Abstract
A combination of staining, light microscopy and SYBR green- and dual-labelled fluorescent probe-based qPCR chemistries with species- and gene-specific primers was employed to evaluate fluctuations in the aerial biomass of Leptosphaeria maculans spores captured by volumetric spore trappings in Poznan, Poland (2006, 2008) and Harpenden, UK (2002, 2006). Arising from these surveys, DNA samples extracted from Burkard spore-trap tapes were screened for fluctuation patterns in the frequencies of AvrLm1 and AvrLm6, the most prominent of the 15 genes that code for avirulence effectors in this Dothideomycete cause of the destructive phoma stem canker disease of oilseed rape worldwide. In Poznan, very low frequencies of AvrLm1 allele were found in the autumn of both 2006 and 2008, reflecting significantly increased cultivation of rape seed with Rlm1-based resistance. In contrast, at least six folds-higher frequencies of AvrLm6, which were also confirmed by end-point PCR bioassays on phoma-infected leaves from the same region of Poland, were obtained during both years. In the UK, however, relatively higher AvrLm1 allele titres were found in L. maculans spores captured in air samples from the autumn of 2002 on the experimental fields of Rothamsted Research, Harpenden, that were historically sown to genetically heterogeneous B. napus cultivars. In the 2006 screen these levels had plummeted, to a 1:4 ratio, in favour of frequencies of the AvrLm6 allele. Patterns of fluctuations in erg11 (CYP51) fragments coding for sterol 14α-demethylase suggest October as the month with the most viable wind-dispersed L. maculans propagules of each season of the screens.
Keywords: Airborne inoculum, Air sampling, Avirulence genes, Disease forecasting, Leptosphaeria maculans, Phoma stem canker
Introduction
Various techniques for spore capture are increasingly being used in supporting decisions on the protection of agricultural crops against important pathogens causing great yield losses. The systems mostly used are volumetric traps that enable a continuous monitoring of fungal spore concentrations in known volumes of air samples following capture. Advice based on spore counts is done routinely in several countries, including USA (Isard et al. 2007), Australia (Blackleg Sporacle, Salam et al. 2003), India (Devi and Singh 2007) and the UK (for the development of PASSWORD; Gladders et al. 2006). Currently in Europe, the biggest monitoring system based on spore counts is the System for Forecasting Disease Epidemics (SPEC; Jedryczka et al. 2004, 2012) which was designed in Poland for monitoring the aerial dynamics of ascospore concentrations of the pathogen Leptosphaeria maculans, the cause of the phoma stem canker disease of oilseed rape. This damaging disease is prevalent on brassicas worldwide (Fitt et al. 2006), except for China and India, where agricultural practices and weather conditions appear to prevent epidemics of this destructive pathogen.
The species L. maculans, usually found in association with the less damaging L. biglobosa (cause of phoma leaf and upper stem necrosis), has become in recent years one of the most widely-studied fungal species (Rouxel and Balesdent 2005; Kaczmarek and Jedryczka 2011). Apart from causing economic yield losses, key features of the biology of this ascomycete fungus make it a candidate for genetic and molecular biological studies. Pseudothecia, the fruiting bodies, of L. maculans are easily formed in natural conditions as well as in controlled environment thereby permitting studies on the prolonged haploid phase of the life cycle and easy separation (by pulsed gel electrophoresis) of its 17 chromosomes (Cozijnsen et al. 2000), including a dispensable minichromosome (Leclair et al. 1996; Balesdent et al. 2013). The pathogen is easily cultured on artificial media and produces numerous conidial spores with identical genetic material. Recent successful transformations of L. maculans with green fluorescent protein (GFP), as well as with DS-Red, reporter gene constructs have made it possible to study the endophytic growth of the pathogen within asymptomatic host tissues (Sexton and Howlett 2001; Eckert et al. 2005). More recently, the genome of this dothideomycete fungus was sequenced and the annotations of fungal avirulence genes have been released publicly (Rouxel et al. 2011).
Knowledge of the distribution of avirulence genes within fungal populations is crucial for choosing the strategies of crop protection against pathogens. As interactions between products of avirulence genes and products of host plant resistance genes determine disease outcome, studies designed to monitor avirulence genes are of great importance in planning both general and specific strategies of disease managements, including the choice of cultivars that are most suitable for resisting pathogen populations as well as planning breeding strategies for future control of the disease. In total, 11 avirulence genes comprising AvrLm1-AvrLm11 (including AvrLm4-7) in L. maculans and corresponding to 11 R genes viz, Rlm1-Rlm11 in brassica (B. napus, B. juncea, B. nigra and B . rapa) host plants (Ansan-Melayah et al. 1995, 1998; Balesdent et al. 2001, 2002, 2005, 2006, 2013; Delourme et al. 2006; Kutcher et al. 2010; Soyer et al. 2014) have been identified so far. Four other resistance genes, LepR1, LepR2, LepR3 and LepR4 were identified in Canadian canola species (Rimmer 2006; Larkan et al. 2013; Yu et al. 2013).
Leptosphaeria maculans has a high evolutionary potential, according to the criteria of McDonald and Linde (2002), arising from a combination of its possession of both sexual and asexual cycles, an effective wind-dispersal mechanism for ascospores and rain-splash method for the infective conidia, as well as its large population size. In the recent past, rapid degradation and loss (‘breakdown’) of qualitative resistance have attended the large scale cropping of oilseed rape cultivars possessing a single vertically-resistant Rlm gene (Gout et al. 2006). For example, the cultivar ‘Surpass 400’ (specific resistance derived from B. rapa spp. sylvestris; Crouch et al. 1994) which was introduced to Australia in 2000 and was widely adopted by rapeseed farmers, became no longer effective by 2003 against L. maculans populations, causing severe losses to the oilseed rape industry (Li and Cowling 2003) thereby. Similarly, the large scale cropping from 1996 to 1999 (43.7 % of total area grown), of oilseed rape with the Rlm1 gene in France, resulted in a pathogen population change (increase in the frequency of avrLm1) that rendered ineffective, by the 2000/2001 growing season, this qualitative resistance/trait (Rouxel et al. 2003). However, Van de Wouw et al. (2009) have suggested that recognition of avirulence in L. maculans towards a Brassica napus cultivar with ‘sylvestris-derived’ resistance involved two resistance genes. Resistance gene Rlm6 currently appears to provide effective resistance to L. maculans. Stachowiak et al. (2006) identified AvrLm6 in 100 % of European isolates examined in 2002, whereas AvrLm1 ranged between 1.2 % in Teendorf (Germany) to 17.6 % in Harpenden (UK, England). The most recent study on AvrLm1 and AvrLm6 in the UK was done in 2006–2009 and found that while AvrLm1 allele frequencies remained low those of AvrLm6 had decreased to 35–66 % (Van de Wouw et al. 2010). In a large-scale survey of French L. maculans in 2001 and 2002 all isolates of L. maculans possessed AvrLm6 (Balesdent et al. 2006). A similar pattern was reported for North America, where AvrLm6 was present in the majority (75–100 %) of isolates, with the exception of a field in Camrose, Alberta (Canada), where it was found to be below 40 % (Dilmaghani et al. 2009). The presence of avirulence gene AvrLm1 in North America appears to depend on both location and popularity and expanse of oilseed rape cultivation; in Mexico the allele was detected in all isolates of L. maculans sampled, whereas in the majority of fields in Alberta and Saskatchewan, Canada, AvrLm1 was completely absent. Strangely, however, this allele was also very low (1.5 %) in Chile, where oilseed rape is not popularly cultivated.
