Abstract
The relative contributions of adaptive selection and neutral drift to observed genetic change are unknown, but likely depend on the inherent abundance of functional genotypes in sequence space and how accessible those genotypes are to one another. To better understand the relative roles of selection and drift in evolution, local fitness landscapes for two different RNA ligase ribozymes were examinedusing a continuous in vitro evolution system under conditions that foster the capacity for neutral drift to mediate genetic change.The exploration of sequence spacewas acceleratedby increasing the mutation rate using mutagenic nucleotide analogs. Drift wasencouraged by carrying out evolution within millions of separate compartments to exploit the founder effect. Deep sequencing of individuals from the evolved populations revealed that the distribution of genotypes did not escape the starting local fitness peak, remaining clustered around the sequenceused to initiate evolution.This is consistent with a fitness landscape where high-fitness genotypes are sparse and well isolated, and suggests, at least in this context, that neutral drift alone is not a primary driver of genetic change. Neutral driftdoes, however, provide a repository of genetic variation upon which adaptive selection can act.
Keywords: Experimental evolution, Fitness landscape, In vitro compartmentalization, Mutagenesis, Neutral drift
Introduction
Evolutionary processesarefundamentally responsible for observed biodiversity, yet a predictive understanding of evolutionary trajectories and the mechanisms that underpin evolution remains elusive. Many approaches to understanding evolutionary outcomes make use of a fitness landscapeheuristic. First conceived by Sewall Wright (1932), a fitness landscape maps the reproductive success of a population of individuals that are connected through mutation.Evolution provides a means to link viable and divergent genotypesto bring about genetic change, but the structure of fitness landscapes for real populations is poorly understood.For a complex phenotype, such as the catalytic activity of an enzyme or the overall ability of an organism to survive, the frequency of high-fitness genotypes out of all possible sequence combinations (sequence space) is undetermined, and the accessibility of these genotypes to one another is unknown (Orr 2009).
Mathematical models of fitness landscapes suggesta highlikelihood that a given high-fitness genotype is directly connected to at least one other high-fitness genotype(Gavrilets and Gravner 1997).If the density of high-fitness genotypes in sequence space is sufficiently high, above a “percolation threshold”, then there will be an extensive neutral network linking these genotypes to one another. Such a network would allow populations to drift across large distances in sequence space without asubstantial reduction in fitness (Gavrilets and Gravner 1997; Kaplan 2008; Pigliucci 2008).
Attempts to enumerate the frequency of high-fitness genotypes in sequence space provide conflicting evidence regarding the extent of neutral networks. Computed distributions of minimum energy secondary structures for either all possible binary RNA sequences or a sampling of quaternary sequences of a given lengthindicate that many different RNA sequencesfold into a smaller set of typical structures, thosesequencesbeing connectable by single mutations (Fontana et al. 1993; Fontana and Schuster 1998; Gruner et al. 1996a; Gruner et al. 1996b).Theseresults suggestthe existence of large neutral networks via redundancy in the map of genotype to structure (reviewed in Schuster and Fontana 1999).Empirical support has come from the construction and testing of intermediate sequences that connecttwo genetically distinctribozymes, a RNAligase and a self-cleaving RNA(Schultes and Bartel 2000). RNAs along a mutational path between these two ribozymes, each intermediate no more than two mutations away from its neighbor, retain partial activity of the closest ribozyme. The central intermediate, with over 40 mutations relative to each starting sequence,isable to catalyze both ribozyme reactions, albeit at a substantially reduced rate.
A more recent empirical study, surveying much broader regions of sequence space by deep sequencing, provides conflicting results (Jiménez et al. 2013). That study assayed all possible sequences of a 24-nucleotide RNA for the ability to bind GTP, and found only 15 unique fitness peaks among the 424 possiblesequences. This low density suggests that peaks are not common in sequence space and that neutral networks are not extensive, even for such a simple phenotype. In fact, it was only possible to trace a functional mutational network connecting just three of the peaks.
Another study used deep sequencing to characterizethe fitness landscape surrounding theclass II ligase ribozyme (Pitt and Ferré-D’Amaré 2010).A mutagenized population was constructed encompassing this neighborhood and was subject to selection for active ribozymes, enriching those individuals with the best catalytic activity. Deep sequencing of the selected variants revealed a fitness landscape with a single high-fitness peak centered on the original sequence, surrounded by closely-related variants that were present at lower frequencies.
Most catalytic RNAscontain too many nucleotides to permit exhaustive sampling of local sequence space. Díaz Arenas and Lehman (2010)instead allowedsmall populations of ligase ribozymes to explore the fitness landscape by undergoing continuous in vitro evolution. Mn2+ ionswere added to the continuous evolution mixture to increase the frequency of mutationto ~3 × 10–4 per nucleotide per cycle of amplification (Vartanian et al. 1999).Aquasispecies distribution emerged, consisting of a set of closely-related genotypes clusteredaroundthe parental sequence (Eigen 1971). However, individuals in the evolved populations typically were separated from each other by only a few mutations, most of which occurred outside the catalytic center of the ribozyme, again suggesting a fitness landscape with few and isolated high-fitness genotypes.
Analyses of protein fitness landscapes also have provided a mixed picture. Studiesassaying the activity of point mutants suggest that proteins are robust to mostsingle mutations(reviewed in Wagner 2013). Evolution experiments in which proteins are subjected to neutral drift have generated viable enzymes that are tolerant of a modest number of non-synonymous mutations (Bershtein et al. 2006; Bloom 2007; Amitai et al. 2007), and some of these variants have novel phenotypic properties with regard to substrate usage. It is not clear, however,whether these examples are indicative ofan overall fitness landscape that has a high degree of neutrality.
Intertwined with questions about the underlying structure of fitness landscapes are issues pertaining to the driving forcesof evolution.If fitness landscapes have isolated high peaks surrounded by genotypes with much lower fitness, thenone might expect classical Darwinian selection to be primarily responsible for evolutionary change. In that case, novel high-fitness genotypes may only emerge when the environment changes and the population is forced to adapt to a new fitness landscape. Alternatively, if fitness landscapes contain extensive neutral networks, then neutral driftmay be achief driver of genetic change.
Neutral drift relies on the stochastic fixation of neutral or nearly neutral sequence variants (Kimura 1984; Ohta 1992). The present study sought to maximize stochastic processes by enforcing a high mutation rate and employing conditions that promote the founder effectdue to population bottlenecks (Soll et al. 2006). Such bottlenecks could provide a means of traversing lower fitness intermediate sequences that connectseparate high-fitness peaks. Isolated from their more fit brethren,the lower-fitness intermediates, provided they are still capable of reproduction, may survive long enough to acquire additional mutations and move to other regions of sequence space. Thus, even if the paths between high-fitness genotypes in a fitness landscape are not perfectly neutral, stochastic evolutionary processes could make those paths accessible.
The structure of local fitness landscapes was investigated using populations of ligase ribozymes undergoing continuous in vitro evolution (Wright and Joyce 1997).The environmental conditions were kept constant during this process to minimize adaptive selection pressure. This work differs in two ways from previous approaches to understanding the structure of fitness landscapes:first, the mutation rate was greatly increased using mutagenic nucleotide analogs (Petrie and Joyce 2010);second, the founder effect was encouraged using compartmentalized water-in-oil emulsions to isolate individual ribozymes (Paegel and Joyce 2010).
