Abstract
The green alga Chlamydomonas reinhardtii is a popular unicellular organism for studying photosynthesis, cilia biogenesis and micronutrient homeostasis. Ten years since its genome project was initiated, an iterative process of improvements to the genome and gene predictions has propelled this organism to the forefront of the “omics” era. Housed at Phytozome, the Joint Genome Institute’s (JGI) plant genomics portal, the most up-to-date genomic data include a genome arranged on chromosomes and high-quality gene models with alternative splice forms supported by an abundance of RNA-Seq data. Here, we present the past, present and future of Chlamydomonas genomics. Specifically, we detail progress on genome assembly and gene model refinement, discuss resources for gene annotations, functional predictions and locus ID mapping between versions and, importantly, outline a standardized framework for naming genes.
Keywords: Chlamydomonas, algae, nomenclature, gene symbols, Phytozome, annotation
Chlamydomonas – a reference green alga
Chlamydomonas reinhardtii (herein referred to as Chlamydomonas) provides an excellent microbial platform for the investigation of fundamental biological functions. Both photosynthesis (a process associated with the plant lineage) and human ciliary disease (associated with the animal lineage) are effectively studied using this organism as a reference system. A decade of work, encompassing the publication of the genome draft sequence [1], has made this organism highly “genome enabled”. Given the substantial recent and on-going genomic improvements, their discussion in this article is timely.
As a unicellular haploid in the vegetative stage of its life cycle, Chlamydomonas shares the experimental advantages associated with microbes. These include: rapid doubling time (~8–12h), well-defined media and growth requirements, the ability to synchronize cultures with periodic light exposure, the capacity for classical genetic crosses to characterize mutant strains and efficient long-term cryopreservation [2]. The Chlamydomonas molecular and genetic toolbox has grown over the years: irradiated or chemically mutagenized lines have been identified with classical genetic screens [3–5], and RNAi-based knock-downs [6, 7]; zinc-finger nuclease-based mutagenesis [8] and efficient protocols for gene-specific mutant screens [9] are now available. A growing collection of laboratory-generated and environmentally-isolated strains is available at the Chlamydomonas resource center (http://chlamy.org/). Complementary to the use of mutants for ascribing gene function, cDNAs [10, 11] and BAC libraries [12] are available for rescuing mutant phenotypes.
Much of the interest in employing Chlamydomonas in the laboratory stems from its unique evolutionary history. Approximately 700 million years separate the Chlorophyte (green algae, including Chlamydomonas) and Streptophyte (non-chlorophyte green algae and land plants) lineages [13], but the photosynthetic apparatus and auxiliary components have remained remarkably similar. In addition, providing acetate as a fixed-carbon source fully overcomes the need to photosynthesize, so that strains with mutations in photosynthesis–related genes can complete the life cycle. This provides an advantage over land plant systems for determining photosynthetic gene function.
Plants and animals diverged prior to the Chlorophyte-Streptophyte split, yet Chlamydomonas and animals have retained many features that were later lost in land plants. In particular, the cilia of Chlamydomonas are highly similar to those in mammals, making this alga an excellent system for studying ciliary disease [14, 15]. Because the cilia of Chlamydomonas are not essential mutants unable to assemble flagella can be isolated and studied, making this system uniquely useful. Furthermore, Chlamydomonas is one of very few model organisms from which it is possible to isolate the basal bodies and flagella, allowing biochemical and proteomic analyses of these organelles [16, 17].
The ability of Chlamydomonas to bridge the plant and animal lineages combined with access to the high quality genome provides a powerful genetic and genomic platform for probing the function of uncharacterized genes, such as the members of the “green cut” [18, 19] and the “cilia cut” [1]. Consequently, hundreds of laboratories around the world exploit Chlamydomonas to address fundamental questions related to photosynthesis, flagella and photoproduction of commercial commodities including biofuels.
Version 3.1: a high-quality draft genome and gene predictions
Following two preliminary versions (reviewed in [20]), a draft Chlamydomonas genome (JGI v3.1) was published in 2007 [1]. CC-503, a cell wall-less strain of mating type +, was selected because the absence of a complete cell wall facilitated cell lysis and high DNA yields. An average of 13x coverage was achieved by sequencing 2.1 million paired-end reads of small insert plasmids, fosmids and BACs on the Sanger platform. The major challenges presented by the high GC content (64%) was overcome with modifications to the sequencing protocols. Reads were assembled (Box 1) with the Joint Genome Institute’s (JGI) JAZZ assembler (Table 1). A typical annotation strategy that combined evidence from ~250k ESTs and de novo prediction tools (Box 2) generated 15,143 gene models on the assembly. The Chlamydomonas community performed unprecedented manual annotation of gene function, gene symbol (gene name), defline and description on 2,973 genes. This version was deposited in Genbank (Accession ABCN01000000). However, gene models in this release were sometimes truncated or missing because supporting expression data was very limited at the time. As discussed below, dramatic improvements in assembly and annotation have taken place and the most up-to-date version is maintained at Phytozome. Many sequence analysis studies were performed using this resource (reviewed in [21]) as well as comparative phylogenomic studies culminating in the creation of the “green cut” and “cilia cut” [1].
