Abstract
Replication inhibitors cause replication fork stalling and double-strand breaks (DSBs) that result from processing of stalled forks. During recovery from replication blocks, the homologous recombination (HR) factor RAD51 mediates fork restart and DSB repair. HR defects therefore sensitise cells to replication inhibitors, with clear implications for cancer therapy. Gemcitabine is a potent replication inhibitor used to treat cancers with mutations in HR genes such as BRCA2. Here we investigate why, paradoxically, mutations in HR genes protect cells from killing by Gemcitabine. Using DNA replication and -damage assays in mammalian cells, we show that even short Gemcitabine treatments cause persistent replication inhibition. BRCA2 and RAD51 are recruited to chromatin early after removal of the drug, actively inhibit replication fork progression and promote the formation of MUS81- and XPF-dependent DSBs that remain unrepaired. Our data suggest that HR intermediates formed at Gemcitabine-stalled forks are converted into DSBs and thus contribute to Gemcitabine-induced cell death, which could have implications for the treatment response of HR-deficient tumours.
Keywords: DNA replication, DNA damage, homologous recombination, nucleoside analogs, tumor suppressors
Introduction
Many cytotoxic anti-cancer treatments target proliferating cells by interfering with DNA replication, thus generating lethal DNA damage. Such treatments exploit the high proliferation rates of cancer cells, and can be further potentiated by cancer-specific defects in DNA repair (1). The mechanisms of action of two replication inhibitors, the ribonucleotide reductase (RNR) inhibitor hydroxyurea (HU) and the DNA polymerase inhibitor aphidicolin, have been studied in detail. Both cause slowing or stalling of replication forks, generating excessive amounts of single-stranded DNA (ssDNA) as DNA polymerases stall but the replicative helicase continues to unwind DNA. Replication inhibition activates the ATR-dependent S phase checkpoint, which stabilises stalled forks and down-regulates new replication initiation (origin firing) to prevent further damage (2). After removal of the inhibitor, replication restarts and the checkpoint is inactivated. Depending on the length of treatment, restart occurs either by resumption of replication fork progression or through new origin firing (2, 3). After a few hours of replication block, structure-specific nucleases such as MUS81-EME1 begin to process the stalled forks into double-strand breaks (DSBs) (3, 4). Accumulation of these DSBs creates a requirement for the DSB repair pathways homologous recombination (HR) and non-homologous end joining (NHEJ) for cellular resistance to replication inhibitors (5). HR depends on the recombinase RAD51 and mediator proteins such as XRCC3 and BRCA2, which promote the loading of RAD51 onto ssDNA. In addition to their roles in DSB repair, BRCA2 and RAD51 also prevent excessive MRE11-dependent resection of the daughter strands at stalled forks (6, 7) and RAD51 promotes restart of stalled forks after release from HU (3). All of these findings are of potential clinical importance as several types of cancer can have genetic defects in HR. This includes breast and pancreatic cancer, where familial and sporadic forms can display inactivating mutations or promoter methylations in BRCA1, BRCA2, PALB2, BRIP1 and other genes of the Fanconi Anaemia pathway (8-11). Breast and pancreatic cancer are treated with the replication inhibitor Gemcitabine (2′,2′-difluorodeoxycytidine). In the cells, Gemcitabine is converted into its di- and triphosphates, which inactivate RNR and inhibit DNA polymerase after incorporation into nascent DNA (12). This strongly inhibits DNA synthesis and causes p53-independent apoptosis. The cytotoxic DNA lesions induced by Gemcitabine and the DNA repair pathways that respond to them are poorly understood. Intriguingly, previous studies found that Chinese hamster cells mutated in BRCA2 or another HR mediator, XRCC3, and the FANCC-mutated pancreatic cancer cell line PL11 were less sensitive to Gemcitabine treatments than their HR-proficient counterparts (13-15).
Here, we investigate the molecular mechanism by which the HR factors BRCA2 and RAD51 promote Gemcitabine-induced cell death. Our data suggest that even after short Gemcitabine treatments, replication forks remain stalled and are converted into DSBs that persist in the cells. BRCA2 and RAD51 are recruited to chromatin, inhibit fork progression and promote the formation of DSBs that are dependent on the structure-specific endonucleases MUS81 and XPF. Our data suggest that HR intermediates formed at stalled forks promote Gemcitabine cytotoxicity, which could have implications for the treatment response of HR-deficient tumours.
