Abstract
OBJECTIVE
To measure the variability of T1ρ relaxation times using CubeQuant, T2 relaxation times using quantitative double echo in steady state (DESS), and normalized sodium signals using 3D cones sodium MRI of knee cartilage in vivo at 3T.
DESIGN
Eight healthy subjects were scanned at 3T at baseline, one day, five months, and one year. Ten regions of interest (ROIs) of knee cartilage were segmented in the medial and lateral compartments of each subject’s knee. T1ρ and T2 relaxation times and normalized sodium signals were measured and the root-mean-square coefficient of variation (CVRMS) was calculated. Intra-subject variability was measured over short, moderate and long-term, as well as intra-observer and inter-observer variability.
RESULTS
The average intra-subject CVRMS measurements over short, moderate, and long-term time periods were 4.6%, 6.1%, and 6.0% for the T1ρ measurements, 6.4%, 9.3%, and 10.7% for the T2 measurements and 11.3%, 11.6%, and 12.9% for the sodium measurements, respectively. The average CVRMS measurements for intra-observer and inter-observer segmentation were 3.8% and 5.7% for the T1ρ measurements, 4.7% and 6.7% for the T2 measurements, and 8.1% and 11.4% for the sodium measurements, respectively.
CONCLUSIONS
These CVRMS measurements are substantially lower than previously measured changes expected in patients with advanced osteoarthritis compared to healthy volunteers, suggesting that CubeQuant T1ρ, quantitative DESS T2 and 3D cones sodium measurements are sufficiently sensitive for in vivo cartilage studies.
Keywords: variability, MRI, knee, cartilage, repeatability, quantitative
INTRODUCTION
Osteoarthritis (OA) causes disability for 10% of people over 60 and costs $60 billion each year (1). OA includes morphological, biochemical and structural changes to the articular cartilage (2). Since the degeneration of articular cartilage is largely irreversible, early detection of OA is critical (3). Quantitative magnetic resonance imaging (MRI) is a promising, noninvasive method for early detection of OA because it can assess joint physiology before morphological changes occur (3–5).
T1ρ and T2 relaxation times and sodium (Na+) MRI are MRI parameters that are correlated with cartilage macromolecules. The two main macromolecules in cartilage are proteoglycans (PG) and collagen. Early stages of OA are associated with a loss of proteoglycans and changes in collagen structure (6). The spin-lattice relaxation time in the rotating frame, T1ρ, is strongly inversely correlated with PG content (R2 = 0.85, p < 0.001) (7). The spin-spin relaxation time, T2, may be correlated with collagen structure (8) and water content (R2 = 0.89, p < 0.001) (9). Sodium is the positive ion correlated with glycosaminoglycan (GAG), which is a negatively charged side chain of proteoglycans (10–13). Sodium signal loss has shown a strong linear correlation with PG loss (R2 = 0.85, p < 0.01) (14).
Changes in T1ρ, T2, and sodium caused by advanced OA can be on the order of 5%–10% (15,16). Previous studies measured increases in T1ρ relaxation times of 14.2% in vivo (17) and 30%–120% ex vivo (15) in osteoarthritic cartilage, suggesting a loss of proteoglycans. Similarly, prior work measured increases in T2 relaxation times of 9.4% (16) to 14.1% in vivo (17) and 5%–50% ex vivo (15). Previous work has measured decreases in sodium signal intensity of 20%–40% in vivo. Since the changes caused by OA can be small, on the order of 5%–10% (15,16), it is important to estimate the variability in quantitative MR parameters..
Many groups have studied the variability of T1ρ, T2, and sodium measurements using a variety of different sequences. For example, three studies recently analyzed T1ρ relaxation times and reported either the coefficients of variation (CV), or the root-mean-square (RMS) CVs (CVRMS), which ranged from CVRMS values of 7%–19% (18), regional CVs of 1.7%–8.7% (19), and CVRMS values of 2.2%–5.9% to 4.8%–8.8% (20). Several groups analyzed T2 measurements and reported CVRMS values of 4%–14% (18), CVRMS average values of 6%–29% (21), and CVRMS values 4.0%–6.2% to 4.2%–8.5% (20). Several studies calculated sodium measurements and found CVRMS values of 7.5%–13.6% (22), within-subjects CVs of 2.2%–13.8% (23), and variability that ranged from 3.0%–8.6% (24). The CVs from these previous studies are typically less than 20% and may vary due to many factors such as the study design, subject population, or sequences.
This paper investigates three novel sequences that confer certain advantages. CubeQuant T1ρ is a novel, SNR-efficient, five minute sequence that can determine 3D T1ρ relaxation times (25). Many groups measure T1ρ relaxation times using 3D T1ρ gradient recalled echo (GRE) sequences (26) or modified GRE sequences, such as a T1ρ-prepared balanced-GRE (27), a spin-lock-based 3D GRE sequence (28), or the Magnetization-Prepared Angle-modulated Partitioned-k-space Spoiled gradient recalled (SPGR) Sequence (MAPSS) (19). Quantitative double echo in steady state (DESS) is a 10 minute steady-state free precession (SSFP) sequence that can provide T2 relaxation times (29,30). A previous in vivo study showed strong correlation between DESS T2 measurement and SE (Pearson product-moment coefficient 0.989; mean absolute difference 1.8%) (30); this version of DESS uses 4 echoes and three-dimensional solution space to fit T2 relaxation times. The Osteoarthritis Initiative (OAI) uses DESS with water excitation (DESSwe); but the data cannot be used to calculate T2 relaxation times and diffusivity (31). In addition to providing morphological images that distinguish fluid and cartilage, quantitative DESS can also calculate T2 relaxation times without diffusion effects and has the potential to calculate apparent diffusion coefficient (ADC) values (30,32). Other groups have measured T2 relaxation times using a 2D multi-section multi-echo (MSME) sequence (18), multi-echo (ME) turbo spin-echo (TSE) sequence (21), or MAPSS (20). We measure sodium signal intensity using a 3D cones k-space trajectory with a short echo time (TE). The 3D cones sequence requires fewer readouts than a Cartesian sequence does because each readout covers a significant portion of k-space rapidly (33,34). Other methods often used for sodium imaging include twisted projection imaging (TPI) (35) or radial trajectories (36).
Since these sequences are novel techniques, there is a need to comprehensively validate the variability of T1ρ relaxation times using CubeQuant T2 relaxation times using quantitative DESS, and sodium signal intensity using 3D cones. Furthermore, previous studies of T1ρ, T2, and sodium measurements typically only calculate short-term variability and intra-subject variability. We investigated these three novel sequences using year-long intra-subject variability studies, and a substantial analysis on the variability of segmentation and post-processing. The purpose of this work was to calculate intra-subject variability over short, moderate, and long-term periods, variability of segmentation due to observer differences, and intra-session variability. We aim to quantify the variability of these techniques to determine if they are sufficiently sensitive for in vivo cartilage studies, such as the progression of OA.
METHODS
Data Acquisition
We scanned the knees of eight healthy subjects (6 males, 2 females, mean age 28.1±4.5 years, mean BMI = 22.1±2.8 kg/m2) at baseline, one day, approximately five months (5.2±2.4 months), and one year (11.7±0.3 months) later, all in the evening. Due to scheduling conflicts, we were not able to scan one subject at one year and two subjects at five months. Informed consent was obtained after the procedure had been explained, and the studies were conducted in accordance with the guidelines of the human ethics committee. All datasets were acquired on two MR750 3T scanners (GE Healthcare, Waukesha, WI). CubeQuant T1ρ and DESS images were acquired with a volume-transmit, 8-channel receive knee coil (Invivo Inc., Gainesville, FL). Cones sodium images and a second set of co-registered proton DESS images were acquired using a custom-built dual-tuned sodium/proton four-ring birdcage knee coil, consisting of a low pass birdcage center section and two half high pass outer proton sections (37). We tightly padded the knees of the subjects in the coils with sponge pads to minimize patient movement.