Box 1. Genome Sequencing.
Current technology cannot sequence entire chromosomes; rather many copies of the chromosomes are randomly fragmented into millions of pieces and these fragments are sequenced. The challenging process of assembly involves recreating the starting chromosomes from millions or even billions of fragment sequences (or reads). Storing all the reads in memory and comparing their sequences to each other can require tens or hundreds of Gb of RAM and assembly software can run for days.
Overlapping identical sequences found on different fragments allow the smallest scale of assembly (known as contigs; contiguous runs with no gaps). Tricks such as sequencing both ends of a piece of DNA of known length help to assemble at the next scale (scaffolds, which link contigs across gaps). By combining sequences from a range of known sized fragments, it is usually possible to recapitulate Mbp-sized runs of the genome sequence. Organizing scaffolds onto complete chromosomes currently requires integrating an optical or genetic map with the scaffold sequences. At this point, the genome sequence is probably a draft. Finishing requires laborious manual experiments to target gaps that need filling, and to correct sequence errors and misassemblies.
Serious problems exist: almost all genomes contain repeats (identical or nearly identical sequences that occur in many locations in the genome). If the sequencing reads are shorter than the repeat sequence, it is not possible to tell which copy of the repeat sequence generated the reads as repeat sequences are identical (to within the limits of sequencing errors). Sequencing errors as well as variation caused by polyploidy can sometimes be corrected, but may interrupt contigs. Further, some regions of the genome (such as high %GC regions, whose DNA forms tight hairpins that cannot be accessed by the sequencing enzyme) are hard to obtain sequence from. This and the random nature of sampling can lead to some regions of the genome that are only covered by a few reads (or, in extreme cases, none at all). Next generation sequencing strategies try to mitigate these problems by sequencing at very high average depth, but even so, poor coverage can generate a stretch of unknown sequence (a gap) in the assembly. There are a few very useful summary statistics for assessing genome quality. The simplest are the percent gaps and the percent of the genome represented in the assembly. More complex are the N/L50: if all the pieces that make up the assembly are ordered from longest to shortest, these are the number (N50) of pieces needed to make up 50% of the assembly (fewer is better) and the length (L50) of the shortest piece in this set (longer is better) (Table 1).
Table 1.
History of C. reinhardtii genome assembliesa
| Genome version | Release date | New data compared to previous releases | Chromosomes | Total Scaffolds | Total sequence (including % gaps) | Scaffold N50/L50 | Contig N50/L50 |
|---|---|---|---|---|---|---|---|
| 3 | 2006 | Sanger sequencing optimized for high %GC genomes | n/a | 1,557 | 120.2 Mb (12.5%) | 24 / 1.7Mb | 603 / 44.6 kb |
| 4 | 2008 | Complete reassembly with targeted Sanger sequencing of poor quality regions, followed by manual finishing and further rounds of targeted genome completion. Repeats resolved with 3kb- to BAC-sized clone sequencing. Genetic map with 349 markers [22] was used to anchor scaffolds on chromosomes. | 17 | 88b | 112.3 Mb (7.5%) | 7 / 6.6Mb | 322 / 90.6 kb |
| 5 | 2012 | New libraries generated at wide range of insert sizes, sequenced with Sanger and 454, with every gap targeted for sequencing. Scaffolds integrated into 957 marker genetic map (pers. comm. Martin Spalding), supported by Rymarquis 2005 [22]. | 17 | 54b | 111.1 Mb (3.6%) | 7 / 7.8 Mb | 140 / 219.4 kb |
initial assemblies consisted of scaffolds (v3). From v4 onwards the scaffolds were mapped to chromosomes using data from genetic maps.
of which 17 are chromosomes (71 and 37 unanchored scaffolds in v4 and v5 respectively)
Box 2. Gene modelling, or finding needles in a haystack.
The raw genome sequence (Box 1) tells us little about biological function. A series of algorithms with varying degrees of accuracy must be employed to tease this information out of the genome. More than half of a typical plant consists of repetitive sequences, i.e. it comprises up to thousands of stretches of sequence that are identical or nearly identical to each other. Repetitive sequences that are similar to each other comprise a repeat family; it is common to have thousands of different repeat families. The presence of many Mb of repetitive sequences greatly increases the computational time it takes to annotate the gene models in the genome (see below) because these regions do not often encode proteins, yet still have to be scanned. Furthermore, some gene finding algorithms will annotate large and spurious families of genes in repetitive sequence. In a process known as repeat masking, the genome is scanned for repetitive sequences and all occurrences are ‘masked’ from further analysis.
