Abstract
Differentiation of CD4+ helper and CD8+ cytotoxic αβ T cells from CD4+CD8+ thymocytes involves up-regulation of lineage-specifying transcription factors and transcriptional silencing of CD8 or CD4 co-receptors, respectively, in major histocompatibility complex (MHC) II or I restricted thymocytes. Here, we demonstrate that inactivation of the Dicer RNA endonuclease in murine thymocytes impairs initiation of Cd4 and Cd8 silencing, leading to development of positively selected, MHCI- and MHCII-restricted mature CD4+CD8+ thymocytes. Expression of the anti-apoptotic BCL2 protein or inactivation of the p53 pro-apoptotic protein rescues these thymocytes from apoptosis, increasing their frequency and permitting accumulation of CD4+CD8+ αβ T cells in the periphery. Dicer-deficient MHCI-restricted αβ T cells fail to normally silence Cd4 and display impaired induction of the CD8-lineage specifying transcription factor Runx3, whereas Dicer-deficient MHCII-restricted αβ T cells show impaired Cd8 silencing and impaired induction of the CD4-lineage specifying transcription factor Thpok. Finally, we show that the Drosha RNA endonuclease, which functions upstream of Dicer in microRNA biogenesis, also regulates Cd4 and Cd8 silencing. Our data demonstrate a previously dismissed function for the microRNA biogenesis machinery in regulating expression of lineage-specifying transcription factors and silencing of Cd4 and Cd8 during αβ T cell differentiation.
Introduction
The generation of distinct cellular lineages from multipotent progenitor cells involves differentiation programs that couple up-regulation of lineage specific genes with silencing of genes expressed in progenitor cells and alternative lineages. The initiation, maintenance, and silencing of gene expression during lineage commitment are regulated by genetic and epigenetic mechanisms. One paradigm for elucidating molecular mechanisms that control gene expression during lineage commitment is the differentiation of CD4+ and CD8+ αβ T cells from CD4+CD8+ (double-positive or DP) thymocytes that have expressed functional αβ TCRs (1, 2). Assembly and expression of T cell receptor (TCR) β genes drives CD4−CD8− (double-negative or DN) thymocytes to differentiate into DP thymocytes (3). This developmental transition initiates rearrangement and expression of TCRα genes, which leads to expression of unique αβ TCRs on immature CD4+CD8+ thymocytes. Specificities of αβ TCRs are selected through interactions of these antigen receptors with self-peptide/MHC complexes expressed on thymic epithelial cells, a process aided by CD4 and CD8 co-receptors (3, 4). Depending on the affinity of such interactions, thymocytes die by “neglect”, are rescued from programmed cell death and further differentiate (positive selection), or are actively deleted (negative selection). Concomitant with positive selection, immature CD4+CD8+ thymocytes up-regulate lineage-specifying transcription factors and silence Cd8 or Cd4 as they differentiate into mature CD4+ or CD8+ (single positive or SP) thymocytes. SP cells exit the thymus and migrate to the spleen, lymph nodes, and other peripheral organs as CD4+ or CD8+ lineage αβ T cells.
Differentiation of CD4+ and CD8+ αβ T cells is regulated by αβ TCR-activated signaling pathways that control downstream transcription factors (2, 5). These factors include Runx3, which is required for CD8 lineage effector functions and Cd4 silencing, and Thpok (encoded by Zbtb7b), which drives differentiation of CD4+ SP thymocytes and facilitates Cd8 silencing (2, 6–10). Runx3 and Thpok are mutually antagonistic as Runx3 represses Zbtb7b expression by binding a silencer upstream of the Zbtb7b promoter (11, 12), while Thpok represses Runx3 expression (13–15) and antagonizes Runx-mediated repression of Cd4, possibly through binding to the Cd4 silencer (14, 16). Despite requirement for Runx3 and the Cd4 silencer in initiation of Cd4 silencing, neither is required to prevent Cd4 re-expression in peripheral CD8+ αβ T cells (17, 18), implying that Cd4 silencing is maintained epigenetically. In contrast to control of Cd4 expression, lineage-specific Cd8 transcription appears to be regulated by developmental stage specific Cd8 enhancers, rather than a cis-acting silencer element (19–22). However, Thpok-mediated recruitment of histone deacetylases to Cd8 enhancers may facilitate Cd8 silencing in CD4+ cells (10). In addition to Runx3 and Thpok, several transcription factors and chromatin modifying enzymes modulate CD4/CD8 lineage commitment and/or co-receptor expression, yet none of these has been shown to directly regulate initiation of Cd4 or Cd8 silencing following positive selection of DP thymocytes (1, 2, 23).
The Dicer and Drosha RNA endonucleases guide cellular differentiation through their ability to control gene expression. Both proteins are required for the biogenesis of microRNAs (miRs), which repress gene expression by binding and destabilizing or blocking translation of mRNAs (24). However, Dicer can also function independently of Drosha to create short-interfering RNAs (siRNAs), which inhibit gene expression by inducing epigenetic changes that block transcription of target loci (25). While inactivation of Dicer or Drosha initiating in mouse DN thymocytes has been shown to increase apoptosis of immature thymocytes, neither was found to affect CD4 and CD8 lineage commitment or Cd4 and Cd8 silencing (26, 27). We demonstrate here that inactivation of Dicer starting in DN thymocytes impairs Cd4 and Cd8 silencing, leading to generation of positively selected, MHCI- and MHCII-restricted mature CD4+CD8+ thymocytes. Expression of the anti-apoptotic BCL2 protein or inactivation of the p53 pro-apoptotic protein rescues these cells from apoptosis, increasing their frequency and permitting accumulation of CD4+CD8+ αβ T cells in the periphery. We demonstrate that Dicer is required for appropriate initiation of Cd4 and Cd8 silencing in thymocytes, but find no evidence for a requirement of Dicer in maintenance of Cd4 and Cd8 silencing in peripheral CD4+ or CD8+ αβ T cells. We also show that Dicer-deficient MHCI-restricted αβ T cells exhibit impaired transcriptional silencing of Cd4 and impaired expression of the Cd4-silencing transcription factor Runx3, while Dicer-deficient MHCII-restricted αβ T cells have reduced expression of Thpok, the master regulator of CD4 lineage commitment. We also show that the Drosha RNA endonuclease also regulates Cd4 and Cd8 silencing, suggesting a role for miRs in this process during lineage-commitment. Our data demonstrate an unexpected role for the miR biogenesis machinery in promoting appropriate co-receptor silencing and lineage commitment during αβ T cell differentiation.
Materials and Methods
Mice
LckCre (28), EμBCL2 (29), Dicerflox/flox (26), Tp53flox/flox (30), Cd4Cre (28), Rag1−/− (31), MHCI−/− (32), MHCII−/− (33), OT-I (34), OT-II (35), and Droshaflox/flox (36) mice have been described. Mice were maintained under specific pathogen-free conditions at the Children's Hospital of Philadelphia (CHOP) or the National Institute of Health. Unless otherwise indicated, studies were conducted on littermate or age-matched mice between 4–8 weeks of age. All studies were performed in accordance with regulations and approved by the CHOP or NIAID/NIH Institutional Animal Care and Use Committees.
Flow Cytometry and Gating
Flow cytometry was as described previously (37). Unless otherwise specified, gating was Forward Scatter × Side Scatter → Singlets → Live cells (Invitrogen LIVE/DEAD) → TCRβhiCD24lo, followed by CD4 and CD8α gating.
Bone marrow chimeras
Single cell suspensions were prepared from tibia and femur bone marrow. CD4+ and CD8+ αβ T cells were removed by magnetic bead depletion (Qiagen). Recipient mice were lethally irradiated (900 rads in two 450 rad doses 4 hours apart) prior to retro-orbital injection. Mice were analyzed 8–10 weeks later.
