Abstract
Eosinophils originate in the bone marrow from an eosinophil lineage–committed, IL-5 receptor alpha (IL-5Rα)–positive, hematopoietic progenitor (eosinophil progenitor). Indeed, IL-5 is recognized as a critical regulator of eosinophilia and has effects on eosinophil progenitors, eosinophil precursors and mature eosinophils. However, substantial levels of eosinophils remain after IL-5 neutralization or genetic deletion, suggesting that there are alternative pathways for promoting eosinophilia. Herein, we investigated the contributory role of IL-5 accessory cytokines on the final stages of eosinophil differentiation. IL-5 stimulation of low-density bone marrow cells resulted in expression of a panel of cytokines and cytokine receptors, including several ligand-receptor pairs. Notably, IL-4 and IL-4Rα were expressed by eosinophil precursors and mature eosinophils. Signaling through IL-4Rα promoted eosinophil maturation when IL-5 was present, but IL-4 stimulation in the absence of IL-5 resulted in impaired eosinophil survival, suggesting that IL-4 cooperates with IL-5 to promote eosinophil differentiation. In contrast, CCL3, an eosinophil precursor–produced chemokine that signals through CCR1, promotes terminal differentiation of CCR1-positive eosinophil precursors in the absence of IL-5, highlighting an autocrine loop capable of sustaining eosinophil differentiation. These findings suggest that brief exposure to IL-5 is sufficient to initiate a cytokine cooperative network that promotes eosinophil differentiation of low-density bone marrow cells independent of further IL-5 stimulation.
Keywords: Allergy, Cell Differentiation, Hematopoiesis, Inflammation
Introduction
Eosinophils are derived from an eosinophil lineage–committed progenitor (EoP) that is derived from the granulocyte/macrophage progenitor in mice and the common myeloid progenitor in humans (1, 2). The EoP is identified via surface expression of CD34, IL-5 receptor alpha (IL-5Rα, a.k.a. CD125) and low levels of c-kit (CD117). EoPs reside in small numbers primarily in the bone marrow (~0.05% of lineage-negative CD34+ cells), with even lower levels found in peripheral blood and in human umbilical cord blood (2). Importantly, EoP numbers in the bone marrow increase substantially with allergen exposure or helminth infection and in patients with eosinophil-associated disorders (2–4). Further, EoPs are found within allergic nasal and pulmonary mucosa and in the sputum from asthmatic patients, suggesting that eosinophil differentiation in situ may contribute to the accumulation of mature eosinophils in tissues (5–7).
EoPs mature into granule-containing eosinophil precursors (preEos) that maintain proliferative capacity and surface expression of IL-5Rα (Figure 1). PreEos are an ill-defined heterogeneous population that matures into fully differentiated eosinophils that have a characteristic segmented nucleus (bilobed in humans and circular in mice) and a cytoplasm rich with eosin-staining granules. IL-5 is recognized as a critical regulator of eosinophilia and has effects on multiple eosinophil-lineage developmental stages (8). Although IL-3, IL-5 and GM-CSF have been shown to induce eosinophil-containing colony formation from bone marrow progenitors, IL-5 stimulation of bone marrow progenitors selectively produces eosinophils (9). Overexpression of IL-5 in mice results in profound eosinophilia, suggesting that IL-5 not only directs differentiation, but also has a proliferative effect on EoPs and/or preEos (10, 11). Yet, IL-5–deficient animals show a near-normal steady-state eosinophil production, suggesting that other unidentified mediators can promote eosinophil development in the absence of IL-5 (12). Notably, although treatment with anti–IL-5 lowers blood and tissue eosinophil levels in mice and man with established disease (13), there are still substantial residual eosinophils, especially in the bone marrow (14–18). Taken together, these studies highlight the need to identify mediators responsible for persistent tissue eosinophilia after IL-5 is neutralized via targeted treatment.
Figure 1. Eosinophil differentiation schematic in low-density bone marrow cultures.
Low-density bone marrow (LDBM) cells include the IL-5 receptor alpha (IL-5Rα)–expressing eosinophil-lineage committed progenitor (EoP), which differentiates into eosinophil precursors (preEos), which expresses IL-5Rα and Siglec-F on its surface. The preEos further differentiates into a mature eosinophil (Eos) identified by surface expression of IL-5Rα, Siglec-F and CCR3.
We utilized an ex-vivo culture system to identify eosinophil-promoting pathways that are initiated by IL-5 and could maintain an eosinophil-promoting environment after IL-5 is withdrawn. We report that IL-5 stimulation of low-density bone marrow (LDBM) cells for 4 days is sufficient to induce a cooperative cytokine network that can promote terminal eosinophil differentiation in the absence of further IL-5 stimulation. This cooperative network includes expression of a panel of cytokines and cytokine receptors, including ligand-receptor pairs that have the potential to influence eosinophil maturation. Notably, IL-4 and its receptor IL-4Rα are expressed during the full spectrum of eosinophil developmental stages. IL-4Rα signal transduction in LDBM cells promoted eosinophil differentiation in the presence of IL-5, whereas IL-4 stimulation of preEos in the absence of IL-5 impaired eosinophil survival, highlighting a cooperative relationship between IL-4 and IL-5 signal transduction. In contrast, preEos express CCL3 and the CCL3 receptor CCR1, which can promote terminal differentiation of preEos in the absence of IL-5. As CCL3 expression is induced in eosinophil-associated disorders (19, 20), our data provide mechanistic insight into the limitations observed in solely targeting IL-5, as other mediators (e.g. CCL3) can promote eosinophil maturation after IL-5 removal.
Materials and Methods
Mice
In-house–bred male and female wild-type (BALB/c), IL-4 knockout (KO) (21), IL-4RαKO (purchased from Taconic, Hudson NY), IL-5KO (12) and IL-4 enhanced green fluorescent protein (eGFP) reporter (22) mice were used as the source of LDBM cells. All mice were housed under specific pathogen-free conditions and handled under approved protocols of the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center (CCHMC).
LDBM cell cultures
LDBM cultures were performed as previously described (23) unless otherwise indicated to differentiate preEos and mature eosinophils (Figure 1). Cell-free culture supernatants were collected after 5, 8 and 11 days of IL-5 stimulation. For the IL-5 withdrawal (IL-5W) model (Figure 2A), LDBM cells were cultured in 24-well plates in media containing IL-5 (10 ng/mL) for 4 days; thereafter, for the remainder of the culture period (6 days), 50–75% of the culture media was removed every 2 days and replaced with media without added IL-5 (3 times total). For the IL-5 removal (IL-5V) model (Figure 2E), LDBM cells were cultured in 24-well plates in media containing IL-5 (10 ng/mL) for 4 or 6 days and then washed with culture media to remove IL-5 and replated and cultured in media without added IL-5 for the remainder of the culture period with media changes every 2 days. For some IL-5V, LDBM cells were washed, replated in media containing the cytokines (Peprotech, Rocky Hill, NJ) CXCL10 (1 ng/mL), CCL3 (50 ng/mL), CCL4 (50 ng/mL), IL-4 (5 ng/mL), GM-CSF (1 ng/mL), IL-9 (1 ng/mL) or IL-5 (10 ng/mL) and collected after 48 hours for flow cytometry analysis, along with cell-free supernatants for ELISA analysis. For some experiments, the neutralizing antibodies anti–IL-5 (5 µg/mL, clone TRFK5, R&D Systems, Minneapolis, MN), anti–GM-CSF (5 µg/mL, clone MP122E9, R&D Systems), anti-CXCL10 (12 µg/mL, R&D Systems), anti-CCL3 (0.2 µg/mL, R&D Systems) or anti-CCL4 (12 µg/mL, R&D Systems) or appropriate isotype IgG control antibodies (rat IgG2a or goat IgG, R&D Systems) were added to media at the indicated time points.
