Abstract
A fragment of the ScHd1 gene derived from eight inbred lines was sequenced and showed homology to other Hd1 genes from different cereals. Sequences were analysed with respect to the presence of a single-nucleotide polymorphism (SNP) difference. A C-T transition at position 312 of the consensus sequence was found, which distinguished two lines from the remaining six. The deduced amino acid sequence revealed a high identity (93%) to a Hd1-like protein from wheat. The identified mutation allowed the localisation of ScHd1 on a genetic map of rye (6RS). A small, statistically significant linkage between ScHd1 and earliness per se (eps) and some morphological traits was also established. The chromosomal region, including the S76 allele for the ScHd1 gene was linked to earlier heading, elongated spikes, a greater number of spikelets per spike and an increased weight of 1000 kernels.
Keywords: Flowering, Heading date, Intrinsic earliness, Secale cereale L, Winter rye
Variability in flowering time in cereals depends on genes controlling photoperiod (Ppd) and vernalisation (Vrn), as well as on other genes that regulate earliness per se (eps). There are many articles concerning cereal eps loci that have been analysed and described as QTLs, but there is little published information relating to the analysis of these genes. One of the best-known eps loci is Eps-A m 1 from Triticum monococcum on chromosome 1Am (Bullrich et al. 2002; Válarik et al. 2006; Lewis et al. 2008; Faricelli et al. 2010). The Eps-3A m gene in Einkorn wheat is also characterised quite well (Hori et al. 2007; Gawroński and Schnurbusch 2012). In rye, many QTLs determining flowering time were detected on all seven chromosomes (Myśków 2012; Myśków et al. 2012).
The object of this study was the Hd1 gene, an orthologue of the Arabidopsis thaliana CONSTANS (CO) gene, involved in photoperiodic flowering. Hd1 gene homologues have been identified in rice, barley and wheat (Yamamoto et al. 1998; Yano et al. 2000; Kojima et al. 2002; Griffiths et al. 2003; Nemoto et al. 2003). Although homologues of Hd1 are key regulators of photoperiodic flowering in plants of short-day (SD) zones (Cockram et al. 2007), they do not cause strong phenotypic effects in temperate cereals. However, transgenic experiments expressing the wheat gene in rice showed that it maintains its strong effect in the genetic background of SD plants (Nemoto et al. 2003).
Although the nucleotide sequence of the Hd1 gene fragment (GU324592.1) is already known, it has not yet been mapped to the rye genome. Additionally, its role in flowering time regulation is unknown. The main purpose of this study was 1) to map Hd1 to the rye genome; 2) to examine its role in the regulation of earliness per se; 3) to examine linkage between ScHd1 and the distribution of morphological traits such as plant height (Ph), spike length (Sl), number of spikelets per spike (Sps), number of kernels per spike (Kps), weight of kernels per spike (Kw), thousand kernel weight (Tkw), pre-harvest sprouting (PHS) and alpha-amylase activity (AA).
Plant material included inbred lines of rye: 541, Ot1-3, Ds2, RXL10, S120, S76, C599 and 620–5, with at least 20 generations of inbreeding. These were the parental breeds used for crossbreeding to obtain high-density genetic maps (Milczarski et al. 2011, Myśków, unpublished data). In addition, the mapping populations were at our disposal (population of recombinant inbred lines: 541×Ot1-3, DS2×RXL10, S120×S76 and C599×620-2).
DNA was isolated from lyophilised leaves using the DNeasy Plant Mini Kit (Qiagen). The DNA was amplified by PCR via standard conditions (detailed methodology upon request) in a DNA Engine Dyad® Thermal Cycler (Bio-Rad Laboratories), using Thermo Scientific (Fermentas) reagents and the primer pair: Hd-F355 (5´-AGCGTGTGCGTGTCTGCGAA-3’) and Hd-R862 (5’-GCAGCCTGCCCTGCTCCTAT-3’) purchased from Genomed. Primers were designed to the conservative region containing the BBOX (B-box-type zinc finger) domain of the wheat TaHd1 gene (AB094491.1, TaHd1-3 mRNA for the Hd1-like protein complete coding sequence). The numbers in the primer names describe the binding positions of the primers.
