to reveal genome-wide patterns of adaptive differentiation. In addition, inferences about adaptive divergence based on high-FST outlier loci can be strengthened by testing for concordance with other ge-nomic signatures of selection (e.g. LD outliers or nucleotide substitution rates; [47]) and ecological, phenotypic, and environmental data, as described below.
Characterizing adaptive differentiation among CUs
There are many different potential approaches for quantifying adaptive differences among ESUs or MUs. This is an important frontier in conservation genomics (see ‘Future directions’ section). Here, we describe one potential approach for characterizing adaptive differentiation among MUs, once ESUs and MUs have already been delineated. First, global FST (among all populations) is calculated among all MUs for each SNP locus. In cases where there is little a priori information about patterns of adaptive differentiation, testing for global FST outliers is most appropriate because this should detect loci that are under strong directional selection in any set of populations. Alternatively, testing for pairwise FST outliers between two predefined groups may be more appropriate when those groups are hypothesized to be adaptively divergent.
Second, high-FST outliers that may be under divergent selection are identified using genome scans [45,48–51]. Third, this outlier SNP data set is used to characterize patterns of adaptive differentiation among MUs using population dendrograms, multivariate statistics, landscape genomics, or another appropriate analysis. The end result of this approach will be quantification of the relative similarity or dissimilarity among MUs at presumably adaptive loci, as shown in Box 2 for threespine sticklebacks (Gasterosteus aculeatus).
Alternatively, population-specific FST values can be calculated [52] using outlier loci to identify the most adaptively divergent populations. For example, consider a set of ten population samples that all show moderate amounts of divergence (global FST ≈0.05) at, say, 10 000 loci. However, one of the ten populations is in a very different environment and is highly divergent for a suite of adaptive loci (say 5%). The signal from these few divergent loci would be difficult to detect if one just looks at global FST for all loci with ten populations. However, these loci would stand out as outliers when one tests for outlier loci using population-specific FST values. This would indicate the presence of adaptive divergence between this individual population and all others.
When possible, ecological, phenotypic, and environmental data should also be used to complement genomic data to strengthen inferences about spatial patterns of adaptation. For one, these data can be used to develop a priori hypotheses about adaptive differentiation. For example, if a species is distributed across multiple environmental axes (e.g. elevation, latitude, and precipitation gradients) and populations vary phenotypically across these axes, this information can be used to develop hypotheses about patterns of adaptive differentiation that can be tested using population genomic data. In addition, these data should be used to back up inferences about adaptive divergence based on outliers, given that some outliers may be false positives. For example, if outlier loci group MUs into two clusters in a PCA, the hypothesis that these clusters represent adaptively divergent sets of populations would be strengthened by phenotypic and environmental data that reveal a similar grouping.
Using information on adaptive differentiation to improve conservation decisions
Patterns of adaptive differentiation can then be used by managers to inform and improve management decisions. First, this information can help with decisions about prioritization of populations and resources. If one goal of management is to maintain adaptive differentiation within a species, then managers should make sure that ESUs or MUs with significant adaptive differences are conserved. For example, managers may want to focus conservation efforts on those CUs with the greatest adaptive differences. Second, information on adaptive differentiation can be used to inform supplementation programs designed to augment declining populations. In particular, managers should try to augment declining populations with source populations that are the most similar at adaptive loci to the target population to minimize the risk of outbreeding depression [20,21,53]. Finally, information on adaptive divergence can be used to inform assisted migration efforts, a management strategy that is likely to become increasingly necessary in the face of climate change. Specifically, an understanding of patterns of adaptive variation and how this variation is related to the landscape and climate can help managers decide the most appropriate sources for colonization of new habitats [54]. Thus, information on spatial patterns of adaptation could greatly improve several management decisions that are currently being made blind with respect to adaptive variation.
Although genomic approaches provide a window into adaptive divergence, genomic results should nonetheless be interpreted with caution [7]. Selection and adaptation are extremely complex, and a complete understanding of adaptive differentiation may never be attained. For example, in cases in which traits are determined by many genes of small effect, there may be insufficient power to detect these loci in genome scans [46]. Moreover, loci that are historically or currently important for adaptation may differ from those loci that will be adaptive in future environments. Below, we suggest future research directions to help determine the best ways to use genomic data for delimiting CUs and quantifying adaptive divergence and the potential, as well as limitations, of using genomic data for this purpose.