Figure 2. IL-5 withdrawal has a mild effect on final eosinophil maturation.
(A) Schematic of in-vitro model for IL-5 withdrawal (IL-5W) from cultured low-density bone marrow (LDBM) cells is shown. (B) Yield of mature eosinophils (mean ± SEM, n = 11 independent experiments with at least 3 wells per condition in each experiment) from LDBM cells cultured in IL-5 continuously (IL-5) or after IL-5W is shown. (C) Representative dot blots showing CCR3 and Siglec-F surface staining of day-matched LDBM cells cultured in IL-5 continuously (IL-5) or after IL-5W are shown. Percentage of gated, live cells is shown in Siglec-F+ quadrants. (D) Kinetics of absolute number (mean ± SEM, n = 3 independent experiments with 3 wells per condition in each experiment) of cells per well from LDBM cells cultured in IL-5 continuously (IL-5) or after IL-5W in the presence of anti–IL-5 antibody or isotype control IgG is shown. ***p < 0.001, ****p < 0.0001 when compared to wells containing isotype control antibody. (E) Schematic of in-vitro model of removal of IL-5 (IL5V) from cultured LDBM cells is shown. (F) Yield of mature eosinophils (mean ± SD, representative of 3 independent experiments with 4 wells per condition per experiment) from LDBM cells cultured following IL-5V after 4 or 6 days compared to cultured with IL-5 added back to culture media is shown. ***p < 0.001, ****p < 0.0001 when compared to wells containing IL-5. (G) Representative dot blots showing CCR3 and Siglec-F surface staining of day-matched LDBM cells cultured following IL-5V or cultured with IL-5 added back to culture media are shown. Percentage of gated, live cells is shown in 3 quadrants. (H) Fraction (mean of 3 independent experiments with 3–6 wells per experiment) of cultured LDBM cells in each quadrant following IL-5V or after being cultured with IL-5 added back to culture media are shown. *p ≤ 0.05 when compared to fraction of LDBM cells when IL-5 is added back to culture media. Eos, mature eosinophil. preEos, eosinophil precursor.
Flow cytometry staining, data acquisition and analysis
Cells were washed twice and resuspended in 100 µL flow buffer (1X PBS containing 1 mM EDTA) at a concentration of 1–100 × 106 cells/mL. Blocking was performed in flow buffer with 0.5–5.0 µg rat anti-mouse CD16/CD32 (BD Bioscience, San Diego, CA) for 5 minutes at 4°C. Cells were incubated with antibodies for 15–30 minutes at 24°C and then stained for 5 minutes with the viability dye described below. Cells were then washed and resuspended in flow buffer. Eosinophils were identified with FITC-CCR3 (83101, R&D Systems), PE-Siglec-F (E50-2440, BD Biosciences, San Diego, CA) and Live/Dead near-IR fixable stain (Molecular Probes, Grand Island, NY). IL-4 eGFP reporter kinetics were determined by staining eosinophils with PE-CCR3 (83101, R&D Systems), Brilliant Violet 421-Siglec-F (BD Biosciences) and Live/Dead near-IR (Molecular Probes). Eosinophils expressing chemokine receptors were identified by staining with PE-CCR1 (R&D Systems), PE-CCR5 (Biolegend, San Diego, CA) or PE-CXCR3 (R&D Systems) and with FITC-CCR3 (83101, R&D Systems), Brilliant Violet 421-Siglec-F (BD Biosciences) and Live/Dead near-IR (Molecular Probes). Heat-killed LDBM (1 minute at 95°C) and live LDBM were used for live/dead staining controls. Cell counts were determined using 5-µm AccuCount particles (Spherotech, Lake Forest, IL) according to manufacturer's instructions. Data were acquired using LSR II, FACS Canto, and LSR Fortessa flow cytometers (BD Bioscience) maintained by the Research Flow Cytometry Core at CCHMC. Data were compensated and analyzed using FlowJo software (Tree Star, Ashland, OR). Compensation was calculated in FlowJo using unstained and stained cells (Live/Dead) and OneComp eBeads (eBioscience, San Diego, CA). Gate boundaries and shapes were determined using fluorescence-minus-one controls including appropriate isotype antibodies.
PreEos isolation and culture
PreEos (SiglecF+CCR3−CD11c−) were purified from Day 8 LDBM cultures via FACS with MoFlo™ XDP (Beckman Coulter, Pasadena, CA) and replated in 24-well plates at 1×106 cells/mL. Cells and cell-free supernatants were collected after 24 and 48 hours. Total RNA was isolated with the Direct-zol RNA miniprep (Zymo Research, Irvine, CA) following the manufacturer’s instructions, and cell-free supernatants were collected for ELISA analysis.
Gene expression
Gene expression in isolated preEos was determined using primers described in Table 1. Reaction mixtures contained 10 ng cDNA template, 100 nM of each primer, and 1X FastStart Universal SYBR Green Master with Rox (Roche Diagnostics, Indianapolis, IN). Real-time quantitative PCR was carried out on an ABI 7900HT (Applied Biosystems, Foster City, CA) thermal cycler using the following program: 95°C for 10 minutes followed by 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds. Expression was calculated using the ΔΔCt method (Livak) and normalized to Gapdh.
Table 1.