The PCR amplicons were separated by electrophoresis in a 1.5% agarose gel with 1×TBE buffer, and were visualised using EtBr. Monomorphic products of amplification of ∼500 bp in length were obtained for eight analysed lines. These fragments were isolated from the gel and cloned into the pCR®II-TOPO® vector (Life Technologies, Invitrogen). A GenomeLab DTCS – Quick Start Kit (Beckman Coulter) and M13 primer were used for PCR sequencing. Sequencing was performed in a Beckman Coulter CEQ 8000 Genetic Analysis System. Each DNA strain was sequenced on both strands and a consensus sequence was obtained using BioEdit (Hall 1999) and Geneious R6 (Biomatters, http://www.geneious.com/).
The obtained sequences, ranging from 370 bp to 520 bp in length, were registered with NCBI and GenBank with the accession numbers from KJ371037 to KJ371044, consecutively. The sequences of the analysed lines and one common consensus sequence were compared with sequences deposited at the NCBI database and homology to GU324592.1 (Secale cereale cultivar Puma Hd1-like protein mRNA, partial cds), AB094488.1 (Triticum aestivum TaHd1-2 gene for Hd1-like protein, complete coding sequence), four homologous wheat sequences and to barley sequences was revealed. The percentage nucleotide identity for rye, wheat and barley was 98%, 95% and 94% (E = 0), respectively.
Sequences were analysed with respect to the presence of SNPs. Single nucleotide differences were detected at 14 sites, but they concerned a single strand, and probably resulted from amplification errors during PCR. Moreover, a single SNP (a C-T transition at position 312 of the consensus sequence) that distinguished lines S76 and C599-5 from the other six lines was detected. Using the predicted amino acid sequence to perform a similarity search using the NCBI database showed a high identity of 99% (one variant amino acid) and 93% (12 different amino acids) to rye (ADR51712.1) and wheat (BAC92733.1) Hd1-like protein, respectively. The observed SNP differences between our lines translate into one amino acid (leucine to phenylalanine) difference at position 169 of the wheat protein of 370 amino acids.
The next step after identifying the sequences of ScHd1 and SNP was the design of new primers to identify the C-T mutation via PCR. The same forward primer was used as previously for amplification, together with one of two new reverse primers: Hd-R1-312R19 (5-´ACATATTGTATCCGACAAA-3´) or Hd-R2-312R19 (5´-ACATATTGTATCCGACAAG-3´) together with hot-start VivaTaq polymerase (Novazym).
The SNP polymorphism in the analysed lines was confirmed using allospecific primers for two variants of the ScHd1 sequence and the analysis was also performed for two mapping populations: S120×S76 (RIL-M) and C599×620-2 (RIL-R). A single gene segregation (χ 2 = 1.29) was obtained for the first population, whereas the allelic polymorphism of the ScHd1 fragment was not found in the second population. The inbred line C599-5 analysed in this experiment was a sister sub-line with respect to that used to produce hybrid C599×620-2. These sub-lines must have differed in terms of mutations in ScHd1. Segregation data for the RIL-M population was added to data from previous analyses, mainly using DarT markers (Myśków 2012; Myśków et al. 2012); mapping of ScHd1 was performed via JoinMap 3.0 software (Van Ooijen and Voorrips 2001) and the marker was associated with loci on chromosome 6R. Mapping was executed for a group consisting of 44 markers most significantly associated with ScHd1 (LOD = 15). The map position of the gene and its nearest loci is presented in Fig. 1.
Fig. 1.
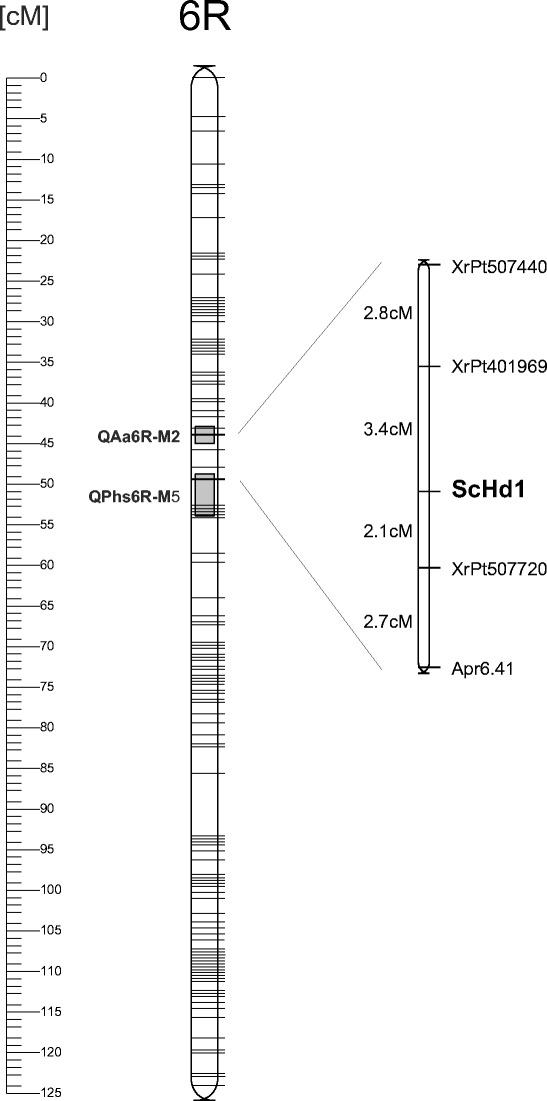
Localisation of ScHd1 on the genetic map of rye on chromosome 6R in the RIL-M population. Grey rectangles mark QTLs mapped to the close vicinity of the gene (QTL for α-amylase activity and pre-harvest sprouting)