Scan Parameters
We acquired the CubeQuant T1ρ, quantitative DESS T2, and sodium image data all in the sagittal plane. T1ρ relaxation times were measured using a magnetization-prepared pseudo-steady-state 3D fast spin echo (FSE) sequence called CubeQuant with a 500 Hz spin-lock frequency pulse, TR = 1228ms, 90° flip angle, partial k-space acquisition using 0.5 averages, a resolution of 0.5mm × 0.625mm × 3mm, four time spin-lock (TSL) durations of 0ms, 10ms, 30ms, and 60ms, and a total scan time of 5:49min (25). T2 relaxation times were measured using the quantitative DESS sequence (38,39), a SSFP sequence with acquisition echoes on either side of an unbalanced gradient. Four magnitude images (2 echoes per acquisition) are obtained by acquiring two 3D quantitative DESS scans: one with a small diffusion gradient and a large flip angle, and the other with a large diffusion gradient and small flip angle. The low diffusion pair had a large flip angle of 35° and gradient of 34.66ms×mT/m on all three axes. The high diffusion pair had a 18° flip angle and gradient of 138.4ms×mT/m on all three axes (30). The first TE was 9 ms and the second effective TE was 43ms, since the second echo was measured from the previous RF pulse (30). The quantitative DESS sequence had a TR = 26ms, 1 average, a resolution of 0.625mm×0.625mm×3mm, and a total scan time of 9:40 min. Sodium images were obtained using a non-Cartesian spoiled gradient-echo sequence with the 3D cones k-space trajectory (34) with TE = 0.6ms, TR = 35ms, a readout duration of 24ms, 70° flip angle, 28 averages, a resolution of 1.25mm×1.25mm×4.0mm, and a total scan time of 21:33min.
Segmentation and Slice Selection
We calculated the MR parameter maps for all three sequences and segmented the knee cartilage on a proton DESS image. T1ρ relaxation times were calculated pixel-wise using a mono-exponential fit in OsiriX. T2 relaxation times were generated from the four DESS images for each pixel by an OsiriX software tool that fits the signals to a three-dimensional solution space created using the Wu-Buxton signal model for a range of T1, T2, and ADC values (30,40). Sodium signal intensities were measured and normalized to the popliteal artery signal. Manual segmentation on a single slice in the lateral (L) and medial (M) compartments of a single DESS image produced the following ROIs: anterior (AF), central (CF), and posterior (PF) femoral cartilage and anterior (AT) and posterior (PT) tibial cartilage. The medial slice was the slice closest to the midline of the leg with continuous medial femoral cartilage. The lateral slice was the slice closest to the midline of the leg with continuous lateral femoral cartilage. The segmentation technique is shown on a DESS image (Figure 1). The femoral cartilage is split into the lateral anterior (LAF), central (LCF) and posterior (LPF) regions (16). The tibial cartilage is bisected into lateral anterior (LAT) and posterior (LPT) tibial cartilage. The same convention was followed for the medial cartilage. If the volunteer moved in between scans, ROIs were moved accordingly. Bone and fluid were excluded in all ROIs.
Figure 1.
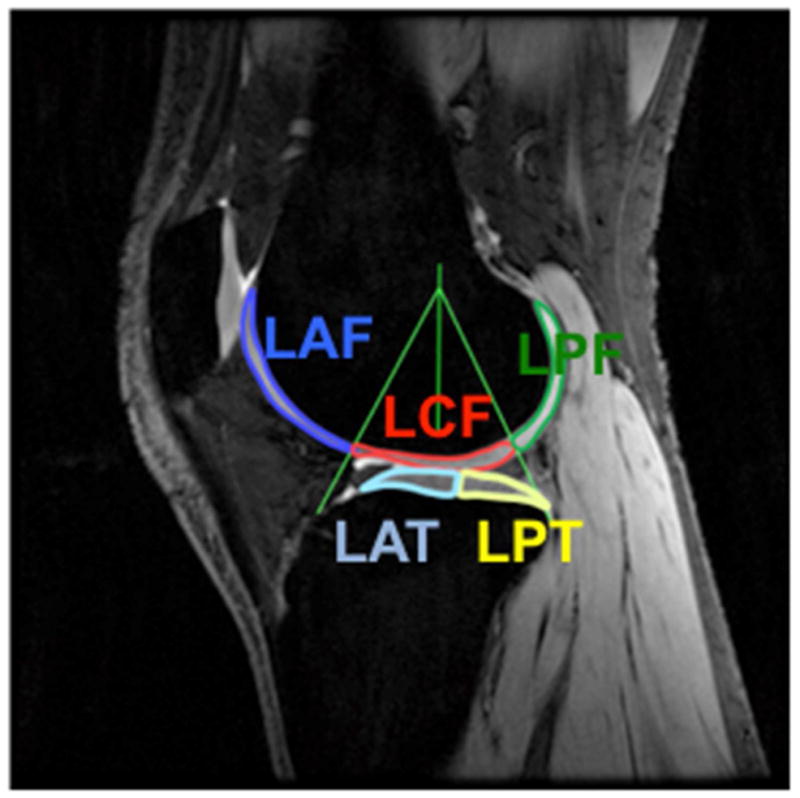
Typical segmentation is shown in the lateral compartment of a healthy subject’s knee cartilage on a DESS image. The femoral cartilage is split into the lateral anterior (LAF), central (LCF) and posterior (LPF) femoral cartilage such that the central cartilage is confined within ±30° of a normal vector to the tibial cartilage. The tibial cartilage is bisected into lateral anterior (LAT) and posterior (LPT) tibial cartilage. The same convention was followed for the medial cartilage.
Data Analysis
The metric of variability is the CV, the standard deviation of the measurements divided by the mean. The mean signal of each ROI for each subject was calculated at each time point. For each subject, we computed the CV for each category of variability for each ROI. The root-mean-square CV (CVRMS) was determined by taking the square root of the mean of the sum of the CV’s squared of the eight subjects for each ROI, CVRMS = Σ√(CV)2/n, where n = number of subjects (41). Finally, we calculated the average, minimum, and maximum CVRMS and the 95% confidence intervals (CI) amongst all 10 ROIs to provide a simplified estimate of the variability of each technique, so that the CVRMS was not specific for any one ROI.
Definitions of Variability
Variability is defined as the precision of results under different conditions (41). We define short-term intra-subject variability to be the variability that results from scanning the same subject on consecutive days. Moderate-term intra-subject variability is the variability of scanning the subject at baseline and approximately five months later. We define long-term intra-subject variability to be the variability of scanning the same subject at baseline and approximately one year later. Intra-observer variability is defined as the variability of the measurements after a single observer segments the same slice twice. Adjacent-slice variability is the variability between the original slice and an adjacent slice in each compartment. Inter-observer variability is the variability after different observers segment the same slices from the same scan. Intra-session variability is the variability within a single session.
Intra-session Variability Calculations
We analyzed three aspects of intra-session variability: 1) consecutive scans without repositioning, 2) consecutive scans with repositioning, and 3) intrinsic MRI noise by simulations. We scanned a ninth subject (34F, left knee) twice consecutively, repositioned the subject, and then scanned a third time, immediately. The same ROIs were used for the consecutive scans, and the third scan was re-segmented by the same observer. Noise simulations were performed on a single scan and ROI by adding increasing levels of complex Gaussian noise to each of the magnitude of the source images. This was done 100 times per technique, and then the magnitude of the summation was taken (42). The T1ρ, T2, and sodium measurements of the noisy images were then calculated and the CV was computed comparing the parameters generated from the noisy source images to the parameters from the original source images. The T1ρ and T2 relaxation times and the normalized sodium signals were calculated in the lateral anterior femoral region. The CV was measured for each parameter as a function of the signal-to-noise ratio (SNR) of the source image with the highest SNR in the set of images.