Primers used for Real-Time Quantitative PCR (RT-qPCR)
| Gene | Gene Accession Number |
Forward Primer | Reverse Primer | Size (bp) |
|---|---|---|---|---|
| Csf2 | NM_009969 | TCGTCTCTAACGAGTTCTCCTT | CGTAGACCCTGCTCGAATATCT | 76 |
| Csf2ra | NM_009970 | CTGCTCTTCTCCACGCTACTG | GAGACTCGCCGGTGTATCC | 200 |
| Csf2rb | NM_007780 | AAAAACAGCCAGTGTCCTGTG | GATGCTGACGTTCTTGGGAAG | 221 |
| Epx | NM_007946 | ATGGAGACAGATTCTGGTGG | CCAGTATTGTCGCATACAATCC | 102 |
| Gapdh | NM_008084 | CTGGTATGACAATGAATACGG | GCAGCGAACTTTATTGATGG | 191 |
| Il3ra | NM_008369 | CTGGCATCCCACTCTTCAGAT | GGTCCCAGCTCAGTGTGTA | 115 |
| Il4 | NM_021283 | CTGTAGGGCTTCCAAGGTGCTTCG | CCATTTGCATGATGCTCTTTAGGC | 209 |
| Il4ra | NM_001008700 | AAT GTG ACC TAC AAG GAA CC | GGA CTC CAC TCA CTC CAG | 124 |
| Il5 | NM_010558 | CTCTGTTGACAAGCAATGAGACG | TCTTCAGTATGTCTAGCCCCTG | 102 |
| Il5ra | NM_008370 | CCACAAGAAGACGAATATGATACC | TTGAGTTCAGCAGAAACCCA | 142 |
| Prg2 | NM_008920 | TTGCAAACTTGACAAGACCC | GTTTCAGAGCTCAGATGAAGAG | 109 |
| Siglec5 | NM_145581 | CTCAGTGTCATCTATGCTCCA | GACTCACCCTCTTGGATCTG | 112 |
Cytokine levels
Cytokine levels were measured in cell-free supernatants from cultured LDBM cells or cultured, isolated preEos via multiplex assay using the Mouse Cytokine/Chemokine Panel I (Millipore) by the Flow Cytometry Core Facility at CCHMC or by ELISA per the manufacturer’s instruction.
Microarray analysis
For RNA extraction, cultured LDBM cells were collected before IL-5 stimulation and after 4 and 10 days of IL-5 stimulation. No selection for any specific cell type was performed prior to RNA extraction. The Affymetrix Exon Chip MoGene-1.0 was used to compare gene expression profiles between LDBM cells before IL-5 stimulation (0 days) and LDBM cells stimulated with IL-5 for 4 or 10 days. Microarray data were analyzed using the GeneSpring software (Agilent Technologies, Santa Clara, CA). Statistical significance was determined at p < 0.05 with one-way ANOVA and Benjamini-Hochberg multiple testing correction. Pathway enrichment analysis was performed with single-experiment analysis in GeneSpring. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (24) and are accessible through GEO Series accession number GSE55386 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ctsfaamehnixdcx&acc=GSE55386).
Statistics
Data were analyzed using either a two-tailed, unpaired Student's t test or a one-way ANOVA with Bonferroni post-hoc test as appropriate (GraphPad Prism). Differences were considered statistically significant when p < 0.05. Data are presented as mean ± SEM except for representative experiments, in which data are presented as mean ± SD.
Results
IL-5 withdrawal has little effect on final eosinophil maturation
To investigate the importance of IL-5 during the later stages (post lineage commitment) of eosinophil differentiation, we used an ex-vivo model of IL-5 withdrawal (IL-5W) in which the culture media initially contains high-dose IL-5 (10 ng/mL) to promote differentiation and expansion of EoPs and preEos for 4 days, after which no further IL-5 was added to the cultures (Figure 2a). Eosinophil yield and maturation as measured by surface expression of Siglec-F and CCR3 were minimally affected by withdrawal of IL-5 for the last 6 days of differentiation (Figure 2b–c). There was an increase in the proportion of Siglec-F+CCR3− cells in the cultures, which suggests that final eosinophil maturation and upregulation of CCR3 are delayed or stalled (Figure 2c). Neutralizing the IL-5 that remained in the cultures resulted in declining yield of mature eosinophils in the cultures with time (Figure 2d). As the number of eosinophils declined with IL-5 neutralization, these data suggest that IL-5 is important for survival of the newly matured eosinophils (Figure 2d).
To further evaluate the contribution of IL-5 on preEos differentiation, we removed IL-5 from the cultures after 4 or 6 days (IL-5V, Figure 2e) and determined the effect on mature eosinophil yield. Stimulating LDBM cells with IL-5 for 4 or 6 days followed by 6 or 4 days of culture with media alone (IL-5V), respectively, still yielded a population of mature eosinophils (Figure 2f–g), albeit significantly lower than in the presence of IL-5. The proportion of mature eosinophils in the cultures increased with longer IL-5 exposure (6 days vs. 4 days, Figure 2h), suggesting that IL-5 is critical for preEos expansion. Together, these data suggest that exposure of preEos to IL-5 resulted in a developmental program capable of driving the final stages of eosinophil differentiation after IL-5 is withdrawn, neutralized or removed.
IL-5 stimulation induces expression of a panel of cytokines by LDBM cells
To identify cytokines that may contribute to the final stages of eosinophil differentiation, we used two unbiased approaches to identify active cytokine signaling pathways in LDBM cells after IL-5 stimulation. Initially, we kinetically measured cytokine levels in the culture supernatants. Cytokines previously associated with eosinophils, including IL-4, IL-9 and GM-CSF, were detected after 5, 8 and 11 days of IL-5 stimulation of LDBM cells (Figure 3a). Notably, in the supernatants, we also detected TNF-α accumulation that declined as the eosinophils matured, and levels of vascular endothelial growth factor increased as the eosinophils matured (Figure 3b). In addition, we detected IL-6 (12 ± 2 pg/mL), IL-10 (7 ± 0.7 pg/mL) and IL-12p40 (16 ± 2 pg/mL) in the supernatants after 5 days of IL-5 stimulation, but these cytokines were undetectable in the supernatants at later time points. Together, these data suggest that IL-5 stimulates LDBM cells to express a panel of cytokines that changes as mature eosinophils become more predominant in the cultures.
Figure 3. IL-5 stimulation induces expression of a panel of cytokines by low-density bone marrow cells.
(A–B) Cytokine level (mean ± SEM, n = 2 experiments with 3 samples per experiment) in the supernatants from low-density bone marrow (LDBM) cells cultured in IL-5 for the indicated number of days. (C) Heat map showing mRNA expression levels for all genes in LDBM cells cultured in IL-5 for the indicated number of days with 2-fold or more change is shown with genes with peak expression after 4 or 10 days of IL-5 are included in box. (D–E) In LDBM cells cultured in IL-5 for the indicated number of days, genes with 2-fold or more increase and that are involved in cytokine-mediated signaling are shown, including a panel of cytokines (D) and chemokine receptors (E). VEGF, vascular endothelial growth factor.