The sequence analysis and the map position suggest that ScHd1 is homologous to the CO/Hd1 gene. TaHd1 was located on the long arm of chromosomes belonging to group 6 in hexaploid wheat (Nemoto et al. 2003). A rye gene was also mapped to the pericentromeric region of chromosome 6, with a genetic distance of 5cM, above the RAPD – APR6.41 marker localised to the short arm on the integrated 6R map (Stojałowski et al. 2009). In the light of new data concerning rearrangements of the rye genome in comparison to wheat (Li et al. 2013), the short arm 6R corresponds not only to the short arm of wheat chromosome 6, which was previously postulated (Devos et al. 1993), but also includes fragment of the long arm in the centromeric region. Therefore, localisation of ScHd1 to the short arm is consistent with present knowledge about genome synteny.
A map used for the map position of Hd1 in rye was previously used for the localisation of the eps loci and of QTLs for different morphological traits such as PHS, AA, Ph, Sl, Sps, Kps, Kw and Tkw (Myśków et al. 2012, 2014). Additionally, the spike density coefficient (Ct) was calculated from the formula: 10 cm × Sps/Sl and collected data were included for further analysis. The results enabled a relationship between ScHd1 segregation and the distribution of the above-mentioned traits in the mapping population to be detected. The Kruskal–Wallis test (STATISTICA 9.0, http://www.statsoft.com) was used to determine inter-relationships and the results are depicted in Table 1. It was also assessed whether the gene co-locates with known QTLs.
Table 1.
Relationship between the segregation of ScHd1 alleles and heading earliness and different morphological traits in the rye mapping population RIL-M (143 inbred lines), identified using the Kruskal-Wallis test (only statistically significant). He – heading earliness (on a nine-point scale), Sl – spike length (cm), Kw – kernel weight per spike (g), Tkw – thousand kernel weight (g), Ct – density of spikelets (number of spikelets per 10 cm of spike). Measurements of earliness were made for four years (2007–2010); other traits were measured over 3 years (2008–2010)
| Trait | N number of lines | H Kruskal-Walis test statistic | Mean value of trait | |||
|---|---|---|---|---|---|---|
| P probability | Allele C (S120) | Allele T (S76) | Difference | |||
| He/07 | 104 | 6.64 | 0.01 | 4.75 | 5.91 | 1.16 |
| He/08 | 131 | 6.56 | 0.01 | 4.33 | 5.06 | 0.73 |
| Sl/08 | 132 | 6.73 | 0.01 | 8.06 | 8.56 | 0.50 |
| Sl/09 | 132 | 4.06 | 0.04 | 6.70 | 6.99 | 0.29 |
| Sl/08–10 | 131 | 5.25 | 0.02 | 7.45 | 7.83 | 0.38 |
| Kw/08–10 | 131 | 5.48 | 0.02 | 0.56 | 0.66 | 0.10 |
| Tkw/08 | 132 | 4.04 | 0.04 | 26.02 | 28.08 | 2.06 |
| Tkw/10 | 124 | 5.01 | 0.03 | 19.37 | 20.47 | 1.10 |
| Tkw/08–10 | 131 | 6.41 | 0.01 | 22.99 | 24.75 | 1.76 |
| Ct/08 | 132 | 11.03 | 0.00 | 26.69 | 28.33 | 1.64 |
| Ct/09 | 132 | 3.75 | 0.05 | 27.36 | 28.31 | 0.95 |
One or two QTLs for earliness per se, depending on the mapping population, were observed on chromosome 6R (Stojałowski and Łapiński 2002; Myśków 2012; Myśków et al. 2012). ScHd1 was not mapped to any of the two QTLs present in the RIL-M population map (Myśków 2012; Myśków et al. 2012). However, the relationship of this marker with the phenotypic segregation of eps was shown to be statistically significant in two years out of a four-year research study, using the Kruskal–Wallis test. The non-parametric Kruskal–Wallis test frequently leads to different results than the CIM analysis (Myśków et al. 2014) and is less accurate with respect to QTL mapping; however, it is a simple method of marker identification that can be used for the selection of an evaluated trait. It also enables the evaluation of a phenotypic effect that can be obtained using a certain selection marker. The Kruskal–Wallis test showed that ScHd1 can affect heading by about one point on a rating scale (according to Masojć and Milczarski 1999), which corresponds to approximately 1 day. Heading earliness was associated with the presence of an allele from line S76. Although the line S76 is later compared to S120, transgression into the RIL-M population and QTL analysis indicates the presence of many eps genes derived from line S76 (Myśków et al. 2012).