RESULTS
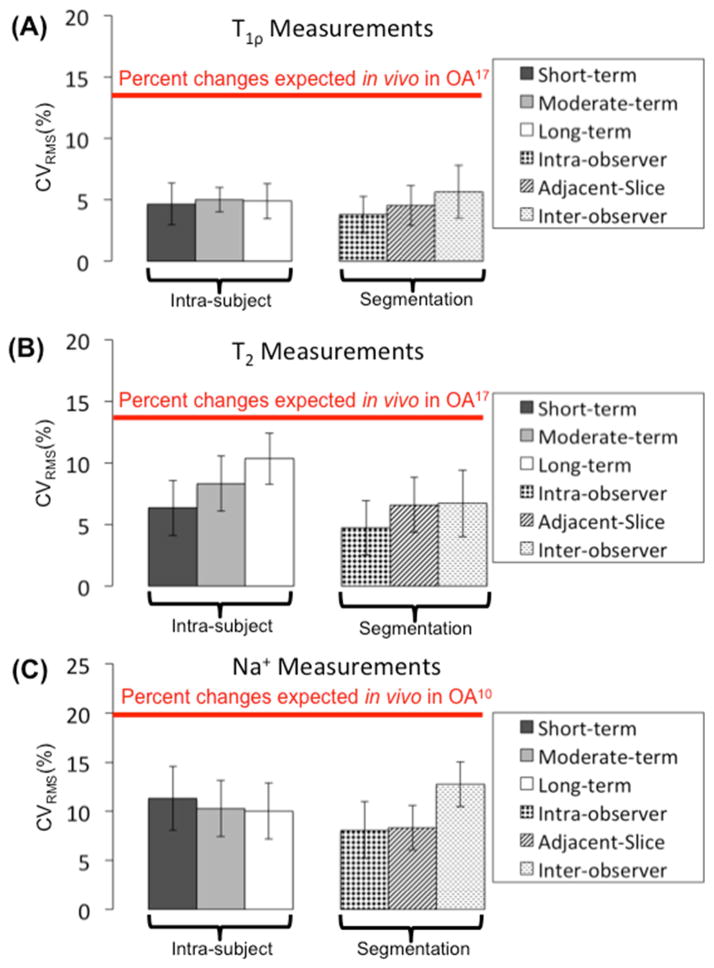
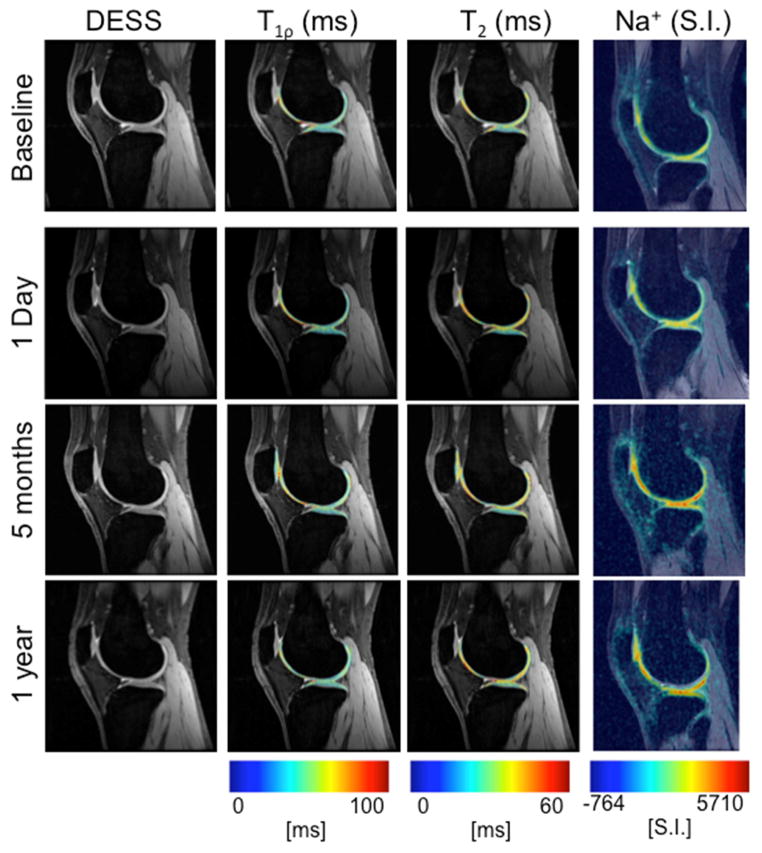
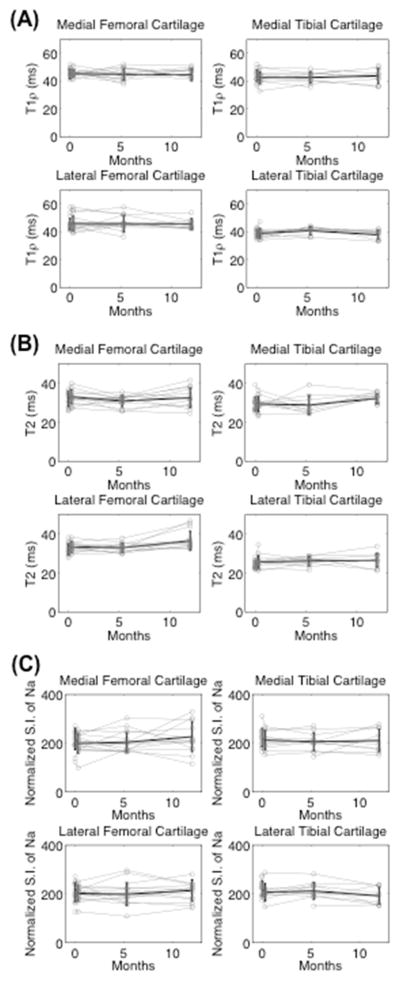
All average CVRMS measurements were substantially lower than changes expected in OA, from prior studies (Figure 2) (10,17). The images shown at baseline, one day later, five months, and one year later demonstrate the consistency of measurements over time (Figure 3). This is true for all subjects, shown by the measurements of all regions of all subjects plotted in gray, and the mean and standard deviation plotted in black (Figure 4).
Figure 2.
The average CVRMS (± standard deviation) for each parameter and technique amongst the 10 ROIs are grouped next to each other in two categories: intra-subject variability due to short, moderate, and long-term time periods and the post-processing segmentation variability due to intra-observer, adjacent-slice, and inter-observer variability for (A) T1ρ, (B) T2, and (C) sodium measurements. All CVRMS measurements are substantially lower than the changes expected due to advanced osteoarthritis (10, 17).
Figure 3.
DESS images and MR parameter maps of T1ρ, T2, and sodium of a single subject are shown at baseline (first row), one day (second row), five months (third row), and one year later (fourth row). The measurements are stable over time with an average CV of 6.0%, 10.7%, and 12.9% for T1ρ, T2, and sodium intra-subject measurements, respectively, over one year.
Figure 4.
The measurements of all regions of the healthy subject’s cartilage are shown in gray. The mean and standard deviation of the eight subjects are shown in black. The 10 ROIs are grouped into the medial and lateral femoral and tibial compartments for simplicity. These graphs show the consistency of the measurements over time for (A) T1ρ (B) T2 and (C) sodium measurements.