Materials and Methods
Cell lines and reagents
Human cell lines were all obtained from ATCC more than two years ago and were therefore authenticated using 8-locus STR profiling (LGC standards). Human U2OS osteosarcoma cells were last authenticated in April 2013. H1299 lung carcinoma cells were last authenticated in March 2011 and have not been cultured since. BxPC3 pancreatic adenocarcinoma cells, MCF7 breast cancer cells and OVCAR3 human ovarian cancer cells were last authenticated in April 2014.
VC8 and VC8-B2 cells were obtained from Malgorzata Z. Zdzienicka (16, authentication not available). Cells were confirmed Mycoplasma-free and grown in DMEM with 10% FCS in a humidified atmosphere containing 5% CO2. OVCAR3 cells were grown in DMEM with 10% foetal bovine serum, 0.01 mg/ml insulin and 1% non-essential amino acids (Sigma). Gemcitabine (Tocris Bioscience) was used at 2 or 5 μM for 2 h. DNA-PK inhibitor NU7441 (Tocris Bioscience) was used at 1 μM. BLM inhibitor ML216 (Sigma-Aldrich) was used at 1.8 μM as previously described (17).
DNA fibre analysis
Cells were labelled with 25μM CldU and 250μM IdU as indicated. For release from Gemcitabine, cells were washed three times with warm PBS. Controls were labelled with CldU and IdU for 20 min each. DNA fibre spreads were prepared as described (3). Acid treated fibre spreads were incubated with rat anti-BrdU (detects CldU, BU1/75, AbD Serotec) and mouse anti-BrdU (detects IdU, B44, Becton Dickinson) for 1h. Slides were fixed with 4% formaldehyde and incubated with anti-rat IgG AlexaFluor 555 and anti-mouse IgG AlexaFluor 488 (Molecular Probes) for 1.5h. Images were acquired on an E600 Nikon microscope using a Plan Apo 60× (1.3NA) oil lens (Nikon), a digital camera (C4742-95, Hamamatsu) and the Volocity acquisition software (Perkin Elmer). Images were analysed using ImageJ (http://rsbweb.nih.gov/ij/). For quantification of replication structures, 60-250 structures were counted per independent experiment.
Immunofluorescence
For phospho-Histone H2AX, 53BP1, Lamin B and phospho-Histone H3, cells were fixed with 4% formaldehyde and permeabilised with 0.2% Triton X-100 for 5 min. For RAD51 foci, cells were pre-extracted with 0.2% Triton X-100 for 1 min. For colocalisation with replication foci, antibodies were fixed with 4% formaldehyde before DNA denaturation with HCl and immunostaining for thymidine analogues. Primary antibodies were rat monoclonal anti-BrdU (BU1/75, AbD Serotec, 1:400) to detect CldU, mouse monoclonal anti-BrdU (B44, Becton Dickinson, 1:50) to detect IdU, mouse monoclonal anti-phospho-Histone H2AX (Ser139) (JBW301, Merck Millipore, 1:1000), rabbit polyclonal anti-RAD51 (H-92, Santa Cruz Biotechnology, 1:500), rabbit polyclonal anti-53BP1 (Bethyl, 1:3000), goat polyclonal anti-Lamin B (Santa Cruz Biotechnology, 1:400) and rabbit polyclonal anti-phospho-Histone H3 (Ser10) (Merck Millipore, 1:500).. Secondary antibodies were anti-Rat IgG AlexaFluor 555, anti-mouse IgG AlexaFluor 488, anti-rabbit IgG AlexaFluor 555 or AlexaFluor 647 and anti-goat IgG Alexafluor 594 (Molecular Probes). DNA was counterstained with DAPI and images acquired as above.
Cell survival assays
For clonogenic survival, defined numbers of cells were plated before treatment with Gemcitabine (0.1 μM – 5 μM) for 2 h. Colonies of >50 cells were allowed to form in fresh medium, fixed and stained with 50% ethanol, 2% methylene blue for 10 min. Apoptosis was quantified by counting fragmented nuclei after DAPI staining and mitotic catastrophe was quantified by counting fragmented nuclei displaying Lamin B staining.
Flow cytometry
5×105 cells per sample were treated as indicated, harvested and fixed with cold 70% ethanol before staining with propidium iodide (10 μg/ml). Cell cycle profiles were gathered using the C6 Flow Cytometer system (Accuri) and analysed with CFlow Plus.