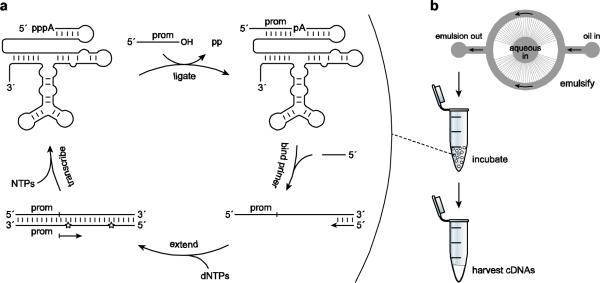
In the continuous evolution system, a population of single-stranded RNA molecules are challenged to ligate a promoter-containing oligonucleotide substrate to their 5′ end (Fig. 1a). All RNA molecules in the population are reverse transcribed via extension of a DNA primer that is complementary to the 3′ end of the ribozyme, but only those RNAs that havealready catalyzed ligation give rise to DNAs that are competent for transcription by a corresponding RNA polymerase. All of the components needed to bring about selective amplification of functional RNAs are contained within a common reaction mixture, and newly-produced progeny RNAs can catalyze new ligation events, all without experimenter intervention. After several hours of selective amplification, a portion of the amplified material can be transferred to a new reaction mixturethat contains a fresh supply of reagents, thus enabling continuous evolution to proceed indefinitely. Because ligation, reverse transcription, and transcription occur rapidly, several logs of selective amplification can occur within a few hours, enabling the study of many generations of evolution in a short amount of time.
Fig. 1.
Compartmentalized, continuous in vitro evolution of ligase ribozymes.a Scheme for continuous evolution. The ribozymes ligate to their 5′ end a chimeric DNA/RNA substrate containing the T7 RNA polymerase promoter sequence (prom). A primer binds to the 3′ end of the ribozymes and is extended by reverse transcriptase to form cDNAs that may contain mutagenic nucleotide analogs (stars). Only cDNAs resulting from ligated RNAs contain a functional double-stranded promoter and are transcribed by T7 RNA polymerase to yieldprogeny RNAs.b Generation of microfluidic water-inoil emulsions. The microfluidic device contains a central aqueous input reservoir with 110 radial nozzles that empty into an annular oil flow channel. An oil-surfactant mixture is supplied at the oil input reservoir and the emulsion is collected at the output reservoir. Aqueous droplets within the emulsion contain all of the components of the continuous evolution mixture, with an average of one founder ribozyme per droplet. Following incubation, the emulsion is broken and the cDNAs are harvested.
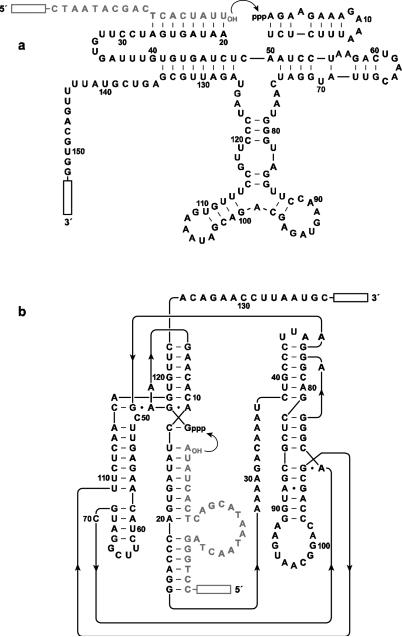
Two distinct ligase ribozymeshave been shown to be capable of undergoing continuous evolution.The DSL ligasewas built on a scaffold of known secondary structural elements that were connected to a randomized region, from which a functional ligase was selected (Ikawa et al. 2004) and subsequently adapted to the conditions of continuous evolution (Voytek and Joyce 2007). The class I ligase ribozyme was originally discovered by in vitro selection starting from random-sequence RNAs (Ekland et al. 1995) and further optimized for the continuous evolution system (Wright and Joyce 1997; Schmitt and Lehman 1999; Paegel and Joyce 2008). The sequences and secondary structures of the two ribozymes used in this studyare shown in Fig.2.
Fig 2.
Sequence and secondary structure of ribozymes used to initiate neutral drift experiments. a DSL ligase variant that wasselected for improved activity under low-Mg2+conditions.b Class I ligase variant that emerged from prior in vitro evolution experiments (Paegel and Joyce 2008). The substrate is shown in gray; curved arrow indicates the ligation junction; boxed regions indicate primer sites for reverse transcription and PCR amplification. Nucleotides that were subject to evolution are numbered.
The mutagenic nucleotide analogs 8-oxo-dGTP and dPTP were added to the continuous evolution mixture to increase the mutation rate to ~10–2per nucleotideper cycle of amplification (Petrie and Joyce 2010), a 30-fold increase over previous efforts(Díaz Arenas and Lehman 2010). These analogs have a characteristic mutation spectrum,with 8-oxo-dGTP promotingA→C changes and dPTP promoting bothA→G and G→A changes. It would be preferable to have a method that causes all possible mutations to occur at high frequency, but no such method currently exists.
Thecomponents of continuous in vitro evolution were emulsified inaqueous droplets dispersed in an oil phase, using a microfluidic device that produces monodisperseemulsions containing millions of discrete droplets (Fig. 1b) (Paegel and Joyce 2010). In each droplet, evolution was initiated with a single ribozyme molecule to achieve the founder effect on a massively parallel scale. The evolution experiments were initiatedwith mutagenized populations of either the DSL or class I ligase, encompassing local sequence space surrounding each high-fitness parental genotype. Because evolution was initiated with a single ribozyme variant in each droplet, competition with the parental wild type was limited and the populations had an unprecedentedopportunity to drift.
After the evolution process was completed, the evolved populations were analyzed bydeep sequencing to characterize rare variants and to determine if neutral drift alone was sufficient to access distant regions of the fitness landscape for this system. This analysis revealed that the evolved populations were dominated by individualshaving a modest number of mutations that occurred at many positions throughout the ribozyme. Ribozymes with larger numbers of mutations were rare and significant divergence from the parental sequence did not occur, suggesting that, at least in the context of the continuouslyevolving ligases, the fitness landscape does not contain readily accessible neutral networks and neutral drift is not the primary driver of genetic change.
Materials and Methods
Materials
Synthetic oligodeoxynucleotides were purchased fromIDT (Coralville, IA). Chimeric DNA/RNA oligonucleotides and DNA libraries were synthesized using standard phosphoramidite chemistry on an Applied Biosystems Expedite 8909 DNA/RNA synthesizer, with amidites and other reagents purchased from Glen Research (Sterling, VA). All oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and desalted prior to use. Histidine-tagged T7 RNA polymerase was purified from E. coli strain BL21 containing plasmid pBH161 (provided by William McAllister, SUNY Brooklyn). Superscript II RNase H– reverse transcriptaseand streptavidin-coated magnetic beads (MyOne C1 Dynabeads) were from Life Technologies (Carlsbad, CA), DNase I was from Roche Applied Science (Indianapolis, IN), and Taq and Phusion DNA polymeraseswere from NEB (Ipswich, MA).Inorganic pyrophosphatase, NTPs, and dNTPs were from Sigma-Aldrich (St. Louis, MO), [α-32P]ATP and [γ-32P]ATP were from Perkin Elmer (Waltham, MA), and mutagenic nucleotide analogs 8-oxo-dGTP and dPTPwere from TriLink (San Diego, CA).
Pre-adaptation of DSL ribozyme to mutagenic conditions
Continuous evolution was initiated with 1 nM of in vitro transcribed RNAcorresponding to the T100-1 variant of the DSL ligase (Voytek and Joyce 2007),which has the sequence 5′-AGAAGAAAGAAAUUUCUCUAAUAGUGAUUCUUGUGAUUUGUGUGGUCUCAAUCCUAAGGCUAAACGCUAUGGAUCAAUGGGUAGGUGCCAAAUAGAGCAGACGAUAAAGUGUUUCCGUUC CCUAGUAGAUUGCGAGUCGUAUUGGCAUAGCUGACUCCGUGCCAUCC-3′. The continuous evolution mixture also contained5μM substratehaving the sequence 5′-CATCGTGCCTTGCTGCTCTAATACGACTCACUAUU-3′ (ribonucleotides in bold, T7 RNA polymerase promoter sequence underlined), 2.5μM cDNA primer having the sequence 5′-GGATGGCACGGAGTCAG-3′, 2.5U/μL T7 RNA polymerase, 4.5U/μl Superscript II RNaseH– reverse transcriptase, 0.001 U/μL inorganic pyrophosphatase, 2mM each NTP, 0.25 mM dPTP, 0.2 mM dATP,0.2 mM dGTP, 0.075 mM dCTP,0.075 mM TTP, 15mM MgCl2,50 mM KCl, 4 mM DTT, and 50 mM EPPS (pH 8.5). The various dNTPs and dNTP analogs were used in unequal proportions to achieve the highest frequency of mutation that did not impair polymerase function, based on a previously optimized protocol (Petrie and Joyce 2010).The continuous evolution mixturewas incubated at 37 °C for 30–60 min, then a small aliquot was transferred to a new mixturecontaining fresh reagents.