Future directions
Many outstanding questions need to be answered to learn how best to take advantage of the power of genomic data to delineate CUs and characterize adaptive differentiation among them. Chief among these are: which analyses are most appropriate and effective for testing for adaptive differentiation using outlier loci? How many SNP loci are needed to delineate different CUs accurately and characterize spatial patterns of adaptation? When will incorporating genomic data change delineation of CUs or change management decisions? How will grouping localities into MUs affect inferred patterns of adaptive differentiation? Many more questions are likely to surface as the use of genomic data to delineate CUs becomes more common.
Both empirical and simulation studies will help address these questions. Case studies using genomic data collected from species with known adaptive differentiation will be particularly useful for testing and comparing different approaches for characterizing adaptive differentiation with outlier loci. For example, Coop et al. [55] looked at patterns of divergence at millions of SNPs in humans to study the geographic distributions of putatively selected alleles at a range of geographic scales. They found that patterns at adaptive loci are predictable from the patterns found at all loci genome wide. They argue that adaptation is constrained by the historical relationships and gene flow between populations. Other species with known patterns of local adaptation that now have population genomic data sets that could be used to test methods for delineating CUs include threespine sticklebacks [26], Trinidadian guppies (Poecilia reticulata [56]), thale cress (Arabidopsis thaliana [57]), and various spruce species (Picea spp. [35,58]).
Simulation studies, which could be parameterized using these or other SNP data sets, would be particularly useful for testing the number of loci needed to characterize accurately adaptive differentiation among ESUs or MUs for different combinations of selection coefficients, gene flow, Ne, and sample sizes (numbers of loci, populations, and individuals; e.g. [59]). Simulation studies would also be useful for testing the effects of grouping localities into MUs on inferred patters of adaptive differentiation.
More work is also needed in developing new analyses to delineate CUs and test for adaptive differentiation. For example, an ideal analysis might simultaneously delineate ESUs, MUs, and quantify adaptive differentiation among these different hierarchical CUs in a single analytical framework, analogous to current Bayesian MCMC approaches developed to infer simultaneously phylogenies, timing of divergence, and historical demographic parameters from sequence data [60].
Genomic data will become increasingly common for species of conservation and management concern. For the first time, these data will enable researchers to understand patterns of adaptive variation in rare and endangered species for which it is impossible to test for adaptation using traditional approaches such as reciprocal transplant experiments. As such, genomic data will play a crucial role in informing management and policy for species of conservation concern. Now is the time to consider carefully how best to take advantage of these data to delineate CUs and characterize adaptive variation and then apply this information to improve conservation decision making.
Figure I.
Grouping of populations of threespine stickleback (Gasterosteus aculeatus) using all, non-outlier, or outlier single nucleotide polymorphisms (SNPs). The data for this analysis are taken from Hohenlohe et al. [26]. (a) Photos of male ocean and lake sticklebacks (courtesy of M. Currey and B. Cresko). (b) Population dendrogram based on all 12 648 SNP loci. (c) Population dendrogram based on 12 016 non-outlier SNP loci (bottom 95% of significance values). (d) Population dendrogram based on 632 outlier SNP loci (top 5% of significance values). This dendrogram is shown at half the scale as the previous two dendrograms. O1–O2 are the two ocean populations and L1–L3 are the three lake populations. (e) Scatterplot of pairwise FST at outlier loci versus pairwise FST at non-outlier loci.
Figure I.
Proposed steps in using genomic data [e.g. single nucleotide polymorphisms (SNPs)] to define conservation units and test for adaptive differentiation among management units (MUs). White circles represent sampling localities, blue outlines are evolutionarily significant units (ESUs), yellow ellipses are MUs, and orange outlines are adaptively similar groups of MUs. The grayscale background is an elevation layer (white is high elevation, black is low elevation).
Acknowledgments
We thank P. Craze, G. Luikart, J.M. Robertson, D.A. Tallmon, R.S. Waples and three anonymous reviewers for providing helpful suggestions that greatly improved this paper. This research was supported by NSF grants DEB 1046408 and DEB 1146489 to W.C.F., DEB 1022196 to J.K.M., and DEB 0742181 to F.W.A. P.A.H. received support from NIH/ NCRR grant P20RR16448 to L. Forney.