CubeQuant T1ρ Variability
The average CVRMS of T1ρ relaxation times using CubeQuant was less than 5.7% for all types of variability. We measured the average CVRMS, 95% CI, and ranges of minimum and maximum CVRMS amongst the 10 cartilage ROIs (Table 2). For intra-subject variability, the average CVRMS [minimum-maximum] of short-term variability was 4.6%[2.2%–8.6%], the average moderate-term CVRMS was 6.1%[4.5%–8.6%], and the average long-term CVRMS was 6.0%[3.1%–8.4%]. For the variability of segmentation, the average intra-observer CVRMS was 3.8%[2.1%–6.3%], the average adjacent-slice CVRMS was 4.5%[3.0%–8.6%], and the average inter-observer CVRMS was 5.7%[2.7%–10.5%].
Table 2.
Variability measurements of T1ρ relaxation times due to intra-subject and post-processing differences.
| ROI | Intra-subject Short-term CVRMS (95% CI) | Intra-subject Moderate-term CVRMS (95% CI) | Intra-subject Long-term CVRMS (95% CI) | Intra-observer CVRMS (95% CI) | Adjacent-slice CVRMS (95% CI) | Inter-observer CVRMS (95% CI) |
|---|---|---|---|---|---|---|
| MEAN | 4.6 (3.6, 5.7) | 6.1 (5.4, 6.9) | 6.0 (4.8, 7.1) | 3.8 (2.9, 4.7) | 4.5 (3.5, 5.5) | 5.7 (4.3, 7.0) |
| MAF | 5.2 (3.1, 7.4) | 8.6 (4.6, 12.5) | 7.2 (4.6, 9.9) | 3.7 (2.4, 5.0) | 3.9 (2.6, 5.3) | 4.7 (2.2, 7.2) |
| MCF | 3.7 (1.9, 5.5) | 6.1 (2.9, 9.3) | 6.4 (4.0, 8.8) | 3.0 (1.7, 4.2) | 4.8 (3.1, 6.6) | 10.5 (4.2, 16.8) |
| MPF | 4.5 (2.1, 7.0) | 5.7 (3.8, 7.6) | 6.0 (3.6, 8.4) | 2.2 (1.0, 3.3) | 4.4 (2.5, 6.3) | 2.7 (1.5, 4.0) |
| MAT | 8.6 (3.8, 13.4) | 5.2 (2.7, 7.7) | 6.9 (4.6, 9.1) | 6.4 (3.7, 9.0) | 4.4 (2.4, 6.4) | 4.0 (2.2, 5.7) |
| MPT | 5.3 (3.1, 7.5) | 5.1 (1.9, 8.3) | 3.2 (1.5, 4.8) | 5.2 (3.0, 7.4) | 8.6 (4.7, 12.6) | 5.0 (2.7, 7.2) |
| LAF | 4.7 (3.3, 6.1) | 6.8 (4.0, 2.7) | 5.3 (2.5, 8.1) | 2.4 (1.2, 3.7) | 5.0 (1.7, 4.6) | 5.9 (3.5, 8.4) |
| LCF | 2.2 (1.3, 3.1) | 5.7 (4.1, 9.5) | 4.8 (2.2, 7.4) | 4.8 (2.4, 7.2) | 3.0 (3.0, 7.0) | 5.6 (3.1, 8.2) |
| LPF | 2.9 (1.5, 4.4) | 7.3 (4.4, 7.3) | 3.0 (2.2, 4.0) | 2.4 (1.3, 3.6) | 4.6 (1.3, 4.7) | 8.0 (3.0, 12.9) |
| LAT | 4.8 (2.5, 7.0) | 6.5 (3.0, 10.2) | 8.2 (4.2, 12.3) | 2.7 (1.3, 4.1) | 3.2 (2.4, 6.8) | 4.9 (2.7, 7.2) |
| LPT | 4.5 (2.1, 6.9) | 4.5 (2.3, 10.1) | 8.4 (4.6, 12.2) | 5.3 (3.0, 7.6) | 2.2 (1.7, 4.7) | 5.2 (3.2, 7.2) |
Quantitative DESS T2 Variability
Using quantitative DESS, the average CVRMS of T2 relaxation times was 12.7% or less for all types of variability. We measured the average CVRMS, 95% CI, and ranges of minimum and maximum CVRMS amongst the 10 cartilage ROIs (Table 3). For intra-subject variability, the average CVRMS [minimum-maximum] of short-term variability was 6.3%[3.6%–10.6%], the average moderate-term CVRMS was 9.3%[5.1%–12.7%], and the average long-term CVRMS was 10.7%[6.7%–12.9%]. For the variability of segmentation, the average intra-observer CVRMS was 4.7%[2.6%–10.1%], the average adjacent-slice CVRMS was 6.6%[3.9%–10.6%], and the average inter-observer CVRMS was 6.7%[2.8%–11.6%].
Table 3.
Variability measurements of T2 relaxation times due to intra-subject and post-processing differences.
| ROI | Intra-subject Short-term CVRMS (95% CI) | Intra-subject Moderate-term CVRMS (95% CI) | Intra-subject Long-term CVRMS (95% CI) | Intra-observer CVRMS (95% CI) | Adjacent-slice CVRMS (95% CI) | Inter-observer CVRMS (95% CI) |
|---|---|---|---|---|---|---|
| MEAN | 6.3 (5.0, 7.7) | 9.3 (7.8, 10.9) | 10.7 (9.6, 11.8) | 4.7 (3.4, 6.1) | 6.6 (5.2, 8.0) | 6.7 (5.0, 8.4) |
| MAF | 5.9 (3.0, 8.9) | 9.8 (5.0, 14.7) | 6.7 (4.3, 9.1) | 3.3 (1.7, 4.9) | 7.6 (4.0, 11.3) | 7.4 (3.4, 11.3) |
| MCF | 6.6 (3.8, 9.4) | 10.8 (6.6, 15.2) | 11.7 (5.8, 17.6) | 5.7 (2.9, 8.6) | 5.7 (4.1, 7.2) | 7.2 (4.4, 10.0) |
| MPF | 5.3 (3.4, 7.3) | 7.0 (5.1, 9.0) | 10.9 (8.0, 13.6) | 2.6 (1.6, 3.7) | 6.9 (4.4, 9.3) | 2.9 (1.5, 4.2) |
| MAT | 3.9 (2.5, 5.3) | 9.0 (3.8, 14.2) | 10.6 (5.9, 15.4) | 5.7 (3.5, 8.0) | 4.0 (2.4, 5.6) | 4.6 (3.0, 6.2) |
| MPT | 7.1 (4.8, 9.3) | 12.7 (5.1, 18.7) | 8.9 (5.6, 12.2) | 10.1 (4.8, 15.3) | 9.7 (5.7, 13.6) | 11.6 (4.7, 18.5) |
| LAF | 3.6 (1.7, 5.5) | 11.1 (3.9, 18.3) | 10.6 (6.5, 14.8) | 3.0 (1.9, 4.2) | 5.3 (3.0, 7.6) | 6.1 (3.7, 8.5) |
| LCF | 6.9 (4.5, 9.4) | 6.5 (6.7, 9.9) | 12.9 (8.6, 17.2) | 3.8 (1.9, 5.6) | 10.6 (5.6, 15.5) | 5.2 (2.5, 8.0) |
| LPF | 4.5 (2.5, 6.4) | 12.0 (4.1, 18.3) | 11.4 (6.0, 16.9) | 5.5 (2.4, 8.5) | 3.9 (2.2, 5.6) | 4.6 (2.3, 7.0) |
| LAT | 9.1 (6.3, 11.8) | 5.1(3.0, 6.9) | 11.1 (7.0, 15.3) | 3.2 (1.9, 4.4) | 5.0 (2.8, 7.3) | 10.7 (5.8, 15.6) |
| LPT | 11.0 (6.3, 14.9) | 9.0 (5.2, 12.8) | 12.2 (6.3, 18.1) | 4.5 (2.7, 6.3) | 7.1 (3.9, 10.4) | 7.0 (2.9, 11.1) |
Cones Sodium Variability
For the 3D cones sodium measurements, the average CVRMS was 12.7% or less for all types of variability. Amongst the 10 cartilage ROIs, we measured the average CVRMS, 95% CI, and ranges of minimum and maximum CVRMS (Table 4). For intra-subject variability, the average CVRMS [minimum-maximum] of short-term variability was 11.3%[8.0%–17.7%], the average moderate-term CVRMS was 11.6%[4.5%–14.9%], and the average long-term CVRMS was 12.9%[7.5%–24.1%]. For the variability due to segmentation, the average intra-observer CVRMS was 8.1%[3.8%–14.0%], the average adjacent-slice CVRMS was 8.3%[5.3%–12.2%], and the average inter-observer CVRMS was 11.4%[4.2%–18.5%].