The next step is gene prediction, which builds “models” of the genes on the genome from statistical algorithms that recognize likely splice sites, translation starts and stops, open reading frames, typical intron and exon numbers and lengths per transcript. Modern algorithms also weave in homology data: regions of the assembly that can be translated into a sequence that is similar to a protein from a different organism are likely to encode a gene. and expression data (to confirm predicted splice junctions, add untranslated regions (UTRs) and putative alternative splice forms to transcript predictions). Toolkits like PASA [24], EVM [41] and MAKER2 [42] are commonly used to integrate expression and homology data into gene models. EST sequences do not usually identify full length mRNAs, so predictive algorithms range from conservative (give a minimum combination of exons) and inclusive (give all possible combinations of exons). A reasonable simple strategy is to generate the “best” model at a locus, at least as a starting point for downstream analysis. Sometimes, the longest model at the locus is used, assuming it is the most complete, however this approach is also subject to errors of locus merging. Finding the beginning and end of transcripts is tricky too, particularly in compact genomes including that of Chlamydomonas. Gene models that split or merge gene loci are the result of errors in predicting transcription starts and ends. Errors in gene models are caused by too little EST information (no transcript evidence is available to help delineate exon-intron structure of the gene model) just as much as from too much EST/RNA-Seq data where noise and inaccuracies in transcription or RNA processing (e.g. intron retention) start to confound what data corresponds to functional transcripts. It is important to note that even with high quality EST data and a good gene prediction, the gene models are just that – i.e. only models.
As genome projects mature, updated (and hopefully improved) assemblies and gene models are generated. It is of great interest to be able to map gene models from previous versions to the new data to leverage published work that references the old data and to new insights from more complete/detailed updated data sets. However, mapping annotations is challenging: previous models can be fragmented or incomplete and resolution of collapsed repeats in the new genome sequence can cause particular problems when trying to map paralogs correctly. Gap filling and assembly rearrangements cause additional problems. That being said, in a typical genome, two-thirds or more of the gene models can be mapped straightforwardly and most of the rest can be mapped to some degree, leaving several percent unmapped.
Tools such as Interproscan [43] are commonly used to do a first pass on predicting function based on sequence similarity or motifs. While having some notion of putative function is desirable, caution must be exercised because inaccuracies are commonplace [39] and computational prediction is no substitute for experimental verification.
Version 4: genome and annotation improvements
Subsequent improvements to the genome assembly and annotation were tackled systematically. Many gaps were filled with targeted sequencing of fragments appropriate to the size of the gap and manual analysis. The genome was completely reassembled and mapped onto a genetic map [22] that recapitulated the 17 chromosomes of Chlamydomonas with only 7.5% of the assembly represented by gaps (Table 1).
Gene models were predicted using a range of tools followed by manual review to reduce errors and increase annotation quality. Initially, gene models were predicted with the JGI pipeline (JGI v4; Table 2). Three annotations were generated with the Augustus algorithm [23], taking advantage of gradual improvements in its methods for integrating EST data. These updates (Aug u5, Aug u9 and Aug u10.2) gradually increased the number of gene models encoding complete proteins from a starting methionine to a terminal stop. This was particularly evident in Aug u10.2 in which expression data from over 7 millions 454 ESTs were incorporated into the gene models, allowing extensive annotation of untranslated regions for the first time (Figure 1; Table 2). The Aug u10.2 update was incorporated into Phytozome v.8 as the official JGI v4.3 annotation for genome assembly v4 (Table 2).
Table 2.