Pulsed-field gel electrophoresis
2×106 cells per sample were treated as indicated, harvested and melted into 1.0% InCert-Agarose (Lonza) inserts. Inserts were digested in 0.5 M EDTA-1% N-laurylsarcosyl-proteinase K (1 mg/ml) at room temperature for 48 h and washed three times in TE buffer. Inserts were loaded onto a separation gel (1.0% chromosomal-grade agarose, Bio-Rad). Separation was performed using a CHEF DR III (BioRad; 120 field angle, 240 s switch time, 4 V cm−1, 14 °C) for 20 h. Images of ethidium bromide-stained gels were acquired using a Syngene G:BOX gel imaging system. DSBs (chromosome fragments >2 Mbp) were quantified by densitometry using ImageJ. Intensity of DNA entering the gel was normalised to total DNA and untreated control was subtracted to obtain final values.
siRNA treatment
siRNA against human RAD51 (14), MUS81 (siGENOME SMARTpool D-016143) and XPF(ERCC4) (OnTARGETplus SMARTpool L-019946-00) were from Thermo Fisher. “Allstars negative control siRNA” (nonT) was from Qiagen. Cells were transfected with 50 nM of each siRNA using Dharmafect 1 (Thermo Fisher) for 24, 48 (RAD51) or 72 h (XPF and MUS81) before treatment with Gemcitabine.
Western blotting
Primary antibodies were rabbit polyclonal anti-RAD51 (H-92, Santa Cruz Biotechnology, 1:500), mouse monoclonal anti-MUS81 (MTA30 2G10/3, Santa Cruz Biotechnology, 1:500), mouse monoclonal anti-XPF (219, Fisher Scientific, 1:200), mouse anti-αTubulin (B512, Sigma, 1:5000), rabbit polyclonal anti-βActin (Cell Signaling, 1:1000) and mouse monoclonal anti-PARP1 (F-2, Santa Cruz Biotechnology, 1:500). For further antibody information see Supplementary Materials and Methods.
Statistical analysis
The means and 1× standard error (SEM) of independent repeats are shown. Statistical significance was determined using the student’s t-test (* p<0.05, ** p<0.01, *** p<0.001).
Results
We used BRCA2-mutated VC8 and BRCA2-complemented VC8-B2 Chinese hamster fibroblasts (p53 mutated), an isogenic model for BRCA2 function that has successfully been used to study the role of BRCA2 in chemotherapy response (18). We tested short Gemcitabine treatments in the micromolar range, similar to clinically relevant concentrations (19, 20). While VC8 cells were hypersensitive to cisplatin as expected (Fig. S1A), they were less sensitive than VC8-B2 cells to higher concentrations of Gemcitabine (Fig. 1A). Similar results were obtained after siRNA-depleting RAD51 in human U2OS osteosarcoma and BxPC3 pancreatic cancer cell lines (both p53 wild type), suggesting that this was not due to secondary mutations acquired in VC8 cells, but to loss of RAD51 function (Fig. 1B, C, Fig. S1B).
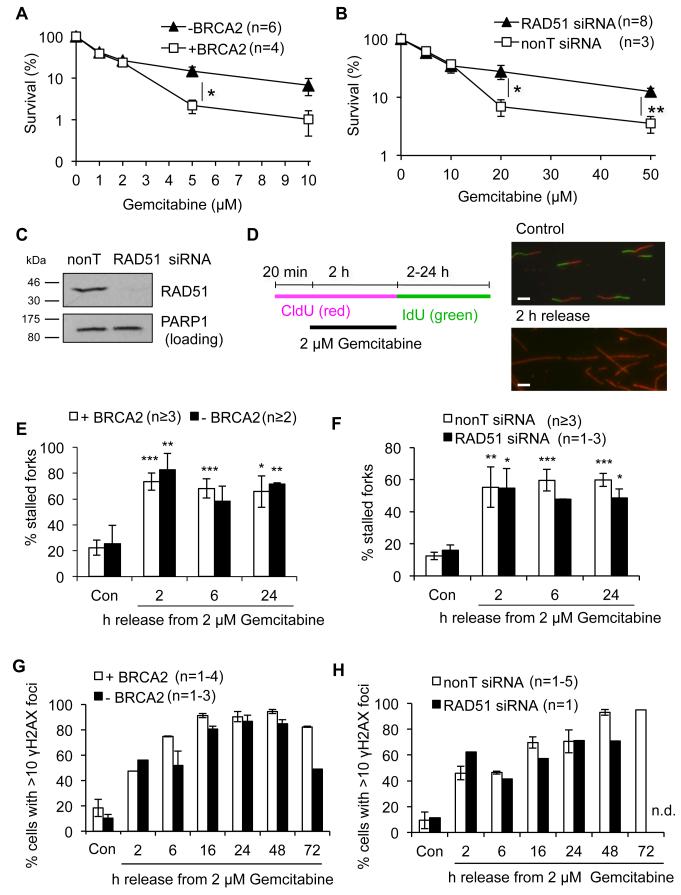
Figure 1. HR defects protect from cell death, but have no effect on irreversible replication fork stalling after release from Gemcitabine.