The kcat and Km of the starting and evolved ribozymeswere determined in a ligation reaction in the presence of either 3.0 or 6.2mM MgCl2, employing eight different concentrations of substrate that spanned the Km,always with substrate in at least 10-fold excess over[α-32P]ATP-labeledribozyme. The reaction mixturesalso contained50mM KCl, 4 mM DTT, and50 mM EPPS (pH 8.5) and were incubated at 37 °C. The reactions were quenchedat various times with EDTA in excess of Mg2+, and the products were separatedby PAGE. Values for kobs wereobtainedfor each concentration of substrate by fitting the data to the equation:
whereFreactedis the fraction reacted at time t,and a is the maximum extent. For reactions that proceeded slowly, kobs was instead determined by a linear fit of the data within the first 15% of the reaction. Values for kcat and Km were obtained by fitting thekobs values to the Michaelis-Menten equation.
Generation of droplets within microfluidic devices
A positive relief mold composed of SU-8 photoresist on a silicon wafer was produced by the Stanford Microfluidics Foundry and provided by Brian Paegel. The mold has microfluidic circuits with the same geometry as described previously (Paegel and Joyce 2010), with a 500 × 150 μm annulus receiving output from110 aqueous emitter nozzles, but with an aqueous layer depth of 10 μm and aqueous channelwidth of 10 μm. Replicates were castfrom the mold in polydimethylsiloxane(Sylgard 184 silicone elastomer kit, Dow Corning, Midland, MI) and bonded to glass microscope slides to produce the microfluidic devices,asdescribedpreviously (Paegel and Joyce 2010).A new device was used for each round of compartmentalized evolution.
The devicewas secured to the stage of a binocular microscope equipped with CCD camera to monitor fluid flow, and connected as described previously (Paegel and Joyce 2010) via Tygonmicrobore tubing (0.01” i.d. × 0.03” o.d.; Saint-Gobain, Valley Forge, PA) to two syringes, one containing the oil phase and the other containing the aqueous phase. The oil phase consisted of 70% DMF-A-6CS silicone oil (Shin-Etsu Silicones, Akron, OH), 26% mineral oil (Sigma-Aldrich), and 4% KF-6038 surfactant (Shin-EtsuSilicones), measured by weight and gently premixed for at least one hour before filling the syringe. The filled syringes were fitted with 30-gauge blunt needles, onto which thetubing was threaded. The tubing was inserted directly into pre-punched reservoirs on the device. Outflow was collected in 1.5-mL Eppendorf tubes, pre-filled with 500 μL of the oil phase and connected to the output reservoir via a 7.5-cm length of tubing. Digital syringe pumps (SP101i, World Precision Instruments, Sarasota, FL) were used to drive the aqueous and oil solutions. Each device was first rinsed with aqueous solution containing continuous evolution buffer and tested for its ability to form good emulsions before loading and emulsifying the actual evolution mixture.
The device was rinsed with buffer-only aqueous phase at a flow rate of 20μL/min for 10min, then at 5 μL/min for 20 min. Flow of the oil phase was started at 70 μL/min, and both phases were allowed to run for a few minutes after droplets had begun to form. In preparation for swapping the aqueous phase, the oil pump was stoppedand the aqueous pump was allowed to continue to flow for 15 min. Then the oilphase tubing was removed, the aqueous pump stopped, both syringes and tubing were discarded, and a new oil-filled syringe was attached. Freshly prepared continuous evolution mixture was kept on ice to prevent preinitiation of the reaction prior to emulsification. This mixture was loaded into a syringe, also kept on ice, which was connected to the device as before, priming at a flow rate of 20μL/min for 0.5 min, then 1 μL/min for 28 min, then 5μL/min for 2min. Oil flow then was initiated at 70μL/minand emulsions were collected, beginning 2 min after stable emulsions had started to form.
Dropletproduction was assayed by includingbromophenol blue in the aqueous phase and visualizing with the binocular microscope. ImageJ (NIH, Bethesda, MD) was used to measure individual dropletsthat had circularity ≥0.75 and area ≥10μm2in the focused regions of the image (Kaltenbach et al. 2012). The average droplet radiuswas 9.5 ± 1.5μm, corresponding to an average volume of 3.5 pL and requiring a seedconcentration of 468 fMtogive, on average, onemolecule per droplet. Due to surface tension of the droplets within the oil phase, there was no appreciable merging of droplets (Paegel and Joyce 2010).
Generation of starting populations
Template DNAs for transcribing the starting pools of RNAs were prepared by PCR cross-extension of two synthetic oligodeoxynucleotides. For the DSL ligase the completed construct had the sequence 5′-CATCGTGCCTTGCTGCTCTAATACGACTCACTATTAGAAGAAAGAAATTTCTCTAATAGTGATCCTTGTGATTTGTGTGATCTCAATCCTAAGACTGAACGTTATGGATCAATGGGTAG GTTCCAAGTAGAGCAGACGATAAAGTGTTTCCGTTCCCTAGTAGATTGCGAGTCGTATTTTGACGTGGCTGACT CCGTGCCATCC-3′ and for the class I ligase had the sequence 5′-GACTAATACGACTCACTATAGGAAGAACACACTATAGTGACCCAGGAAAA-GACAAATCTGCCCTTAGAGCTTGAGAACATCTTCGGATGCACGGGAGGCAGCTCGCGATGGAAGTAACGGAC CCAGCGTTCTCAACAGTGTTCACAGAACCTTAATGCACGAGCATGGAGGGACT-3′ (T7 RNA polymerase promoter sequence and region of overlap for cross-extension underlined; nucleotides randomized at 10% degeneracy italicized). Cross-extension was carried out by 10 cycles of PCR, using primers for the DSL ligase having the sequence 5′-CATCGTGCCTTGCTGCT-3′ and 5′-GGATGGCACGGAGTCAG-3′ and for the class I ligase having the sequence 5′-GACTAATACGACTCACTATA-3′ and 5′-AGTCCCTCCATGCTCGT-3′. The cross-extension products were used as input for in vitro transcription, and the resulting RNAs were purified by PAGE.
Prior to continuous evolution of the class I ligase, the population was pre-cleared of inactive variants by mixing 3 nmol (15 μM) [α-32P]ATP-labeled RNA with 30 μM substrate, 15 mM MgCl2, 50 mM KCl, and 50mM EPPS (pH 7.5), then incubatingat 37°C for 1 h. The ligated RNAswere purified by PAGE, reverse transcribed, PCR amplified, and transcribed to generate RNAs that were used to initiate evolution.