Quantitative real-time PCR
RNA isolation and cDNA generation were as described previously (37). Primers: Cd4 Exon 1F: 5’-GCAGAGTGAAGGAAGGACTGG-3’, Cd4 Intron 1R: 5’- CAGAACATTCCGGCACATTAGC-3’. Primers for Zbtb7b were previously described (38). Primers for Rorc and Foxo1 were purchased from Life Technologies (murine Taqman assays).
Western Blot
Cell pellets were lysed in 1% (vol/vol) SDS buffer under reducing conditions, separated on a 10% SDS-PAGE gel, transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore) and analyzed by immunoblot and chemiluminescence. The anti-Runx antibody was from Epitomics (# 2593-1).
Quantification of miRNA levels
Sorted cells were resuspended in TRIzol (Ambion) and RNA isolated using miRNeasy Mini kit (Qiagen). Reverse transcription was performed using the Taqman microRNA Reverse Transcription Kit (Applied Biosystems) and probe-specific primers. qRT-PCR was performed on a ViiA 7 (Applied Biosystems) instrument using Taqman Universal Master Mix (Applied Biosystems) according to manufacturer’s instructions. Matched reverse transcription and qPCR primers for miR-181a, let-7c, and snoRNA202 were from Life Technologies.
Statistical Analyses
Unless otherwise indicated, Student’s t-test was utilized for statistical analyses. Error bars are standard error of the mean (SEM).
Results
Suppressing apoptosis of Dicer-deficient thymocytes results in generation of CD4+CD8+ mature αβ T cells
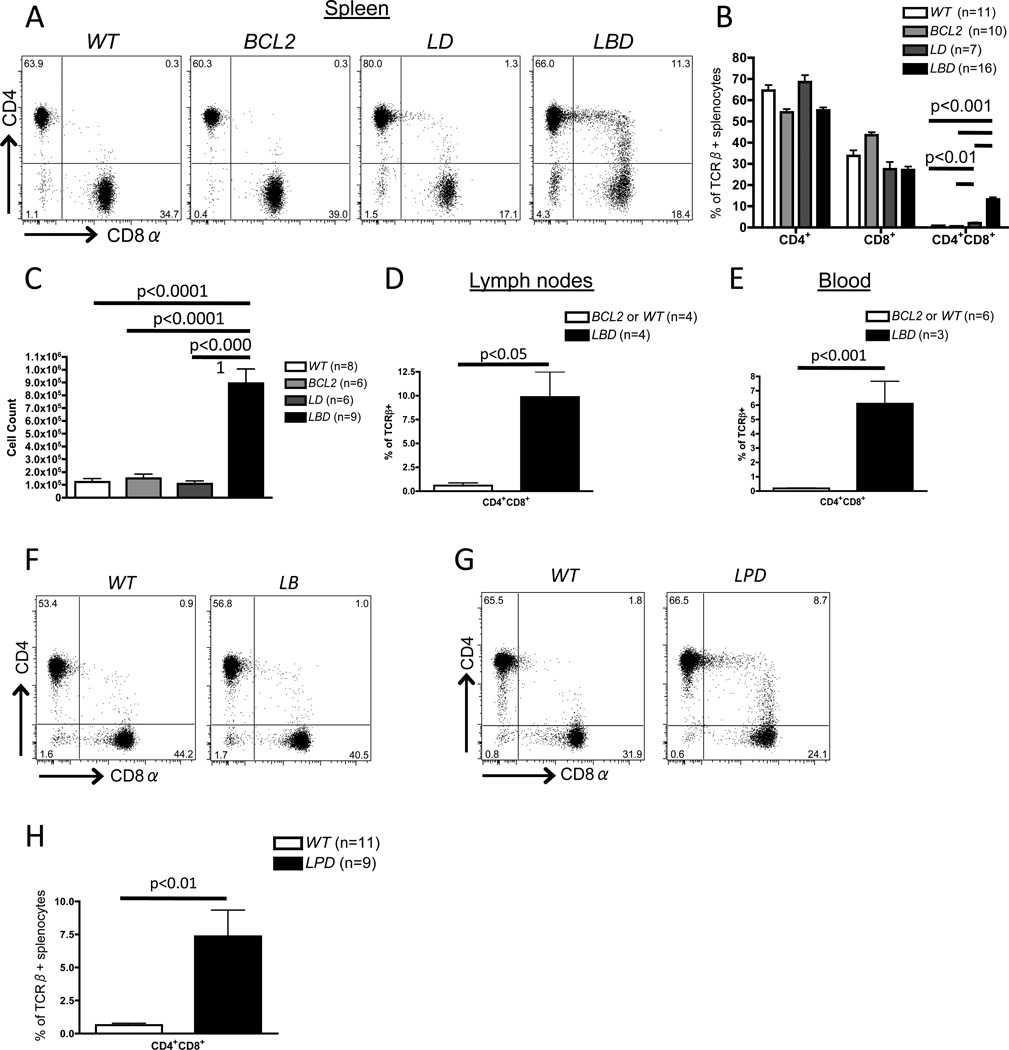
While previous reports suggested that Dicer does not control lineage commitment of CD4+ and CD8+ αβ T cells (26, 27, 39), we considered the possibility that apoptosis of Dicer-deficient thymocytes might mask a role for Dicer in CD4/CD8 lineage commitment. To address this possibility, we analyzed LckCre:EμBCL2:Dicerflox/flox (LBD), LckCre:Dicerflox/flox (LD), EμBCL2:Dicerflox/flox (BCL2), and Dicerflox/flox (wild-type or WT) mice. We and others have shown that the LckCre transgene drives efficient Dicer deletion in LD and LBD DN thymocytes (26, 27, 37). The EμBCL2 transgene drives expression of the anti-apoptotic BCL2 protein throughout αβ T cell development (29) and inhibits apoptosis of LD thymocytes, which results in rescue of thymic DP cellularity as we have previously reported (37). We show here that SP thymocyte numbers are also rescued in LBD mice (Supplemental Fig. S1A and S1B), although peripheral αβ T cell numbers are reduced in LBD mice relative to controls (Supplemental Fig. S1C and S1D), likely because Eμ activity (and therefore transgenic BCL2 expression) declines as αβ T cells mature (Supplemental Fig. S1E). Notably, BCL2 expression does not alter the extent of Dicer deletion in DN thymocytes of LBD mice relative to LD mice (37), making it unlikely that phenotypic differences between LBD and LD mice arise from different efficiencies of Dicer deletion and resultant miR loss. Flow cytometry analyses in LBD mice showed large numbers of splenic αβ T cells aberrantly expressing both CD4 and CD8 at varying levels (hereafter referred to as CD4+CD8+ cells; Fig. 1A,B), corresponding to ~15% of splenic αβ T cells in 4–8 week old animals. This population was absent from WT and BCL2 mice (Fig. 1A,B). The increased frequency of CD4+CD8+ αβ T cells was not simply due to loss of CD4+ or CD8+ cells since LBD mice exhibited an ~10-fold increase in total number of splenic CD4+CD8+ cells as compared to control mice (Fig. 1C). We also observed increased frequencies of CD4+CD8+ αβ T cells in the lymph nodes and blood of LBD mice compared to WT and BCL2 mice (Fig. 1D,E). In addition, CD4+CD8+ αβ T cells were absent in Dicer-sufficient, LckCre:EμBCL2 (LB mice (Fig. 1F), indicating that this phenotype of LBD mice is not due to combined expression of Cre and BCL2.
Figure 1. Generation of aberrant peripheral CD4+CD8+ cells in Dicer deficient mice expressing a BCL2 transgene or lacking Trp53.