Table 4.
Variability measurements of Na+ due to intra-subject and post-processing differences.
| ROI | Intra-subject Short-term CVRMS (95% CI) | Intra-subject Moderate-term CVRMS (95% CI) | Intra-subject Long-term CVRMS (95% CI) | Intra-observer CVRMS (95% CI) | Adjacent-slice CVRMS (95% CI) | Inter-observer CVRMS (95% CI) |
|---|---|---|---|---|---|---|
| MEAN | 11.3 (9.3, 13.4) | 11.6 (9.7, 13.6) | 12.9 (9.5, 16.3) | 8.1 (6.3, 9.9) | 8.3 (6.9, 9.7) | 11.4 (8.6, 14.3) |
| MAF | 14.4 (9.8, 19.2) | 14.9 (9.5, 20.3) | 20.3 (10.0, 30.6) | 11.1 (8.5, 13.7) | 10.7 (6.0, 15.4) | 16.0 (11.4, 20.7) |
| MCF | 12.8 (7.9, 17.8) | 12.8 (5.0, 20.6) | 9.9 (6.2, 13.6) | 6.2 (3.4, 9.0) | 6.8 (3.5, 10.0) | 7.8 (4.7, 10.9) |
| MPF | 8.6 (4.7, 12.6) | 11.6 (7.0, 16.3) | 9.2 (4.4, 14.0) | 6.1 (3.6, 8.6) | 8.8 (6.7, 10.9) | 12.9 (7.1, 18.8) |
| MAT | 8.0 (4.8, 11.2) | 10.2 (5.8, 14.8) | 12.2 (8.9, 15.5) | 8.9 (4.4, 13.5) | 12.2 (5.9, 18.0) | 18.5 (9.7, 27.4) |
| MPT | 10.4 (5.5, 15.3) | 12.7 (5.9, 19.5) | 11.8 (7.0, 16.6) | 7.9 (4.0, 11.8) | 10.4 (3.9, 14.9) | 16.9 (10.5, 23.2) |
| LAF | 17.7 (7.8, 27.6) | 4.5 (2.8, 6.3) | 24.1 (10.6, 37.5) | 3.8 (2.4, 5.3) | 5.3 (3.9, 6.9) | 10.5 (6.0, 15.0) |
| LCF | 13.9 (8.3, 19.6) | 12.7 (6.6, 18.8) | 7.5 (4.6, 10.3) | 8.3 (5.4, 11.2) | 6.8 (3.8, 9.9) | 9.6 (5.9, 13.3) |
| LPF | 9.6 (4.9, 14.2) | 14.3 (5.8, 22.8) | 10.6 (6.4, 14.8) | 14.0 (7.1, 20.9) | 5.7 (4.0, 7.5) | 8.6 (5.5, 11.8) |
| LAT | 9.6 (5.5, 13.8) | 8.6 (4.2, 13.1) | 8.2 (5.0, 11.5) | 5.9 (4.2, 7.6) | 7.4 (3.3, 11.4) | 4.2 (2.3, 6.1) |
| LPT | 8.1 (3.8, 12.5) | 13.8 (4.5, 23.1) | 15.4 (8.5, 22.2) | 8.6 (4.8, 12.3) | 9.1 (4.6, 13.6) | 9.3 (5.1, 13.5) |
Intra-session Variability Results
We calculated the average CV of the intra-session variability measurements over all 10 ROIs to be 9.2% or less. The CV of the single subject who was scanned twice, repositioned, and scanned a third time, demonstrates a remaining source of variability (Figure 5). The average CV [minimum-maximum] amongst the 10 ROIs after scanning the same subject consecutively was 1.9%[0.0%–4.6%], 2.8%[0.2%–5.6%], and 7.3%[1.1%–19.9%] for T1ρ, T2, and sodium measurements, respectively. After repositioning the subject, the average CV amongst the 10 ROIs was 3.0%[0.7%–6.8%], 5.2%[1.0%–14.0%], and 9.2%[0.2%–22.2%] for T1ρ, T2, and sodium measurements, respectively. The Monte Carlo simulations demonstrate that variability decreases as SNR of the source images decreases (Fig. 6). The CV of the measurements from the noisy source images compared to the measurements from the original source images is calculated as a function of the SNR of the source image with the highest SNR. The SNR of the original source image with the highest SNR is marked along the curve to demonstrate the expected CV for a typical scan. The CVs were 2.3% for the T1ρ measurements, 0.7% for the T2 measurements, and 1.5% for the sodium sequence.
Figure 5.
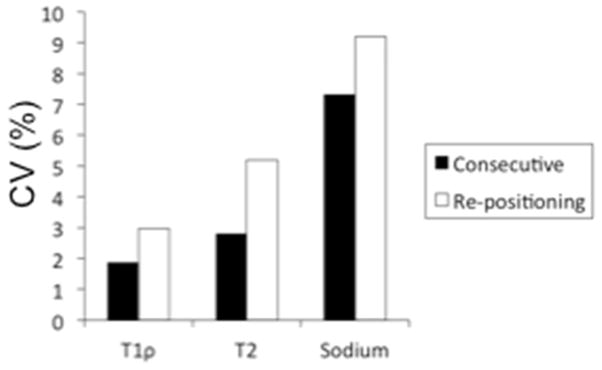
The average CV amongst 10 ROIs of a single subject who was scanned twice consecutively, repositioned, and then scanned a third time demonstrates a remaining baseline source of variability.
Figure 6.
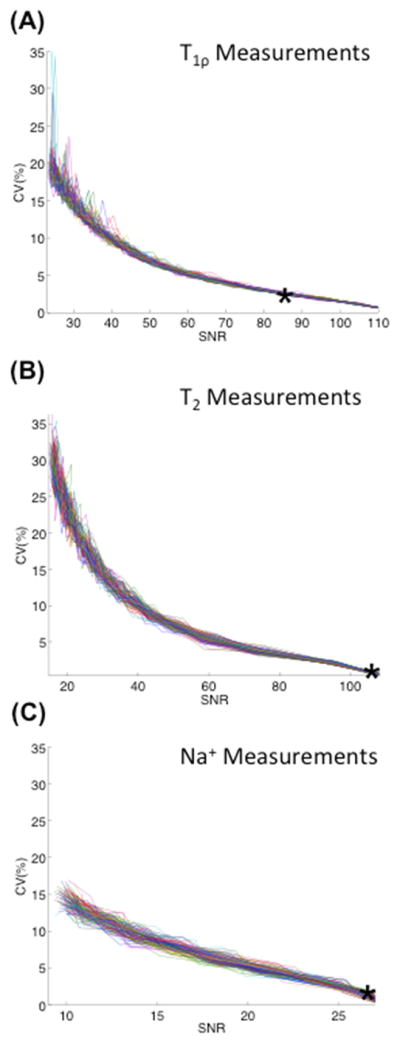
Monte Carlo simulations were performed by adding increasing factors of complex Gaussian noise to each of the magnitude source images for all sequences and then taking the magnitude of the summation. Each color represents one of the 100 simulations. The T1ρ, T2, and sodium measurements were calculated and the CV was computed for the (A) T1ρ, (B) T2, and (C) sodium measurements. These graphs demonstrate that the variability increases as the noise levels increase and the SNR of the source images decreases. The typical SNR of the source image with the highest SNR is plotted along the curve, as indicated by the asterisk.