Compartmentalized continuous evolution
To prevent premature initiation, the continuous evolution reactions were pre-assembled on iceas two separate mixtures: one containing the population of ribozymes or corresponding cDNAs, 5′-carboxyfluorescein-labeled cDNA primer, and NTPs; the other containing substrate, T7 RNA polymerase, reverse transcriptase, and inorganic pyrophosphatase; and both containingdNTPS, MgCl2 in 6.2 mM excess over the total concentration of NTPs anddNTPs, 50 mM KCl, 4 mM DTT, and 50 mM EPPS (either pH 8.5 for the DSL ligase or pH 7.5 for the class I ligase). The substrate for the DSL ligase had the sequenceshown above and the substrate for the class I ligase had the sequence 5′-CCGAAGCCTGGGATCAATAATACGACTCACUAUA-3′ (ribonucleotides in bold, T7 RNA polymerase promoter sequence underlined). The two mixtures were combined to give a final concentration of 468 fM ribozymes or corresponding cDNAs, 2.5μM cDNA primer, 2mM each NTP, either 0.2 mM each dNTP (for non-mutagenic rounds) or 8 mM 8-oxo-dGTP, 0.25 mM dPTP, 0.2 mM dATP, 0.05 mM dGTP, 0.075 mM dCTP, and 0.075 mM TTP (for mutagenic rounds), 2.5U/μL T7 RNA polymerase, 4.5U/μl Superscript II RNaseH– reverse transcriptase, and 0.001 U/μL inorganic pyrophosphatase. The combined mixture was loaded into a chilled syringe and used to produce the emulsion, as described above.
Emulsified material (~120 μL aqueous component) was collected in Eppendorf tubes, then warmed to 37 °C to initiate continuous evolution.Following6 h incubation, 0.2 M NaOH was added to prevent further amplification, the top oil layer was discarded, the remaining oil was extracted five times with hexane, andthe residual hexane was removed using a vacuum concentrator. The RNA was hydrolyzed by incubating at 90°C for 20 min, then the mixture was neutralized with HCl and the remaining cDNAs were purified by ethanol precipitation and quantitated by PAGE analysis, comparing to known amounts of 5′-carboxyfluorescein-labeled cDNA primer. The cDNAs were either used directly or PCR amplified, then transcribed to generate material to begin the next round of continuous evolution. All PCR amplifications were carried out together with negative control reactions that contained no added cDNA, confirming that no detectable amount of amplified material was produced due to contamination by extraneous DNA molecules.
Analysis of catalytic activity
The catalytic activity of the wild-type ribozyme and final evolved population was determinedin a ligation reaction employing 0.5 μM ribozyme, 5 μM [5′-32P]-labeled substrate, 15 mM MgCl2, 50 mM KCl, 4 mM DTT, and 50 mM EPPS (either pH 8.5 for the DSL ligase or pH 7.5 for the class I ligase), which was incubated at 37 °C for various times. The reaction products were separated by PAGE and values for kobs were obtained as described above.
Sequencing analysis
Conventional sequencingemployedcDNAs that werepurified by PAGE, PCR amplified,cloned, and sequenced. For deep sequencing, the cDNAs were PCR amplified if necessary and transcribed to corresponding RNAs, then the mixture was treatedwithDNase I, and the remaining RNAs were allowed to undergo ligation with a 5′-biotinylated substrate. The reacted RNAs were captured onstreptavidin-coated magnetic beads and unreacted RNAs were removed by repeated washing: once with 1 M NaCl, 10 mM Tris(pH 7.5), and 0.05% Tween20;twice with 8 M urea, 10 mM Tris(pH 7.5), and 0.05% Tween20; twice with 20mM NaOH and 0.05% Tween20;and twice with 10 mM Tris(pH 7.5). The biotinylatedRNAsthen wereeluted by incubating with 95% formamide and 10 mM EDTA at 95°C for 10 min to denature the streptavidin, then reverse transcribed and PCR amplified usingPhusionhigh-fidelity DNA polymerase. Deep sequencing was carried out on anIlluminaMiSeq machine, using ~200 ng of DNA that had first been purified byagarose gel electrophoresis.
The samples were prepared for deep sequencing using the NEBNext Ultra DNA Library Prep Kit for Illumina. Each sample was end-repaired, kinased, and A-tailed with Taq polymerase, then ligated toIlluminaTruSeq barcoded adapters according to NEB protocols. The samples then were pooled, PCR amplified, gel purified, and sequenced in a 250-cycle, paired-end run, carried out concurrently with unrelated material.The samples were demultiplexed with Illumina Pipelineand CASAVA (version 1.8.2)to generate aFASTQ filefor each sample.
All files were processed in a Unix BASH environment, using custom shellscripts that are available upon request.Adaptor sequences were removed usingTrimmomatic (Lohse et al. 2012), with four tolerated mismatches in the seed, palindrome clip threshold of 30, simple clip threshold of seven, and minimum adaptor length to clip of one. After trimming, FLASH (Mago and Salzberg 2011) was used to merge the paired forward and reverse reads, retaining only those with perfect complementarity. Based on FLASH-assigned quality scores, reads were discarded if they contained any position with aPhred score less than 36.Reads that differed in length compared to the wild typeby more than two nucleotides also were discarded. Otherwise all reads were retained, with no restriction on the number of substitution mutations.
Reads were aligned to the wild-type sequence using Bowtie2 (Langmead and Salzberg 2012)in end-to-end mode with default parameters.From thealignment file, reads with particular edit distances were extracted usingSAMtools (Li et al. 2009). A module from BreSeq(Barrick et al. 2009; Barrick and Lenski 2013) was used to generate an ensemble gapped alignment for all aligned reads, from which sub-sequences were extracted, corresponding to regions of interest in the ribozyme. The gapped alignment also was used to tabulate the base identity or gaps at every nucleotide position, using a custom Python scriptthat is available upon request, from which the frequency of observed mutation was calculated. Insertions were collapsed into the upstream nucleotideposition.
Results
Pre-adaptation of DSL ribozyme to mutagenic conditions
Before carrying out neutral drift experiments with the DSL ligase, it first was necessary to adapt the ribozyme to the lower Mg2+ concentration employed with the mutagenic nucleotide analogs.Clonally pure ribozyme was used to initiate continuous evolution, carried out in bulk solutionin the presence of15 mM MgCl2and 0.25 mMdPTP. The population was maintained via serial transferfor an overall amplification of 10100-fold. During this time the population acquired 11 consensus mutations relative to the starting ribozyme. A consensus clone was identified(Fig.2a) thathas an improved ligation rate at the lower Mg2+ concentration. Although the totalMg2+ concentration during continuous evolution was 15 mM, the effective concentration was6.2 mM, accounting for stoichiometric chelation of Mg2+by NTPs and dNTPs in the mixture. In the presence of 6.2 mM MgCl2, the evolved ligase has a kcat of 0.57 min–1 and Km of 0.81μM, which is only slightly improved compared to the starting ribozyme. However, the evolved variant retains good activity in the presence of only 3 mM MgCl2, witha catalytic efficiency (kcat/Km) of 7.2 × 105 M–1 min–1, compared to 6.8 × 103 M–1 min–1 for the starting ribozyme. Thispre-adapted DSL ligasevariant was used to initiate subsequent experiments and is hereafter referred to as the wild-type or parent ribozyme.
Neutral drift of ligase ribozymes within compartments
Two separate lines of compartmentalized evolution were initiated with libraries based on either the DSL or class I ligase ribozyme (Fig. 2). The parent ribozymes were randomly mutagenized at afrequency of 10% per nucleotide position to introduce variation in the neighborhood of the wild-type sequence.The resulting pools of RNAsweremixed with the other components of the continuous in vitro evolution system and emulsified as the aqueous phase of a water-in-oil emulsion (Fig. 1b). The concentration of RNA was chosen toensure, on average, one seed molecule per aqueous droplet in the emulsion. Given this concentration and the total volume of the aqueous phase, the startingpopulation size was 3 × 107molecules. Among these,all single-error mutants relative to the parental sequence were represented, with 12-error mutants being the most common error class. The continuous evolution mixture was kept on ice prior to emulsification so that no reactions could take place until the RNA molecules were isolated within individual droplets.
Once emulsified, the compartmentalized materials were incubated for 6 h to allow selective amplification to occur within the seededcompartments. This ample incubation time was provided to ensure that amplification would reach a common ceiling for droplets that had been seeded with a viable ribozyme.Following incubation, theribozymes were digested by adding 0.2 M NaOH, thus preventing any further reactions. Then the oil phase was extracted with hexane and the aqueous portion was neutralized with HCl. ThecDNAsthat remained in the mixture were analyzed by PAGE to determine the amount of full-length material, corresponding to ligated ribozymes. This material was used to prepare the next emulsion, again with an average of one seed molecule per compartment. This procedure was repeated for several rounds to provide an opportunity for the population to drift away from the parental sequence.