Glossary
- Adaptive locus
a polymorphic locus with one or more alleles that increase fitness in the local environment.
- Conservation unit (CU)
a population of organisms that is considered distinct for purposes of conservation, such as a MU, DPS, or ESU.
- Divergent selection
a form of selection in which different alleles are favored in different populations, resulting in divergence in allele frequencies among these populations.
- Effective population size (Ne)
the size of the ideal, panmictic population that would experience the same loss of genetic variation, through genetic drift, as the observed population.
- Evolutionarily significant unit (ESU)
a classification of populations that have substantial reproductive isolation, which has led to adaptive differences so that the population represents a significant evolutionary component of the species. Maintenance of different ESUs is important for maximizing the potential for adapting to future environmental change. The original term used was ‘evolu-tionarily’ [11]. However, both evolutionarily and evolutionary are currently used in the literature.
- Gene flow
exchange of genetic information between demes (randomly mating populations) through migration. Gene flow is often more narrowly defined as the absolute number of migrant individuals per generation (Nem).
- Genetic drift
random changes in allele frequencies in a population between generations due to sampling individuals that become parents and binomial sampling of alleles during meiosis. Genetic drift is more pronounced in small populations.
- Genomic data
genetic information (e.g. SNP genotypes, DNA sequences, etc.) at thousands to millions of loci across the genome of a sample of organisms, gathered using next-generation sequencing or other high-throughput techniques.
- Linkage disequilibrium (LD)
non-random association of alleles at different loci within a population. Also known as gametic disequilibrium.
- Management unit (MU)
a local population that is managed as a distinct unit because of its demographic independence. Maintaining multiple MUs is important for ensuring long-term persistence of species.
- Migration rate (m)
the proportion of exchange among demes. More specifically, m is the probability of an individual breeding in a deme other than that of his or her birth.
- Outlier loci
loci that might be under selection (or in gametic disequilibrium with loci under selection) that are detected because they fall outside the range of the expected distribution for some summary statistic compared with that of neutral loci in a sample (e.g. extremely high or low values of FST).
- Population genomics
the study of numerous loci to understand the roles of evolutionary processes (genetic drift, gene flow, selection, and mutation) that shape variation across genomes and populations [34, 70].
- Single nucleotide polymorphism (SNP)
a nucleotide site (base pair) in a DNA sequence that is polymorphic in a population and can be used as a marker to assess genetic variation within and among populations [71]. Usually only two alleles exist for a SNP in a population.
References
- 1.Rokas A, Abbot P. Harnessing genomics for evolutionary insights. Trends Ecol. Evol. 2009;24:192–200. doi: 10.1016/j.tree.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Rice AM, et al. A guide to the genomics of ecological speciation in natural animal populations. Ecol. Lett. 2011;14:9–18. doi: 10.1111/j.1461-0248.2010.01546.x. [DOI] [PubMed] [Google Scholar]
- 3.Stapley J, et al. Adaptation genomics: the next generation. Trends Ecol. Evol. 2010;25:705–712. doi: 10.1016/j.tree.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Elmer KR, Meyer A. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol. Evol. 2011;26:298–306. doi: 10.1016/j.tree.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Thomas MA, Klapper R. Genomics for the ecological toolbox. Trends Ecol. Evol. 2004;19:439–445. doi: 10.1016/j.tree.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Kohn MH, et al. Genomics and conservation genetics. Trends Ecol. Evol. 2006;21:629–637. doi: 10.1016/j.tree.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Allendorf FW, et al. Genomics and the future of conservation genetics. Nat. Rev. Genet. 2010;11:697–709. doi: 10.1038/nrg2844. [DOI] [PubMed] [Google Scholar]
- 8.Frankham R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 2010;143:1919–1927. [Google Scholar]
- 9.Allendorf FW, Luikart G. Genetics and the Conservation of Populations. Blackwell Publishing; 2007. [Google Scholar]