(A) Clonogenic survival of VC8 (−BRCA2) and VC8-B2 (+ BRCA2) cells treated with Gemcitabine for 2 h and released into fresh medium. (B) Clonogenic survival of U2OS cells ± RAD51 treated as in (A). (C) Protein levels of RAD51 and PARP1 (loading control) in U2OS cells 24 h after transfection with RAD51 or nonT siRNA. (D) Schematic and representative images for DNA fibre labelling. CldU-only labelled tracks (stalled forks) were normalised to all CldU-containing tracks. Bars: 10 μm. (E) Quantification of stalled forks in VC8 and VC8-B2 cells (asterisks compare to Con). (F) Quantification of stalled forks in U2OS cells ± RAD51 siRNA (asterisks compare to Con). (G) Percentages of cells displaying more than 10 γH2AX foci after release from Gemcitabine. (H) Percentages of U2OS cells ± RAD51 siRNA displaying more than 10 γH2AX foci after release from Gemcitabine. Error bars: SEM; Asterisks: * p<0.05, ** p<0.01, *** p<0.001, Student’s t-test.
We initially used 2 μM Gemcitabine, which has been shown to inhibit fork progression and allowed about 50% survival in our cell lines, and measured replication restart using DNA fibre analyses (Fig. 1D). Even at this low concentration, most forks remained stalled and did not resume progression for at least 24 h after release from 2 h Gemcitabine (Fig. 1D). Levels of fork stalling were comparable between BRCA2-proficient and deficient cells and similar results were obtained using RAD51-depleted U2OS cells (Fig. 1E, F). Levels of phospho-S139-H2AX (γH2AX), a marker of stalled forks (3), increased after Gemcitabine release and remained high for at least 72 h, suggesting that stalled forks persisted for several days (Fig. 1G, H). The induction of γH2AX was comparable in BRCA2-proficient and -deficient as well as control- and RAD51-depleted cells (Fig. 1G, H). However, BRCA2-deficient cells displayed lower γH2AX staining after 72 h release, suggesting a quicker recovery from Gemcitabine-induced DNA damage (Fig. 1G). Overall these data did not suggest that promotion of fork restart by BRCA2 or RAD51 plays a role in response to cytotoxic Gemcitabine treatments.
Despite persistent fork stalling, cells resumed replication between 6-24 h release, firing new origins and resuming slow progression through S phase (Fig. 2A, B). Nevertheless, markers of S phase checkpoint signalling remained active during replication restart (Fig. S2). Cell cycle progression was accompanied by apoptosis and mitotic catastrophe (MC), which peaked after 2-3 days release (Fig. 2B, C). The appearance of MC suggests that some cell death did result from aberrant mitotic entry in presence of unrepaired DNA damage (21). VC8 cells displayed lower induction of MC and apoptosis after 5 μM Gemcitabine (Fig. 2D), which was not due to VC8 cells being prevented from cycling and mitotic entry. Instead, VC8 cells displayed higher percentages of cells positive for phospho-histone H3 (Fig. 2E) and faster progression into the next G1 phase 1 day after release compared to VC8-B2 cells (Fig. 2F). Initial accumulation in S phase was also not lower in BRCA2-deficient cells (Fig. 2F), confirming that reduced Gemcitabine sensitivity was not due to fewer cells entering S phase. Interestingly, VC8 cells displayed fewer γH2AX-positive cells and a lower percentage of S phase cells at 3 days after release, suggesting a quicker recovery from Gemcitabine in absence of BRCA2 (Fig. 1G, Fig. 2F).