In recent reports (Kaczmarek et al. 2009, 2012; Jedryczka et al. 2010b; Karolewski et al. 2012; Atkins et al. 2013) we have presented studies that demonstrated by quantitative PCR techniques the detection, discrimination and quantification of seasonally captured propagules of Leptosphaeria spp, Pyrenopeziza brassicae and Sclerotinia sclerotiorum in air samples from oilseed rape fields of Poland and southern UK. These studies utilized Burkard 7-day air samplers, primers based on internal transcribed spacer (ITS) region or β-tubulin gene fragment sequences and SYBR-green or dual-labelled fluorescent probe chemistry to monitor seasonal fungal abundance in air spora ahead of the onset of damaging symptoms of diseases caused by these pathogens. In the current study, we seek to further demonstrate the possibilities of monitoring, screening and characterizing these spores for viability and the presence of virulence and pathogenicity effector genes. A report (Van de Wouw et al. 2010) has demonstrated the determination of frequencies of avirulence alleles in airborne inoculum of L. maculans.
The aim of this study was to screen for the proportions of AvrLm1 and Avrlm6 avirulence genes in L. maculans propagule populations captured in autumnal air samples collected during the 2006/2007 and 2008/2009 seasons from Great Poland, one of the main oilseed rape growing areas of Poland. The relative abundance of AvrLm1 and Avrlm6 avirulence alleles in L. maculans aerial propagule populations sampled in Hertfordshire, UK during the autumns of the 2002/2003 and 2006/2007 oilseed rape seasons was also determined for comparison. These studies were undertaken primarily to assess the minimum detection limits for concentrations of fungal DNA in a sample that would enable the detection of avirulence gene fragments and whether this resolution might be put to practical use. The proportions of avirulence alleles in spores were compared to the ratio of these alleles in leaf explants that were sampled from field-sown plants after natural phoma lesion infection. In addition, L. maculans spore populations were evaluated by light microscopic counts and abundance determination was assessed by real-time, quantitative PCR employing either SYBR green or dual-labelled fluorescent dye chemistry. Azoles, the largest group of sterol 14α-demethylation inhibiting fungicides (Cools et al. 2013), are extensively used in European rapeseed cultivation, particularly in Poland (Jedryczka et al. 2012) and the UK (Fitt et al. 2006), for the field control of phoma stem canker and foliar diseases (Carter et al. 2013). The sterol enzyme (14α-demethylase; ERG11, a cytochrome P450) inhibited by these durable fungicides (imidazoles and triazoles) is pivotal to ergosterol biosynthesis, essential for fungal plasma membrane function and a determinant of fungal growth and viability (Weete et al. 2010). To further evaluate fungal biomass and ascertain the potential for fungal viability and active growth, spores were also screened for the presence of fragments of Lmac erg11 (Griffiths and Howlett 2002) the sterol 14α-demethylase (=CYP51) gene from L. maculans.
Materials and methods
Ascospore sampling
Sampling of the spores of Leptosphaeria spp. was done in the autumns of 2006 and 2008 in Poznan, Poland using a Hirst type seven-day volumetric spore sampler (Burkard Manufacturing Co., Rickmansworth, UK). The trap was located at the grounds of the Institute of Plant Genetics, the Polish Academy of Sciences in Poznan in Poland. The spore trap was surrounded by oilseed rape stubble infected with stem canker which had been collected from the local fields after harvest in July 2006 and 2008. The spore sampler was operated according to the instructions of Lacey and West (2006). Weekly strips of Melinex tape were divided into daily pieces (14 × 48 mm). These were then cut longitudinally, with one piece used for extraction of DNA whilst the other was mounted onto a microscope slide, stained with 0.1 % (w/v) trypan blue in lactophenol and examined with a light microscope under 400× magnification (Zeiss Axiostar, Germany). The numbers of spores present on tapes were re-calculated to daily ascospore numbers per 1 m3 of air. A similar sampling and detection protocol was done in the autumn of the 2002/2003 and of the 2006/2007 oilseed rape seasons at Rothamsted Research in Harpenden, Hertfordshire, UK.
Sampling and isolation of pathogens from plant tissues
The isolates characterized in this study originated from field experiments established in autumn 2008 at four sites in Poland in Great Poland (Cerekwica, Gora, Nowa Wies Ujska, Pawlowice). The field plots were sown with the winter cultivar of oilseed rape: Bosman (Plant Breeding Strzelce) in Cerekwica and Winna Gora, PR46W10 (Pioneer Hi-Breed) in Nowa Wies Ujska and Pawlowice. In total one hundred leaves with visible symptoms of phoma leaf spotting were collected from rapeseed fields located in the region of study. Fragments of leaves with disease symptoms were surface-sterilized with 1 % sodium hypochlorite for 2 min followed by rinsing (3 times, 2 min) in sterile distilled water. Small sections of sterilized leaves were placed on potato dextrose agar (PDA, Sigma) medium supplemented with streptomycin sulphate (0.02 % w/v). Fungal isolates were subcultured until they were free of contaminants. A hyphal tip of each isolate was excised with a sterile needle and a binocular microscope and subcultured on to fresh PDA medium. The taxonomic identification was based on culture morphology and size of ITS products from end-point PCR amplification, as proposed by Williams and Fitt (1999).
Axenic cultures were kept at 22 °C for 12–14 days under alternating 12 h white/12 h near-UV light. Conidia of individual isolates of L. maculans were suspended in sterile double-distilled water; the suspensions were adjusted to a concentration of 107 conidia ml−1 and stored at−20 °C for further analysis.
Cotyledon test
Fifty one isolates obtained from naturally-infected plants of oilseed rape in the region of Great Poland were studied using the cotyledon test, according to the method described by Stachowiak et al. (2006). Three differential cultivars/lines were used to perform the test: Columbus possessing Rlm1 and Rlm3 resistance genes, line 22-1-1 with Rlm3 and Goéland with Rlm9 resistance gene (Balesdent et al. 2005). Moreover, the experiment included cv. Darmor-MX, with the Rlm6 resistance gene, but with unknown Rlm9 locus (Stachowiak et al. 2006). The avirulence genes were characterized based on the reaction of differential cultivars/line to artificial inoculation using the tested isolates. No reaction to inoculation or the formation of small dark ring around the infection site was recorded as a resistant reaction, identifying the isolate as avirulent, whereas formation of light green or pale beige leaf spots with pycnidia was recorded as susceptible reaction, resulting from the virulence of the isolate (Balesdent et al. 2001).
The test was done using 12-day old seedlings, cultivated at controlled environment room with 12 h photoperiod, with 20 °C at the light phase and 16 °C in darkness. Prior to inoculation the plants were watered to maintain high humidity. Each half of the cotyledon was punctured with a needle and then inoculated with a 10 μL droplet of inoculum suspension of 1 × 107 conidia ml−1. Such a droplet was placed on each half of the cotyledon, so that one plant contained four inoculation sites. Each isolate was screened on all four cultivars with eight to ten plants. After inoculation, trays with plants were covered with a plastic propagator lid and kept in darkness for 60 h, following which plants were kept at alternating 12 h photoperiod and the temperature conditions described above. The humidity in the chamber was increased to 70 % by using a humidifier HumiDisk (Carel Deutschland GmbH, Germany). Host response was scored 14 days after inoculation, using a 0–6 scale, as established (Balesdent et al. 2001) and described by Stachowiak et al. (2006).