- 10.Fraser DJ, Bernatchez L. Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol. Ecol. 2001;10:2741–2752. [PubMed] [Google Scholar]
- 11.Ryder OA. Species conservation and systematics: the dilemma of subspecies. Trends Ecol. Evol. 1986;1:9–10. [Google Scholar]
- 12.Moritz C. Defining evolutionarily significant units for conservation. Trends Ecol. Evol. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 13.Waples RS. Pacific salmon, Oncorhynchus spp., the definition of ‘species’ under the Endangered Species Act. Mar. Fish. Rev. 1991;53:11–22. [Google Scholar]
- 14.Crandall KA, et al. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- 15.Palsbøll PJ, et al. Identification of management units using population genetic data. Trends Ecol. Evol. 2007;22:11–16. doi: 10.1016/j.tree.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Hanski IA, Gilpin ME. Metapopulation Biology: Ecology, Genetics, and Evolution. Academic Press; 1997. [Google Scholar]
- 17.McKay JK, Latta RG. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 2002;17:285–291. [Google Scholar]
- 18.Holderegger R, et al. Adaptive vs. neutral genetic diversity: implications for landscape genetics. Landsc. Ecol. 2006;21:797–807. [Google Scholar]
- 19.de Guia APO, Saitoh T. The gap between the concept and definitions in the Evolutionarily Significant Unit: the need to integrate neutral genetic variation and adaptive variation. Ecol. Res. 2007;22:604–612. [Google Scholar]
- 20.Moritz C. Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas. 1999;130:217–228. [Google Scholar]
- 21.Frankham R, et al. Predicting the probability of outbreeding depression. Conserv. Biol. 2011;25:465–475. doi: 10.1111/j.1523-1739.2011.01662.x. [DOI] [PubMed] [Google Scholar]
- 22.Glenn TC. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 23.Hohenlohe PA, et al. Next-generation RAD sequencing identifies thousands of SNPs for assessing hybridization between rainbow and westslope cutthroat trout. Mol. Ecol. Resour. 2011;11:117–122. doi: 10.1111/j.1755-0998.2010.02967.x. [DOI] [PubMed] [Google Scholar]
- 24.Rowe HC, et al. RAD in the realm of next-generation sequencing technologies. Mol. Ecol. 2011;20:3499–3502. doi: 10.1111/j.1365-294x.2011.05197.x. [DOI] [PubMed] [Google Scholar]
- 25.Reznick D, Endler JA. The impact of predation on life-history evolution in Trinidadian guppies (Poecilia reticulata) Evolution. 1982;36:160–177. doi: 10.1111/j.1558-5646.1982.tb05021.x. [DOI] [PubMed] [Google Scholar]
- 26.Hohenlohe PA, et al. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li WH. Maintenance of genetic variability under joint effect of mutation, selection and random drift. Genetics. 1978;90:349–382. doi: 10.1093/genetics/90.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haldane JBS. A mathematical theory of natural and artificial selection. VI. Isolation. Proc. Camb. Philos. Soc. 1930;26:220–230. [Google Scholar]
- 29.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slatkin M. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 1985;16:393–430. [Google Scholar]
- 31.Pritchard JK, et al. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goss EM, Bergelson J. Variation in resistance and virulence in the interaction between Arabidopsis thaliana and a bacterial pathogen. Evolution. 2006;60:1562–1573. [PubMed] [Google Scholar]
- 33.Sambatti JBM, Rice KJ. Local adaptation, patterns of selection, and gene flow in the Californian serpentine sunflower (Helianthus exilis) Evolution. 2006;60:696–710. [PubMed] [Google Scholar]
- 34.Luikart G, et al. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- 35.Prunier J, et al. Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Mol. Ecol. 2011;20:1702–1716. doi: 10.1111/j.1365-294X.2011.05045.x. [DOI] [PubMed] [Google Scholar]
- 36.Bonin A, et al. Population adaptive index: a new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conserv. Biol. 2007;21:697–708. doi: 10.1111/j.1523-1739.2007.00685.x. [DOI] [PubMed] [Google Scholar]
- 37.Salemi M, Vandamme A-M. The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny. Cambridge: University Press; 2003. [Google Scholar]
- 38.Pritchard JK, et al. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guillot G, et al. GENELAND: a computer package for landscape genetics. Mol. Ecol. Notes. 2005;5:712–715. [Google Scholar]