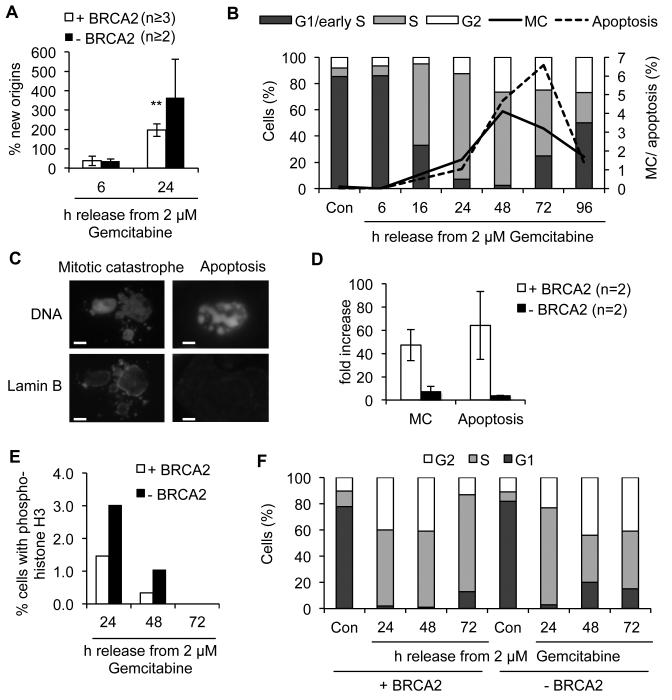
Figure 2. BRCA2-deficient and –proficient cells display aberrant cell cycle progression after release from Gemcitabine.
(A) New origin firing in VC8 and VC8-B2 cells after release from Gemcitabine. DNA fibre labelling was performed as in Fig. 1D and IdU-only labelled tracks (new origins) were normalised to all CldU-containing tracks. (B) FACS analysis of cell cycle progression and time course of mitotic catastrophe (MC) and apoptosis in VC8-B2 cells after release from Gemcitabine. (C) Representative images of DAPI- and Lamin B1-stained VC8-B2 cells with mitotic catastrophe or apoptotic phenotypes after 48 h release from Gemcitabine. Bars: 10 μm. (D) Increase in MC and apoptosis in VC8 and VC8-B2 cells after release from 5 μM Gemcitabine. (E) Percentages of VC8-B2 and VC8 cells positive for phospho-histone H3 staining following release from Gemcitabine in the presence of 1.5 μM Nocodazole. (F) Cell cycle progression in VC8 and VC8-B2 cells after release from 5 μM Gemcitabine for 24-72 h. Error bars: SEM; Asterisks: ** p<0.01, Student’s t-test.
We decided to further investigate the role of BRCA2 and RAD51 at Gemcitabine-stalled replication forks. In addition to promoting fork restart, RAD51 and BRCA2 also prevent shortening of daughter strands at stalled forks (6, 7), and RAD51 inhibits fork progression during cisplatin and camptothecin treatments (22, 23). To investigate if either of these processes occurs after release from Gemcitabine, we compared the length of DNA replicated during 2 h Gemcitabine treatment and after 4 h release from 5 μM Gemcitabine in VC8 and VC8-B2 cells (Fig. 3A). Tracks replicated during the 2 h Gemcitabine treatment were longer in presence of BRCA2, as has been described before (6). However, after release from Gemcitabine replicated tracks in BRCA2-proficient cells remained the same length, while tracks in BRCA2-deficient cells further increased in length, suggesting that some forks were still progressing (Fig. 3B, C, F). Similar results were obtained using RAD51-depleted U2OS cells (Fig. 3D, E, G). Our data suggest that after release from Gemcitabine, BRCA2 and RAD51 are recruited to forks where RAD51 promotes transactions that inhibit further fork progression. In HR-proficient cells, RAD51 foci indeed accumulated and persisted for 72 h after release (Fig. 3H, I), suggesting that HR was initiated but not completed during that time.