History of gene models and locus identifiers
| Gene model version a |
Release date |
Transcripts (alternative forms) |
New data compared to previous releases |
Locus ID format and example |
Transcript ID example | Data available at: |
|---|---|---|---|---|---|---|
| JGI v3 | 2005 | 15,143 (82b) | 204k Sanger ESTs | protein ID, unique number | 196029 | http://genome.jgi-psf.org/Chlre3/Chlre3.home.html |
| JGI v4 | 2008 | 16,709 (0) | New v4 assembly | protein ID, unique number | 334127 | http://genome.jgi-psf.org/Chlre4/Chlre4.home.html |
| Aug u5 | 2008 | 15,818 (1,070) | Includes alternate transcript predictions. Transcriptional starts and stops inferred from EST data [44] and trained on a set of manually inspected 5′ and 3′ UTR regions. | au5.gYYYYY_t1; YYYYY is a serial number along the assembly starting at 1 at the beginning of chromosome 1. | au5.g5896_t1 | http://augustus.gobics.de/predictions/chlamydomonas/ |
| Aug u9 | 2009 | 15,935 (0) | Augustus algorithm improvements | Au9.CreXX.gZZZZZZZ.t1; XX is the chromosome or scaffold number and ZZZZZZZ is a serial number along the assembly, increasing by 50. | Au9.Cre01.g003650.t1 |
http://augustus.gobics.de/predictions/chlamydomonas/ http://www.phytozome.net/chlamy |
| JGI v4.3 (Phytozome 8) | 2012 | 17,114 (0) | Based on Augustus u10.2. Incorporates 6.32M JGI and 0.69M Genoscope 454 ESTs, homology to Volvox carteri, proteomics data. | CreXX.gZZZZZZZ.t1.B; XX and ZZZZZZ as for Aug u9, B is the version number of this transcript sequence. | Cre01.g042500.t1.2 | http://genomes.mcdb.ucla.edu/cgi-bin/hgGateway |
| JGI v5.3.1 (Phytozome 9.1) | 2012 | 17,737 (1,789) | New v5 assembly. Based on Augustus u11.6. Incorporates 1.03 M 454 ESTs and 239M 2×100bp Illumina read pairsc and other Illumina data totalling 1.03 B reads. Alternate splice forms included in prediction. Initial partial mapping forwards of v4.3 locus IDs. | CreXX.gZZZZZZ.tA.B; XX and ZZZZZZ as for Aug u9, A is the number of the splice form, B is the version number of this splice form sequence. 13,448 models have stable IDs of this form. The remaining 6,078 models are of the form gYYYYY.tA where YYYYY is a serial number along the assembly and A is the number of the splice form. | Cre01.g006450.t2.1 or g200.t1 | http://www.phytozome.net/chlamy |
| JGI 5.5 (Phytozome 10) | 2014 | 17,741 (1,785) | Based on Augustus u11.6. Improved mapping forwards from v4.3. All loci have stable locus ID. | CreXX.gZZZZZZ.tA.B | Cre08.g386100.t3.1 | http://www.phytozome.net/chlamy |
All previous versions are mapped forward and can be browsed at http://www.phytozome.net/chlamy
Alternative transcripts annotated by hand
Of these four sequencing runs (116M reads) used strand specific sequencing.
Figure 1.
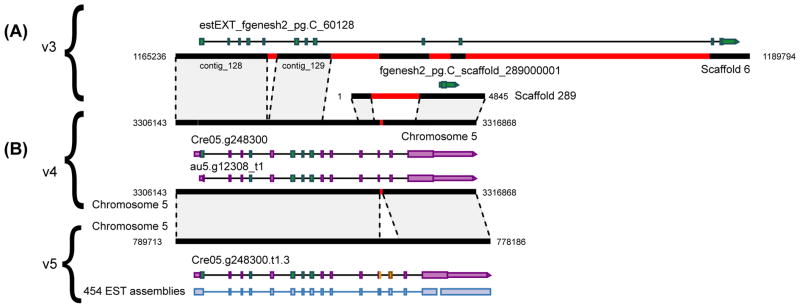
Refinement of the NRAMP4 gene model. Black and red boxes represent genome sequence and gaps respectively on portions of scaffolds or chromosomes (coordinates in bp indicated at the edges), for genome assembly versions as labelled on the left. Gene models are depicted as filled boxes (exons) along horizontal lines (introns). Box fill color indicates the first assembly version an exon was predicted in (green is v3, mauve is v4, orange is v5); wide and narrow sections represent coding sequence and untranslated regions respectively) and an arrowhead indicates the direction of transcription. Shading between dotted lines represents identical nucleic acid sequence between genome assemblies. (A) Comparing assembly v3 to v4, note the amount of gap sequence (red) that was filled, allowing more accurate gene loci to be predicted. The sequence from contig_128 and contig_129 from scaffold 6 were placed on chromosome 5, as was all of scaffold 289. The gap between contig_128 and contig_129 was filled (by addition of 17bp) in v4, while the gap in scaffold 289 was partially filled (by addition of a further 1178bp). (B) The gap in v4 was filled in the v5 assembly (899bp), which is near-finished quality, allowing the extension of exon 12 and prediction of a new exon (both represented by orange boxes) and a gene model that is completely consistent with assembled 454 EST evidence (lilac track at the bottom).
Version 5: further improvements
Version 5 of the genome assembly, released in 2012, improved on v4 by targeting remaining gaps and using new Sanger- and 454-based sequencing from a wide range of library sizes. This approach successfully filled approximately half of the gaps (Table 1), and combined with a 957 genetic marker map (Martin Spalding pers. comm.) allowed 34 of the 71 unanchored scaffolds in v4 to be incorporated into chromosomes (Table 1), leaving just 37 unanchored scaffolds in the v5 assembly.
The v5 gene models were generated by integrating new expression data from 59 RNA-Seq experiments totalling 1.03B reads. These included 239M read pairs from JGI, roughly a quarter of which were strand-specific, allowing the direction of transcription and hence the strand of the gene model to be inferred. Gene models were based on Augustus update 11.6 (Aug u11.6) predictions. However, these predictions were made without repeat masking (because the 67% GC content of Chlamydomonas coding regions [1] leads to excessive repeat masking (Box 2)). They were filtered to remove gene models with ≥30% overlap to known transposable elements, open reading frames <50 amino acids or internal stop codons.