DNA extraction from Melinex tape and isolates
Daily, Melinex tape sections (48 mm) were cut into sections using sterile forceps and scissors and placed into a 2 ml microfuge tube. To each of these tubes was added one scoop (approx. 150 mg) of sterile acid-washed Ballotini beads (425–600 μm diameter). Fungi were grown in 50-ml Czapek-Dox broth (Sigma) with yeast extract (Oxoid) for 7–14 days at 25 °C. Fresh mycelium was freeze-dried and homogenized. DNA was extracted from the tapes and from individual isolates grown in culture media using a method described by Kaczmarek et al. (2009). The intial homogenate was incubated at 70 °C and partitioned against an equal volume of a 24:1 mixture of chloroform and isoamyl alcohol. DNA was precipitated by chilling incubation at −20 °C with ethanol and sodium acetate (3 M, pH 5). DNA pellets were washed with ice-cold 70 % ethanol, dried, dissolved in 100 μL 1 mM TE (Tris HCl pH 7.5, 1 mM EDTA) buffer and stored at −20 °C.
Quantitative PCR to assess proportions of DNA of each Leptosphaeria maculans in the spore samples
For quantitative PCR, a standard 20-μL reaction contained 5 μL template DNA, 200 nM forward primer, 180 nM reverse primer (Mahuku et al. 1996; Liu et al. 2006), 10 μL SYBR Green JumpStart Taq ReadyMix (Sigma, UK), 0.08 μL ROX internal reference dye (Sigma, UK) and 3.78 μL nuclease-free, sterile water. For this study, duplicate 10-μL assays were routinely done. Thermal cycling parameters for detection were 95 ºC for 2 min followed by 38 cycles of 95 ºC for 15 s, 60 ºC for 30 s and 72 ºC for 45 s. To ascertain the specificity of the procedure, a dissociation (melting) curve was done after the final amplification cycle by heating samples at 95 °C for 15 s, cooling to 60 °C for 1 min and then heating to 95 °C for 15 s. Fluorescence was measured continuously and standard curves were developed from the Ct values of the amplification of standard genomic DNA concentrations (10 ng μL−1 to 1 pg μL−1) from L. maculans mycelial cultures. Leptosphaeria maculans DNA from these tapes was also assessed and quantified by real-time PCR utilizing dual-labelled probes and species-specific primers based on the nucleotides sequences of β-tubulin gene fragments from L. maculans (Kaczmarek et al. 2012). For this chemistry, a 20 μL reaction volume contained 5 μL template DNA, 10 μL qPCR supermix (Invitrogen, UK), 200 nM forward primer and 200 nM reverse primer and 100 nM of the dual-labelled fluorescent probe that was targeted at the β-tubulin gene fragment of L. maculans, 0.08 μL 50× ROX reference dye (Invitrogen, UK) and 2.5 μL autoclaved distilled water. Amplification and product detection were done at 50 °C for 2 min, 1 cycle at 95 °C for 2 min, followed by 45 cycles at 95 °C for 15 s and 60 °C for 45 s, while recording changes in fluorescence at 60 °C during each cycle and using nuclease-free water (Sigma, UK) as the no-template control. A standard curve was generated by plotting the Ct value for each sample of a standard series of genomic DNA concentrations (10 ng uL−1 to 100 fg uL−1) extracted from mycelial cultures of L. maculans. All samples were collected in duplicate.
Quantitative PCR to screen for alleles of the AvrLm1, AvrLm6 and erg11 genes in captured spore samples
Real-time PCR quantitation of AvrLm1, AvrLm6 (Gout et al. 2006; Fudal et al. 2007) and erg11 (Griffiths and Howlett 2002) alleles in L. maculans spore samples was done by running uniplex diagnostic reactions based on the SYBR Green chemistry. Primers AvrLm1F (5′-ATTTCCAGACGTTCCGAGTG-3′), AvrLm1R (5′-ACGTTGTAATGAGCGGAACC-3′), AvrLm6F (5′-CGCACGAAGGCAACTATGTA-3′), AvrLm6R (5′-GCTTTTGGAGTTGGTCATGG-3′), LmacERG11F (5′-AAACCCGCACACATAATCGAAG-3′) and LmacERG11R (5′-TTCTCGTCGTCCTTTTCGTC-3′) were designed and used. Each standard 20-μL reaction contained 5 μL template DNA, 0.6 μL forward primer, 0.54 μL reverse primer, 10 μL SYBR Green JumpStart Taq ReadyMix (Sigma, UK), 0.08 μL ROX internal reference dye (Sigma, UK) and 3.78 μL nuclease-free, sterile water. For this study, duplicate 10-μL assays were routinely done. Thermal cycling parameters for detection were 95 ºC for 2 min followed by 38 cycles of 95 ºC for 15 s, 60 ºC for 30 s and 72 ºC for 45 s. To ascertain the specificity of the procedure, a dissociation (melting) curve was done after the final amplification cycle by heating samples at 95 °C for 15 s, cooling to 60 °C for 1 min and then heating to 95 °C for 15 s. Fluorescence was measured continuously and standard curves were developed from the Ct values of the amplification of standard genomic DNA concentrations (10 ng μL−1 to 1 pg μL−1) from L. maculans mycelial cultures. The primer pairs were tested (Ta = 60 °C) for cross reactivity and specificity by end-point PCR against genomic DNA (10–100 ng uL−1) from mycelial samples of a panel of fungal species comprising L. biglobosa, Pyrenopeziza brassicae, Botrytis cinerea, Sclerotinia sclerotiorum, Alternaria brassicae, A. brassicicola, Verticillium longisporum and Zymoseptoria tritici (=Mycosphaerella graminicola).
PCR to screen for alleles of the AvrLm1 and AvrLm6 genes in mycelial samples
End-point PCR amplifications of avirulent allele AvrLm1 and AvrLm6 of L. maculans were carried out by the methods described above. The PCR primers (Van de Wouw et al. 2010) used were AvrLm1qF (5′-GGGTGTTTACTTCGCCTCAC-3′), AvrLm1qR (5′-ACGTTGTAATGAGCGGAACC-3′), AvrLm6qF (5′- TATTGGACAAAAGCCGAAGG-3′) and AvrLm6qR (5′-GCGAGAAGCAAGTGGAATGT-3′). PCR amplification was done in MJ Research PTC 200 Thermal Cycler (MJ Research Inc., Canada) in the total volume of 10 μL which contained 1 μL extracted DNA solution (10–100 ng uL−1), 0.2 mM dNTPs, 1 μM forward primer 1 μM 1 μM reverse primer, 0.5 U DNA DreamTaq polymerase (Thermo Fisher Scientific Inc. USA), 1 μL PCR buffer (10×, with Mg2+ 20 mM) and sterilized water was added to the total volume of 10 μL. PCR mixture was covered with 20 μL mineral oil. PCR conditions were as follows: 2 min at 94 ºC, followed by 45 cycles of 30 s at 94 ºC, 30 s at 58 ºC and 60 s at 72 ºC, with a final extension of 5 min at 72 ºC. PCR products were separated on a 2.0 % agarose gel, stained with ethidium bromide and visualized with UV light. To preclude negative detections being regarded as arising from PCR inhibition, duplex reactions were done in all cases along with the detection of minisatellite MinLm2452, which yielded two sizes of amplicons; a 103 bp product as the more prominent variant and a second less frequent 82 bp amplicon (Jedryczka et al. 2010a). The primers that were used for this amplification were MinLm2452F (5′-GTACATGGGCGGACAGGC-3′) and MinLm2452R (5′-CATTTACACTGCACACCTGCTCA-3′). The positive control was DNA from L. maculans isolate POL85 containing both AvrLm1 and AvrLm6 alleles. The negative control was water with no target DNA. A second negative control with target DNA template from L. maculans isolate PAW10 containing avrLm1 and avrLm6 alleles was included. This isolate lacked the expected avirulence allele PCR products but had minisatellite MinLm2452.