To screen for mediators and receptors that are induced by IL-5 and may contribute to eosinophil differentiation, we performed gene expression profiling on LDBM cells cultured with IL-5 for 4 or 10 days. Expression of 681 or 1055 genes was significantly (p < 0.05) induced ≥2 fold after 4 or 10 days of IL-5 stimulation, respectively, when compared to expression levels prior to IL-5 stimulation (Figure 3c and Supplementary Table 1). As expected, the genes with the greatest induction in LDBM cells after 4 days of IL-5 stimulation encoded constituents of eosinophil granules, including eosinophil ribonucleases and eosinophil peroxidase. Two genes, Ccr3 and Alox15, that encode for proteins important for mature eosinophil effector functions were the most induced genes in LDBM cells stimulated with IL-5 for 10 days. We also identified distinct subsets of genes that could be delineated on the basis of peak expression level after 4 or 10 days of IL-5 stimulation, when preEos and mature eosinophils predominate in the cultures (Figure 3c). Pathway analysis revealed significant enrichment for cytokine signaling pathways, including IL-5, chemokines, IL-3 and IL-4 (Table 2). We further explored changes in gene expression specific to cytokine-mediated signaling (Figure 3d and Supplementary Table 2) and chemokine receptors (Figure 3e). Together, this analysis revealed panels of genes with IL-5–mediated differential expression during eosinophil development from LDBM cells, suggesting that eosinophilopoiesis could be differentially influenced by mediators depending upon the developmental stage.
Table 2.
Pathway Analysis of Differentially Expressed Genes
| Pathway | P value | Genes in Pathway |
|---|---|---|
| Mm_IL-5_Signaling_Pathway _WP151_69175 | 9.36E-08 | 25 |
| Mm_Chemokine_signaling_pathway_WP2292_53116 | 1.84E-06 | 44 |
| Mm_IL-3_Signaling_Pathway_WP373_69196 | 6.42E-06 | 28 |
| Mm_IL-4_signaling_Pathway_WP93_41293 | 1.28E-03 | 16 |
PreEos express a specific panel of cytokines
As the LDBM cultures used for our gene expression profiling comprise a heterogeneous population of cells that include preEos until late in the cultures, when mature eosinophils predominate, we evaluated the specific expression of cytokines and cytokine receptors by preEos (Siglec-F+CCR3−) sorted from IL-5–stimulated LDBM cultures. The mRNA for the receptors for IL-3, IL-5 and GM-CSF, their shared β chain and the receptor for IL-4 was detected in preEos sorted from cultures after 4 days of IL-5 stimulation (Figure 4a), suggesting that preEos may be responsive to these cytokines. Levels of mRNA for these cytokine receptors, with the exception of the β chain, were similar to that of Siglec-F, which is also expressed on the surface of preEos. We investigated IL-5-stimulated cytokine expression by the sorted preEos and detected the cytokines IL-4, IL-6 and IL-13 in the supernatants (Figure 4b). Notably, no IL-9 or GM-CSF was detected in the supernatants from the purified preEos cultured with IL-5, suggesting that the IL-9 and GM-CSF detected in the supernatants from the LDBM cultures is produced and secreted by other bone marrow–derived cells in those cultures (Figure 3a). No significant difference in levels of mRNA for IL-5Rα, CCL3, IL-4, or the granule proteins major basic protein and eosinophil peroxidase were observed in preEos sorted from LDBM cells stimulated with IL-5 at 10, 1 or 0.1 ng/mL (Figure 4c and data not shown), suggesting that IL-5 stimulates an expression profile in preEos that is independent of the concentration of IL-5. We next evaluated the kinetic expression of IL-4 mRNA by preEos during maturation using an IL-4 fluorescent reporter (4get) mouse line (22). The absolute number of cells that express both IL-4 and Siglec-F increased after 7 days of IL-5 stimulation (Figure 4d), and very few cells that expressed Siglec-F did not express IL-4 (Figure 4d). Together, these data reveal that mRNA for IL-4 is expressed by both preEos and mature eosinophils and that IL-5–stimulated preEos produce and secrete multiple chemokines and cytokines, including IL-4 and IL-13.
Figure 4. IL-5 stimulation of eosinophil precursors induces cytokine expression.
Normalized expression (mean ± SD) of cytokine receptors (A) in sorted eosinophil precursors (PreEos) after 4 days of IL-5 stimulation is shown from a representative of 3 independent experiments with 3 wells per time point per experiment. (B) Cytokine level (mean ± SD) in the supernatants from sorted PreEos after stimulation with IL-5 for 24 or 48 hours is shown from a representative of 3 independent experiments with 3 wells per time point per experiment. (C) Normalized gene expression (mean ± SEM, n = 3 independent experiments) in PreEos sorted from LDBM cells after 4 days of IL-5 at 10, 1 or 0.1ng/mL is shown. (D) Absolute number (mean ± SD) of Siglec-F+ (solid line) and Siglec-F+GFP+ (dashed line) cells per well from low-density bone marrow (LDBM) cells cultured in IL-5 for the indicated number of days is shown from a representative of 3 independent experiments with 3 wells per time point in each condition. βc, β chain; R, receptor.
IL-4Rα signaling cooperates with IL-5 to promote eosinophil differentiation
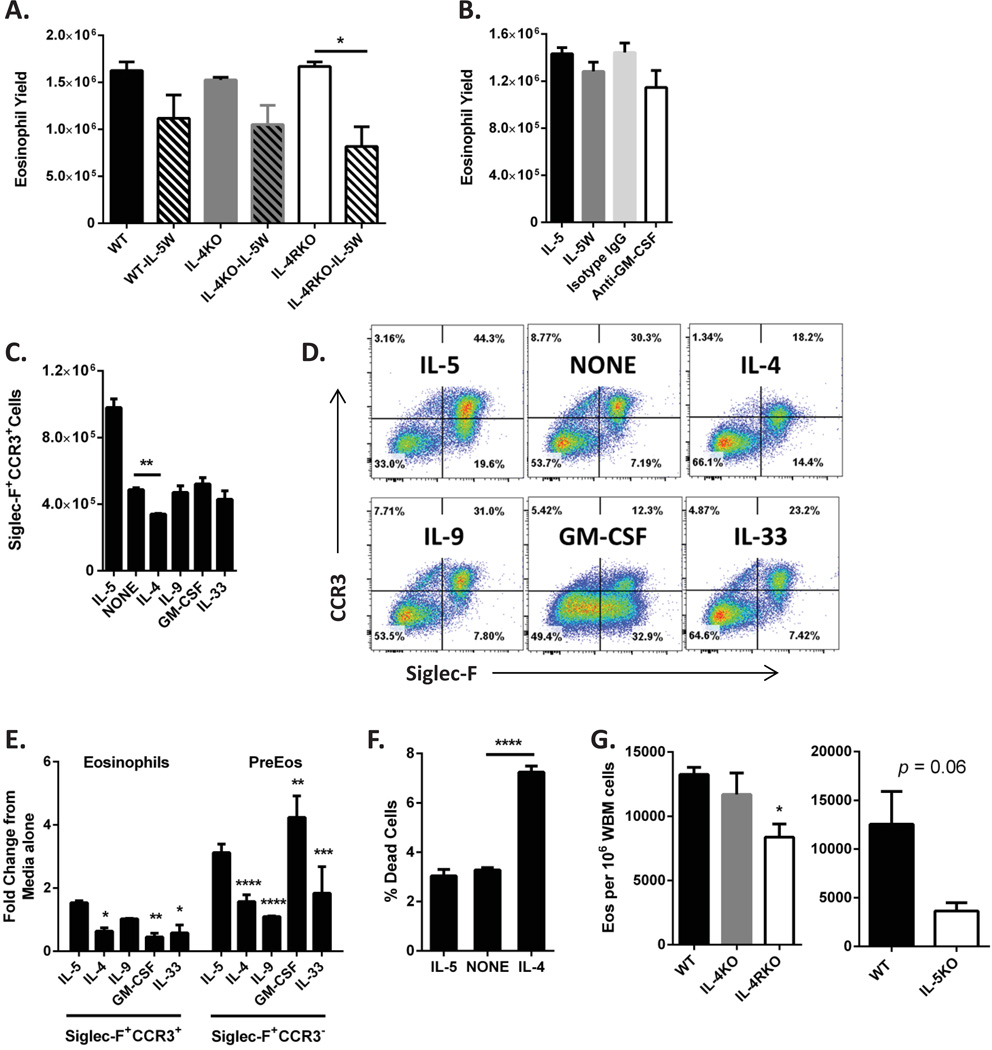