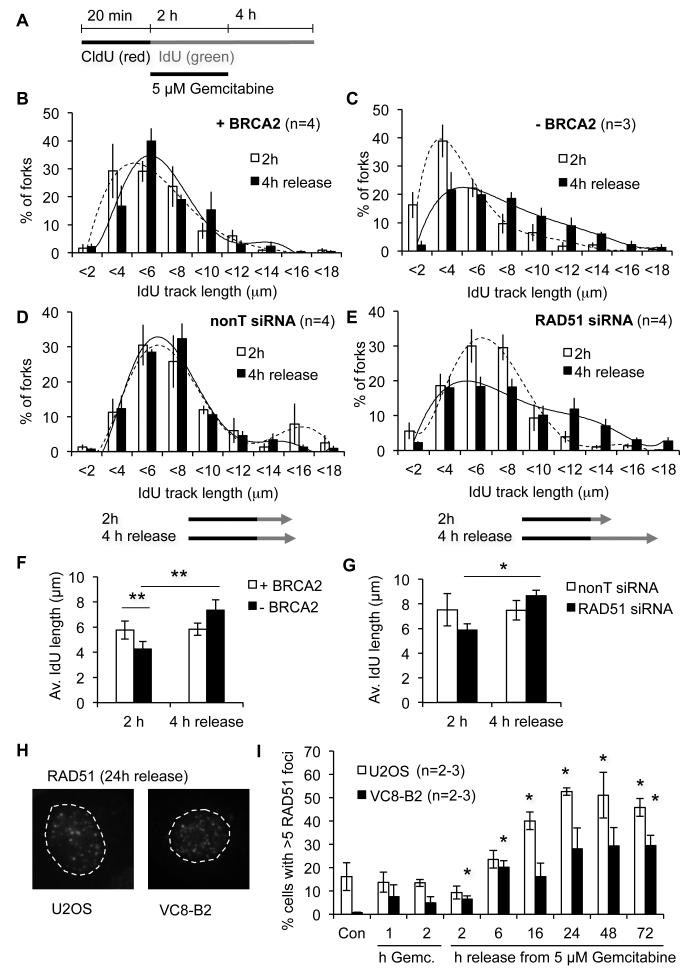
Figure 3. BRCA2 and RAD51 inhibit replication fork progression after release from Gemcitabine.
(A) Labelling protocol for DNA fibre analyses. Cells were labelled with CldU, treated with IdU and 5 μM Gemcitabine for 2 h and released into IdU for 4 h. (B) Length distributions of IdU-labelled tracks from VC8-B2 cells (+ BRCA2). (C) Length distributions of IdU-labelled tracks from VC8 cells (−BRCA2). (D) Length distributions of IdU-labelled tracks from U2OS cells treated with nonT siRNA. (E) Length distributions of IdU-labelled tracks from U2OS cells treated with RAD51 siRNA. (F) Average lengths of IdU tracks in VC8 and VC8-B2 cells treated as in (A). (G) Average lengths of IdU tracks in U2OS cells +/− RAD51. (H) Representative images of RAD51 foci in cells released from 5 μM Gemcitabine for 24 h. (I) Percentages of cells displaying more than 5 RAD51 foci during 1 and 2 h Gemcitabine treatment and after release from Gemcitabine (asterisks compare to Con). Error bars: SEM; Asterisks: * p<0.05, ** p<0.01, Student’s t-test.
Next we tested whether Gemcitabine-stalled forks were processed into DSBs. We first measured accumulation of nuclear 53BP1 foci, which mark sites of DSBs (24). High numbers of 53BP1 foci that co-localised with replication foci accumulated after 2 and 16 h release in U2OS and VC8-B2 cells, respectively (Fig. 4A, B). Compared to γH2AX foci (Fig. 1G, H), 53BP1 foci formation was delayed and only around half of γH2AX-positive cells also contained 53BP1 foci. This supports the idea that γH2AX marks all stalled replication forks as well as DSBs, while 53BP1 only accumulates at the subset of forks that have been processed into DSBs. Pulsed-field gel electrophoresis of genomic DNA confirmed that the increase in 53BP1 foci correlated with an increase in DSB levels (Fig. 4C, Fig. S3A). As with RAD51 foci, DSB levels remained high for 2-3 days after release, suggesting that little DSB repair was occurring.
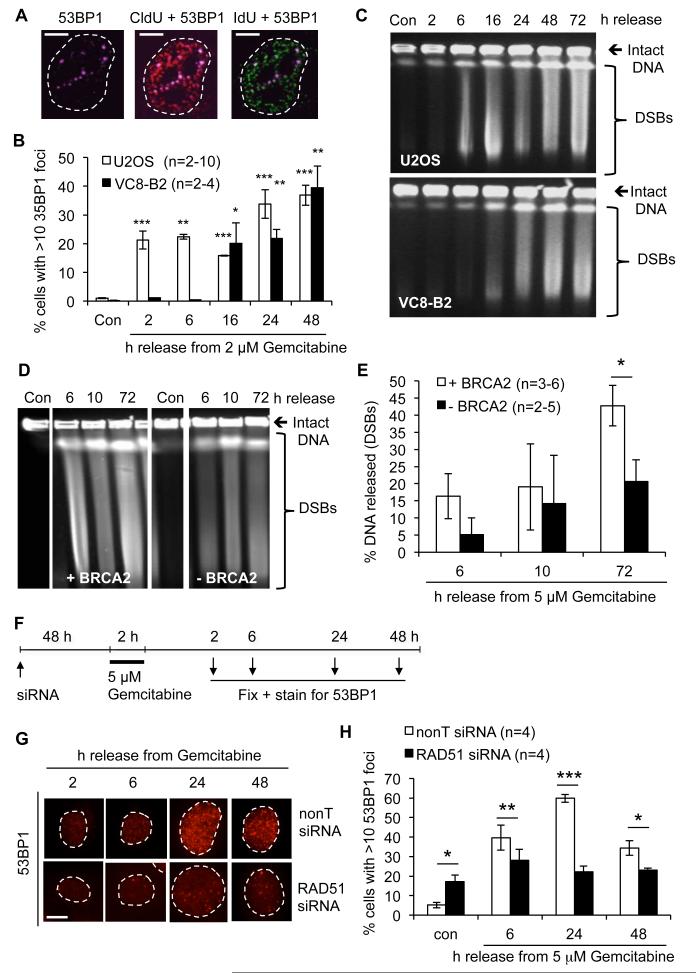
Figure 4. Gemcitabine causes double strand breaks that depend on BRCA2 and RAD51.