Statistical analyses
DNA yields were correlated against the corresponding quantities of avirulence alleles of AvrLm1 and AvrLm6, and also of erg11, with extracted spore tapes using Statistica version 9.0 (StatSoft Inc.). Due to non-linearity and non-normality of data for amounts of DNA of L. maculans a Spearman’s rank correlation was used to examine the relationships between the two measurement parameters.
Results
Quantitative PCR analyses enabled the detection, separation and evaluation of avirulence alleles AvrLm1 and AvrLm6 in captured propagule samples thereby permitting a visualization of the patterns of the seasonal fluctuations of these fungal effectors across different years and experimental sites. In Poznan, Poland (Fig. 1) AvrLm6 was predominant while AvrLm1 was detected only at comparatively low levels in both 2006 and 2008. In the UK, in the autumn of 2002, however, AvrLm1 allele predominated over AvrLm6 in Harpenden (Fig. 2a). As shown in Table 1, in that year, the AvrLm1 titre was at least 1.2 times greater than that for AvrLm6. Four years later, in 2006, the AvrLm6 allele became over two times more prominent than AvrLm1 in the qPCR assays of UK samples (Fig. 2c). The correlation coefficient between DNA concentrations of AvrLm1 and AvrLm6 was 0.712 (Table 1), and it ranged from 0.268 for data obtained in Poznan in 2006 to 0.859 for Rothamsted Research 2002. The relatively low correlation for data obtained in Poznan in 2006 appeared to arise from the very low AvrLm1 allele titres, but corresponded nevertheless to the much higher frequency of AvrLm6 DNA in these autumnal Polish air samples as compared to AvrLm1. In Poland AvrLm6 allele was on average 8.5 times more frequent than AvrLm1 in the 2 years studied, and the ratio ranged from 7:1 to 15:1 in favour of AvrLm6. It is noteworthy that the correlation between the qPCR-based detection of avirulence genes and the visual spore count was highly significant (r = 0.954; P > 0.001).
Fig. 1.
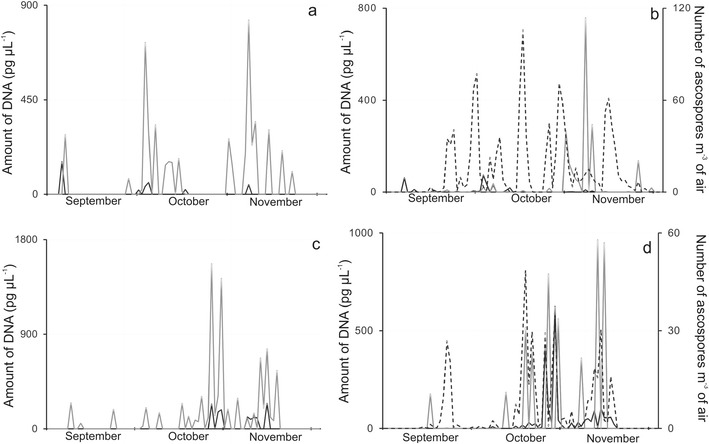
Seasonal fluctuations in DNA quantities or ascospore counts obtained by sampling airborne propagules collected over two autumns (a, b; 2006 and c, d; 2008) periods on Melinex tapes of a Burkard 7-day volumetric air sampler located in Poznan, Poland. Quantitative PCR assays detected avirulence alleles AvrLm1 (a, c; black line), AvrLm6 (a, c; grey line) or with primers based on β-tubulin (b, d; black line) or erg11 (b, d; grey line) fragments in the propagules of Leptosphaeria maculans. Ascospore release patterns (b, d; dotted line) were determined by light microscopic counts of Leptosphaeria-like ascospores per m3 of air
Fig. 2.
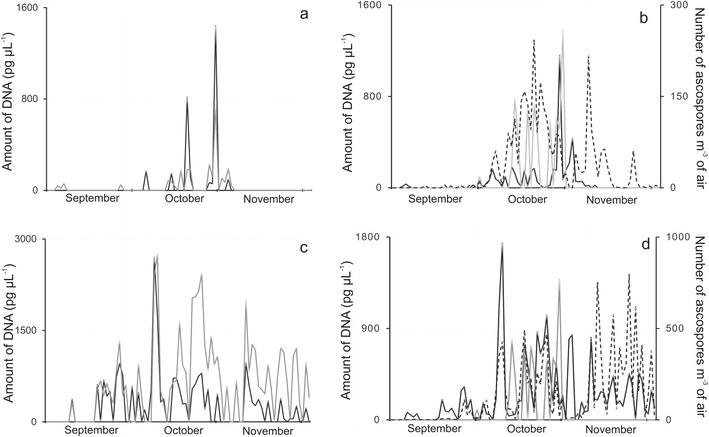
Seasonal fluctuations in DNA quantities or ascospore counts obtained by sampling airborne propagules collected over two autumns (a, b; 2002 and c, d; 2006) periods on Melinex tapes of a Burkard 7-day volumetric air sampler located in Rothamsted Research, Harpenden, UK. Quantitative PCR assays detected avirulence alleles AvrLm1 (a, c; black line), AvrLm6 (a, c; grey line) or with primers based on β-tubulin (b, d; black line) or erg11 (b, d; grey line) fragments in the propagules of Leptosphaeria maculans. Ascospore release patterns (b, d; dotted line) were determined by light microscopic counts of Leptosphaeria-like ascospores per m3 of air
Table 1.
Comparison of scores from light microscopic counts of Leptosphaeria-like ascospores, qPCR detection of DNA from L. maculans spores and of avirulence alleles in air samples from Poland or the UK
| Location x year | Spore count | DNA variant detected in air sample (pg) | Correlation AvrLm6:AvrLm1 | Ratio AvrLm6:AvrLm1 | |
|---|---|---|---|---|---|
| AvrLm1 allele | AvrLm6 allele | ||||
| Rothamsted 2002 | 2942 | 2911.2 | 2399.9 | 0.859 | 0.82 |
| Rothamsted 2006 | 11327 | 22536.5 | 51040.0 | 0.684 | 2.26 |
| Poznan 2006 | 1224 | 338.6 | 4989.6 | 0.268 | 14.74 |
| Poznan 2008 | 421 | 1228.2 | 8306.0 | 0.724 | 6.76 |
| Mean | 3978.5 | 6753.6 | 16683.9 | 0.634 | 6.15 |
In both Poland (Figs. 1a–d) and the UK (Figs. 2a–d) fluctuations in the abundance of these L. maculans–specific avirulence alleles occurred in consonance with the seasonal patterns of ascospore release as judged by light microscopy and were also in agreement with patterns of fluctuations in the quantities of L. maculans DNA determined by qPCR assays. Judging by the titres of erg11 from L. maculans, ergosterol biosynthetic activity was more easily measurable in airborne propagules from Poznan in 2008 than in 2006 (Figs. 1b and d). As shown in Fig. 2b and d, however, this activity was more easily quantified in captured air spora from the UK in both 2002 and 2006 and the fluctuation patterns had maximal prominence in the October of both years.
For avirulence alleles, a similar data set (Table 2) was obtained in planta, from bioassays done on infected cotyledonary leaves of Brassica napus employing molecular detection of either AvrLm1 or AvrLm6 by end-point PCR with allele-specific primers (Fig. 3). Out of the 51 isolates of L. maculans that were sampled randomly from fields in Great Poland (within which the Burkard volumetric spore trap was located) only one isolate, obtained from an oilseed rape field in Pawlowice (Fig. 3a, sample 11), was avirulent on the oilseed rape differential cultivar (cv. Columbus) with the corresponding Rlm1 resistance gene. All the remainder were virulent on this cultivar, i.e. possessed the avrlm1 allele. In contrast, all tested isolates possessed AvrLm6 avirulence allele (Fig. 3b).