As preEos express the mRNA for IL-4Rα (Figure 4a) and secrete the IL-4Rα ligands IL-4 and IL-13 (Figure 4b), we investigated the contribution of IL-4Rα signal transduction in promoting preEos differentiation. Deficiency in IL-4Rα, but not IL-4, resulted in decreased eosinophil yield under IL-5W conditions (Figure 5a), suggesting that signaling through IL-4Rα (via IL-4 and/or IL-13) can contribute to preEos maturation. In contrast, although preEos also express mRNA for GM-CSF receptor (Figure 4a), neutralizing GM-CSF after IL-5W did not result in a significant change in eosinophil yield, suggesting that GM-CSF does not promote preEos differentiation (Figure 5b). We next investigated the potential for IL-4, GM-CSF, IL-9 and IL-33 to promote preEos maturation when IL-5 was completely removed from the cultures. The addition of IL-9, GM-CSF or IL-33 to the culture media for 2 days in the absence of IL-5 did not result in enhanced preEos maturation (as measured by upregulation of CCR3 on the surface of the Siglec-F+ preEos) compared to media alone (Figure 5c), but the frequency of preEos (Siglec-F+CCR3−) in the cultures increased with the addition of GM-CSF (Figure 5d). Notably, the increase in preEos with GM-CSF stimulation was greater than IL-5 (Figure 5e), suggesting that GM-CSF promotes preEos expansion. The addition of IL-4 resulted in decreased preEos maturation compared to media alone (Figure 5c) with an increase in the frequency of preEos (Figure 5d–e), suggesting that IL-4 signaling in the absence of IL-5 results in decreased survival of the newly matured eosinophils. Indeed, the percentage of non-viable cells after 48 hours of IL-4 stimulation in the absence of IL-5 was significantly greater compared to media alone (Figure 5f). In addition, deficiency in IL-4Rα resulted in a significant reduction in the baseline frequency of mature eosinophils in the bone marrow compared to bone marrow from wild-type animals (Figure 5g). IL-4 or IL-5 deficiency had no significant effect on baseline eosinophil production in vivo (Figure 5g). Together, these data suggest that IL-4Rα signaling cooperates with IL-5 to promote preEos maturation and survival but that IL-4Rα signaling in the absence of IL-5 results in impaired survival of newly matured eosinophils.
Figure 5. Effects of IL-4Rα signaling on eosinophil differentiation in the absence of IL-5.
(A) Eosinophil yield (mean ± SEM, n = 3 independent experiment with 3–4 wells per condition in each experiment) from wild-type (WT), IL-4–deficient (IL-4KO) and IL-4Rα–deficient (IL-4RKO) low-density bone marrow (LDBM) cells cultured with IL-5 continuously (solid bars) or IL-5 withdrawal (IL-5W, hatched bars) is shown. *p < 0.05. (B) Eosinophil yield (mean ± SEM, n = 3 independent experiment with 4–6 wells per condition in each experiment) from LDBM cells cultured with IL-5 continuously (IL-5) or IL-5W with anti–GM-CSF antibody or isotype control antibody (IgG). Absolute number (C, mean ± SD) of mature eosinophils (CCR3+Siglec-F+), dot blots (D) showing surface CCR3 and Siglec-F expression, fold change (E, mean ± SEM) in percentage of mature eosinophils (Siglec-F+CCR3+) and PreEos (Sigelc-F+CCR3−) and percentage of non-viable cells (F) from LDBM cells cultured for 8 days in IL-5 and then washed and replated in media alone (NONE) or with the indicated cytokines for 2 additional days is shown. (C), (D) and (E) are from a representative of 3 independent experiments with 3 wells for each condition in each experiment; (F) is the mean ± SEM of the 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 when compared to fold change when IL-5 is added back to culture media. (G) Frequency (mean ± SEM, n = 4 independent experiments with 3–4 mice per group per experiment) of mature eosinophils (Eos) in the whole bone marrow (WBM) of WT, IL-4KO, IL-4RKO and IL-5KO mice is shown. *p < 0.05.
CCL3 promotes eosinophil differentiation
In addition to cytokine expression (Figure 3a), we also investigated IL-5–mediated expression of chemokines by cultured LDBM cells. Accumulation of a subset of chemokines (CCL2, CCL5 and CXCL10) in the culture supernatants changed significantly during IL-5–induced eosinophil differentiation (Figure 6a), whereas protein levels of another subset of chemokines (CCL3, CCL4, CXCL2 and CXCL9) remained relatively constant throughout the culture period (Figure 6b). To determine which chemokines are specifically expressed by preEos within the LDBM cultures, we sorted preEos and replated the purified cells to measure cytokine levels in the supernatants after 24 and 48 hours of IL-5 stimulation. CCL2, CCL3 and CXCL10 protein were detected in the supernatants from purified preEos (Figure 6c). We next evaluated expression of chemokine receptors by preEos to identify potential chemokine receptor-ligand pairs that may influence preEos maturation. Surface expression for CCR1 (receptor for CCL3), CCR5 (receptor for CCL3 and CCL4) and CXCR3 (receptor for CXCL10 and CXCL9) on preEos (Siglec-F+CCR3−) and mature eosinophils (Siglec-F+CCR3+) was measured by flow cytometry on cultured LDBM cells. We detected CCR1 on the surface of a subset (~one-third) of the preEos after 4 days of IL-5 stimulation, whereas CCR5 and CXCR3 were detected on much smaller subsets of preEos (Figure 6d). No significant expression of CCR1, CCR5 or CXCR3 was detected on the surface of mature eosinophils (Figure 6d). Together, these data suggest that surface expression of chemokine receptors changes during IL-5–mediated eosinophil development and that preEos express ligands for chemokine receptors on their surface, which may result in developmental stage–specific responses to chemokines.
Figure 6. CCL3 promotes eosinophil differentiation.