Annotation version JGI v4.3 consisted of 17,114 gene loci (Table 1). A preliminary mapping of 12,263 (72)% of the stable locus identifiers from v4 (see below) was released (JGI v5.3.1, Table 2). The latest version (JGI v5.5) used a more robust mapping algorithm that used local synteny to map loci (12,647 loci, 74%). In addition, genes on the 34 scaffolds that were integrated into chromosomes were given a new locus updated to reflect their new location (2,487 loci, 15%). Due to large changes in the gene models between versions, the remaining loci (1,980, 12%) could not be mapped from v4 to v5 in a straightforward manner, and new loci were generated. Expert annotation of gene symbols, deflines and descriptions was carried forwards during the mapping process.
Thanks to the high quality genome sequence and the substantial amount of expression data available, as well as the functional annotation efforts of the community, gene models in the JGI flagship genome of Chlamydomonas represent the most highly curated genomic data for any alga.
Future work
Developments in the Chlamydomonas genome project will continue. A systematic review of gene symbols is nearing completion and will form the basis of an updated Chlamydomonas GenBank submission. A more involved update of deflines (see glossary) and gene descriptions with genes will come later in 2014 together with methods for a user to contribute new information to the database.
As sequencing technologies develop, new kinds of data on e.g. chromatin state will become available and incorporating them into the Chlamydomonas genome project will enable novel and exciting analyses on gene regulation.
Resources for gene identifier conversion and bulk annotations
Gene identifier conversion
As Chlamydomonas assembly versions and gene models are refined, updated annotations with new locus and transcript identifiers have been generated. This necessitates the ability to convert between versions. For instance, if an RNA-Seq experiment was published with JGI v4 transcript IDs, a researcher would need to convert the old IDs for comparison to present work being performed using the new Aug u11.6 IDs. For small tasks, this can be done manually with BLAT [25] searches of transcripts against the genome. However, for longer lists of genes, The Algal Functional Annotation Tool offers a Batch Identifier Conversion tool (Table 3). Currently, the tool can convert between JGI v3, JGI v4, Augustus u5, u9 u10.2 (JGI v4.3) and u11.6 (JGI v5.3.1 and v5.5). The Program to Assemble Spliced Alignments (PASA) tool [24] was used to map previous gene models to the v5 assembly; this was aided by a BLAT [25] and BLASTP [26] based approach that used neighbouring genes to help map loci. Future releases of Chlamydomonas gene models will be integrated into the tool.
Table 3.
Online Chlamydomonas resources
| Database | URL | Summary |
|---|---|---|
| Phytozome [27] | http://www.phytozome.net | Primary repository of Chlamydomonas genome/gene models. Bulk retrieval of annotation data. Structured to enable comparative genomics with other plants and algae. Contains user validated annotations, and PFAM, Panther and GO predicted annotations. |
| UCLA algal genomics portal | http://genomes.mcdb.ucla.edu/ | Chlamydomonas genome browser. Repository for multiple transcriptomic datasets. |
| Algal Annotation Tool [45] | http://pathways.mcdb.ucla.edu/algal/index.html | Batch conversion of gene identifiers. Bulk annotation prediction via Kegg, MapMan, GO, Panther, Metacyc. |
| GIAVAP | https://giavap-genomes.ibpc.fr/chlamydomonas | Comparison of v5.5 gene predictions with previous versions, browser with BAC and fosmid ends. |
| Iomiqs [29] | http://iomiqsweb1.bio.uni-kl.de | Bulk annotation prediction via MapMan with visual output. |
| Predalgo [31] | https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main | Green algal-specific protein localization predictions. |
| BioCyc [46] | http://biocyc.org/CHLAMY/organism-summary | Maps gene products onto metabolic pathways. |
| Chlamydomonas Connection | http://www.chlamy.org/ | A Gateway to Resources for Chlamydomonas Research: news, methods, jobs, gene nomenclature etc. |
| Chloroplast genome [47] | http://www.chlamy.org/chloro | Map and gene lists. |
| Flagellar proteome [17] | http://labs.umassmed.edu/chlamyfp/index.php | Based on version 3, but lists JGIv4 equivalence; UMASS Amherst. |
| Kazusa Institute [10] [11] | http://est.kazusa.or.jp/en/plant/chlamy/EST | Distributes cDNA clones corresponding to their EST collection. |
| Chlamydomonas Resource Center | http://chlamycollection.org/ | Distributes strains, plasmids, cDNA libraries, kits etc. |
| ChlamyStation | http://chlamystation.free.fr/ | Paris (IBPC) Collection of photosynthesis mutants. |
| Transcription factors | http://plntfdb.bio.uni-potsdam.de/v3.0/index.php?sp_id=CRE4 | Part of the Plant Transcription Factor Database, University of Potsdam. |
| Silencing RNAs [48] | http://cresirna.cmp.uea.ac.uk/ | from the Sainsbury Laboratory, D.C.Baulcombe group. |
| Green Genie2 [49] | http://stormo.wustl.edu/GreenGenie2/ | Green genie gene models. |
| Plant TFDB [50] | http://planttfdb.cbi.pku.edu.cn/index.php?sp=Cre | Database of Chlamydomonas transcription factors |
However, automated mapping is impossible or misleading if the underlying genomic sequence (and hence the gene model and, potentially, the protein sequence) for a particular locus has changed drastically between versions such as in split/merged genes (Box 2) or filling of large exon encoding gaps.