In the evolutionary lineage with the DSL ligase, the mutagenic nucleotide analogs were included throughout the course of compartmentalized evolution. The yield of full-lengthcDNA was low, so this materialwas PCR amplified between rounds. Six rounds of compartmentalized evolution were carried out, for an overall amplification of 1.2 × 105-fold. Following this process, individualswere cloned from the population and sequenced, which revealed that, on average, 3.4 ± 1.2% of the nucleotideswere mutated relative to the wild-typeDSL ligase.
In the evolutionary lineage with theclass I ligase, the starting population first was pre-cleared of low-activity variants by removing RNA molecules thatdid not undergo ligation in one hour. Additionally, standard dNTPs were used in the first two rounds of compartmentalized evolution.As a result of these procedures, the frequency of mutations decreased from 9.0 ± 2.3% for the starting population, to 5.8 ± 1.5% for the pre-cleared population, and then to 2.2 ± 1.4% for the population after the second round of compartmentalized evolution. Sequencediversity then was allowed to increase by carrying out ten additional roundsin the presence of the mutagens, causingmutations to occurat a rate of ~10–2 per nucleotide per 10-fold amplification(Petrie and Joyce 2010), throughoutan overall amplification of 1043-fold. Sequence analysis of the final population revealed that, on average, 4.6 ± 1.7% of the nucleotides were mutated relative to the wild-type class I ligase.
The catalytic activity of the DSL and class I ligase populations was monitored throughout the course of compartmentalized evolution by determining the yield of full-length cDNA after each round, corresponding to the amount of ligated RNA. Following the initial rounds, which removed low-activity variants that were present in the starting pool, the yield of full-length cDNA remained unchanged (4.4 ± 1.4 pM for the DSL lineage; 8.8 ± 1.5 nM for the class I lineage), indicating that adaptive evolution had not occurred. In addition, assays were carried out to compare the catalytic activity of the wild-type ribozyme and final evolved population. For the DSL ligase, the wild type has a kobs of 0.6 min–1 and the final population has a kobs of 0.4 min–1;for the class I ligase, the wild type has a kobs of 1.0 min–1 and the final population has a kobs of 0.1 min–1;again demonstrating that adaptive evolution had not occurred.
Deep sequencing of evolved populations
Next-generation sequencing was used to examinesequence variation among the evolvedpopulation of ribozymesforeach of the two lineages. Prior to sequencing, full-length cDNAswereharvested from the continuous evolution mixtureand used as templates for in vitro transcription. The resulting RNAs were allowed to ligate themselves to a 5′-biotinylated substrate and then captured on streptavidinto removethe mutagen-containing cDNAs, mutagenic nucleotide analogs, and any unreacted ribozymes. The isolated RNAs were reverse transcribed and PCR amplified to generate mutagen-free DNA corresponding to functional RNAs in the population. Two control samples were prepared for each ribozyme, employing either wild-type DNAthat was simply amplified or wild-type RNA that was treated in the same manner as for the evolved populations.
All six samples were analyzed on an IlluminaMiSeq instrument in a 250-cycle paired-end run, which generated >350,000 sequence reads per sample, each read corresponding to a distinct molecule in the sample. Because the read length was longer than the ribozyme sequence, all molecules were sequenced completely in both the forward and reverse directions. Only read pairs with perfect complementarity in the forward and reverse directions were included in the analysis, then merged into a single sequence (Chen-Harris et al. 2013). Additionally, quality score and length filters were applied: only reads for which all positions had a Phredscore of ≥36 (0.025% errorprobability) were included, and any readsthat differed in length by more than two nucleotides relative to the wild type werediscarded.Table 1 shows the percentage of reads that passed each step of data filtering.
Table 1.
Number of raw and filteredreads from deep sequencing analysis. Controls using either the wild-type DSL or class I ribozyme were sequenced after either PCR amplification or processing in the same manner as for the evolved populations.
| Raw | Paired filter | Quality filter | Length filter | Aligned | |
|---|---|---|---|---|---|
| DSL control, amplified only | 386,156 | 362,828 (94.0%) | 316,257 (81.9%) | 314,917 (81.6%) | 314,917 (81.6%) |
| DSL control, processed | 406,100 | 373,641 (92.0%) | 322,770 (79.5%) | 316,221 (77.9%) | 316,214 (77.9%) |
| DSL evolved, processed | 648,489 | 581,460 (89.7%) | 488,451 (75.3%) | 476,849 (73.5%) | 476,836 (73.5%) |
| Class I control, amplified only | 404,358 | 370,018 (91.5%) | 321,868 (79.6%) | 319,617 (79.0%) | 319,616 (79.0%) |
| Class I control, processed | 501,786 | 444,887 (88.7%) | 381,689 (76.1%) | 374,565 (74.6%) | 374,522 (74.6%) |
| Class I evolved, processed | 708,308 | 600,623 (84.8%) | 505,094 (71.3%) | 475,050 (67.1%) | 474,772 (67.0%) |
Bowtie2 (Langmead and Salzberg 2012) was used to align the reads to the reference sequence. Nearly all reads (>99.9%) were successfully aligned. A gapped alignment was generated to determine the frequency of observedmutation at each nucleotide position, and these positional frequencies were averaged to determine the overall frequency of mutation (Table 2).
Table 2.
Observed mutation frequenciesin the wild-type controls and evolved populations of DSL and class I ribozymes. Mutations attributable to the mutagenic nucleotide analogs are shown in bold.
| Mutation frequency (%) |
||||||||
|---|---|---|---|---|---|---|---|---|
| wt | A | C | G | U | insert | delete | all | |
| DSL control | A | – | 0.00 | 0.05 | 0.01 | 0.01 | 0.06 | 0.13 |
| C | 0.07 | – | 0.00 | 0.03 | 0.00 | 0.01 | 0.11 | |
| G | 0.04 | 0.01 | – | 0.08 | 0.01 | 0.02 | 0.15 | |
| U | 0.01 | 0.02 | 0.00 | – | 0.00 | 0.01 | 0.05 | |
| N | 0.11 | |||||||
| DSL evolved | A | – | 0.93 | 4.97 | 0.08 | 0.08 | 0.05 | 6.07 |
| C | 0.01 | – | 0.00 | 0.23 | 0.02 | 0.00 | 0.27 | |
| G | 3.79 | 0.37 | – | 0.03 | 0.02 | 0.01 | 4.21 | |
| U | 0.03 | 0.23 | 0.12 | – | 0.05 | 0.02 | 0.44 | |
| N | 2.95 | |||||||
| Class I control | A | – | 0.01 | 0.03 | 0.02 | 0.00 | 0.02 | 0.07 |
| C | 0.01 | – | 0.00 | 0.04 | 0.00 | 0.01 | 0.07 | |
| G | 0.03 | 0.02 | – | 0.03 | 0.01 | 0.02 | 0.11 | |
| U | 0.01 | 0.02 | 0.00 | – | 0.00 | 0.01 | 0.04 | |
| N | 0.07 | |||||||
| Class I evolved | A | – | 2.74 | 8.95 | 0.26 | 0.03 | 0.14 | 12.11 |
| C | 0.13 | – | 0.56 | 1.61 | 0.02 | 0.07 | 2.39 | |
| G | 2.25 | 0.45 | – | 0.24 | 0.03 | 0.11 | 3.07 | |
| U | 0.04 | 0.19 | 0.10 | – | 0.07 | 0.09 | 0.46 | |
| N | 5.31 | |||||||
For the evolved DSL and class I ribozymes, the overallfrequency of mutation was3.0% and 5.3% per nucleotide position, respectively, albeit with a high degree of positional variation. These numbers are comparable to what was seen with the limited amount of conventional sequencing, as described above.The spectrum of mutations coincided with what would be expected from the mutagens,mostly involving A→C transversions and both A→G and G→A transitions (Petrie and Joyce 2010). For the untreated controls withthe wild-type DSL and class I ribozymes, the frequency of mutation was 0.04%and 0.02%, respectively, reflecting mutations that aroseduring PCR amplification. For theprocessed controls,these frequencies were 0.11% and 0.07%, respectively, reflecting the additional opportunity for mutation during the transcription and reverse transcription steps. The latterfrequenciesindicate the background level of mutation and were used to establish a threshold for significance in subsequentanalyses of individual haplotypes in the populations.