The presence of pleiotropic effects is an interesting phenomenon with respect to genes that regulate flowering time. Numerous studies concerning Ppd and Vrn genes have confirmed their simultaneous effect on other vital plant traits. Pleiotropic effects of Ppd genes on plant height, spike morphology, and the number and weight of kernels has been confirmed in wheat (Worland et al. 1998) and barley (Laurie et al. 1994; Karsai et al. 1999; Wang et al. 2010).
Several studies have also suggested a relationship between the activity of intrinsic earliness genes and other important crop traits. For example, it was established that the best-characterised gene of earliness per se (locus Eps-A m 1) also exhibits pleiotropic effects and affected the number of spikelets per spike of the diploid wheat via regulation of the length of the spike development stages (Lewis et al. 2008). In diploid wheat, it was also observed that the QTL for the heading date from 3A overlapped with a QTL for the length of spike and number of spikelets. The co-localisation of some QTLs of earliness per se with QTLs for pre-harvest sprouting was observed in rye (Myśków 2012; Myśków et al. 2012), which might indirectly provide evidence concerning the influence of flowering time genes on the germination process in subsequent progeny.
The results presented here show that similar to other wheat and barley eps genes, ScHd1 might be related to morphological traits, including spike morphology, such as the length of the spike, spike density and thousand kernel weight (Table 1). Plants with the allele from line S76 were also characterised by earlier heading, an elongated spike, an increase in the number of spikelets per spike, increased weight of kernels per spike and thousand kernel weight. This might indicate that the confidence interval of an average QTL usually contains several dozens of polymorphic genes for different features.
Throughout the three years of study, the Kruskal–Wallis test showed no correlation of ScHd1 segregation with α-amylase activity or susceptibility to pre-harvest sprouting; however, this gene is linked with the XrPt507440 and XrPt507720 markers that localise within QTLs responsible for these traits.
Acknowledgments
The study was financially supported by the National Science Centre of Poland, under grant NN310 067639.
References
- Bullrich L, Appendino ML, Tranquilli G, Lewis S, Dubcovsky J. Mapping of a thermo-sensitive earliness per se gene on Triticum monococcum chromosome 1Am. Theor Appl Genet. 2002;105:585–593. doi: 10.1007/s00122-002-0982-5. [DOI] [PubMed] [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA, Greenland AJ. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J Exp Bot. 2007;58(6):1231–1244. doi: 10.1093/jxb/erm042. [DOI] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RL, Liu CJ, Masojć P, Xie DX, Gale MD. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet. 1993;85:673–680. doi: 10.1007/BF00225004. [DOI] [PubMed] [Google Scholar]
- Faricelli ME, Válarik M, Dubkovsky J. Control of flowering time and spike development in cereals: the earliness per se Eps-1 region in wheat, rice, and Brachy podium. Funct Integr Genomics. 2010;10:293–306. doi: 10.1007/s10142-009-0146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawroński P, Schnurbusch T. High-density mapping of the earliness per se-3Am (Eps-3Am) locus in diploid einkorn wheat and its relation to the syntenic regions in rice and Brachypodium distachyon L. Mol Breeding. 2012;30:1097–1108. doi: 10.1007/s11032-011-9697-0. [DOI] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003;131:1855–67. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hori K, Takehara S, Nankaku N, Sato K, Sasakuma T, Takeda K. Barley EST markers enhance map saturation and QTL mapping in diploid wheat. Breeding Sci. 2007;57:39–45. doi: 10.1270/jsbbs.57.39. [DOI] [Google Scholar]
- Karsai I, Mészáros K, Szücs P, Hayes PM, Láng L, Bedö Z. Effect of loci determining photoperiod sensitivity (Ppd-H1) and vernalization response (Sh2) on agronomic traits in the “Dictoo”דMorex” barley mapping population. Plant Breed. 1999;118:399–403. doi: 10.1046/j.1439-0523.1999.00408.x. [DOI] [Google Scholar]