Table 2.
Frequency (%) of avirulence alleles in populations of Leptosphaeria maculans isolates from infected cotyledonary leaves of oilseed rape from Great Poland region in central-west Poland
| No. | Location | Cotyledon test | Molecular detection | ||||
|---|---|---|---|---|---|---|---|
| No. of isolates | AvrLm1 allele | AvrLm6 allele | AvrLm9 allele | AvrLm1 allele | AvrLm6 allele | ||
| 1 | Cerekwica | 11 | 0 | 100 | 0 | 0 | 100 |
| 2 | Gora | 4 | 0 | 100 | 0 | 0 | 100 |
| 3 | Nowa Wies Ujska | 20 | 0 | 100 | 0 | 0 | 100 |
| 4 | Pawlowice | 16 | 6.25 | 100 | 0 | 6.25 | 100 |
| Mean | 51 | 1.96 | 100 | 0 | 1.96 | 100 | |
Fig. 3.
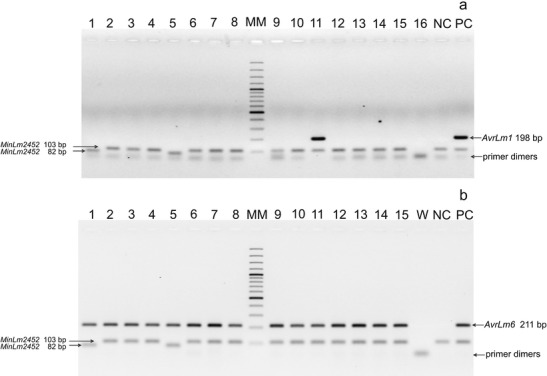
Detection of avirulence alleles AvrLm1 (a) and AvrLm6 (b) in isolates (lanes 1 to 15) of Leptosphaeria maculans obtained from infected plants of oilseed rape grown in Pawlowice, central-west Poland. W – water, NC – negative control (DNA of the isolate PAW10 containing avrLm1 and avrLm6), PC – positive control (DNA of the isolate POL85 containing AvrLm1 and AvrLm6)
Discussion
The results of this study demonstrate a successful derivation of biological information, beyond earlier description and evaluation of numerical abundance, from season-long screening and monitoring of air borne fungal pathogen spores. Quantitative PCR techniques were employed for the detection of the L. maculans avirulence alleles AvrLm1 and AvrLm6 in DNA extracted from airborne particles on tapes from spore traps that were operated in Poland and the UK within oilseed rape fields over the autumn of 2002, 2006 and 2008. The patterns of Leptosphaeria-like ascospore release were also assessed on stained Melinex tapes by light microscopy and fluctuations in pre-harvest aerial biomass of L. maculans inoculum were determined by real-time qPCR. In this study, the primers that were designed for AvrLm1and AvrLm6 detection recognized, respectively, a 301 bp (nucleotides 109–409) and a 307 bp (nucleotides 353–659) fragment of the open reading frames of these avirulence effectors. Primers recognizing a 198 bp (nucleotides 212–409, for AvrLm1) and a 211 bp (nucleotides 328–479, for AvrLm6) gene fragment were used instead by Van de Wouw et al. (2010) to determine and compare by qPCR the proportions of AvrLm1 and AvrLm6 alleles in airborne ascospores of L. maculans that were captured from Rothamsted Research with their ratio on infected plants sampled from farmer’s fields of oilseed rape in the same area. The latter primers found application in the current study in the end-point PCR screening of infected oilseed rape plants for AvrLm1 and AvrLm6 alleles.
The correlations between the ascospore counts and concentrations of L. maculans DNA in captured air spora in this study were highly significant, suggesting the possibility of replacing the laborious and time consuming visual assessments done by staining and light microscopic counts with molecular biological detection. The latter approach furthermore provides opportunities for generating additional informative data. Evidence for ergosterol biosynthetic activity (as judged by fluctuations in the titre of fragments of erg11 from L. maculans) was conceived in the current study as a test for spore viability potential that could also assist in future detection of and screens for mutations affecting sensitivity to azole fungicides. Evidence is currently lacking in L. maculans (Huang et al. 2011) for mutations in this Cyp51 gene encoding the azole target sterol 14α-demethylase.
Avirulence alleles of L. maculans code for small secreted protein effectors which play a putative role in modulating innate immunity in oilseed rape, the major brassica host plants (Rouxel and Balesdent 2010). Genes coding for AvrLm1, AvrLm4-7 and AvrLm6 have been cloned (Gout et al. 2006; Fudal et al. 2007). When constitutively or inducibly expressed at the onset of infection, recognition in the host plant by Rlm1 and Rlm6 of these products of AvrLm1 and AvrLm6 results in phoma disease resistance. Continual and widespread cultivation of oilseed rape cultivars with monogenic Rlm1-mediated resistance precipitated in France and Australia the rapid evolution of avrLm1-based virulence (through deletion mutation events within the AvrLm1 locus; Gout et al. 2007) in L. maculans populations, epiphytotic phoma stem canker disease spread and massive oilseed rape yield losses. Consequently, oilseed rape breeders and farmers in continental Europe and Australia switched to the cultivation of oilseed rape cultivars with multigenic and more durable resistance. In the current study, the avirulence gene AvrLm1 was rarely detected in Polish air samples and only sporadically in plants, apparently subsequent to the ineffectiveness and impractical usefulness of Rlm1 as resistance gene source. In contrast, AvrLm6 allele was detected in most airborne L. maculans propagules and phoma-infected field-sown oilseed rape plants from Poland implying the sustained effectiveness of Rlm6-based stem canker resistance. This was evinced by a minimum of 8:1 average ratio in the frequencies of AvrLm6:AvrLm1 alleles across the autumn of 2006 and 2008. In the UK, however, the frequency ratios of AvrLm6:AvrLm1 alleles was less polarized, and even though AvrLm6 was more frequently detected, the proportion between these avirulence alleles was about 2:1. It may be concluded that in the UK, the AvrLm1 allele is present in air samples approximately four times more often than in Poland. Annually, a varied number of genetically heterogeneous oilseed rape cultivars and lines are sown to field experimental plots at Rothamsted Research; it is reasonable that DNA from L. maculans propagules captured by traps operated at this UK site around the debris of these cultivars would reflect the greater diversity obtained in the current study. In general, data obtained in Poland established that AvrLm6 allele was present in all or most DNA samples obtained from ascospores of L. maculans, whereas AvrLm1 was nearly non existent. It is clear from our study that no spores with avrLm6 in the air resulted from no plants infected with isolates of L. maculans possessing avrLm6 allele, what in turn resulted in no inoculum with avrlm6.