Distribution of mutations in the evolved populations
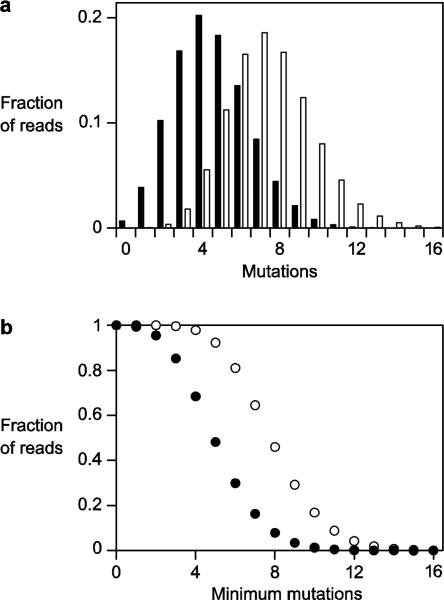
For each evolved population, the sequence reads were binned according to their edit distance (number of substitutions, insertions, or deletions relative to the wild-type sequence) (Fig. 3a). Reads that differed in length by more than two nucleotides compared to the wild type were discarded to avoid including amplification artifacts, these reads constituting only ~4% of the total. For the DSL and class I ligase lineages, the most abundant edit distance is four andseven mutations, respectively. There are numerous reads with more divergent sequences, but as the number of mutations increases, there are progressively fewer reads for the most abundant unique sequence in each edit class.
Fig. 3.
Number of observed mutations among individuals isolated from the evolved populationof DSL (black) or class I (white) ribozymes. a Fraction of population with a particular number of mutations. b Fraction of population with at least a given number of mutations.
The distribution of edit distances suggests that neitherlineage contains a substantially divergent ribozyme motif, at least based on overall sequence. To the extent that read abundance can be used as a proxy for fitness, the broad distribution of edit distances suggests that both ribozymes are tolerant of mutation, keeping in mind that the sequences were obtained from molecules that were the product of the RNA-catalyzed ligation reaction. The distribution does not tail off for the DSL and class I ribozymes until these molecules contain more than seven or ten mutations, respectively (Fig. 3b). Thesemutations are broadly scattered throughout the molecules.
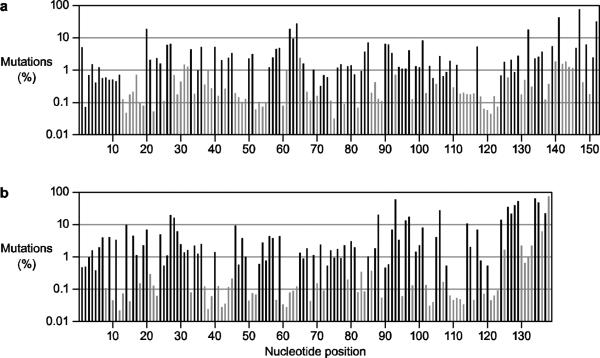
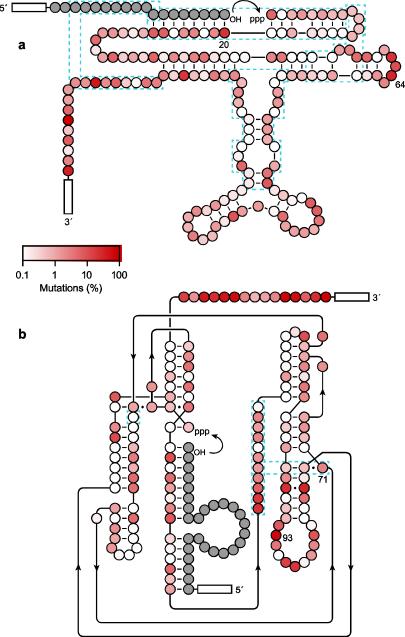
In both evolved populations, the frequency of observedmutation varies at different positions (Fig. 4). This is true for all positions, whether or not they were targeted by themutagens. Some positions have mutations levelsexceeding50%, while othersarehighly conserved relative to the wild-type sequence.The latter likely reflect positions that play a significant role in ribozyme function. The frequency of observed mutation at various positions was mapped onto the secondary structure of the ribozymes (Fig. 5). For the DSL ligase, the connecting loop at positions 61–65 is highly tolerant of mutation, whereas the region of the catalytic center at positions 75–85 and 115–126is much more conserved. For the class I ligase, the flexible tail at positions 124–138 has a high level of mutation, whereas most of the intricately folded catalytic center is more conserved. Even within the conserved regions, however, nearly every position that was targeted by the mutagensexhibits a mutation frequency exceeding the significance threshold established by the control samples. Again, this suggests that both ribozymes are broadly tolerant of mutation, especially mutations that have only a modest effecton catalytic function.
Fig. 4.
Frequency of observed mutations at each nucleotide position in the evolved population of (a) DSLor(b) class I ribozymes. Black bars indicate positions subject to mutation by 8-oxo-dGTP and dPTP.
Fig. 5.
Frequency of observed mutations at each nucleotide position in the evolved population of (a) DSL or (b) class I ribozymes, mapped onto their corresponding secondary structure. Color intensity is proportional to the frequency of mutation; dashed blue lines indicate local structural motifs that were studied for haplotype variation; index mutations for haplotype analysis are numbered; gray circles indicate substrate nucleotides; boxed regions indicate primer sites.
Haplotype analysis revealsscant divergence from parental sequences
Regional haplotypes were extracted from the sequences of the evolved ribozymes to search for novel local motifs that might represent alterationof the canonical structure. Haplotypes were considered only if they exceeded the significance threshold. For the DSL ligase lineage, haplotypes were extracted from regions of well-known secondary structure and from the catalytic center (Fig.5a). Various combinations of mutations occur throughout the stem that encompasses the ligation junction, but nearly all of these preserve the overall base-pairing interaction and none are suggestive of any structural rearrangement. There isnever more than a single mutation in the region comprising the tetraloop-receptor interaction thatprovides a structural scaffold for the ribozyme (Ikawa et al. 2004). Mutations are scattered throughout the catalytic center, but again none are suggestive of any structural rearrangement. Finally, mutations occur throughout the accessory substrate-binding domain near the 3′ end of the ribozyme(Voytek and Joyce 2007), but none disrupt basepairingwith the substrate.
For the class I ligase, the X-ray crystal structure (Shechner et al. 2009)was used to guide the search for haplotypes that might represent a local structural rearrangement (Fig.5b). The overall structure of the ribozyme is definedby three extended stems, arranged as a tripod. Two of these stems are connected by an unpaired region that contains two adenosine stacks which dock into the minor grooves of the connected stems, thus helping to anchor the tripod. The 5′ adenosine stack at positions 27–30 is more tolerant of mutation, perhaps because its role in helping to dock the substrate-containing stem is less critical for the ligase variant employed in this study, whichalso contains an accessory substrate-binding domain (Paegel and Joyce 2008). The 3′ adenosine stack at positions 34–36 is more conserved, never with more than one residue being mutated. In addition to forming a minor-groove docking interaction, these adenosines help coordinate a Mg2+ion that is thought to be essential for catalysis (Shechner et al. 2009). No more than a single mutation was observed within the cluster of seven nucleotides (31–33, 50, 71, 86, and 108) that helpto structure the active site of the ribozyme at the ligation junction.