DISCUSSION
In summary, we found that CubeQuant T1ρ, quantitative DESS T2, and cones sodium measurements have similar variability as previous studies using different sequences, and that the average CVRMS for each technique is substantially lower than the percentage changes previously measured in several in vivo studies in osteoarthritic cartilage (10,17). We calculated nine types of variability, which include the short, moderate and long-term intra-subject variability and the segmentation variability from intra-observer, adjacent-slice and inter-observer differences. Intra-session variability was calculated due to the effects of repositioning after scanning consecutively, and the effect of variation due to noise alone.
Our average CVRMS measurements are comparable to other published literature CVRMS and CV measurements. Our average CVRMS measurements of T1ρ ranged from 4.6%–6.1% for intra-subject variability and 3.8%–5.7% for post-processing segmentation variability. Our average CVRMS measurements are within the range of previously reported measurements of short-term intra-subject variability, which have CVs of 7%–19% in a multi-center trial (18), 1.7%–8.7% in a healthy volunteer study (19) and 2.2%–5.9% in a morning scan to 4.8%–8.8% in an evening scan of healthy volunteers (20). Since we scanned healthy subjects instead of patients, and we scanned at the same center for all scans, these factors may contribute to the substantially high variability of our T1ρ results. A previous study found no significant difference due to diurnal variation in T1ρ or T2 measurements between morning and evening scans, so although we might expect somewhat more variation, the time of day might not be a significant factor (20). The average CVRMS measurements of T2 ranged from 6.3%–10.7% for intra-subject variability and 4.7%–6.7% for post-processing segmentation variability. Our measurements are again similar to previously reported measurements of the short-term intra-subject variability of T2, which ranged from 4%–14% (18), regional CVs ranged from 6%–29% (21) and CVs of 4.0%–6.2% for a morning scan to 4.2%–8.5% for evening scans, respectively (20). Our average CVRMS measurements of sodium ranged from 11.3%–12.9% for intra-subject variability and 8.1%–11.4% for post-processing segmentation variability. Our results are similar to or somewhat higher than previously reported CVRMS ranges of 7.5%–13.6% for six healthy volunteers (22). Two studies that also used the cones sodium MRI sequence for a short-term variability study reported CVs of 2.2%–13.8% (23) and 3.0%–8.6% (24); however, our study additionally measures long-term variability and post-processing variability in healthy volunteers only (23,24).
By gathering these nine types of variability, we discovered several trends that provide insight into the potential sources of variability for in vivo cartilage studies and the regions of cartilage that may have the greatest variation. For example, long-term intra-subject variability trended towards being worse than moderate-term or short-term intra-subject variability. The inter-observer variability also trended towards being worse than intra-observer or adjacent-slice variability. Comparing the short-term intra-subject CVRMS values, the medial anterior tibial region was the most variable ROI for the T1ρ measurements, the medial posterior tibial region was the most variable ROI for the T2 measurements, and the lateral anterior femoral region was the most variable ROI for the sodium measurements. The medial anterior femoral region is typically the smallest ROI and may include partial volume artifacts due to nearby joint fluid, so these factors may account for the increased variability. T1ρ measurements were the least variable, followed by the T2 measurements, and then finally the sodium measurements.
For the intra-session variability, the low CVs for T1ρ and T2 were expected; however, we did observe that sodium was more variable and had higher CVs than predicted. Consecutive scans were the least variable, and when we repositioned the subject, we observed greater variation. For the single ninth volunteer, the measurements were relatively consistent, as expected for a short-term intra-subject consecutive scan, and the CVs are about half that of the average numbers for the group of eight volunteers for T1ρ and T2 (Figure 5). For the sodium measurements, the CVs for the single volunteer were surprisingly about the same as the CVs for the group of volunteers over the short term. Therefore, it appears that intrinsic MRI noise is a substantial source of variation in sodium MRI. Possible reasons for high noise for the single volunteer is that the subject’s knee was smaller than most other subjects, so the knee may not have loaded the sodium coil as well as the larger volunteers, thereby reducing SNR and affecting the CV more than the proton measurements with the 8-channel knee coil.
We performed Monte Carlo simulations because we found during the consecutive scans, with no repositioning or other external influences, that there are remaining variations of about 1%–2%. Therefore, we felt it was important to explore the potential source of the remaining variation that may be inevitable with MRI. While some of this could possibly be attributed to patient movement, the Monte Carlo simulation results suggest that MRI noise may limit the lowest CV measurements, for these techniques, given the current typical SNRs of the source images to ranges of 0.7%–2.3%. Noise may be more dominant with the sodium measurements, compared to the T1ρ and T2 measurements, as a source of variation. Fig. 6 demonstrates that the remaining variation could be considered an approximate lower bound of variability.
Many studies have experimentally fit equations that mathematically relate the MR parameters with the macromolecules. For example, one group fit experimental data for T1ρ and T2 at 8.5T, and found T1ρ = 1.7e−0.06[GAG], T1ρ = 1.6e−0.17[Collagen] and T2 = 1.1e−0.07[GAG], T2 = 1.1e−0.17[Collagen], demonstrating the inverse association between T1ρ or T2, and GAG or collagen (43). As the GAG or collagen content decreases, T1ρ and T2 should increase. The sodium concentration has been shown to be mathematically related to GAG by the equation, , where FCD is the fixed charged density of the negatively charged GAG, counterbalanced by the positively charged Na+ ions (44). The FCD is demonstrated to be related to the sodium concentration by the equation, , where SF is the synovial fluid sodium concentration in the image near cartilage (44). There is a high correlation between the FCD from the sodium MRI and PG loss (slope = 0.89 and R2 = 0.81), showing the direct correlation between sodium MRI and GAG (13).
While this study only considered healthy volunteers, these techniques are designed for patients with OA and other musculoskeletal disorders, and the variability is likely to increase. A multi-center study of T1ρ and T2 measurements, which included 18 healthy volunteers, 16 mild OA patients, and 16 moderate OA patients scanned four times over a two month period, demonstrated that the reproducibility of T1ρ was somewhat lower with increasing grades of OA, and the reproducibility was more consistent for T2 measurements across the three grades of cartilage health (18). A recent study of sodium MRI showed that the test-retest variability in sodium MRI was similar between 15 patients with OA and five age and sex matched healthy controls for two MRI sessions on the same day (23). However, over a longer period of several years, patients with OA may have more substantial degradation of the cartilage, and the variability of the measurements would likely worsen.
Technical factors may be one source of variability. For example, the T2 measurements from quantitative DESS are particularly sensitive to noise, as the second echo of the high diffusion/low flip angle acquisition typically has a low SNR. For sodium measurements, subjects of different sizes may have different SNR. Standard deviations may be large in an individual ROI if there are any partial volume artifacts including fluid or bone, although we attempted to avoid this by manually reshaping the ROI to avoid such artifacts. Magic angle effects are present, but knee position was similar between timpeoints.
This study was designed to estimate the variability of quantitative MRI measurements at commonly collected timepoints in longitudinal OA studies. A limitation of this study is that we could not assess the effect of time on the quantitative data. Another limitation is the lack of direct comparison between these novel techniques presented and published sequences. A final limitation of this work is that we did not include phantom experiments that may help estimate the underlying variability due to human subjects. Future studies with more subjects could be designed to assess the change in quantitative metrics over time in both healthy volunteers and OA patients and comparisons to standard MRI techniques.