- 40.Joost S, et al. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol. Ecol. 2007;16:3955–3969. doi: 10.1111/j.1365-294X.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- 41.Joost S, et al. Spatial analysis method (SAM): a software tool combining molecular and environmental data to identify candidate loci for selection. Mol. Ecol. Resour. 2008;8:957–960. doi: 10.1111/j.1755-0998.2008.02162.x. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz MK, et al. Landscape genomics: a brief perspective. In: Cushman SA, Huettmann F, editors. Spatial Complexity, Informatics, and Wildlife Conservation. Springer; 2009. pp. 165–174. [Google Scholar]
- 43.Manel S, et al. Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol. Ecol. 2010;19:3760–3772. doi: 10.1111/j.1365-294X.2010.04717.x. [DOI] [PubMed] [Google Scholar]
- 44.Manel S, et al. Common factors drive adaptive genetic variation at different spatial scales in Arabis alpina. Mol. Ecol. 2010;19:3824–3835. doi: 10.1111/j.1365-294X.2010.04716.x. [DOI] [PubMed] [Google Scholar]
- 45.Narum SR, Hess JE. Comparison of FST outlier tests for SNP loci under selection. Mol. Ecol. Resour. 2011;11:184–194. doi: 10.1111/j.1755-0998.2011.02987.x. [DOI] [PubMed] [Google Scholar]
- 46.Rockman MV. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution. 2012;66:1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hohenlohe PA, et al. Using population genomics to detect selection in natural populations: key concepts and methodological considerations. Int J. Plant Sci. 2010;171:1059–1071. doi: 10.1086/656306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genomescans. Mol. Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- 49.Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 50.Bonin A. Population genomics: a new generation of genome scans to bridge the gap with functional genomics. Mol. Ecol. 2008;17:3583–3584. doi: 10.1111/j.1365-294X.2008.03854.x. [DOI] [PubMed] [Google Scholar]
- 51.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaggiotti OE, Foll M. Quantifying population structure using the F-model. Mol. Ecol. Resour. 2010;10:821–830. doi: 10.1111/j.1755-0998.2010.02873.x. [DOI] [PubMed] [Google Scholar]
- 53.Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- 54.McKay JK, et al. ‘How local is local?’ A review of practical and conceptual issues in the genetics of restoration. Restor. Ecol. 2005;13:432–440. [Google Scholar]
- 55.Coop G, et al. The role of geography in human adaptation. PLoS Genet. 2009;6:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willing E-M, et al. Genome-wide single nucleotide polymorphisms reveal population history and adaptive divergence in wild guppies. Mol. Ecol. 2010;19:968–984. doi: 10.1111/j.1365-294X.2010.04528.x. [DOI] [PubMed] [Google Scholar]
- 57.Hancock AM, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- 58.Namroud M-C, et al. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol. Ecol. 2008;17:3599–3613. doi: 10.1111/j.1365-294X.2008.03840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waples RS, Gaggiotti O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 2006;15:1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- 60.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dizon AE, et al. Rethinking the stock concept – a phylogeographic approach. Conserv. Biol. 1992;6:24–36. [Google Scholar]
- 62.Avise JC. Molecular Markers, Natural History, and Evolution. Chapman & Hall; 1994. [Google Scholar]
- 63.Vogler AP, Desalle R. Diagnosing units of conservation management. Conserv. Biol. 1994;8:354–363. [Google Scholar]
- 64.USFWS (U.S. Fish and Wildlife Service) and NMFS (National Marine Fisheries Service) Policy regarding the recognition of distinct vertebrate population segments under the Endangered Species Act. Fed. Regist. 1996;61:4721–4725. [Google Scholar]
- 65.Wilding CS, et al. Differential gene exchange between parapatric morphs of Littorina saxatilis detected using AFLP markers. J. Evol. Biol. 2001;14:611–619. [Google Scholar]
- 66.Cresko WA, et al. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goudet J, et al. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 69.Kalinowski ST. How well do evolutionary trees describe genetic relationships between populations? Heredity. 2009;102:506–513. doi: 10.1038/hdy.2008.136. [DOI] [PubMed] [Google Scholar]
- 70.Black WC, IV, et al. Population genomics: genome-wide sampling of insect populations. Annu. Rev. Entom. 2001;46:441–469. doi: 10.1146/annurev.ento.46.1.441. [DOI] [PubMed] [Google Scholar]
- 71.Morin PA, et al. SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 2004;19:208–216. [Google Scholar]