A, Representative CD4 and CD8 staining on CD24loTCRβ+ splenocytes of WTBCL2LD, or LBD mice. B, Average percentages of CD4+CD8+, and CD4+CD8+ cells amongst CD24loTCRβ+ splenocytes of WTBCL2LD, or LBD mice. C, Total numbers of CD4+CD8+CD24loTCRβ+ splenocytes in mice of the indicated genotypes. D and E, Frequency of CD4+CD8+ cells among TCRβ+ cells in lymph nodes (D) or blood (E) of LBD or control mice. F, Representative CD4 and CD8 staining on CD24loTCRβ+ splenocytes of WT or LckCre:EμBCL2 mice. G, Representative CD4 and CD8 staining on CD24loTCRβ+ splenocytes of WT or LPD mice. H, Average frequency of CD4+CD8+ cells amongst CD24loTCRβ+ splenocytes of WT or LPD mice. B, C, D, E, and H, The total numbers of mice analyzed are indicated. B, C, D, E, F, and H, each experiment was performed at least 3 independent times.
Normally, pre-selection DP thymocytes are CD24hiTCRβlo cells, become CD24hiTCRβhi cells after up-regulation of TCRβ expression during positive selection, and then down-regulate CD24 expression to become CD24loTCRβhi SP mature thymocytes (40); mature peripheral αβ T cells are similarly CD24loTCRβhi. We found that CD4+CD8+ αβ T cells were CD24loTCRβhi (Fig. 1A,B), suggesting that they are mature, post-selection αβ T cells. To further clarify their nature, we performed qPCR for mRNAs that are differentially expressed between thymocytes and mature αβ T cells. Similar to normal splenic CD4+ and CD8+ αβ T cells, splenic CD4+CD8+CD24loTCRβhi αβ T cells did not express Rag1 or Rorc, two markers of immature DP thymocytes (Fig. S2A and S2B). Splenic CD4+CD8+CD24loTCRβhi αβ T cells also exhibited higher expression of Foxo1 (Fig. S2C), which is up-regulated in mature αβ T cells. These data indicate that the CD4+CD8+ αβ T cells that develop in LBD mice exhibit multiple features of mature, post-selection αβ T cells and are not simply immature, pre-selection DP thymocytes that exited the thymus. Therefore, our data demonstrate that expression of the pro-survival BCL2 protein throughout development of Dicer-deficient αβ T lineage cells permits generation of mature, post-selection peripheral CD4+CD8+ αβ T cells.
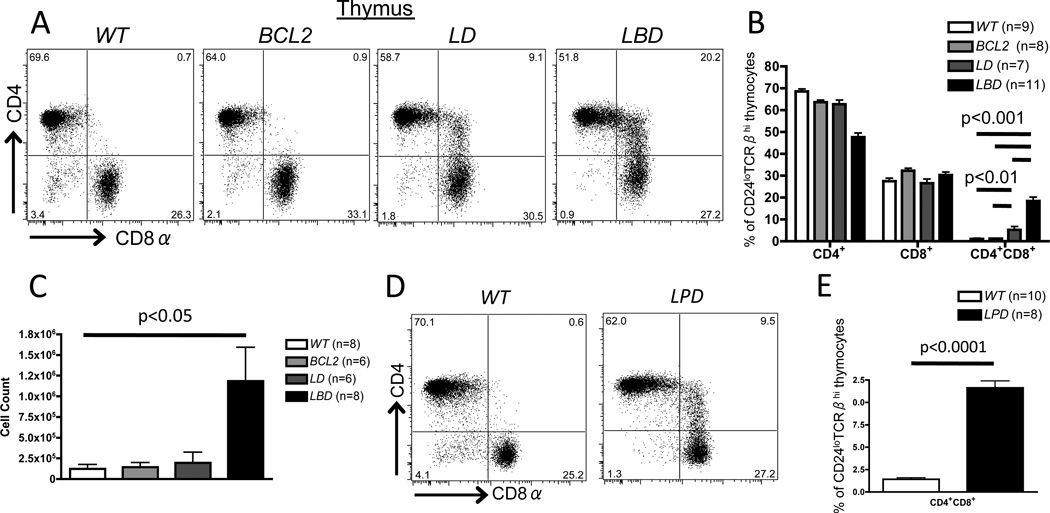
In addition to promoting survival, ectopic BCL2 expression affects other pathways and processes that regulate αβ T lymphocyte differentiation, including NFAT signaling and αβ TCR selection (41, 42). Thus, to rule out the possibility that peripheral CD4+CD8+ αβ T cells in LBD mice arise from effects of BCL2 expression other than promoting survival of Dicer-deficient cells, we generated and analyzed LckCre:p53flox/flox:Dicerflox/flox (LPD) mice with combined inactivation of Dicer and p53 initiating in DN thymocytes. The p53 protein activates cell cycle checkpoints in response to DNA damage and other cellular stresses, and induces apoptosis when such stresses are too severe (43). Similar to the case for LBD mice, we found higher frequencies of mature splenic CD4+CD8+ αβ T cells in LPD mice relative to WT mice (Fig. 1G,H). This finding indicates that inactivation of the pro-apoptotic p53 protein in Dicer-deficient thymocytes also permits accumulation of splenic CD4+CD8+ αβ T cells. Consequently, we conclude that inhibiting apoptosis of Dicer-deficient αβ T lineage cells unmasks a requirement for Dicer in appropriate CD4/CD8 silencing in mature αβ T cells.
Dicer is required for normal initiation of CD4/CD8 silencing following αβ TCR selection
The splenic CD4+CD8+ αβ T cells in LBD mice could result from impaired initiation of CD4/CD8 silencing upon selection of thymocytes and/or impaired maintenance of CD4/CD8 silencing following thymic egress of mature post-selection thymocytes. To determine whether initiation of CD4/CD8 silencing is impaired in LBD mice, we analyzed CD4 and CD8 expression on pre- and post-selection LBD and control thymocytes. We found that 18% of CD24loTCRβhi mature thymocytes in LBD mice aberrantly expressed both CD4 and CD8, whereas WT and BCL2 mice contained essentially no CD4+CD8+ mature thymocytes (Fig. 2A,B). The increased frequency of CD4+CD8+ cells was not simply due to loss of CD4+ or CD8+ cells since LBD mice exhibited an ~5-fold increase in the total number of thymic CD4+CD8+ cells relative to controls (Fig. 2C). We found similar increased frequencies of CD4+CD8+CD24loTCRβhi thymocytes in LPD mice relative to WT mice (Fig. 2D,E). In addition, we found that 5% of CD24loTCRβhi mature thymocytes in LD mice express both CD4 and CD8. While this frequency is above the levels observed in mature thymocytes of WT and BCL2 mice (Fig. 2A,B), the limited numbers of these thymocytes (<1% of total thymocytes in LD mice) could explain why they were not observed in previous analyses of LD mice (26, 27). To evaluate whether incomplete deletion and inactivation of Dicer could account for impaired initiation of CD4/CD8 silencing in only a subset of LBD and LD thymocytes, we quantified the expression of two miRs that are highly expressed in thymocytes (miR-181a and let-7c) (44). Post-selection (CD24loTCRβhi) thymocytes from LD and LBD mice exhibited >80% reduction in miR-181a and let-7c levels (Fig. S3A, B), indicating comparable and substantial inactivation of Dicer in post-selection thymocytes of both LD and LBD mice. Furthermore, the levels of each miR were reduced similarly among post-selection CD4+CD8+, or CD4+CD8+ thymocytes from LBD and LD mice (Fig. S3A, B), indicating that Dicer is required for normal initiation of CD4/CD8 silencing in only a subset of post-selection thymocytes. Regardless, our data show that CD4+CD8+ mature thymocytes arise after positive selection of Dicer-deficient thymocytes, and that expression of BCL2 or inactivation of p53 is not required for development of these cells. Therefore, we conclude that Dicer is required for appropriate initiation of Cd4 and/or Cd8 silencing during intrathymic αβ T cell differentiation.