In conclusion, our CVRMS measurements are lower than most previously measured changes in OA. CubeQuant T1ρ, quantitative DESS T2, and 3D cones sodium MRI may therefore be sufficiently sensitive for detecting changes resulting from OA.
Table 1.
Summary of Subject Demographics
| Demographics | |||||
|---|---|---|---|---|---|
| n = 8 | Age | Sex | Height (m) | Weight (kg) | BMI (kg/m2) |
| Subjects | 28.1±4.5 | M=6 | 1.8±0.1 | 68.2±11.9 | 22.1±2.8 |
Acknowledgments
The authors gratefully acknowledge Brady Quist for his assistance with the Monte Carlo simulations, Ernesto Staroswiecki for his expertise on sodium MRI and the DESS sequence, and Bragi Sveinsson for his assistance with the DESS computations.
Funding sources
This project was supported by the following: the National Institutes of Health (Grant Numbers EB002524, AR062068, and R01-AR0063643), the Arthritis Foundation, the Richard M. Lucas Foundation, the Natural Sciences and Engineering Research Council of Canada (NSERC) Post-Doctoral Fellowship, and GE Healthcare. The study sponsors had no involvement in the study design, collection, analysis, or interpretation of data, nor in the decision to submit the manuscript for publication.
Footnotes
Author Contributions
The authors contributed to this manuscript in the following ways: Conception and design (CDJ, EJM BAH, GEG); Analysis and interpretation of data (CDJ, EJM, UDM, BAH, GEG); Drafting of the article (CDJ, EJM, UDM); Obtaining of funding (NKB, BAH, GEG); Technical support (RDW, WC, NKB); Collection and assembly of data (CDJ, UDM). All authors critically revised the manuscript and gave final approval of the article. CDJ and GEG take responsibility for the integrity of the manuscript as a whole (gold@stanford.edu).
Competing interest statement
Caroline D. Jordan is employed by St. Jude Medical. Emily J. McWalter is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Post-Doctoral Fellowship. Uchechukwuka D. Monu has no competing interests. Ronald D. Watkins has no competing interests. Weitian Chen is employed by GE Healthcare. Neal K. Bangerter has no competing interests. Brian A. Hargreaves receives research or institutional support from GE Healthcare and the NIH, and receives patent royalties from GE Healthcare, Siemens, and Philips. Garry E. Gold receives research or institutional support from GE Healthcare, the NIH, the Arthritis Foundation, and provides consulting services to Boston Scientific.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Caroline D. Jordan, Email: cjordan@alumni.stanford.edu.
Uchechukwuka D. Monu, Email: udmonu@stanford.edu.
Emily J. McWalter, Email: mcwalter@stanford.edu.
Ronald D. Watkins, Email: watkinsr@stanford.edu.
Weitian Chen, Email: weitian.chen@ge.com.
Neal K. Bangerter, Email: nealb@ee.byu.edu.
Brian A. Hargreaves, Email: bah@stanford.edu.
Garry E. Gold, Email: gold@stanford.edu.
References
- 1.Buckwalter JA, Martin JA. Osteoarthritis. Adv Drug Deliv Rev. 2006;58:150–67. doi: 10.1016/j.addr.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Felson DT, Lawrence RC, Dieppe PA, Hirsch R, Helmick CG, Jordan JM, et al. Osteoarthritis: new insights. Part 1: the disease and its risk factors. Ann Intern Med. 2000;133:635–46. doi: 10.7326/0003-4819-133-8-200010170-00016. [DOI] [PubMed] [Google Scholar]
- 3.Burstein D. Tracking longitudinal changes in knee degeneration and repair. J Bone Joint Surg Am. 2009;91 (Suppl 1):51–3. doi: 10.2106/JBJS.H.01412. [DOI] [PubMed] [Google Scholar]
- 4.Conaghan P. Is MRI useful in osteoarthritis? Best Pract Res Clin Rheumatol. 2006;20:57–68. doi: 10.1016/j.berh.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Hunter DJ, Felson DT. Osteoarthritis. BMJ. 2006;332:639–42. doi: 10.1136/bmj.332.7542.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maroudas A, Venn M. Chemical composition and swelling of normal and osteoarthrotic femoral head cartilage. II. Swelling. Ann Rheum Dis. (1977/10/01) 1977;36(5):399–406. doi: 10.1136/ard.36.5.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regatte RR, Akella SVS, Borthakur A, Kneeland JB, Reddy R. Proteoglycan depletion-induced changes in transverse relaxation maps of cartilage: comparison of T2 and T1rho. Acad Radiol. 2002;9:1388–94. doi: 10.1016/s1076-6332(03)80666-9. [DOI] [PubMed] [Google Scholar]
- 8.Dardzinski BJ, Mosher TJ, Li S, Van Slyke MA, Smith MB. Spatial variation of T2 in human articular cartilage. Radiology. 1997;205:546–50. doi: 10.1148/radiology.205.2.9356643. [DOI] [PubMed] [Google Scholar]
- 9.Chou M-C, Tsai P-H, Huang G-S, Lee H-S, Lee C-H, Lin M-H, et al. Osteoarthr Cartil. 4. Vol. 17. Elsevier Ltd; 2009. Apr, Correlation between the MR T2 value at 4.7 T and relative water content in articular cartilage in experimental osteoarthritis induced by ACL transection; pp. 441–7. [DOI] [PubMed] [Google Scholar]
- 10.Wheaton AJ, Borthakur A, Shapiro EM, Regatte RR, Akella SVS, Kneeland JB, et al. Proteoglycan loss in human knee cartilage: quantitation with sodium MR imaging--feasibility study. Radiology. 2004;231:900–5. doi: 10.1148/radiol.2313030521. [DOI] [PubMed] [Google Scholar]
- 11.Bashir A, Gray ML, Hartke J, Burstein D. Nondestructive imaging of human cartilage glycosaminoglycan concentration by MRI. Magn Reson Med. 1999;41:857–65. doi: 10.1002/(sici)1522-2594(199905)41:5<857::aid-mrm1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 12.Reddy R, Insko EK, Noyszewski EA, Dandora R, Kneeland JB, Leigh JS. Sodium MRI of human articular cartilage in vivo. Magn Reson Med. 1998;39:697–701. doi: 10.1002/mrm.1910390505. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro EM, Borthakur A, Gougoutas A, Reddy R. 23Na MRI accurately measures fixed charge density in articular cartilage. Magn Reson Med. 2002;47:284–91. doi: 10.1002/mrm.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borthakur A, Shapiro EM, Beers J, Kudchodkar S, Kneeland JB, Reddy R. Sensitivity of MRI to proteoglycan depletion in cartilage: comparison of sodium and proton MRI. Osteoarthritis Cartilage. 2000;8:288–93. doi: 10.1053/joca.1999.0303. [DOI] [PubMed] [Google Scholar]
- 15.Regatte RR, Akella SVS, Lonner JH, Kneeland JB, Reddy R. T1rho relaxation mapping in human osteoarthritis (OA) cartilage: comparison of T1rho with T2. J Magn Reson Imaging. 2006;23:547–53. doi: 10.1002/jmri.20536. [DOI] [PubMed] [Google Scholar]