(A) 53BP1 foci (far-red) and colocalisation with replication foci (CldU, red and IdU, green) in VC8-B2 cells 6 h after release from 2 μM Gemcitabine. Bars: 10 μm. (B) Percentages of cells displaying more than 10 53BP1 foci after release from Gemcitabine (asterisks compare to Con). (C) Pulsed-field gel electrophoresis showing DSB induction after release from 2 μM Gemcitabine in U2OS and VC8-B2 cells (see Fig. S3A for quantification). (D) PFGE of DSB induction in VC8-B2 and VC8 cells after release from 5 μM Gemcitabine (cropped lanes are from one gel, see Fig. S3B). (E) Percentages of DNA released from plugs in DSB in VC8-B2 and VC8 cells. (F) Outline of experimental design for 53BP1 foci quantification. 24 h after transfection with RAD51 or nonT siRNA, U2OS cells were treated with 5 μM Gemcitabine for 2 h, released for the times indicated, fixed and stained for 53BP1. (G) Representative images of 53BP1 foci in U2OS cells ± RAD51 siRNA released from 5 μM Gemcitabine. Bar: 10 μm. (H) Percentages of U2OS cells ± RAD51 siRNA displaying more than 10 53BP1 foci after release from 5 μM Gemcitabine. Error bars: SEM; Asterisks: * p<0.05, ** p<0.01, *** p<0.001, Student’s t-test.
As DSBs are highly toxic DNA lesions and likely to contribute to Gemcitabine toxicity, we next analysed whether Gemcitabine-induced DSB formation depended on BRCA2 and RAD51. Indeed, PFGE showed that while VC8 cells displayed higher background levels of unrepaired DSBs, the additional increase in DSBs after Gemcitabine was higher in the presence of BRCA2 (Fig. 4D, E, Fig. S3B). Similarly, RAD51-depleted U2OS cells accumulated fewer 53BP1 foci than control cells after release from Gemcitabine (Fig. 4F-H). Comparable results were obtained in RAD51-depleted human cancer cell lines derived form pancreatic (BxPC3, p53 wild type), breast (MCF7, p53 wild type) and ovarian cancer (OVCAR3, p53 mutated) (Fig. S4, S5, S6). HR-deficient cells thus accumulate fewer DSBs after Gemcitabine treatment.
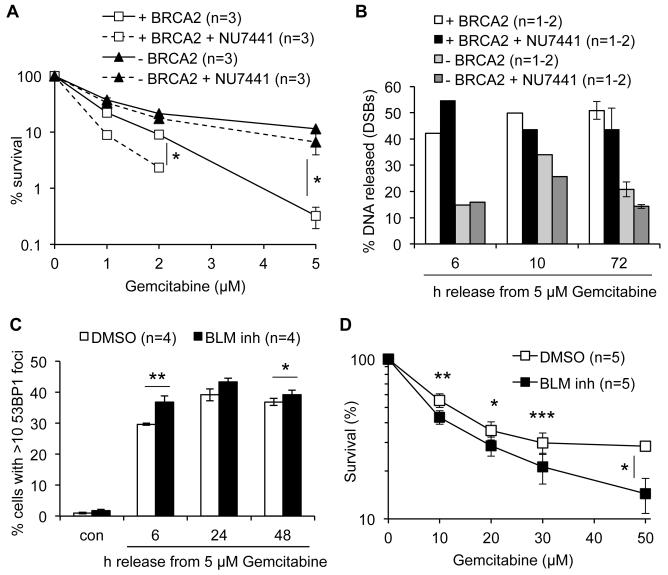
In addition to HR, NHEJ acts as an alternative and competing repair pathway for DSBs. Although NHEJ may not be able to faithfully repair one-DSBs at collapsed replication forks, it can promote resistance to replication inhibitors such as HU, suggesting that some replication-dependent breaks are substrates for NHEJ (5). We considered that reduced Gemcitabine sensitivity in HR-deficient cells might result from DSBs being more efficiently repaired by NHEJ in absence of HR. We inhibited NHEJ using DNA-PK inhibitor NU7441 to test whether this could sensitise HR-deficient cells to Gemcitabine. NU7441 alone increased the background levels of unrepaired DSBs (Fig. S3B). However, co-treatment with NU7441 did not increase Gemcitabine toxicity (Fig. 5A) or Gemcitabine-induced DSB levels in BRCA2-deficient cells (Fig. 5B, Fig. S3B). In contrast, BRCA2-proficient cells treated with NU7441 were more sensitive to Gemcitabine and accumulated more DSBs early after release from Gemcitabine (Fig. 5A, B), suggesting that some BRCA2-dependent DSBs are repaired by NHEJ.