Although no alternative regional haplotypeswere observed among the evolved populations of DSL and class I ribozymes, it is possible that novel discontinuous haplotypes arose througheither neutral drift or adaptive change. To search for evidence of such discontinuous haplotypes, index nucleotide positions were identified that have a high level of mutations, and mutationswere talliedacross all other positions, conditional upon either the presence or the absence of the index mutation. For both the DSL and class I ligases, two index mutations were chosen, one in a region that was expected to be neutral with regard to function and the other in a region that was expected to have significant functional consequences.
Within the DSL ligase, nucleotide position A64 is located in the loop that closes the stem of the tetraloop receptor. This position has been shown to be readily substitutable in the context of the wild-type ribozyme (Ikawa et al. 2004)and was mutated in 28% of individuals in the evolved population. In contrast, nucleotide position A20 is located immediately adjacent to the ligation junction, forming a Watson-Crick pair with the 3′-terminal uridine residue of the substrate. Position A20 was mutated to G in 18% of the sequenced individuals, retaining the ability to form a wobble pair with the substrate. Remarkably, for both of these index mutations there was no correlated difference in the distribution of mutations at all other nucleotide positions in the ribozyme (Fig. 6a,b).
Fig. 6.
Scan for mutations that co-occur with certain index mutations. The set of evolvedsequences were divided based on either the presence (black) or absence (gray) of the index mutation.a DSL ligase position A20. b DSL ligase position A64. c Class I ligase position A71. dClass I ligase position A93. The absence of the index mutation is represented by a gray dot on the horizontal axis.
Within the class I ligase, nucleotide position A93 is located in the loop at the distal end of one of the stems that forms the structural tripod, far from the active site and in a region that can be readily modified without perturbing the function of the wild-type ribozyme (Ekland et al. 1995). This position was mutated in 60% of individuals in the evolved population. In contrast, nucleotide position A71 forms part of a base triple that provides the “floor” of the active site of the ribozyme and is important for catalysis (Shechner et al. 2009).Again, for both these index mutations there was no correlated difference in the distribution of mutations at all other nucleotide positions in the ribozyme (Fig. 6c,d).
Discussion
This study sought tomaximize the opportunity for neutral driftto occur during the continuous in vitro evolution of two different ribozymes:the DSL ligase, which contains152 nucleotides, and the class I ligase, which contains 138 nucleotides. Mutagenic nucleotide analogs were used to achieve an error rate of ~1% per nucleotide per cycle of replication, approaching the error threshold for the propagation of heritable information for molecules of this length (Eigen 1971). Evolution was carried out within millions of separate compartments, each seeded by a single ribozyme to maximize the founder effect and to allow stochastic processes the opportunity to dominate evolution. Despite these efforts, neutral drift failed todiscoversignificantly divergent genotypes or even novelregional haplotypes within thetwo catalytic motifs. These observationsare consistent with an evolutionary fitness landscape that is surprisingly sparse. There do not appear to be extensive, easily accessed neutral networks for either of the two ribozymes, suggesting that adaptation may be the primary mechanism for theirevolutionary change.The haplotype analysiswas limited to mutations with a frequencyexceeding the significance threshold of the sequencing method (~0.1%). Thus it is possible that there are even rarer high-fitness variants in the population that represent neutrally connected divergent genotypes. However, if thesegenotypes areof such low abundance,they would not provide an easily traversable routefor neutral drift to drive evolutionary change.
It is possible that the evolving populations did not explore a sufficient fraction of sequence space to capture a neutral network. Yetat nucleotide positions that were targeted by the mutagens and are known to have little functional consequence, the observed mutation frequency washigh, likelyreachingsaturation. Perhaps if all nucleotide positions could be targeted by high-level mutagenesisa neutral network could be discovered. Currentexperimental techniques do not make this possible while still retaining the other desirable features of the continuous in vitro evolution system. However, with all purine nucleotides being targeted, and with the opportunity for both transition and transversion mutations, ample sequence space was available to the evolving populations over which drift could have occurred.
Highly parallelized compartmentalization was used to relax the selection pressure that results from competition within a population. When individuals are isolated, theirrelative fitness is less critical for reproductive success.It is possible, however, that this relaxation of selection pressure was not fully achieved. The relaxation of competition requires that, no matter the amplification rate of a founding individual in a droplet, it will reach the samefixed ceiling of copy number, limited only by the supply of reagents in the droplet. However, some droplets that were seeded with individuals having an especially slow amplification rate may not have had sufficient time to reach the ceiling before the materials were harvested to begin the next round of evolution. Even given this possibility, compartmentalization greatly reduced the opportunity for a high-fitness genotype to sweep the population. Such individuals would have reached the amplification ceiling within a fraction of the allotted incubation time, and most other variants would have reached the same ceilingduring the remainder of the time. The fact that the catalytic activity of the population did not increase over the course of the two lineages strongly suggests that adaptive evolution did not occur.
Deep sequencing analysis did reveal that single mutations are tolerated at almost every nucleotide position that had beentargeted by the mutagens (Fig. 4). This suggests that the two ligase ribozymes are highly robust to point mutation.However, the co-occurrence of multiple mutationswithin a given individual resulted in a precipitous decline in abundance (Fig. 3). Thus the picture of the fitness landscape is one of a narrow plateau of neutral mutations, tightly clustered about the wild-type sequence andsurrounded by higher-error variants of substantially lower fitness. This agrees with the results ofmutational drift experiments with TEM-1 β-lactamase in E. coli, where a “threshold robustness” was observed that demonstrated tolerance to a limited number of neutral mutations, but beyond which fitness declined sharply (Bershtein et al. 2006).
How then does evolution escape a steep and narrow local fitness peak, and can neutral drift play any role in this regard? One possibility is that neutral drift provides a reservoir of variation upon which adaptive selection can act if the environment were to change. In the evolved populations of ribozymes, there are many distinct sequences having a small number of mutations. Such tolerance to sequence variation has been proposed as a correlate of evolvability in changing environments (Wagner 2008; Wagner 2013).Neutral drift has been used to create libraries of functional protein variants that are especially capable of subsequent adaptive evolution (Bershtein et al. 2008; Gupta and Tawfik 2008).Another recent study examinedevolving populations of the Azoarcusgroup I ribozyme that first were subject to stabilizing selection with the opportunity for neutral mutations, followed by directional selectionthat resulted in adaptation to an alternativesubstrate (Hayden et al. 2011; Hayden et al. 2014).Conditionally neutral, cryptic variation that accumulated in the first phase was found to facilitate adaptation in the second phase.
A greater role for neutral drift may emerge when there is greater capacity for redundancy. For example, a gene duplication event allows one copy of the gene to drift while the other copy maintainsfunction and overall neutrality (Ohno 1970; Zhang 2003). The two ligase ribozymes have compact, interwoven structures that provide little capacity for redundancy. Furthermore, for these molecules to become amplified in the continuous evolution mixture, they must have some minimum rate of ligation that outpaces the rate of reverse transcription,which would otherwise render them inactive. Under such a threshold condition, the survival of substantially less fit variants may be more limited. For more complex evolving systems, where the threshold behavior of a particular enzyme is not absolutely critical for survival, there may be redundancy elsewhere in the system that allows neutral drift to play a larger role.
Despite these limitations, continuous in vitro evolution remains a powerful tool for testingevolutionary hypotheses. As demonstrated here,continuous evolution can be combined with high-level mutagenesis and compartmentalization to accelerate the exploration of sequence space and to explore the structure of fitness landscapes, subject to a minimum threshold for viability. Studies such as these contribute to a fundamental understanding of the nature of fitness landscapes and the factors that influence evolutionary outcomes.
Acknowledgments
The authors are gratefulto Brian Paegel for advice on microfluidics technology, the TSRI Next Generation Sequencing Core for library preparation, sequencing, and de-multiplexing, and Jeffrey Barrickfor advice on sequence alignment and analysis. This work was supported by the NSF (grant no. MCB-0948161) and the NIH Predoctoral Training Program in Molecular Evolution (grant no. T32GM080209).