(A–B) Chemokine levels (mean ± SD) in the supernatants from low-density bone marrow (LDBM) cells stimulated with IL-5 for 5, 8 or 11 days is shown from a representative of 2 independent experiments with 3 wells per time point per experiment. *p < 0.05, ***p < 0.001, ****p < 0.0001 when compared to levels after 5 days of IL-5 stimulation. (C) Cytokine level (mean ± SD) in the supernatants from eosinophil precursors (PreEos) sorted from LDBM cells cultured in IL-5 for 4 days and subsequently stimulated with IL-5 for 24 and 48 hours is shown from a representative of 2 independent experiments with 3 wells per time point per experiment. ****p < 0.0001 when compared to level present after 24 hours. (D) Percentage of eosinophil precursors (Siglec-F+; PreEos) and mature eosinophils (Siglec-F+CCR3+) with surface expression of indicated chemokine receptors in LDBM cells stimulated for 4 days with IL-5 is shown from a representative of 3 independent experiments. (E) Eosinophil yield (mean ± SD) from LDBM cells cultured with IL-5 continuously (IL-5) or IL-5 withdrawal (IL-5W) with anti-CXCL10, anti-CCL3, anti-CCL4 or isotype control antibody (Goat IgG) or no antibody (no Ab) is shown from a representative of 3 independent experiments with 3 wells per condition in each experiment. Absolute number (F, mean ± SD) of mature eosinophils (Siglec-F+CCR3+) and dot blots (G) showing surface expression of Siglec-F and CCR3 from LDBM cells cultured for 8 days in IL-5 and then washed and replated in media alone (NONE) or with the indicated chemokines for 3 additional days is shown. ***p < 0.001. (F) and (G) are from a representative of 3 independent experiments with 3 wells for each condition in each experiment.
Although preEos express the receptor and ligand pairs, individually neutralizing the chemokines CCL3, CCL4 and CXCL10 resulted in no significant change in eosinophil yield following IL-5W (Figure 6e). We next investigated the potential for CCL3, CCL4 and CXCL10 to promote preEos maturation after the removal of IL-5. Although there was no enhanced preEos maturation above media alone with the addition of CCL4 or CXCL10, adding CCL3 to the culture media after IL-5 removal modestly promoted preEos maturation greater than media alone (Figure 6f–g), suggesting that CCL3 can promote eosinophil differentiation in the absence of IL-5.
Discussion
The aim of IL-5–targeted therapy has been to safely reduce blood and tissue eosinophilia to prevent eosinophil-mediated tissue damage. In a range of eosinophil-associated disorders, anti–IL-5 treatment results in a partial reduction in tissue eosinophilia (14–18) and a greater inhibitory effect on the later eosinophil developmental stages in the bone marrow (25). Similar to observations in IL-5–deficient mice (12), IL-5–targeted treatments do not reduce homeostatic levels of tissue eosinophils (26). Together, these data highlight the presence of clinically relevant IL-5–independent pathways that promote eosinophil development and tissue accumulation. As disease-associated eosinophilia primarily results from IL-5–mediated enhancement of eosinophil production, we aimed to identify cytokines that could promote persistent eosinophil differentiation after IL-5 is withdrawn. We demonstrate that IL-5 stimulation of LDBM cells results in expression of stage-specific subsets of cytokines and chemokines that can influence eosinophilopoiesis and survival of developing and newly formed eosinophils. We also demonstrate differences in chemokine receptor surface expression between preEos and eosinophils, suggesting that stage-specific chemokine responses could influence eosinophil development and mobilization of preEos vs. eosinophils from the bone marrow and into inflamed tissues. Our study highlights an IL-5–initiated cooperative cytokine pathway that contributes to persistent eosinophil differentiation and survival even after IL-5 is completely withdrawn.
In our culture system, in which progenitor-enriched LDBM cells are stimulated with high-dose IL-5 for 4 days, we demonstrate that preEos are still capable of expanding and differentiating for at least another 48 hours after IL-5 is removed from the culture media without the addition of any further cytokines. This finding suggests that IL-5 stimulation initiates a developmental program that can continue in its absence. Our data further highlight that IL-5 is critical to promote the survival of the mature eosinophils that result from the differentiating preEos. In this study, we pursued the identification of cytokines and cytokine receptors that are components of the IL-5–initiated developmental program using unbiased approaches to measure cytokines produced and genes expressed by the LDBM cells. We report the IL-5–mediated gene expression profile from LDBM cells (GEO Series GSE55386). The kinetic analysis reveals an expression profile that changed as the preEos mature into eosinophils. After 4 days of IL-5 stimulation, the genes with the greatest induction of expression are granule proteins, including several eosinophil-associated ribonucleases and eosinophil peroxidase (Supplementary Table 1), which is consistent with a prior study investigating IL-5–dependent changes in gene expression in an infection model (27). We focused our expression analysis on cytokines and cytokine receptors produced by the LDBM cells and investigated their ability to influence eosinophilopoiesis. We show that the level of mRNA that encode for multiple cytokine receptors, including CCR1, CCR3 and IL-5Rα, was increased in LDBM cells stimulated with IL-5 for 4 days and further increased after 10 days of IL-5 stimulation. Importantly, expression of the CCR1 ligand CCL3 was also induced by IL-5. Interestingly, we show that expression of mRNA that encodes for CXCR3 and its ligand CXCL10 peaked after 4 days of IL-5 but then declined with further IL-5–mediated differentiation. The expression analysis suggests that IL-5 stimulation of progenitor-enriched LDBM cells results in a developmental stage–specific transcriptome that ultimately leads to differential cytokine responsiveness between preEos and mature eosinophils.
We investigated the role of IL-4 in promoting preEos expansion and differentiation in the presence and absence of IL-5, and we demonstrate that IL-4 and IL-4Rα are expressed during eosinophil development from preEos to mature eosinophils. In addition, preEos express and secrete IL-13, which also signals through IL-4Rα, highlighting the potential for this signaling pathway to influence eosinophilopoiesis. Deficiency of IL-4Rα, but not IL-4, resulted in decreased eosinophil yield from LDBM cells after IL-5 was withdrawn and in decreased numbers of baseline mature eosinophils in the bone marrow, suggesting that IL-13–mediated signaling through IL-4Rα can promote preEos differentiation and expansion. Our studies also propose a cooperative signaling pathway between IL-4Rα and IL-5Rα, as the addition of IL-4 to the culture media when IL-5 is absent results in decreased eosinophil survival.