Figure 2. Impaired initiation of co-receptor silencing in mature thymocytes of Dicer deficient mice expressing a BCL2 transgene or lacking Trp53.
A, Representative CD4 and CD8 staining on CD24loTCRβhi mature thymocytes of WTBCL2LD, or LBD mice. B, Average percentages of CD4+CD8+and CD4+CD8+ cells amongst CD24loTCRβhi mature thymocytes of WTBCL2LD, or LBD mice. C, Total numbers of CD4+CD8+CD24loTCRβhi mature thymocytes in mice of the indicated genotypes. D, Representative CD4 and CD8 staining on CD24loTCRβhi mature thymocytes of WT or LPD mice. E, Average frequency of CD4+CD8+ cells amongst CD24loTCRβ+ mature thymocytes of WT or LPD mice. B, C, and E, the numbers of mice analyzed are indicated; each experiment was performed at least 3 independent times.
Cd4 and Cd8 silencing is maintained in Dicer-deficient αβ T cells
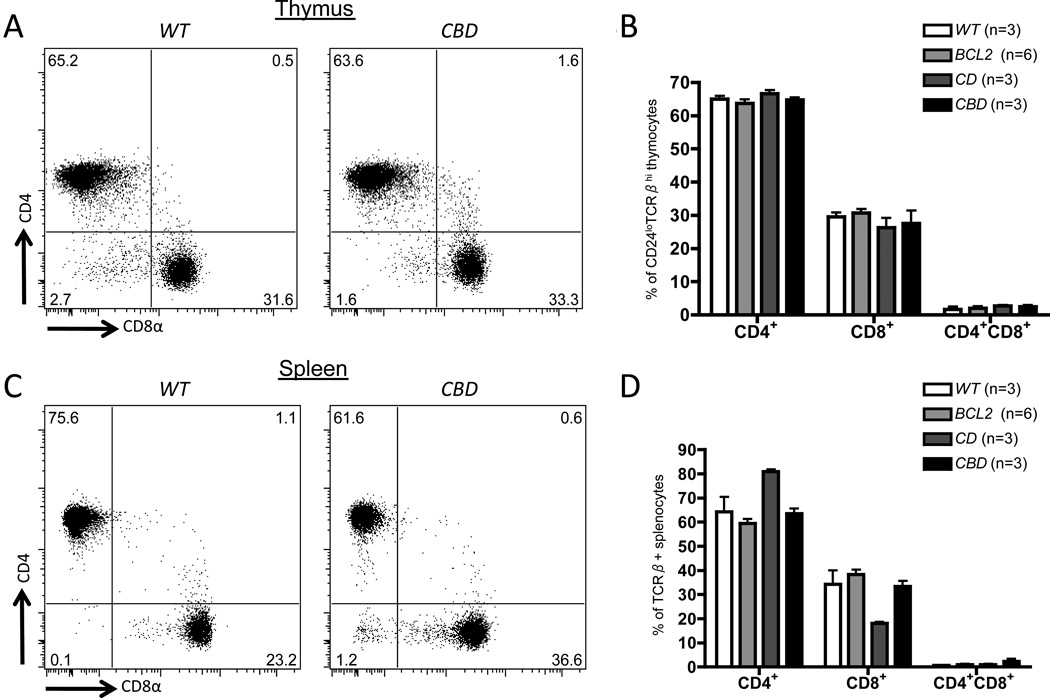
A requirement for Dicer in initiation of Cd4 and/or Cd8 silencing does not preclude a role for Dicer in maintenance of Cd4 and Cd8 silencing in mature αβ T cells. To determine whether Dicer is also required for appropriate maintenance of Cd4 and Cd8 silencing, we employed a genetic approach. The Cd4Cre transgene drives expression of Cre recombinase and deletion of Dicerflox alleles initiating in DP thymocytes (27, 39). However, published reports have shown that Cd4Cre-mediated Dicer deletion does not lead to appreciable loss of Dicer-dependent miRs until after initiation of Cd4 and Cd8 silencing and CD4/CD8 lineage commitment (27, 39, 45). In contrast, peripheral αβ T cells of Cd4Cre:Dicerflox/flox mice exhibit near complete deletion of Dicerflox alleles, low expression of miRs, and phenotypes indicative of Dicer inactivation (27, 39, 45). Based on these observations, we reasoned that Cd4Cre-mediated deletion of Dicer starting in CD4+CD8+ thymocytes would allow initiation of Cd4 and Cd8 silencing before substantial loss of Dicer-dependent miRs, and thereby permit evaluation of whether Dicer has a role in maintenance of co-receptor silencing in CD4+ and CD8+ αβ T cells. Thus, we generated and analyzed Cd4Cre:EμBCL2:Dicerflox/flox (CBD) mice. In striking contrast to LBD mice, CBD mice had neither mature CD4+CD8+ thymocytes (Fig. 3A,B) nor CD4+CD8+ splenic αβ T cells (Fig. 3C,D). We conclude that Dicer and miRs are not required for the maintenance of Cd4 and Cd8 silencing in mature splenic αβ T cells.
Figure 3. Normal initiation of Cd4 and Cd8 silencing in thymocytes and T cells with Dicer deletion initiating in DP thymocytes.
A and C, Representative CD4 and CD8 staining on mature thymocytes (A) or CD24loTCRβ+ splenocytes (C) of WT or CBD mice. B and D, Average percentages of CD4+CD8+and CD4+CD8+ cells amongst mature thymocytes (B) or CD24loTCRβ+ splenocytes (D) of WTBCL2CD, and CBD mice. B and D, the numbers of mice analyzed are indicated. Each experiment was performed at least 3 independent times.
Dicer controls initiation of both Cd4 and Cd8 silencing
The CD4+CD8+ αβ T cells in LBD mice could comprise MHCI-restricted cells with impaired Cd4 silencing, MHCII-restricted cells with impaired Cd8 silencing, or both. To address this issue we restricted the ability of thymocytes to develop on MHCI or MHCII by transferring bone marrow cells from LBD or BCL2 mice into irradiated MHCI−/− or MHCII−/− recipient mice.