- Kojima S, Takahashi Y, KobayashiY ML, Sasaki T, Araki T, Yano M. Hd3a, a rice ortolog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. Genetic analysis of a photoperiod response gene on the short arm of chromosome 2 (2H) of barley (Hordeum vulgare L.) Heredity. 1994;72:619–627. doi: 10.1038/hdy.1994.85. [DOI] [Google Scholar]
- Lewis S, Faricelli ME, Appendino ML, Válarik M, Dubkovsky J. The chromosome region including the earliness per se locus Eps-Am1 affects the duration of early developmental phases and spikelet number in diploid wheat. J Exp Bot. 2008;59:3595–3607. doi: 10.1093/jxb/ern209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Endo TR, Saito M, Ishikawa G, Nakamura T, Nasuda S. Homoeologous relationship of rye chromosome arms as detected with wheat PLUG markers. Chromosoma. 2013;122:555–564. doi: 10.1007/s00412-013-0428-7. [DOI] [PubMed] [Google Scholar]
- Masojć P, Milczarski P. The use of molecular map of the rye genome and QTL analysis for the identification of genes for earliness earing. Biul IHAR. 1999;211:205–210. [Google Scholar]
- Milczarski P, Bolibok-Brągoszewska H, Myśków B, Stojałowski S, Heller-Uszyńska K, Góralska M, Brągoszewski P, Uszyński G, Kilian A, Rakoczy-Trojanowska M. A high density consensus map of rye (Secale cereale L.) based on DArT markers. PLoS One. 2011;6(12):e28495. doi: 10.1371/journal.pone.0028495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myśków B (2012) Identification of quantitative trait loci (QTL) that control earliness and susceptibility to preharvest sprouting of rye (Secale cereale L.) using dense genetic maps for recombinant inbred lines (RILs) populations. Dissertation, West-Pomeranian University of Technology in Szczecin (in Polish)
- Myśków B, Stojałowski S, Łań A, Bolibok-Brągoszewska H, Rakoczy-Trojanowska M, Kilian A. Detection of the QTL for α-amylase activity on high-density genetic map of rye and comparing their localization to loci controlling preharvest sprouting and earliness. Mol Breed. 2012;30(1):367–376. doi: 10.1007/s11032-011-9627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myśków B, Hanek M, Banek-Tabor A, Maciorowski R, Stojałowski S. The application of high-density genetic maps of rye for detection of QTLs controlling morphological traits. J Appl Genetics. 2014;55:15–26. doi: 10.1007/s13353-013-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y. Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 2003;36:82–93. doi: 10.1046/j.1365-313X.2003.01859.x. [DOI] [PubMed] [Google Scholar]
- Stojałowski S, Łapiński M. The use of RAPD technique to construct a linkage map of rye interline hybrid. Zesz Probl Postep Nauk Rol. 2002;488:153–159. [Google Scholar]
- Stojałowski S, Myśków B, Milczarski P, Masojć P. A consensus map of chromosome 6R in rye (Secale cereale L.) Cell Mol Biol Lett. 2009;14(2):190–198. doi: 10.2478/s11658-008-0042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válarik M, Linkiewicz AM, Dubkovsky J (2006) A microlinearity study at the earliness per se gene Eps-Am1 region reveals an ancient duplication that preceded the wheat-rice divergence. Theor Appl Genet 112:945–957 [DOI] [PubMed]
- Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0, Software for the calculation of genetic linkage maps. Plant Res Intern, Wageningen, the Netherlands
- Wang G, Schmalenbach I, von Korff M, Léon J, Kilian B, Rode J, Pillen K. Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor Appl Genet. 2010;120:1559–1574. doi: 10.1007/s00122-010-1276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland AJ, Börner A, Korzun V, Li WM, Petrovic S, Sayers EJ. The influence of photoperiod genes on the adaptability of European winter wheats. Euphytica. 1998;100:385–394. doi: 10.1023/A:1018327700985. [DOI] [Google Scholar]
- Yamamoto T, Kuboki Y, Lin SY, Sasaki T, Yano M. Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theor Appl Genet. 1998;97:37–44. doi: 10.1007/s001220050864. [DOI] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]