CCR1 expression on eosinophils varies between and within human donors (28, 29), is expressed by murine eosinophils as well (30, 31) and has been shown to be important for recruitment of mature eosinophils into tissues (32, 33). In addition, the CCR1/CCL3 axis has been shown to enhance proliferation of lineage-committed myeloid progenitors (34). In this study, we demonstrate expression of CCR1 on the surface of preEos, an eosinophil lineage–committed precursor and further demonstrate that CCL3 activation of preEos modestly enhanced the expansion and differentiation of preEos in the absence of IL-5. Our data focused on the single eosinophil lineage and are consistent with a prior study demonstrating that CCL3 stimulation of LDBM increases the number of GM-CSF–induced colonies containing granulocytes and macrophages (34). As CCL3 (19) and preEos (5) have been noted to be in the airways of patients with atopic disorders, it is possible that activation of CCR1-expressing preEos by CCL3 in the airways induces in-situ eosinophil differentiation in the inflamed tissue and contributes to the persistent tissue eosinophilia seen in anti–IL-5–treated asthmatics (15). Thus, although the heightened preEos differentiation and expansion mediated by CCL3 is modest, we propose that it is likely clinically relevant in inflamed tissues where small numbers of eosinophils have been associated with persistent disease and clinical symptoms even after IL-5–targeted therapy (15, 35).
After the removal of IL-5 from the culture media, the percentage of LDBM-derived cells that expressed neither CCR3 nor Siglec-F increased in the cultures. We propose that the increased frequency of Siglec-F−CCR3− cells results from the combination of cessation of preEos expansion and persistence of preEos maturation via differentiation-promoting pathways, such as CCL3/CCR1 as described above. However, we cannot rule out the possibility that the removal of IL-5 results in the down-regulation of Siglec-F, thereby increasing the double-negative population. Our studies also revealed a significant increase in the percentage of preEos after the addition of GM-CSF to cultures in which IL-5 had been removed, with this increase being greater than the increase in preEos observed when IL-5 was added back to the media. As the absolute number of mature eosinophils that resulted in the presence of GM-CSF was similar to media alone, GM-CSF likely strongly promotes preEos proliferation but not differentiation.
Herein, we describe a novel, eosinophil-promoting cooperative cytokine network that is initiated by IL-5 stimulation of bone marrow progenitors and propagates eosinophilia by an autocrine mechanism in the absence of further IL-5 exposure. Importantly, expression of the mediators and receptors identified, specifically IL-4/IL-13/IL-4Rα and CCL3/CCR1, has been noted in eosinophil-associated disorders. Together, our data provide mechanistic insight into the potential limitations observed in solely targeting IL-5 in eosinophil-associated disorders.
Supplementary Material
Acknowledgements
We wish to thank Shawna Hottinger for her editorial assistance.
This work was supported by the NIH grants R37 AI045898 (M.E.R.), R01 AI083450 (M.E.R.) and K08 AI093673 (P.C.F.); the Buckeye Foundation; the Food Allergy Research & Education (FARE); and the Campaign Urging Research for Eosinophilic Disease (CURED). This project was also supported in part by the NIH grant P30 DK078392 (Gene and Protein Expression and Flow Cytometry Cores of the Digestive Disease Research Core Center in Cincinnati).
Abbreviations used in this article
- L-5Rα
IL-5 receptor alpha
- EoP
eosinophil lineage–committed progenitor
- preEos
eosinophil precursors
- LDBM
low-density bone marrow
- KO
knockout
- eGFP
enhanced green fluorescent protein
- CCHMC
Cincinnati Children's Hospital Medical Center
- IL-5W
IL-5 withdrawal protocol
- IL-5V
IL-5 removal protocol
References
- 1.Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. The Journal of experimental medicine. 2005;201:1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. The Journal of experimental medicine. 2009;206:183–193. doi: 10.1084/jem.20081756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inman MD, Ellis R, Wattie J, Denburg JA, O'Byrne PM. Allergen-induced increase in airway responsiveness, airway eosinophilia, and bone-marrow eosinophil progenitors in mice. Am.J.Respir.Cell Mol.Biol. 1999;21:473–479. doi: 10.1165/ajrcmb.21.4.3622. [DOI] [PubMed] [Google Scholar]
- 4.Gaspar Elsas MI, Joseph D, Elsas PX, Vargaftig BB. Rapid increase in bone-marrow eosinophil production and responses to eosinopoietic interleukins triggered by intranasal allergen challenge. Am.J.Respir.Cell Mol.Biol. 1997;17:404–413. doi: 10.1165/ajrcmb.17.4.2691. [DOI] [PubMed] [Google Scholar]
- 5.Robinson DS, Damia R, Zeibecoglou K, Molet S, North J, Yamada T, Kay AB, Hamid Q. CD34(+)/interleukin-5Ralpha messenger RNA+ cells in the bronchial mucosa in asthma: potential airway eosinophil progenitors. Am.J.Respir.Cell Mol.Biol. 1999;20:9–13. doi: 10.1165/ajrcmb.20.1.3449. [DOI] [PubMed] [Google Scholar]
- 6.Cameron L, Christodoulopoulos P, Lavigne F, Nakamura Y, Eidelman D, McEuen A, Walls A, Tavernier J, Minshall E, Moqbel R, Hamid Q. Evidence for local eosinophil differentiation within allergic nasal mucosa: Inhibition with soluble IL-5 receptor. Journal of Immunology. 2000;164:1538–1545. doi: 10.4049/jimmunol.164.3.1538. [DOI] [PubMed] [Google Scholar]
- 7.Allakhverdi Z, Comeau MR, Smith DE, Toy D, Endam LM, Desrosiers M, Liu YJ, Howie KJ, Denburg JA, Gauvreau GM, Delespesse G. CD34+ hemopoietic progenitor cells are potent effectors of allergic inflammation. The Journal of allergy and clinical immunology. 2009;123:472–478. doi: 10.1016/j.jaci.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Kouro T, Takatsu K. IL-5- and eosinophil-mediated inflammation: from discovery to therapy. Int.Immunol. 2009;21:1303–1309. doi: 10.1093/intimm/dxp102. [DOI] [PubMed] [Google Scholar]
- 9.Clutterbuck EJ, Hirst EM, Sanderson CJ. Human interleukin-5 (IL-5) regulates the production of eosinophils in human bone marrow cultures: comparison and interaction with IL-1, IL-3, IL-6, and GMCSF. Blood. 1989;73:1504–1512. [PubMed] [Google Scholar]