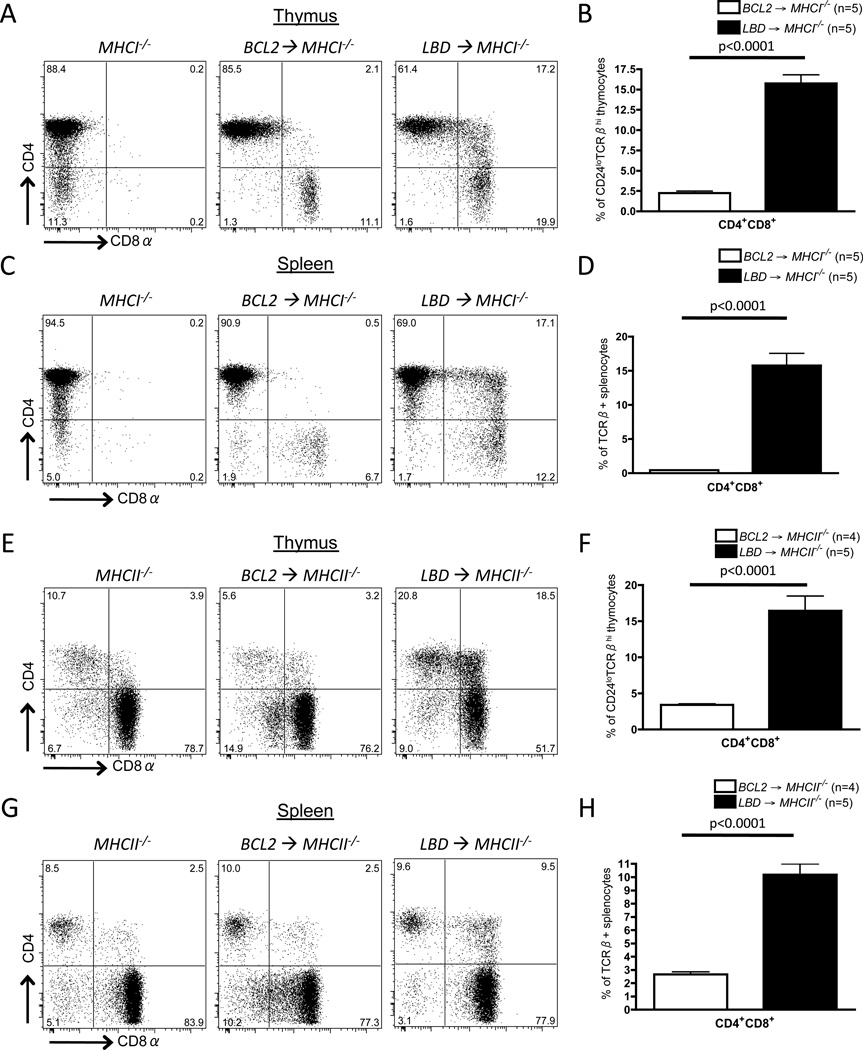
To determine whether Dicer is required for appropriate initiation of Cd8 silencing during development of MHCII-restricted CD4+ αβ T cells, we analyzed irradiated MHCI−/− mice (32) reconstituted with LBD or BCL2 bone marrow cells. It has been shown that BCL2 expression in MHCI−/− mice allows development of small numbers of splenic CD8+ T cells (46). We found the same in MHCI−/− mice reconstituted with BCL2 or LBD bone marrow cells (Fig. 4C). However, we also observed that ~15% of CD24loTCRβhi mature thymocytes aberrantly expressed both CD4 and CD8 in MHCI−/− mice reconstituted with LBD bone marrow, while only 2% of CD24loTCRβhi mature thymocytes were CD4+CD8+ in MHCI−/− mice reconstituted from BCL2 cells (Fig. 4A,B). We also found that ~15% of splenic αβ T cells were CD4+CD8+ in MHCI−/− mice reconstituted with LBD bone marrow cells, while only 0.5% of splenic αβ T cells expressed both CD4 and CD8 in MHCI−/− mice reconstituted with BCL2 bone marrow (Fig. 4C,D). While we cannot rule out that the CD4+CD8+ αβ T cells in MHCI−/− mice reconstituted with LBD cells developed from CD8+ cells that failed to silence Cd4, the substantial population of CD4+CD8+ αβ T cells after transfer of LBD versus BCL2 cells is more consistent with Dicer inactivation leading to impaired initiation of Cd8 silencing during intrathymic differentiation of MHCII-restricted αβ T cells.
Figure 4. Dicer is required for appropriate initiation of Cd4 silencing in MHCI-restricted cells and Cd8 silencing in MHCII-restricted cells.
A-D, Representative CD4 and CD8 staining on mature thymocytes (A) or CD24loTCRβ+ splenocytes (C) of MHCI−/− mice reconstituted with BCL2 or LBD bone marrow. Average frequencies of CD4+CD8+ cells amongst mature thymocytes (B) or CD24loTCRβ+ splenocytes (D) of MHCI−/− mice reconstituted with BCL2 or LBD bone marrow. E-H, Representative CD4 and CD8 staining on mature thymocytes (E) or CD24loTCRβ+ splenocytes (G) of MHCII−/− mice reconstituted with BCL2 or LBD bone marrow. Average frequencies of CD4+CD8+ cells among mature thymocytes (F) or CD24loTCRβ+ splenocytes (H) of MHCII−/− mice reconstituted with BCL2 or LBD bone marrow. B, D, F and H, The numbers of mice analyzed are shown. The experiment was performed twice with at least 4 recipient mice per group; a representative experiment is shown.
To determine whether Dicer is required for appropriate initiation of Cd4 silencing during development of MHCI-restricted CD8+ αβ T cells, we analyzed irradiated MHCII−/− mice (33) reconstituted with bone marrow from LBD or BCL2 mice. As previously shown (33, 46), a small population of CD4+ αβ T cells does develop in MHCII−/− mice, although the vast majority of cells are CD8+ (Fig. 4E). We found that ~16% of CD24loTCRβhi mature thymocytes expressed both CD4 and CD8 in MHCII−/− mice reconstituted with LBD bone marrow, but only ~4% of these cells were CD4+CD8+ in MHCII−/− mice reconstituted with BCL2 cells (Fig. 4E,F). We also found that ~10% of splenic αβ T cells were CD4+CD8+ in MHCII−/− mice reconstituted from LBD bone marrow, but only ~2.5% of splenic αβ T cells expressed both CD4 and CD8 in MHCII−/− mice reconstituted from BCL2 cells (Fig. 4G,H). Although we cannot rule out that the CD4+CD8+ αβ T cells in MHCII−/− mice reconstituted with LBD cells developed from CD4+ cells that failed to silence Cd8, the substantial population of CD4+CD8+ αβ T cells after transfer of LBD versus BCL2 cells is more consistent with Dicer inactivation causing impaired initiation of Cd4 silencing during intrathymic differentiation of MHCI-restricted αβ T cells.
Based on our analyses of MHCI−/− and MHCII−/− mice reconstituted with LBD or BCL2 cells, we conclude that Dicer expression in immature DP thymocytes is needed for appropriate initiation of Cd4 and Cd8 silencing in MHCI and MHCII-restricted cells, respectively.
Dicer regulates Cd4 and Cd8 silencing and expression of Runx3 and Thpok in positively-selected αβ T cells
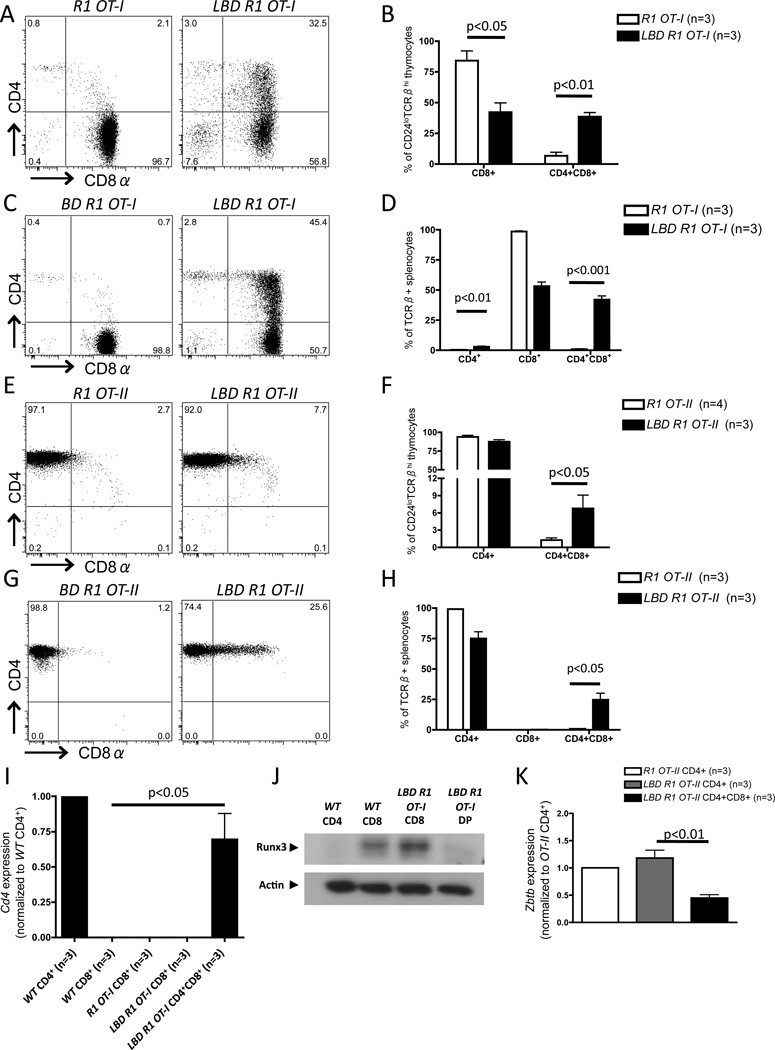
To gain further support for our conclusion that Dicer is required for appropriate initiation of both Cd4 and Cd8 silencing, we generated LBD mice that express the MHCI-restricted OT-I αβ TCR transgene, which normally promotes positive selection of only CD8+ T cells (34), or the MHCII-restricted OT-II αβ TCR transgene, which normally promotes positive selection of only CD4+ T cells (35). We generated these mice on a Rag1−/− background (LBD R1 OT-I and LBD RI OT-II mice) to prevent TCRβ and TCRα gene rearrangements that could subvert the ability of these αβ TCR transgenes to restrict MHC specificity. We found that positively-selected LBD R1 OT-I mature thymocytes (Figs. 5A, B) and αβ T cells (Figs. 5C, D) exhibited impaired Cd4 silencing, with ~35–45% of cells aberrantly expressing both CD4 and CD8, indicating that Dicer is required for appropriate initiation of Cd4 silencing in cells expressing an MHCI-restricted αβ TCR transgene. Similarly, we found that positively-selected LBD R1 OT-II mature thymocytes (Figs. 5E, F) and αβ T cells (Figs. 5G, H) exhibited impaired Cd8 silencing, with ~5% of mature thymocytes and ~25% of mature splenic αβ T cells expressing both CD4 and CD8, indicating that Dicer is also required for normal Cd8 silencing in cells expressing an MHCII-restricted αβ TCR transgene. Collectively, these data demonstrate that Dicer ensures appropriate silencing of both Cd4 and Cd8 in positively-selected αβ T cells.