Knowledge on variation of avirulence genes in L. maculans populations in Poland dates back to 2000–2001, when a population of 92 isolates originating from different parts of the country was studied using a cotyledon test, followed by PCR with Vir1 marker (Jedryczka 2007). The population in those years was found to be monomorphic with respect to AvrLm7 (avirulent genotype) and avrLm2, avrLm3 and avrLm9 (virulent genotypes) but polymorphic for AvrLm1 (58.7 %) and AvrLm4 (3.3 %). The interaction phenotype agreed with molecular study (90.2 %). The most recent Europe-wide monitoring, concerning the occurrence of avirulence genes was done by Stachowiak et al. (2006) at six experimental sites across the main oilseed rape growing regions embracing Poland (two sites), the UK (two), Germany (one) and Sweden (one). In total, 603 isolates were collected during the autumn of 2002 (Germany and the UK) and 2003 (Poland and Sweden). No isolates had the virulence allele avrLm6 and three virulence alleles (avrLm2, avrLm3 and avrLm9) were present in all isolates. The isolates were polymorphic for AvrLm1, AvrLm4, AvrLm5 and AvrLm7 alleles, with virulence alleles at AvrLm1 and AvrLm4 loci and avirulence alleles at AvrLm7 and AvrLm5 loci predominant in populations. Virulent avrLm7 isolates were found at only one site in Sweden. Approximately 90 % of all isolates belonged to one of two races, Av5–6–7 (77 % of isolates) or Av6–7 (12 %). The results were comparable to those of a similar survey done in France in autumn 2000 and 2001 (Balesdent et al. 2005) which reported that resistant genes Rlm6 and Rlm7 were still effective against L. maculans in oilseed rape in Europe.
The current study on avirulence alleles showed that the situation in the Poznan area of Poland had not deviated in the 3–5 years from a previous monitoring work that was done in 2003, and that AvrLm6 is still present in isolates of L. maculans, whereas AvrLm1 is nearly absent in the pathogen’s population. Moreover the situation in the UK with AvrLm1 avirulence gene was similar to that reported by Stachowiak et al. (2006), with nearly 20 % of the isolates avirulent on cultivars possessing Rlm1 resistance gene. In the current work we have also established that AvrLm1 isolates are on average four times more frequent in the UK, in comparison to Poland. This screen suggested, however, a higher proportion of isolates virulent to oilseed rape lines with Rlm6 resistance gene than found in Rothamsted in 2001 by Stachowiak et al. (2006). A large survey of L. maculans isolates from Australia, Canada and South America has also shown that most isolates possessed the AvrLm6 avirulence allele (Dilmaghani et al. 2009). The annual, seasonal and regional variations might originate in selective pressures dictated by cultivar choice as well as other unforeseen reasons, such as the composition of brassicaeous weeds, with different resistance genes from those found in crucifers. The study suggests that Rlm6 resistance gene could provide effective resistance to L. maculans in Poland. However, in the UK, the resistance appears to be under threat. The deployment of Rlm6 resistance cultivars of oilseed rape should when effected be carefully managed by supplementation with qualitative resistance that will prevent rapid stem canker resistance erosion that was witnessed with Rlm1 in France (Rouxel et al. 2003) and in Australian B. napus cultivar Surpass 400 (Li et al. 2003) with ‘sylvestris-derived’ resistance.
The results obtained in the current study confirm quantitative real-time PCR as an accurate and reliable technique due to its enormous sensitivity, possibility of detection of reaction progress, speed of analyses and precision permitting the examination of pico- and even femto-gram differences between spore and effector samples. Additional use of reference genes, such as actin in plant stress-related studies makes it even more reliable (Kozera and Rapacz 2013). The current study further demonstrates that methods employed for screening avirulence genes in air samples (Van de Wouw et al. 2010) could be successfully employed for routine larger-scale and epidemiological studies of plant pathogen populations (Heard and West 2014; West 2014), yielding deeper insights into the avirulence allele compositions and race structures of wind- and rain-dispersed pathogens in particular. At the limits of the powers of the application of qPCR, opportunities exist with other approaches (such as high throughput pyrosequencing; Van de Wouw and Howlett 2012) for molecular biological analyses of other avirulence and effector genes in L. maculans propagules captured on spore tapes. Integrated plant protection guidelines, obligatory for all European Union Member States since early 2014 (Directive 2009/128/EC), stipulate the restriction of chemical sprays of agricultural crops to real needs in specific seasons and locations, based on monitoring results and threshold levels. Taken together, molecular biological detection of gene variants, when combined with various early detection techniques such as those based of aerobiology, are a powerful tool that can be employed for low-, as well as, non-chemical, early and regular interventions in integrated pest management.
Acknowledgments
This work was supported by research projects from Polish Ministry of Science and Higher Education (NN310 086436) and from the National Research Centre of Poland (N N310 298439). Rothamsted Research receives funding from the UK Biotechnology and Biological Sciences Research Council. The authors acknowledge DuPont Poland for funding the SPEC project and the UK Department for Environment, Food and Rural Affairs OREGIN Programme for financial support with analytical work. Funding was also received from the British-Polish Young Scientists Programme of the British Council (WAR/342/88), AHDB-HGCA, The Arable Group, The Chadacre Trust and the Morley Agricultural Foundation. We thank Marie-Helene Balesdent (INRA-Thiverval Grignon, France) for the gift of seeds of differential oilseed rape cultivars and lines.
Footnotes
Joanna Kaczmarek and Akinwunmi O. Latunde-Dada are to be regarded as joint first authors of this paper
References
- Ansan-Melayah D, Balesdent MH, Buée M, Rouxel T. Genetic characterization of AvrLm1, the first avirulence gene of Leptosphaeria maculans. Phytopathology. 1995;85:1525–1529. doi: 10.1094/Phyto-85-1525. [DOI] [Google Scholar]
- Ansan-Melayah D, Balesdent MH, Delourme R, Pilet ML, Renard M, Tanguy X, Rouxel T. Genes for race-specific resistance against blackleg disease in Brassica napus L. Plant Breed. 1998;117:373–378. doi: 10.1111/j.1439-0523.1998.tb01956.x. [DOI] [Google Scholar]
- Atkins SL, Atkins SD, Latunde-Dada AO, Stonard JF, West JS. Detection and quantification of airborne ascospores of Sclerotinia sclerotiorum by quantitative-PCR. Int Organ Biol Integr Control (IOBC) Bull. 2013;92:173–178. [Google Scholar]