- 10.Dent LA, Strath M, Mellor AL, Sanderson CJ. Eosinophilia in transgenic mice expressing interleukin 5. The Journal of experimental medicine. 1990;172:1425–1431. doi: 10.1084/jem.172.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee NA, McGarry MP, Larson KA, Horton MA, Kristensen AB, Lee JJ. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J Immunol. 1997;158:1332–1344. [PubMed] [Google Scholar]
- 12.Kopf M, Brombacher F, Hodgkin PD, Ramsay AJ, Milbourne EA, Dai WJ, Ovington KS, Behm CA, Kohler G, Young IG, Matthaei KI. IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity. 1996;4:15–24. doi: 10.1016/s1074-7613(00)80294-0. [DOI] [PubMed] [Google Scholar]
- 13.Busse WW, Ring J, Huss-Marp J, Kahn JE. A review of treatment with mepolizumab, an anti-IL-5 mAb, in hypereosinophilic syndromes and asthma. J.Allergy Clin.Immunol. 2010;125:803–813. doi: 10.1016/j.jaci.2009.11.048. [DOI] [PubMed] [Google Scholar]
- 14.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, Pavord ID. Mepolizumab and exacerbations of refractory eosinophilic asthma. The New England journal of medicine. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS. Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am.J Respir.Crit Care Med. 2003;167:199–204. doi: 10.1164/rccm.200208-789OC. [DOI] [PubMed] [Google Scholar]
- 16.Plotz SG, Simon HU, Darsow U, Simon D, Vassina E, Yousefi S, Hein R, Smith T, Behrendt H, Ring J. Use of an anti-interleukin-5 antibody in the hypereosinophilic syndrome with eosinophilic dermatitis. The New England journal of medicine. 2003;349:2334–2339. doi: 10.1056/NEJMoa031261. [DOI] [PubMed] [Google Scholar]
- 17.Straumann A, Conus S, Grzonka P, Kita H, Kephart G, Bussmann C, Beglinger C, Smith DA, Patel J, Byrne M, Simon HU. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut. 2010;59:21–30. doi: 10.1136/gut.2009.178558. [DOI] [PubMed] [Google Scholar]
- 18.Assa'ad AH, Gupta SK, Collins MH, Thomson M, Heath AT, Smith DA, Perschy TL, Jurgensen CH, Ortega HG, Aceves SS. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology. 2011;141:1593–1604. doi: 10.1053/j.gastro.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 19.Alam R, York J, Boyars M, Stafford S, Grant JA, Lee J, Forsythe P, Sim T, Ida N. Increased MCP-1, RANTES, and MIP-1alpha in bronchoalveolar lavage fluid of allergic asthmatic patients. Am J Respir Crit Care Med. 1996;153:1398–1404. doi: 10.1164/ajrccm.153.4.8616572. [DOI] [PubMed] [Google Scholar]
- 20.Fulkerson PC, Zimmermann N, Hassman LM, Finkelman FD, Rothenberg ME. Pulmonary chemokine expression is coordinately regulated by STAT1, STAT6, and IFN-gamma. The Journal of Immunology. 2004;173:7565–7574. doi: 10.4049/jimmunol.173.12.7565. [DOI] [PubMed] [Google Scholar]
- 21.McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. The Journal of experimental medicine. 1999;189:1565–1572. doi: 10.1084/jem.189.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 23.Bouffi C, Rochman M, Zust CB, Stucke EM, Kartashov A, Fulkerson PC, Barski A, Rothenberg ME. IL-33 markedly activates murine eosinophils by an NF-kappaB-dependent mechanism differentially dependent upon an IL-4-driven autoinflammatory loop. J Immunol. 2013;191:4317–4325. doi: 10.4049/jimmunol.1301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menzies-Gow A, Flood-Page P, Sehmi R, Burman J, Hamid Q, Robinson DS, Kay AB, Denburg J. Anti-IL-5 (mepolizumab) therapy induces bone marrow eosinophil maturational arrest and decreases eosinophil progenitors in the bronchial mucosa of atopic asthmatics. J.Allergy Clin.Immunol. 2003;111:714–719. doi: 10.1067/mai.2003.1382. [DOI] [PubMed] [Google Scholar]
- 26.Conus S, Straumann A, Bettler E, Simon HU. Mepolizumab does not alter levels of eosinophils, T cells, and mast cells in the duodenal mucosa in eosinophilic esophagitis. J.Allergy Clin.Immunol. 2010;126:175–177. doi: 10.1016/j.jaci.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 27.Bystrom J, Wynn TA, Domachowske JB, Rosenberg HF. Gene microarray analysis reveals interleukin-5-dependent transcriptional targets in mouse bone marrow. Blood. 2004;103:868–877. doi: 10.1182/blood-2003-08-2778. [DOI] [PubMed] [Google Scholar]
- 28.Elsner J, Dulkys Y, Gupta S, Escher SE, Forssmann WG, Kapp A, Forssmann U. Differential pattern of CCR1 internalization in human eosinophils: prolonged internalization by CCL5 in contrast to CCL3. Allergy. 2005;60:1386–1393. doi: 10.1111/j.1398-9995.2005.00893.x. [DOI] [PubMed] [Google Scholar]
- 29.Phillips RM, Stubbs VE, Henson MR, Williams TJ, Pease JE, Sabroe I. Variations in eosinophil chemokine responses: an investigation of CCR1 and CCR3 function, expression in atopy, and identification of a functional CCR1 promoter. J Immunol. 2003;170:6190–6201. doi: 10.4049/jimmunol.170.12.6190. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira SH, Lira S, Martinez AC, Wiekowski M, Sullivan L, Lukacs NW. Increased responsiveness of murine eosinophils to MIP-1beta (CCL4) and TCA-3 (CCL1) is mediated by their specific receptors, CCR5 and CCR8. J Leukoc Biol. 2002;71:1019–1025. [PubMed] [Google Scholar]
- 31.Borchers MT, Ansay T, DeSalle R, Daugherty BL, Shen H, Metzger M, Lee NA, Lee JJ. In vitro assessment of chemokine receptor-ligand interactions mediating mouse eosinophil migration. J Leukoc Biol. 2002;71:1033–1041. [PubMed] [Google Scholar]
- 32.Gaga M, Ong YE, Benyahia F, Aizen M, Barkans J, Kay AB. Skin reactivity and local cell recruitment in human atopic and nonatopic subjects by CCL2/MCP-1 and CCL3/MIP-1alpha. Allergy. 2008;63:703–711. doi: 10.1111/j.1398-9995.2007.01578.x. [DOI] [PubMed] [Google Scholar]
- 33.Domachowske JB, Bonville CA, Gao JL, Murphy PM, Easton AJ, Rosenberg HF. The chemokine macrophage-inflammatory protein-1 alpha and its receptor CCR1 control pulmonary inflammation and antiviral host defense in paramyxovirus infection. J Immunol. 2000;165:2677–2682. doi: 10.4049/jimmunol.165.5.2677. [DOI] [PubMed] [Google Scholar]
- 34.Broxmeyer HE, Cooper S, Hangoc G, Gao JL, Murphy PM. Dominant myelopoietic effector functions mediated by chemokine receptor CCR1. The Journal of experimental medicine. 1999;189:1987–1992. doi: 10.1084/jem.189.12.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leckie MJ, ten Brinke A, Khan J, Diamant Z, O'Connor BJ, Walls CM, Mathur AK, Cowley HC, Chung KF, Djukanovic R, Hansel TT, Holgate ST, Sterk PJ, Barnes PJ. Effects of an interleukin-5 blocking monoclonal antibody on eosinophils, airway hyper-responsiveness, and the late asthmatic response. Lancet. 2000;356:2144–2148. doi: 10.1016/s0140-6736(00)03496-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.