Bulk retrieval of gene function annotation
Whole-genome scale datasets of gene function annotations must be downloaded to perform global -omics studies. Several online resources provide this functionality (Table 3). The Phytozome database [27] has integrated the Intermine tool [28] for bulk download of sequence and annotation information. Phytozome maintains the gold standard, experimentally validated, user annotations, descriptions and deflines and in silico functional predictions. Alternatively, the Iomiqs database [29] utilizes MapMan ontologies to provide a visual output that “bins” genes into various metabolic groupings. More specific types of annotation can be found on the Chlamydomonas section of BioCyc, which maps genes onto metabolic pathways, the cis-regulatory element prediction database [30], and PredAlgo [31], providing green algae-specific protein localization predictions (Table 3).
Uniform and stable gene names for Chlamydomonas
Following in the footsteps of the reference plant, Arabidopsis, once the Chlamydomonas assembly was mapped to chromosomes in version 4, every genetic locus in the genome was given a permanent address or locus identifier (e.g. Cre01.g123450, Table 2). These identifiers ensure continuity in nomenclature going forwards. Such frameworks are widespread for other commonly used organisms and have undoubtedly contributed to their adoption as model systems [32–38].
In addition to the following guidelines, we recommend that researchers use Phytozome as the primary repository for name and annotation data. A mechanism for manual annotation of genes is under active development.
To name or not to name?
Over-annotation in databases, whether of an automated origin, or user-initiated, is common and detrimental: errors can proliferate as computer algorithms map data to new genomes [39]. We therefore propose that genes should only be named (i.e. given what geneticists formally call a gene symbol, such as ODA11 or RBCS2) if one of the following is true: (1) A function or involvement in a specific biological process is associated with a publication. In this case, a pubmed ID (PMID) or other citation should accompany the gene symbol, which should be included in the Phytozome Description. (2) A gene is associated with a high-throughput screen or global study, e.g. proteomes of flagella resulting in the naming of flagellar associated proteins (FAP) or the conserved green-lineage (CGL) associated genes. (3) The gene function is confidently predicted by a rigorous bioinformatic study. Indeed, annotation by investigators with extensive knowledge of particular pathway has been very valuable [40].
If the above criteria are not met, then a gene symbol should not be created. This includes genes encoding proteins with poor similarity to sequences in other organisms (forcing an annotation) or for which the naming is only based on a single conserved domain. In a similar vein, genes should not be named on the basis of homology to proteins involved in a process that does not (or has not been shown to) exist in Chlamydomonas. For example, the protein encoded by Cre02.g116900 displays high similarity to small hydrophilic plant seed proteins in Arabidopsis. In the absence of seed production, this protein clearly cannot perform this function in Chlamydomonas, and therefore should not be named after the Arabidopsis gene ATEM1. Genes without an assigned symbol should be referred to by their locus ID, since every locus has a unique and stable ID. To distinguish between a gene and an encoded protein, we suggest italicizing locus IDs (Crex.gyyyyyy) and non-italicizing proteins (Crex.gyyyyyy).
How to devise a gene symbol
Gene nomenclature guidelines have been established by the Chlamydomonas community (http://www.chlamy.org/nomenclature.html), but are not always strictly followed. We hereafter recall the basic rules, and when it is accepted to depart from them.
-
The preferred format for gene symbols in C. reinhardtii is a 3–5 letter root, in uppercase for nuclear genes, or lower case for organelle genes; this is followed by a number denoting isoform, or occasionally subunits (although for historically named genes, a combination of letters or numbers has been used and can denote numbered mutants recovered in a genetic screen. Alternatively, the gene symbol, including a number, has on occasion been maintained exactly from the orthologous gene of another organism). In general, 3 letters is preferred, but may not always be possible (for example when using an Arabidopsis gene name, which does not conform to a 3-letter standard, the name should not be abbreviated). The root should indicate or abbreviate some aspect of function or phenotype. For example GPD1-GPD4 encode 4 isoforms of glycerol-3-phosphate dehydrogenase, ASA1-ASA9 encode the 9 Chlorophyceae-specific subunits of the mitochondrial ATP synthase and ACLA1 and ACLB1 encode ATP citrate lyase subunits A and B). For historical reasons, some names depart from this scheme, for example HSP70A, HSP70B, HSP70C encode three isoforms of HSP70. Nuclear genes for photosynthesis will retain their cyanobacterial name, followed by a number to denote isoform, unless several isoforms exist (for example RBSCS1-RBCS2, PSBP1-PSBP9)
To make nomenclature more intuitive, gene symbols can be adapted from those of orthologs in other organisms where characterized orthologs exist. This will ensure related gene symbols across organisms, simplifying comparisons between organisms and retrieval of associated literature.