- Balesdent MH, Attard A, Ansan-Melayah D, Delourme M, Renard M. Genetic control and host range of avirulence towards Brassica napus cultivars Quinta and Jet neuf in Leptosphaeria maculans. Phytopathology. 2001;91:70–76. doi: 10.1094/PHYTO.2001.91.1.70. [DOI] [PubMed] [Google Scholar]
- Balesdent MH, Attard A, Kuhn AL, Rouxel T. New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans. Phytopathology. 2002;92:1122–1133. doi: 10.1094/PHYTO.2002.92.10.1122. [DOI] [PubMed] [Google Scholar]
- Balesdent MH, Barbetti MJ, Li H, Sivasithamparam K, Gout L, Rouxel T. Analysis of Leptosphaeria maculans race structure in a world-wide collection of isolates. Phytopathology. 2005;85:1061–1071. doi: 10.1094/PHYTO-95-1061. [DOI] [PubMed] [Google Scholar]
- Balesdent MH, Louvard K, Pinochet X, Rouxel T. A large-scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France. Eur J Plant Pathol. 2006;114:53–65. doi: 10.1007/s10658-005-2104-0. [DOI] [Google Scholar]
- Balesdent MH, Fudal I, Ollivier B, Bally P, Granaubert J, Chevre A, Leflon M, Rouxel T. The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa. New Phytol. 2013;198:889–898. doi: 10.1111/nph.12178. [DOI] [PubMed] [Google Scholar]
- Carter HE, Fraaije BA, West JS, Kelly SL, Mehl A, Shaw MW, Cools HJ. Alterations in the predicted regulatory and coding regions of the sterol 14α-demethylase gene (CYP51) confer decreased azole sensitivity in the oilseed rape pathogen Pyrenopeziza brassicae. Mol Plant Pathol. 2013 doi: 10.1111/mpp.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools HJ, Hawkins NJ, Fraaije BA. Constraints on the evolution of azole resistance in plant pathogenic fungi. Plant Pathol. 2013;62:36–42. doi: 10.1111/ppa.12128. [DOI] [Google Scholar]
- Cozijnsen AJ, Popa KM, Purwantara A, Rolls BD, Howlett BJ. Genomic analysis of the plant pathogenic ascomycete Leptosphaeria maculans; mapping mating type and host specificity loci. Mol Plant Pathol. 2000;1:293–302. doi: 10.1046/j.1364-3703.2000.00033.x. [DOI] [PubMed] [Google Scholar]
- Crouch JH, Lewis BG, Mithen RF. The effect of a genome substitution on the resistance of Brassica napus to infection by Leptosphaeria maculans. Plant Breed. 1994;112:265–278. doi: 10.1111/j.1439-0523.1994.tb00684.x. [DOI] [Google Scholar]
- Delourme R, Chevre AM, Brun H, Rouxel T, Balesdent MH, Dias JS, Salisbury P, Renard M, Rimmer SR. Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus) Eur J Plant Pathol. 2006;114:41–52. doi: 10.1007/s10658-005-2108-9. [DOI] [Google Scholar]
- Devi TK, Singh NI. Aerobiology and epidemiology of false smut disease of rice by Ustilagnoidea virens (Syn. Claviceps oryzae sativae) in thoubal district. Manipur. J Mycopathol Res. 2007;45:107–108. [Google Scholar]
- Dilmaghani A, Balesdent MH, Didier JP, Wu C, Davey J, Barbetti MJ, Li H, Moreno-Rico O, Phillips D, Despeghel JP, Vincenot L, Gout L, Rouxel T. The Leptosphaeria maculans-Leptosphaeria biglobosa species complex in the American continent. Plant Pathol. 2009;58:1044–1058. doi: 10.1111/j.1365-3059.2009.02149.x. [DOI] [Google Scholar]
- Eckert M, Maguire K, Urban M, Foster S, Fitt BDL, Lucas J, Hammond-Kosack K. Agrobacterium tumefaciens-mediated transformation of Leptosphaeria spp. and Oculimacula spp. with the reef coral reef DsRed and the jellyfish gene. FEMS Microbiol Lett. 2005;253:67–74. doi: 10.1016/j.femsle.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Fitt BDL, Brun H, Barbetti MJ, Rimmer RS. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus) Eur J Plant Pathol. 2006;114:3–15. doi: 10.1007/s10658-005-2233-5. [DOI] [Google Scholar]
- Fudal I, Ross S, Gout L, Blaise F, Kuhn ML, Eckert MR, Cattolico L, Bernard-Samain S, Balesdent MH, Rouxel T. Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map-based cloning of AvrLm6. Mol Plant Microbe Int. 2007;20:457–470. doi: 10.1094/MPMI-20-4-0459. [DOI] [PubMed] [Google Scholar]
- Gladders P, et al. Pest and disease management system for supporting winter oilseed rape decisions (PASSWORD) – validation phase. HGCA Proj Rep Number. 2006;390:1–163. [Google Scholar]
- Gout L, Fudal I, Kuhn ML, Blaise F, Eckert M, Cattolico L, Balesdent MH, Rouxel T. Lost in the middle of nowhere: the AvrLm1 avirulence gene of the dothideomycete Leptosphaeria maculans. Mol Microbiol. 2006;60:67–80. doi: 10.1111/j.1365-2958.2006.05076.x. [DOI] [PubMed] [Google Scholar]
- Gout L, Kuhn ML, Vincenot L, Berbard-Samain S, Cattolico L, Berbetti M, Moreno-Rico O, Balesdent MH, Rouxel T. Genomic structure impacts molecular evolution at the AvrLm1 avirulence locus of the plant pathogen Leptosphaeria maculans. Environ Microbiol. 2007;9:2978–2992. doi: 10.1111/j.1462-2920.2007.01408.x. [DOI] [PubMed] [Google Scholar]
- Griffiths KM, Howlett BJ. Transcription of sterol ∆5,6-desaturase and sterol 14α-demethylase is induced in the plant pathogenic ascomycete, Leptosphaeria maculans, during treatment with a triazole fungicide. FEMS Microbiol Lett. 2002;217:81–87. doi: 10.1111/j.1574-6968.2002.tb11459.x. [DOI] [PubMed] [Google Scholar]
- Heard S, West JS. New developments in identification and quantification of airborne inoculum chapter. In: Gullino ML, Bonants P, editors. Plant pathology in the 21st century: detection and diagnosis of plant pathogens. Dordrecht: Springer; 2014. [Google Scholar]
- Huang YJ, Hood JR, Eckert MR, Stonard JF, Cools HJ, King GJ, Rossall S, Ashworth M, Fitt BDL. Effects of fungicide on growth of Leptosphaeria maculans and L. biglobosa in relation to development of phoma stem canker on oilseed rape (Brassica napus) Plant Pathol. 2011;60:607–620. doi: 10.1111/j.1365-3059.2010.02416.x. [DOI] [Google Scholar]
- Isard SA, Russo JM, Ariatti A. The integrated aerobiology modelling system applied to the spread of soybean rust into the Ohio river valley during September 2006. Aerobiologia. 2007;23:271–282. doi: 10.1007/s10453-007-9073-z. [DOI] [Google Scholar]
- Jedryczka M. Epidemiology and damage caused by stem canker of oilseed rape in Poland. Phytopathol Pol. 2007;45:73–75. doi: 10.1146/annurev.phyto.45.062806.094401. [DOI] [Google Scholar]
- Jedryczka M, Matysiak R, Graham K. Lepto-net and SPEC – new projects to control the stem canker of oilseed rape in Poland. Int Organ Biol Control Bull. 2004;27:125–130. [Google Scholar]
- Jedryczka M, Irzykowski W, Jajor E, Korbas M. Polymorphism of ten new minisatellite markers in subpopulations of phytopathogenic fungus Leptosphaeria maculans differing with metconazole treatment. J Plant Prot Res. 2010;50:103–109. doi: 10.2478/v10045-010-0018-0. [DOI] [Google Scholar]
- Jedryczka M, Plachka E, Kaczmarek J, Poslusna J, Latunde-Dada AO, Maczynska A. Monitoring Leptosphaeria maculans and L. biglobosa ascospores around East Sudethian mountains – a joint initiative of Poland and the Czech Republic. Oilseed Crops. 2010;31:49–66. [Google Scholar]
- Jedryczka M, Brachaczek A, Kaczmarek J, Dawidziuk A, Kasprzyk I, Maczynska A, Karolewski Z, Podlesna A, Sulborska A. System for forecasting disease epidemics (SPEC) – decision support system in polish agriculture, based on aerobiology. Alergol Immunol. 2012;9:89–91. [Google Scholar]