- 16.Stahl R, Blumenkrantz G, Carballido-Gamio J, Zhao S, Munoz T, Hellio Le Graverand-Gastineau MP, et al. MRI-derived T2 relaxation times and cartilage morphometry of the tibio-femoral joint in subjects with and without osteoarthritis during a 1-year follow-up. Osteoarthritis Cartilage. 2007;15:1225–34. doi: 10.1016/j.joca.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Benjamin Ma C, Link TM, Castillo D-D, Blumenkrantz G, Lozano J, et al. In vivo T(1rho) and T(2) mapping of articular cartilage in osteoarthritis of the knee using 3 T MRI. Osteoarthritis Cartilage. 2007;15:789–97. doi: 10.1016/j.joca.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosher TJ, Zhang Z, Reddy R, Boudhar S, Milestone BN, Morrison WB, et al. Knee articular cartilage damage in osteoarthritis: analysis of MR image biomarker reproducibility in ACRIN-PA 4001 multicenter trial. Radiology. 2011;258(3):832–42. doi: 10.1148/radiol.10101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Han ET, Busse RF, Majumdar S. In vivo T(1rho) mapping in cartilage using 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS) Magn Reson Med. 2008;59:298–307. doi: 10.1002/mrm.21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Wyatt C, Rivoire J, Han ET, Chen W, Schooler J, et al. Simultaneous Acquisition of T1r and T2 Quantification in Knee Cartilage: Repeatability and Diurnal Variation. J Magn Reson Imaging. 2013 doi: 10.1002/jmri.24253. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser C, Mendlik T, Dinges J, Weber J, Stahl R, Trumm C, et al. Global and regional reproducibility of T2 relaxation time measurements in human patellar cartilage. Magnetic Resonance in Medicine. 2006:527–34. doi: 10.1002/mrm.21005. [DOI] [PubMed] [Google Scholar]
- 22.Madelin G, Babb JS, Xia D, Chang G, Jerschow A, Regatte RR. Reproducibility and repeatability of quantitative sodium magnetic resonance imaging in vivo in articular cartilage at 3 T and 7 T. Magn Reson Med. 2012;68:841–9. doi: 10.1002/mrm.23307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newbould RRD, Miller SRS, Tielbeek JAW, Toms LD, Rao AW, Gold GE, et al. Reproducibility of sodium MRI measures of articular cartilage of the knee in Osteoarthritis. Osteoarthr …. 2012;20(1):29–35. doi: 10.1016/j.joca.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo S, Staroswiecki E, Bangerter NK, Hargreaves BA, Gold GE. Repeatability and age-related change of sodium in the knee articular cartilage measured with sodium MRI (Abstract). Proc Annual Meeting ISMRM; 2011. p. 506. [Google Scholar]
- 25.Chen W, Takahashi A, Han ET. 3D Quantitative Imaging of T1rho and T2 (Abstract). Proc Annu Meet ISMRM; 2011. p. 231. [Google Scholar]
- 26.Borthakur A, Wheaton A, Charagundla SR, Shapiro EM, Regatte RR, Akella SVS, et al. Three-dimensional T1rho-weighted MRI at 1.5 Tesla. J Magn Reson Imaging. 2003;17:730–6. doi: 10.1002/jmri.10296. [DOI] [PubMed] [Google Scholar]
- 27.Witschey WRT, Borthakur A, Elliott MA, Fenty M, Sochor MA, Wang C, et al. T1rho-prepared balanced gradient echo for rapid 3D T1rho MRI. J Magn Reson Imaging. 2008;28:744–54. doi: 10.1002/jmri.21444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Chang G, Xu J, Vieira RLR, Krasnokutsky S, Abramson S, et al. T1rho MRI of menisci and cartilage in patients with osteoarthritis at 3T. European Journal of Radiology. 2012:2329–36. doi: 10.1016/j.ejrad.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welsch GH, Scheffler K, Mamisch TC, Hughes T, Millington S, Deimling M, et al. Rapid estimation of cartilage T2 based on double echo at steady state (DESS) with 3 Tesla. Magn Reson Med. 2009;62:544–9. doi: 10.1002/mrm.22036. [DOI] [PubMed] [Google Scholar]
- 30.Staroswiecki E, Granlund KL, Alley MT, Gold GE, Hargreaves Ba. Simultaneous estimation of T(2) and apparent diffusion coefficient in human articular cartilage in vivo with a modified three-dimensional double echo steady state (DESS) sequence at 3 T. Magn Reson Med. 2012;67:1086–96. doi: 10.1002/mrm.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckstein F, Hudelmaier M, Wirth W, Kiefer B, Jackson R, Yu J, et al. Double echo steady state magnetic resonance imaging of knee articular cartilage at 3 Tesla: a pilot study for the Osteoarthritis Initiative. Ann Rheum Dis. (2005/08/30) 2006;65(4):433–41. doi: 10.1136/ard.2005.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bieri O, Ganter C, Welsch GH, Trattnig S, Mamisch TC, Scheffler K. Fast diffusion-weighted steady state free precession imaging of in vivo knee cartilage. Magn Reson Med. 2012;67:691–700. doi: 10.1002/mrm.23061. [DOI] [PubMed] [Google Scholar]
- 33.Gurney PT, Hargreaves BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med. (2006/02/02) 2006;55(3):575–82. doi: 10.1002/mrm.20796. [DOI] [PubMed] [Google Scholar]
- 34.Staroswiecki E, Bangerter NK, Gurney PT, Grafendorfer T, Gold GE, Hargreaves BA. In vivo sodium imaging of human patellar cartilage with a 3D cones sequence at 3 T and 7 T. J Magn Reson Imaging. 2010;32(2):446–51. doi: 10.1002/jmri.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian Y, Stenger VA, Boada FE. Parallel imaging with 3D TPI trajectory: SNR and acceleration benefits. Magn Reson Imaging. 2009;27:656–63. doi: 10.1016/j.mri.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konstandin S, Nagel AM, Heiler PM, Schad LR. Two-dimensional radial acquisition technique with density adaption in sodium MRI. Magn Reson Med. 2011;65:1090–6. doi: 10.1002/mrm.22684. [DOI] [PubMed] [Google Scholar]
- 37.Watkins RD, Staroswiecki E, Bangerter NK, Hargreaves BA, Gold GE. High SNR Dual-Tuned Sodium/Proton Knee Coil (Abstract). Proc Annual Meeting ISMRM; 2010. p. 1503. [Google Scholar]
- 38.Bruder H, Fischer H, Graumann R, Deimling M. A new steady-state imaging sequence for simultaneous acquisition of two MR images with clearly different contrasts. Magn Reson Med. 1988;7:35–42. doi: 10.1002/mrm.1910070105. [DOI] [PubMed] [Google Scholar]
- 39.Redpath TW, Jones RA. FADE--a new fast imaging sequence. Magn Reson Med. 1988;6:224–34. doi: 10.1002/mrm.1910060211. [DOI] [PubMed] [Google Scholar]
- 40.Sveinsson B, Staroswiecki E, Gold GE, Hargreaves BA. Quantitative 3D Diffusion Musculoskeletal Imaging (Abstract) Orthopedic Research Society; 2012. p. 0457. [Google Scholar]
- 41.Glüer CC, Blake G, Lu Y, Blunt BA, Jergas M, Genant HK. Accurate assessment of precision errors: how to measure the reproducibility of bone densitometry techniques. Osteoporos Int. 1995;5:262–70. doi: 10.1007/BF01774016. [DOI] [PubMed] [Google Scholar]
- 42.Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magn Reson Med. 1995;34:910–4. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menezes NM, Gray ML, Hartke JR, Burstein D. T2 and T1rho MRI in articular cartilage systems. Magn Reson Med. 2004;51:503–9. doi: 10.1002/mrm.10710. [DOI] [PubMed] [Google Scholar]
- 44.Borthakur A, Mellon E, Niyogi S, Witschey W, Kneeland JB, Reddy R. Sodium and T 1r MRI for molecular and diagnostic imaging of articular cartilage y. NMR Biomed. 2006;19:781–821. doi: 10.1002/nbm.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]