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Amitai G, Gupta RD, Tawfik DS. Latent evolutionary potentials under the neutral mutational drift of an enzyme. HFSP J. 2007;1:67–78. doi: 10.2976/1.2739115. doi: 10.2976/1.2739115/10.2976/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nat Rev Genetics. 2013;14:827–839. doi: 10.1038/nrg3564. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J MolBiol. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- Chen-Harris H, Borucki MK, Torres C, Slezak TR, Allen JE. Ultra-deep mutant spectrum profiling: improving sequencing accuracy using overlapping read pairs. BMC Genomics. 2013;14:96. doi: 10.1186/1471-2164-14-96. doi: 10.1186/1471-2164-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz Arenas C, Lehman N. Quasispecies-like behavior observed in catalytic RNA populations evolving in a test tube. BMC EvolBiol. 2010;10:80. doi: 10.1186/1471-2148-10-80. doi: 10.1186/1471-2148-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. doi: 10.1007/bf00623322. [DOI] [PubMed] [Google Scholar]
- Ekland EH, Szostak JW, Bartel DP. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- Fontana W, Stadler PF, Bornberg-Bauer EG, et al. RNA folding and combinatory landscapes. Phys Rev E. 1993;47:2083–2099. doi: 10.1103/physreve.47.2083. doi: 10.1103/physreve.47.2083. [DOI] [PubMed] [Google Scholar]
- Fontana W, Schuster P. Continuity in evolution: on the nature of transitions. Science. 1998;280:1451–1455. doi: 10.1126/science.280.5368.1451. doi: 10.1126/science.280.5368.1451. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Gravner J. Percolation on the fitness hypercube and the evolution of reproductive isolation. J TheorBiol. 1997;184:51–64. doi: 10.1006/jtbi.1996.0242. doi: 10.1006/jtbi.1996.0242. [DOI] [PubMed] [Google Scholar]
- Grüner W, Giegerich R, Strothmann D, et al. Analysis of RNA sequence structure maps by exhaustive enumeration 1. Neutral networks. MonatshChem. 1996a;127:355–374. doi: 10.1007/bf00810881. [Google Scholar]
- Grüner W, Giegerich R, Strothmann D, et al. Analysis of RNA sequence structure maps by exhaustive enumeration 2. Structures of neutral networks and shape space covering. MonatshChem. 1996b;127:375–389. doi: 10.1007/bf00810882. [Google Scholar]
- Gupta RD, Tawfik DS. Directed enzyme evolution via small and effective neutral drift libraries. Nat Methods. 2008;5:939–942. doi: 10.1038/nmeth.1262. doi: 10.1038/nmeth.1262. [DOI] [PubMed] [Google Scholar]
- Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. doi: 10.1007/s00239-013-9604-x. [DOI] [PubMed] [Google Scholar]
- Hayden EJ, Bratulic S, Koenig I, Ferrada E, Wagner A. The effects of stabilizing and directional selection on phenotypic and genotypic variation in a population of RNA enzymes. J MolE. 2014;78:101–108. doi: 10.1007/s00239-013-9604-x. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- Ikawa Y, Tsuda K, Matsumura S, Inoue T. De novo synthesis and development of an RNA enzyme. PNAS. 2004;101:13750–13755. doi: 10.1073/pnas.0405886101. doi: 10.1073/pnas.0405886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Lu Z, Chen D, Raval A, Venturelli OS, Arnold FH. Evolution favors protein mutational robustness in sufficiently large populations. BMC Biology. 2007;5:29. doi: 10.1186/1741-7007-5-29. doi: 10.1186/1741-7007-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez JI, Xulvi-Brunet R, Campbell GW, Turk-MacLeod R, Chen IA. Comprehensive experimental fitness landscape and evolutionary network for small RNA. PNAS. 2013;110:14984–14989. doi: 10.1073/pnas.1307604110. doi: 10.1073/pnas.1307604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach M, Devenish SR, Hollfelder F. A simple method to evaluate the biochemical compatibility of oil/surfactant mixtures for experiments in microdroplets. Lab Chip. 2012;12:4185–4192. doi: 10.1039/c2lc40281e. doi: 10.1039/c2lc40281e. [DOI] [PubMed] [Google Scholar]
- Kaplan J. The end of the adaptive landscape? BiolPhilos. 2008;23:625–638. doi: 10.1007/s10539-008-9116-z. [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge UK: 1983. [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer-Verlag; New York: 1970. [Google Scholar]
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev EcolSyst. 1992;23:263–286. doi: 10.1146/annurev.es.23.110192.001403. [Google Scholar]
- Orr HA. Fitness and its role in evolutionary genetics. Nat Rev Genetics. 2009;10:531–539. doi: 10.1038/nrg2603. doi: 10.1038/nrg2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paegel BM, Joyce GF. Darwinian evolution on a chip. PLoSBiol. 2008;6:900–906. doi: 10.1371/journal.pbio.0060085. doi: 10.1371/journal.pbio.0060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paegel BM, Joyce GF. Microfluidic compartmentalized directed evolution. ChemBiol. 2010;17:717–724. doi: 10.1016/j.chembiol.2010.05.021. doi: 10.1016/j.chembiol.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie KL, Joyce GF. Deep sequencing analysis of mutations resulting from the incorporation of dNTP analogs. Nucleic Acids Res. 2010;38:8095–8104. doi: 10.1093/nar/gkq685. doi: 10.1093/nar/gkq685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. Sewall Wright's adaptive landscapes: 1932 vs. 1988. BiolPhilos. 2008;23:591–603. doi: 10.1007/s10539-008-9124-z. [Google Scholar]
- Pitt JN, Ferré-D'Amaré AR. Rapid construction of empirical RNA fitness landscapes. Science. 2010;330:376–379. doi: 10.1126/science.1192001. doi: 10.1126/science.1192001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt T, Lehman N. Non-unity molecular heritability demonstrated by continuous evolution in vitro. ChemBiol. 1999;6:857–869. doi: 10.1016/s1074-5521(00)80005-8. doi: 10.1016/S1074-5521(00)80005-8. [DOI] [PubMed] [Google Scholar]
- Schultes EA, Bartel DP. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- Schuster P, Fontana W. Chance and necessity in evolution: lessons from RNA. Physica D: Nonlinear Phenomena. 1999;133:427–452. doi: 10.1016/S0167-2789(99)00076-7. [Google Scholar]
- Shechner DM, Grant RA, Bagby SC, Koldobskaya Y, Piccirilli JA, Bartel DP. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science. 2009;326:1271–1275. doi: 10.1126/science.1174676. doi: 10.1126/science.1174676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll SJ, Arenas CD, Lehman N. Accumulation of deleterious mutations in small abiotic populations of RNA. Genetics. 2006;175:267–275. doi: 10.1534/genetics.106.066142. doi: 10.1534/genetics.106.066142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Sala M, Henry M, Wain-Hobson S, Meyerhans A. Manganese cations increase the mutation rate of human immunodeficiency virus type 1 ex vivo. J Gen Virol. 1999;80:1983–1986. doi: 10.1099/0022-1317-80-8-1983. [DOI] [PubMed] [Google Scholar]
- Voytek SB, Joyce GF. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. PNAS. 2007;104:15288–15293. doi: 10.1073/pnas.0707490104. doi: 10.1073/pnas.0707490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Robustness and evolvability in living systems. Princeton University Press; Princeton NJ: 2005. [Google Scholar]
- Wagner A. Robustness and evolvability: a paradox resolved. ProcBiolSci. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. In: Jones DF, editor. Proceedings of the sixth international congress of genetics. Vol. 1. Brooklyn Botanical Gardens; Ithaca, NY: 1932. pp. 356–366. [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends EcolEvol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [Google Scholar]