Figure 5. Dicer regulates Cd4 and Cd8 silencing and expression of Runx3 and Zbtb7b in positively-selected αβ T cells.
A, Representative CD4 and CD8 staining on CD24loTCRβhi mature thymocytes of R1 OT-I and LBD R1 OT-I mice. B, Average percentages of CD4+CD8+, and CD4+CD8+ cells amongst CD24loTCRβhi mature thymocytes of R1 OT-I and LBD R1 OT-I mice. C, Representative CD4 and CD8 staining on CD24loTCRβ+ splenocytes of BD R1 OT-I and LBD R1 OT-I mice. D, Average percentages of CD4+CD8+, and CD4+CD8+ cells amongst CD24loTCRβ+ splenocytes of R1 OT-I and LBD R1 OT-I mice. E, Representative CD4 and CD8 staining on CD24loTCRβhi mature thymocytes of R1 OT-II and LBD R1 OT-II mice. F, Average percentages of CD4+CD8+, and CD4+CD8+ cells amongst CD24loTCRβhi mature thymocytes of R1 OT-II and LBD R1 OT-II mice. G, Representative CD4 and CD8 staining on CD24loTCRβ+ splenocytes of BD R1 OT-II and LBD R1 OT-II mice. H, Average percentages of CD4+CD8+, and CD4+CD8+ cells amongst CD24loTCRβ+ splenocytes of R1 OT-II and LBD R1 OT-II mice. I, qRT-PCR for primary (un-spliced) Cd4 transcripts in sorted splenic populations from WTR1 OT-I, or LBD R1 OT-I mice. J, Runx3 Western blot in sorted splenic CD4+ or CD8+ cells from WT mice and CD8+ or CD4+CD8+ DP) cells from LBD R1 OT-I. Three independent replicates were performed; a representative blot is shown. K, Zbtb7b qRT-PCR in sorted populations from R1 OT-II or LBD R1 OT-II mice. B, D, F, H, I, and K, The numbers of mice analyzed are indicated. At least 3 independent experiments were performed in each case.
The expression of both CD4 and CD8 on Dicer-deficient αβ T cells could result from impaired transcriptional or translational silencing of Cd4 and Cd8. To evaluate the role of Dicer in control of co-receptor transcriptional silencing, we conducted qRT-PCR analyses of primary (un-spliced) Cd4 transcripts in mature αβ T cells sorted from spleens of LBD R1 OT-I and control mice. We detected similarly high levels of primary Cd4 transcripts in LBD R1 OT-I CD4+CD8+ cells and WT CD4+ cells (Fig. 5I). In contrast, we were unable to detect primary Cd4 transcripts in CD8+ cells of WTR1 OT-1, or LBD R1 OT-I mice (Fig. 5I). These results demonstrate that Dicer is required for appropriate transcriptional silencing of Cd4 in MHCI-restricted αβ T cells.
Following positive selection, Runx3 expression is up-regulated in MHCI-restricted thymocytes and drives appropriate initiation of Cd4 transcriptional silencing in CD8 lineage cells (6, 7). To determine whether Dicer controls expression of Runx3 in positively selected, MHCI-restricted αβ T cells, we conducted Western blot analyses of Runx3 protein in mature αβ T cells sorted from spleens of LBD R1 OT-I and control mice. We detected a decreased level of Runx3 protein in CD4+CD8+ cells of LBD R1 OT-I mice as compared to CD8+ cells of WT and LBD R1 OT-I mice (Fig. 5J), indicating that Dicer is required for appropriate expression of Runx3 in positively-selected, MHCI-restricted αβ T cells.
Analogous to Runx3 up-regulation in MHCI-restricted cells, positive selection of MHCII-restricted cells induces Thpok expression, which drives CD4 lineage commitment and facilitates Cd8 silencing (8–10). Given that Runx3 expression was impaired in Dicer-deficient MHCI-restricted αβ T cells, we hypothesized that Dicer might similarly control expression of Thpok in MHCII-restricted cells. To test this hypothesis, we performed qRT-PCR analyses for Zbtb7b mRNA (since we were unable to isolate enough cells for Western blot with available Thpok antibodies) in sorted cells from LBD R1 OT-II and control mice. We found a lower level of Zbtb7b mRNA in CD4+CD8+ cells from LBD R1 OT-II mice relative to CD4+ cells from control R1 OT-II mice (Fig. 5K), revealing that Dicer is also required for normal expression of Zbtb7b in positively-selected, MHCII-restricted αβ T cells. Collectively, these data demonstrate that Dicer promotes appropriate expression of “master” transcriptional regulators of the CD4 and CD8 αβ T cell lineages in MHC II- or I-restricted cells, respectively, following positive selection.