- Kaczmarek J, Jedryczka M. Characterization of two coexisting pathogen populations of Leptosphaeria spp., the cause of stem canker of brassicas. Acta Agrobot. 2011;64:3–14. doi: 10.5586/aa.2011.012. [DOI] [Google Scholar]
- Kaczmarek J, Jedryczka M, Fitt BDL, Lucas JA, Latunde-Dada AO. Analyses of air samples for ascospores of Leptosphaeria maculans and L. biglobosa with light microscopic and molecular techniques. J Appl Genet. 2009;50:411–419. doi: 10.1007/BF03195702. [DOI] [PubMed] [Google Scholar]
- Kaczmarek J, Jedryczka M, Cools HJ, Fitt BDL, Lucas JA, Latunde-Dada AO. Quantitative PCR analysis of abundance of airborne propagules of Leptosphaeria species in air samples from different regions of Poland. Aerobiologia. 2012;28:199–212. doi: 10.1007/s10453-011-9228-9. [DOI] [Google Scholar]
- Karolewski Z, Kaczmarek J, Jedryczka M, Cools HJ, Fraaije BA, Lucas JA, Latunde-Dada AO. Detection and quantification of airborne inoculum of Pyrenopeziza brassicae in polish and UK winter oilseed rape crops by real-time PCR assays. Grana. 2012;51:270–279. doi: 10.1080/00173134.2011.653401. [DOI] [Google Scholar]
- Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutcher R, Balesdent MH, Rimmer SR, Rouxel T, Chevre AM, Delourme R, Brun H. Frequency of avirulence genes in Leptosphaeria maculans in western Canada. Can J Plant Pathol. 2010;32:77–85. doi: 10.1080/07060661003594109. [DOI] [Google Scholar]
- Lacey M, West JS (2006) The air spora: a manual for catching and identifying airborne biological particles. Springer, Heidelberg p 156
- Larkan NJ, Lydiate DJ, Parkin IAP, Nelson MN, Epp DJ, Cowling WA, Rimmer SR, Borhan MH. The Brassica napus blackleg resistance LepR3 enclodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013;197:595–607. doi: 10.1111/nph.12043. [DOI] [PubMed] [Google Scholar]
- Leclair S, Ansan-Melayah D, Rouxel T, Balesdent MH. Meiotic behavior of the minichromosome in the pathogenic ascomycete Leptosphaeria maculans. Curr Genet. 1996;30:541–548. doi: 10.1007/s002940050167. [DOI] [PubMed] [Google Scholar]
- Li CX, Cowling WA. Identification of a single dominant allele for resistance to blackleg in Brassica napus ‘Surpass 400’. Plant Breed. 2003;122:485–488. doi: 10.1111/j.1439-0523.2003.00863.x. [DOI] [Google Scholar]
- Li H, Sivasithamparam K, Barbetti MJ (2003) Breakdown of a B. rapa ssp. sylvestris single dominant resistance gene in B. napus by L. maculans field isolates. Plant Dis 87:752 [DOI] [PubMed]
- Liu SY, Liu Z, Fitt BDL, Evans N, Foster SJ, Huang YJ, Latunde-Dada AO, Lucas JA. Resistance to Leptosphaeria maculans (phoma stem canker) in Brassica napus (oilseed rape) induced by L. biglobosa and chemical defence activators in field and controlled environments. Plant Pathol. 2006;55:401–412. doi: 10.1111/j.1365-3059.2006.01354.x. [DOI] [Google Scholar]
- Mahuku GS, Hall R, Goodwin PH. Co-infection and induction of systemic acquired resistance by weakly and highly virulent isolates of Leptosphaeria maculans in oilseed rape. Physiol Mol Plant Pathol. 1996;49:61–72. doi: 10.1006/pmpp.1996.0039. [DOI] [Google Scholar]
- McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol. 2002;40:349. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- Rimmer SR. Resistance genes to Leptosphaeria maculans in Brassica napus. Can J Plant Pathol. 2006;28:288–297. doi: 10.1080/07060660609507386. [DOI] [Google Scholar]
- Rouxel T, Balesdent MH. The stem canker (blackleg) fungus, Leptosphaeria maculans, enters the genomic era. Mol Plant Pathol. 2005;6:225–241. doi: 10.1111/j.1364-3703.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- Rouxel T, Balesdent MH (2010) Avirulence genes. In: Encyclopedia of life sciences (ELS). Wiley, Ltd: Chichester, UK. doi:10.1002/9780470015902.a0021267
- Rouxel T, Penaud A, Pinochet X, Brun H, Gout L, Delourme R, Schmit J, Balesdent MH. A ten-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene in oilseed rape. Eur J Plant Pathol. 2003;109:871–881. doi: 10.1023/A:1026189225466. [DOI] [Google Scholar]
- Rouxel T, Grandaubert J, Hane JK, Hoede C, van de Wouw AP, et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nat Commun. 2011;2:202. doi: 10.1038/ncomms1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam MU, Khangura RK, Diggle AJ, Barbetti MJ. Blackleg sporacle: a model for predicting onset of pseudothecia maturity and seasonal ascospore showers in relation to blackleg of canola. Phytopathology. 2003;93:1073–1081. doi: 10.1094/PHYTO.2003.93.9.1073. [DOI] [PubMed] [Google Scholar]
- Sexton A, Howlett BJ. Green fluorescent protein as a reporter in the Brassica-Leptosphaeria maculans interaction. Physiol Mol Plant Pathol. 2001;58:13–21. doi: 10.1006/pmpp.2000.0307. [DOI] [Google Scholar]
- Soyer JL, El Ghalid M, Glasser N, Ollivier B, Linglin J, Grandaubert J, Balesdent MH, Connolly LR, Freitag M, Rouxel T, Fudal I. Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans. PLoS Genet. 2014;10(3):e1004227. doi: 10.1371/journal.pgen.1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak A, Olechnowicz J, Jedryczka M, Rouxel T, Balesdent MH, Happstadius I, Gladders P, Latunde-Dada A, Evans N. Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe. Eur J Plant Pathol. 2006;114:67–75. doi: 10.1007/s10658-005-2931-z. [DOI] [Google Scholar]
- Van de Wouw AP, Howlett BJ. Estimating frequencies of virulent isolates in field populations of a plant pathogenic fungus, Leptosphaeria maculans, using high-throughput pyrosequencing. J Appl Microbiol. 2012;113:1145–1153. doi: 10.1111/j.1365-2672.2012.05413.x. [DOI] [PubMed] [Google Scholar]
- Van de Wouw AP, Marcroft SJ, Barbetti MJ, Li H, Salisbury PA, Gout L, Rouxel T, Howlett BJ, Balesdent MH. Dual control of avirulence in Leptosphaeria maculans towards a Brassica napus cultivar with ‘sylvestris-derived’ resistance suggests involvement of two resistance genes. Plant Pathol. 2009;58:305–313. doi: 10.1111/j.1365-3059.2008.01982.x. [DOI] [Google Scholar]
- Van de Wouw AP, Stonard JF, Howlett BJ, West JS, Fitt BDL, Atkins SD. Determining frequencies of avirulent alleles in airborne Leptosphaeria maculans inoculum using quantitative PCR. Plant Pathol. 2010;59:809–818. doi: 10.1111/j.1365-3059.2010.02311.x. [DOI] [Google Scholar]
- Weete JD, Abril M, Blackwell M. Phylogenetic distribution of fungal sterols. PLoS ONE. 2010;5:e10899. doi: 10.1371/journal.pone.0010899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JS (2014) Plant pathogen dispersal. In: eLS Wiley, Ltd; Chichester, UK. doi:10.1002/9780470015902.a0021272
- Williams RH, Fitt BDL. Differentiating A and B groups of Leptosphaeria maculans, causal agent of stem canker (blackleg) of oilseed rape. Plant Pathol. 1999;48:161–175. doi: 10.1046/j.1365-3059.1999.00333.x. [DOI] [Google Scholar]
- Yu F, Gugel RK, Kutcher HR, Peng G, Rimmer SR. Identification and mapping of a novel blackleg resistance locus LepR4 in the progenies from Brassica napus x B. rapa subsp. sylvestris. Theor Appl Genet. 2013;126:307–315. doi: 10.1007/s00122-012-1919-2. [DOI] [PubMed] [Google Scholar]