Potential confusion should be avoided by confirming the proposed gene symbol is not already in use in Chlamydomonas. The authors of this manuscript are available to help researchers verify this. Ideally, it should also not be used in another organism for a different function. The global gene hunter tool (http://www.yeastgenome.org/help/community/global-gene-hunter) enables six databases to be searched simultaneously for this purpose. The Gene database (http://www.ncbi.nlm.nih.gov/gene), at the National Center for Biotechnology Information (NCBI), is also useful for this purpose and can be used to trace gene name roots across different organisms.
Historically, many genes were discovered following genetic studies of mutants named on the basis of a phenotype, or expression or localization studies (e.g. LF5 mutants have long flagella, LCI5 is low-CO2 inducible). Whenever informative of function, these names are preferred as the primary gene symbol over names describing molecular functions. Alternative gene symbols are stored as aliases in Phytozome, allowing the gene to be found if any of its symbols is used as a search term. This effectively links genes to all related literature and vice versa.
Concluding remarks
The culmination of the substantial efforts over a decade is a near-finished Chlamydomonas assembly at the scale of complete chromosomes annotated with high-confidence gene models (JGI v5.5), and mappings from previous versions [24]. In addition, our gene naming guidelines provide an empirical framework in which gene names are both likely to reflect function and searchable. If future gene naming follows the policy outlined above, this will help maximize the benefits that the Chlamydomonas community derives from its genome project, particularly as refinements and developments continue into the future.
Highlights.
Chlamydomonas is a model algal system with a mature genome project.
Substantial improvements to the genome assembly and gene models have been made
Diverse ‘omics data are publicly available, centered at Phytozome.net
A uniform gene symbol and stable gene locus nomenclature aids researchers
Acknowledgments
This work was supported by the National Institutes of Health R24 GM092473 to S.M. I.K.B. and C.B.-H are supported by training grants from the National Institutes of Health (T32ES015457 and GM100753 respectively). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We thank Stefan Schmollinger, Alizée Malnoë, Patrice Salomé and Ursula Goodenough for critical reading of the manuscript.
Glossary
- Defline
A short (2–6 word) description of the encoded protein. For example, for LAO1, the description is Periplasmic L-amino acid oxidase, catalytic subunit
- Description
A lengthier, yet concise, description of the encoded protein with supporting evidence. For example, for LAO1, the defline is L-amino acid oxidase, catalytic subunit M[alpha]; induced by nitrogen starvation [PMID: 8344302]
- Gene name
also known as gene symbol. A series of letters and/or numbers assigned to a gene of known function or with known involvement in a biological process. The gene name is unique within Chlamydomonas, and for non-historically named genes, it should be identical to orthologous gene names from other model organisms. E.g. FTR1 in Chlamydomonas and FTR1 in Saccharomyces cereviase
- Locus ID
Defines the genomic region (nuclear, mitochondrial or plastid) of a feature (typically a gene). In the absence of a gene name, the locus ID should be used to refer to a specific gene. Nuclear loci have the form Cre01.g123450
- Transcript ID
Typically one or more transcripts are transcribed from a locus. These have .t1, t2 etc. appended to the locus name e.g. a locus that expresses two alternative spliceforms might be described by the following transcript IDs: Cre01.g123450.t1 and Cre01.g123450.t2. Strictly, a complete transcript ID ends with a version number that increases whenever the sequence of the transcript model changes e.g. Cre01.g123450.t1.1. In everyday usage, the version number is often omitted for clarity
- User annotation
the “gold standard” in gene function annotation. Applied to a gene by an expert in the relevant biological process and supported by experimental or non-automated informatic evidence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science (New York, NY) 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kropat J, et al. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. The Plant journal : for cell and molecular biology. 2011;66:770–780. doi: 10.1111/j.1365-313X.2011.04537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neupert J, et al. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. The Plant journal : for cell and molecular biology. 2009;57:1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 4.Barbieri MR, et al. A forward genetic screen identifies mutants deficient for mitochondrial complex I assembly in Chlamydomonas reinhardtii. Genetics. 2011;188:349–358. doi: 10.1534/genetics.111.128827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tunçay H, et al. A forward genetic approach in Chlamydomonas reinhardtii as a strategy for exploring starch catabolism. PLoS One. 2013;8:e74763. doi: 10.1371/journal.pone.0074763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerutti H, et al. RNA-mediated silencing in Algae: biological roles and tools for analysis of gene function. Eukaryot Cell. 2011;10:1164–1172. doi: 10.1128/EC.05106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroda M. RNA silencing in Chlamydomonas: mechanisms and tools. Curr Genet. 2006;49:69–84. doi: 10.1007/s00294-005-0042-1. [DOI] [PubMed] [Google Scholar]