Drosha is also required for normal Cd4 and Cd8 silencing during αβ T cell development
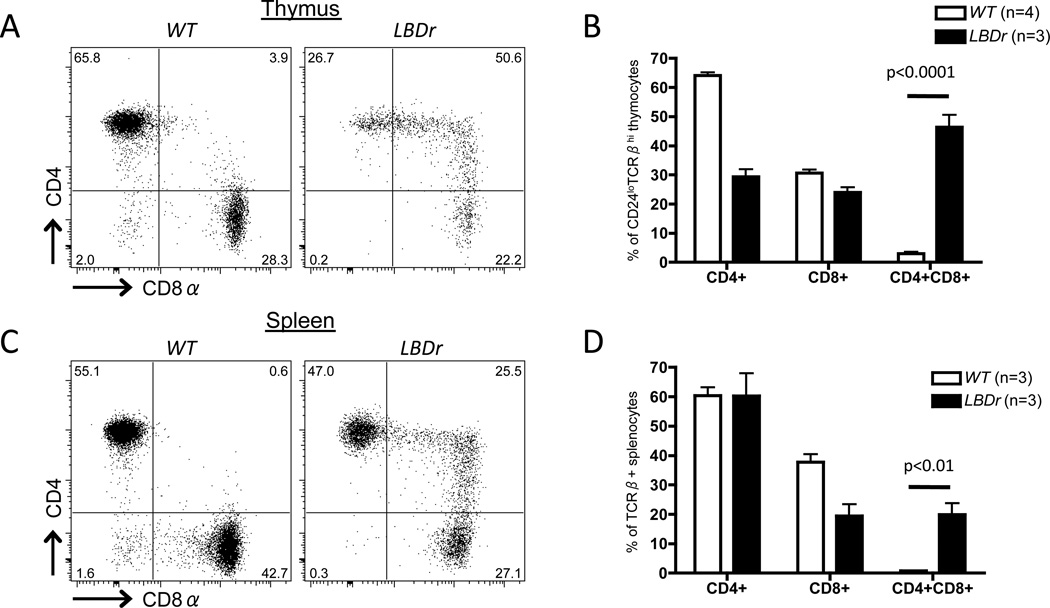
Dicer could regulate Cd4 and Cd8 silencing through generation of siRNAs that directly halt transcription of these loci and/or via biogenesis of miRs that indirectly regulate expression of Runx3, Thpok, or other factors that control CD4 and CD8 expression. To determine if Dicer-dependent siRNAs and/or miRs regulate initiation of Cd4 and Cd8 silencing, we generated and analyzed LckCre:EμBCL2:Droshaflox/flox (LBDr) mice because the Drosha RNA endonuclease is required for production of miRs, but not siRNAs (47). We observed that ~ 40% of CD24loTCRβhi mature thymocytes in LBDr mice were CD4+CD8+ (Fig. 6A,B), revealing that Drosha is required for appropriate initiation of Cd4 and/or Cd8 silencing. We also found that ~20% of mature splenic αβ T cells in LBDr mice were CD4+CD8+ (Fig. 6C,D), indicating that ectopic expression of BCL2 throughout development of Drosha-deficient αβ T cells permits the accumulation of mature post-selection splenic CD4+CD8+ αβ T cells. Since these data demonstrate that both Drosha and Dicer are required for regulation of CD4 and CD8 expression in mature αβ T cells, we conclude that miRs) likely control the appropriate initiation of Cd4 and Cd8 silencing during αβ T cell differentiation.
Figure 6. Drosha is required for appropriate initiation of Cd4 and Cd8 silencing after positive selection.
Representative CD4 and CD8 staining on CD24loTCRβhi mature thymocytes (A) or TCRβ+ splenocytes (C) of WT or LBDr mice. Average percentages of CD4+CD8+, and CD4+CD8+ cells among mature thymocytes (B) or TCRβ+ splenocytes (D) of WT and LBDr mice. B and D, The numbers of mice analyzed are indicated; each experiment was performed at least 3 independent times.
Discussion
We have demonstrated that expression of the Dicer and Drosha proteins in thymocytes is required for appropriate initiation of Cd4 and Cd8 silencing during intrathymic differentiation of CD8+ and CD4+ αβ T cells, respectively. The positive selection of CD24hiTCRβloCD4+CD8+ immature thymocytes activates intracellular signals that up-regulate TCRβ expression and down-regulate CD24 expression as these cells differentiate into lineage-committed CD24loTCRβhiCD4+ or CD24loTCRβhiCD8+ mature thymocytes that exit the thymus as mature CD4+ or CD8+ αβ T cells (40). Our detection of CD24loTCRβhiCD4+CD8+ mature thymocytes in mice with Dicer or Drosha inactivation starting in DN thymocytes demonstrates that a Dicer- and Drosha-dependent mechanism(s) is required for the appropriate initiation of Cd4 and Cd8 silencing. The increased presence of these cells and the generation of peripheral CD4+CD8+ αβ T cells following BCL2 expression (or p53 inactivation) indicates that apoptosis of Dicer- and Drosha-deficient αβ T cells after thymic emigration obscures the critical role of Dicer and Drosha in Cd4 and Cd8 silencing. For this reason, previous analyses of splenic αβ T cells in mice with Dicer or Drosha inactivation starting in DN thymocytes failed to discover that Dicer and Drosha control Cd4 and Cd8 silencing (26, 27). We previously showed that ectopic BCL2 expression in Dicer-deficient thymocytes similarly unmasks a requirement for Dicer in promoting survival of DN thymocytes that attempt TCRβ gene rearrangements (37). Thus, our observations indicate that suppressing apoptosis should be standard practice when analyzing and interpreting phenotypes of Dicer- or Drosha-deficient cells.
Our discovery that Dicer- and Drosha-deficient αβ T cells exhibit similar defects in CD4 and CD8 expression provides strong support for our conclusion that miRs regulate Cd4 and Cd8 silencing during CD4/CD8 lineage commitment. In this context, while our data do not exclude another mechanism, miR biogenesis is the only known process for which both Drosha and Dicer are required. How might miRs control the initiation of Cd4 and Cd8 silencing? TCR-activated signaling pathways regulate expression of Runx3, Thpok, and possibly other proteins that modulate transcription of Cd4 and Cd8 loci (2). Our finding that Dicer promotes expression of Runx3 and Thpok in MHC I- or II-selected αβ T cells, respectively, suggests that miRs likely are required for appropriate up-regulation of these transcription factors, and thus proper initiation of Cd4 and/or Cd8 silencing. Loss of miRs that bind Runx3 and/or Thpok mRNAs should increase expression of their encoded proteins. Accordingly, Dicer likely controls expression of these lineage-specifying factors through miRs that enhance signaling pathways or transcriptional networks that induce Runx3 and Thpok expression, and/or through miRs that inhibit repressors of Runx3 and Thpok expression. Alternatively, it is possible that the development of CD4+CD8+ αβ T cells results from indirect effects of pro-apoptotic signals generated upon miR loss, rather than direct roles of miRs in Cd4 and Cd8 silencing.
While we have demonstrated that Dicer deficiency impairs Cd4 transcriptional silencing, our results do not rule out additional roles for Dicer (or Drosha) in regulating post-transcriptional silencing of Cd4 and/or Cd8. For example, miRs could bind to and induce degradation or block translation of Cd4 or Cd8 transcripts during initiation of CD4 and CD8 silencing, respectively. Consistent with this notion, positive selection decreases the half-lives of Cd4 and Cd8α mRNAs (48). Yet, these changes depend on protein synthesis (48), Cd8α and Cd8β mRNAs lack conserved miR seed sequences (49), and Cd4 reporter genes that lack the Cd4 3’UTR exhibit normal silencing (50), which together argue against a role for miRs in control of Cd4 and Cd8 silencing via direct inhibition of Cd4 and Cd8 mRNAs.
Notably, inactivation of Dicer or Drosha initiating in DN thymocytes leads to the failure of Cd4 or Cd8 silencing in less than half of positively selected CD4+CD8+ αβ T lineage cells. This could be due in part to the timing of Dicer or Drosha deletion relative to positive selection of CD4+CD8+ thymocytes and concomitant Cd4 or Cd8 silencing. LckCre-mediated deletion of Dicer starting in DN thymocytes does not lead to a complete absence of miRs in total thymocytes (26), raising the possibility that a significant fraction of Dicer-deficient CD4+CD8+ thymocytes might undergo positive selection and CD4/CD8 lineage-commitment in the presence of miRs that promote Cd4 and/or Cd8 silencing. This model would be consistent with our data that Cd4Cre-mediated Dicer deletion starting in DP thymocytes is not sufficient to generate CD4+CD8+ mature thymocytes or αβ T cells. Alternatively, our data that most positively-selected αβ T lineage cells that lack Dicer or Drosha are capable of normal Cd4 or Cd8 silencing may indicate that miRs serve to facilitate efficient co-receptor silencing, rather than being absolutely required. For example, miRs could function to increase the recruitment or “on-rate” of transcriptional repressors or chromatin modifiers such as histone deacetylases that mediate CD4 and CD8 co-receptor silencing. In the absence of miRs, these factors may still bind to co-receptor loci and silence transcription, but the kinetic delay in recruitment would manifest as impaired co-receptor silencing in a fraction of cells as observed. In future studies it will be critical to determine whether aberrant peripheral CD4+CD8+ cells exhibit lineage plasticity or are instead fully committed to either the CD4 or CD8 lineage but simply delayed in their differentiation (e.g., could sustained TCR signals that normally promote CD4 lineage commitment re-direct MHCI-restricted splenic CD4+CD8+ cells to the CD4 lineage?).