- 8.Sizova I, et al. Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. The Plant journal : for cell and molecular biology. 2012:873–882. doi: 10.1111/tpj.12066. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Ballester D, et al. Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods. 2011;7:24. doi: 10.1186/1746-4811-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asamizu E, et al. A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. 1999 doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- 11.Asamizu E, et al. Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 2000;7:305–307. doi: 10.1093/dnares/7.5.305. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, et al. Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol Biol. 1994;24:663–672. doi: 10.1007/BF00023562. [DOI] [PubMed] [Google Scholar]
- 13.Becker B. Snow ball earth and the split of Streptophyta and Chlorophyta. Trends Plant Sci. 2013;18:180–183. doi: 10.1016/j.tplants.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Silflow CD, Lefebvre PA. Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol. 2001;127:1500–1507. [PMC free article] [PubMed] [Google Scholar]
- 15.Pazour GJ, Witman GB. The Chlamydomonas Sourcebook. Vol. 3. Elsevier; New York, NY: 2009. The Chlamydomonas flagellum as a model for human ciliary disease. [Google Scholar]
- 16.Keller LC, et al. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 17.Pazour GJ, et al. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]; DNA research : an international journal for rapid publication of reports on genes and genomes. 6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- 18.Heinnickel ML, Grossman AR. The GreenCut: re-evaluation of physiological role of previously studied proteins and potential novel protein functions. Photosynth Res. 2013;116:427–436. doi: 10.1007/s11120-013-9882-6. [DOI] [PubMed] [Google Scholar]
- 19.Karpowicz SJ, et al. The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem. 2011;286:21427–21439. doi: 10.1074/jbc.M111.233734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman AR, et al. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryotic cell. 2003;2:1137–1150. doi: 10.1128/EC.2.6.1137-1150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallon O, Dutcher S. Treasure hunting in the Chlamydomonas genome. Genetics. 2008;179:3–6. doi: 10.1534/genetics.104.179101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rymarquis LA, et al. Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 2005;137:557–566. doi: 10.1104/pp.104.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanke M, et al. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 24.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic acids research. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic acids research. 2012;40:D1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smedley D, et al. BioMart--biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mühlhaus T, et al. Quantitative shotgun proteomics using a uniform 15N-labeled standard to monitor proteome dynamics in time course experiments reveals new insights into the heat stress response of Chlamydomonas reinhardtii. Mol Cell Proteomics. 2011;10:M110.004739. doi: 10.1074/mcp.M110.004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding J, et al. Systematic prediction of cis-regulatory elements in the Chlamydomonas reinhardtii genome using comparative genomics. Plant Physiol. 2012;160:613–623. doi: 10.1104/pp.112.200840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tardif M, et al. PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol. 2012;29:3625–3639. doi: 10.1093/molbev/mss178. [DOI] [PubMed] [Google Scholar]
- 32.Wain HM, et al. Guidelines for human gene nomenclature. Genomics. 2002;79:464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- 33.Eppig JT, et al. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee SY, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marygold SJ, et al. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demerec M, et al. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966;54:61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demerec M, et al. A proposal for a uniform nomenclature in bacterial genetics. J Gen Microbiol. 1968;50:1–14. doi: 10.1099/00221287-50-1-1. [DOI] [PubMed] [Google Scholar]
- 39.Schnoes AM, et al. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS computational biology. 2009;5:e1000605–e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anton BP, et al. The COMBREX Project: Design, Methodology, and Initial Results. PLoS Biol. 2013;11:e1001638. doi: 10.1371/journal.pbio.1001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome biology. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang C, et al. Expressed sequence tags with cDNA termini: previously overlooked resources for gene annotation and transcriptome exploration in Chlamydomonas reinhardtii. Genetics. 2008;179:83–93. doi: 10.1534/genetics.107.085605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez D, et al. Algal Functional Annotation Tool: a web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC bioinformatics. 2011;12:282–282. doi: 10.1186/1471-2105-12-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maul JE, et al. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell. 2002;14:2659–2679. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molnár A, et al. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 49.Kwan, et al. Improving gene-finding in Chlamydomonas reinhardtii:GreenGenie2. BMC Genomics. 2009;10:210. doi: 10.1186/1471-2164-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin, et al. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42:D1182–1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]