Identification and characterization of the Cd4 silencer and failure to discover a Cd8 silencer have led to acceptance in the field that distinct mechanisms control initiation of Cd4 and Cd8 silencing (2). Our data that the miR biogenesis machinery controls appropriate silencing of both Cd4 and Cd8 challenges this dogmatic view. Furthermore, our results suggest that miRs are required for appropriate expression of “master regulators” of CD4 and CD8 lineage commitment following positive selection. Specifically, Dicer deficiency uncouples the regulatory modules that mediate lineage commitment and migration out of the thymus, with phenotypically mature Dicer deficient cells capable of exiting the thymus prior to normal up-regulation of lineage-specifying factors. It will be important to determine whether miRs regulate CD4 and CD8 lineage commitment via a shared pathway, such as TCR signaling, or via distinct mechanisms. Given the role of CD4+ and CD8+ αβ T cell development as a paradigm for elucidating genetic and epigenetic mechanisms that control gene expression changes during cellular differentiation, elucidating Dicer- and Drosha-dependent mechanisms that control CD4 and CD8 lineage commitment should have broad relevance.
Supplementary Material
Acknowledgements
We thank Katherine Yang-Iott and Qi Xiao for technical assistance.
This work was supported by the University of Pennsylvania Training Grants in Computational Genomics HG-000046 (L.J.R.), Cell and Molecular Biology GM-07229 (B.L.B., M.E.D.) and Immunobiology of Normal and Neoplastic Lymphocytes CA-009140 (M.E.D.); NIH Intramural Research Programs of the Center for Cancer Research, NCI (A.C.C., R.B.) and NIAID (B.L.B., S.A.M.); NIH grant AI059621 (A.B.); and Leukemia and Lymphoma Society Scholar Award and NIH grants CA125195 and CA136470 (C.H.B.).
References
- 1.Singer A, Adoro S, Park J-H. Lineage fate and intense debate: myths, models and mechanisms of CD4− versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taniuchi I, Ellmeier W. Transcriptional and Epigenetic Regulation of CD4/CD8 Lineage Choice. Adv. Immunol. 2011;110:71–110. doi: 10.1016/B978-0-12-387663-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 3.Boehmer von H. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv. Immunol. 2004;84:201–238. doi: 10.1016/S0065-2776(04)84006-9. [DOI] [PubMed] [Google Scholar]
- 4.Starr TK, Jameson SC, Hogquist KA. Positive and Negative Selection of T Cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Xiong Y, Bosselut R. Tenuous paths in unexplored territory: From T cell receptor signaling to effector gene expression during thymocyte selection. Semin Immunol. 2010;22:294–302. doi: 10.1016/j.smim.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 7.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, Levanon D, Groner Y. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci USA. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galéra P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 9.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 10.Rui J, Liu H, Zhu X, Cui Y, Liu X. Epigenetic silencing of CD8 genes by ThPOK-mediated deacetylation during CD4 T cell differentiation. J Immunol. 2012;189:1380–1390. doi: 10.4049/jimmunol.1201077. [DOI] [PubMed] [Google Scholar]
- 11.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 12.He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ. CD4-CD8 Lineage Commitment Is Regulated by a Silencer Element at the ThPOK Transcription-Factor Locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R. The Zinc Finger Transcription Factor Zbtb7b Represses CD8-Lineage Gene Expression in Peripheral CD4+ T Cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 15.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, Bosselut R. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J Immunol. 2007;179:4405–4414. doi: 10.4049/jimmunol.179.7.4405. [DOI] [PubMed] [Google Scholar]
- 17.Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 18.Chong MMW, Simpson N, Ciofani M, Chen G, Collins A, Littman DR. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellmeier W, Sunshine MJ, Losos K, Hatam F, Littman DR. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- 20.Ellmeier W, Sunshine MJ, Losos K, Littman DR. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- 21.Hostert A, Tolaini M, Roderick K, Harker N, Norton T, Kioussis D. A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity. 1997;7:525–536. doi: 10.1016/s1074-7613(00)80374-x. [DOI] [PubMed] [Google Scholar]
- 22.Hostert A, Garefalaki A, Mavria G, Tolaini M, Roderick K, Norton T, Mee PJ, Tybulewicz VL, Coles M, Kioussis D. Hierarchical interactions of control elements determine CD8α gene expression in subsets of thymocytes and peripheral T cells. Immunity. 1998;9:497–508. doi: 10.1016/s1074-7613(00)80633-0. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y, Bosselut R. CD4–CD8 differentiation in the thymus: connecting circuits and building memories. Current opinion in immunology. 2012;24:139–145. doi: 10.1016/j.coi.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong MMW, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 29.Strasser A, Harris AW, Cory S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 30.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 31.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 32.Koller BH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in β2M, MHC class I proteins, CD8+ T cells. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- 33.Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, Mathis D, Fugger L. Mice lacking all conventional MHC class II genes. Proc Natl Acad Sci USA. 1999;96:10338–10343. doi: 10.1073/pnas.96.18.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 35.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunology and Cell Biology. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 36.Chong MMW, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brady BL, Rupp LJ, Bassing CH. Requirement for Dicer in Survival of Proliferating Thymocytes Experiencing DNA Double-Strand Breaks. J Immunol. 2013;190:3256–3266. doi: 10.4049/jimmunol.1200957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilic I, Koesters C, Unger B, Sekimata M, Hertweck A, Maschek R, Wilson CB, Ellmeier W. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bendelac A, Matzinger P, Seder RA, Paul WE, Schwartz RH. Activation events during thymic selection. J Exp Med. 1992;175:731–742. doi: 10.1084/jem.175.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linette GP, Li Y, Roth K, Korsmeyer SJ. Cross talk between cell death and cell cycle progression: BCL-2 regulates NFAT-mediated activation. Proc Natl Acad Sci USA. 1996;93:9545–9552. doi: 10.1073/pnas.93.18.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villunger A, Marsden VS, Zhan Y, Erlacher M, Lew AM, Bouillet P, Berzins S, Godfrey DI, Heath WR, Strasser A. Negative selection of semimature CD4+8-HSA+ thymocytes requires the BH3-only protein Bim but is independent of death receptor signaling. Proc Natl Acad Sci USA. 2004;101:7052–7057. doi: 10.1073/pnas.0305757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirigin FF, Lindstedt K, Sellars M, Ciofani M, Low SL, Jones L, Bell F, Pauli F, Bonneau R, Myers RM, Littman DR, Chong MMW. Dynamic microRNA gene transcription and processing during T cell development. J Immunol. 2012;188:3257–3267. doi: 10.4049/jimmunol.1103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linette GP, Grusby MJ, Hedrick SM, Hansen TH, Glimcher LH, Korsmeyer SJ. Bcl-2 is upregulated at the CD4+CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity. 1994;1:197–205. doi: 10.1016/1074-7613(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 47.Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci USA. 2011;108:13159–13164. doi: 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cibotti R, Bhandoola A, Guinter TI, Sharrow SO, Singer A. CD8 coreceptor extinction in signaled CD4+CD8+ thymocytes: coordinate roles for both transcriptional and posttranscriptional regulatory mechanisms in developing thymocytes. Mol Cell Biol. 2000;20:3852–3859. doi: 10.1128/mcb.20.11.3852-3859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 50.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.