Figure 5. Roles of non-homologous end joining and BLM helicase in the response to Gemcitabine.
(A) Clonogenic survival of VC8-B2 and VC8 cells treated with Gemcitabine and 1 μM NU7441 for 2 h and released into fresh medium containing 1 μM NU7441, compared to survival without NU7441 (Fig. 1A). (B) Percentages of DNA released from plugs in DSB in VC8-B2 and VC8 cells released from treatment with 5 μM Gemcitabine in presence or absence of 1 μM NU7441 (see Fig. S3B for gel). Error bars: SD. (C) Percentages of U2OS cells ± BLM inhibitor displaying more than 10 53BP1 foci after release from 5 μM Gemcitabine. Cells were pre-incubated with 1.8 μM BLM inhibitor for 1 h before Gemcitabine treatment and released in fresh medium containing BLM inhibitor. (D) Clonogenic survival of U2OS cells ± BLM inhibitor treated with Gemcitabine for 2 h as in (C). Error bars: SEM; Asterisks: * p<0.05, ** p<0.01, *** p<0.001, Student’s t-test.
To further support a role for HR in Gemcitabine-induced DSB formation and cell death, we used a small molecule inhibitor of the BLM helicase, ML216 (17). BLM counteracts RAD51 function in the initiation of HR by resolving D-loop structures (25). If RAD51-mediated HR is responsible for Gemcitabine-induced DSBs and cell death, then BLM inhibition should exacerbate both DSB formation and cell death. Indeed we observed a small but reproducible increase in DSBs and cell death in cells treated with BLM inhibitor (Fig. 5C, D).
Finally, we used siRNA depletion in U2OS cells to test which enzymes were responsible for converting stalled forks into DSBs, focussing on the MUS81-EME1 and XPF-ERCC1 structure-specific endonucleases. In vitro, only MUS81-EME1 cleaves stalled replication fork structures, but in vivo XPF-ERCC1 and MUS81-EME1 can act in parallel pathways to process joint molecule recombination intermediates such as Holliday junctions (26, 27). Cells were transfected with XPF or MUS81 siRNA for 72 h, treated with 5 μM Gemcitabine for 2 h, and released for up to 72 h. Proteins remained depleted for at least 2 days after treatment (Fig. 6A-C). Depletion of MUS81 or XPF could prevent Gemcitabine-induced DSB formation, with co-depletion of both proteins being more effective (Fig. 6D,E). DSBs in Gemcitabine-treated cells thus depend on BRCA2 and RAD51, and therefore likely on RAD51 loading and filament formation for the initiation of HR, and on endonucleases that cleave HR intermediates. These data suggest that these DSBs arise not simply through endonucleolytic cleavage of stalled replication forks but also through processing of recombination intermediates.
Figure 6. Gemcitabine-induced double-strand breaks depend on MUS81 and XPF.
(A) Protein levels of XPF and βActin (loading control) after transfection with nonT or XPF siRNAs for 72 h, treated with 5 μM Gemcitabine for 2 h and release for the times indicated. (B) Protein levels of MUS81 after transfection with nonT or MUS81 siRNAs as in (A). (C) Protein levels of XPF, MUS81 and βActin (loading control) after transfection with nonT or MUS81 and XPF siRNAs as in (A). (D) Percentage of U2OS cells ± XPF and MUS81 displaying more than 10 53BP1 foci after release from Gemcitabine. (E) Quantification of increase in cells displaying more than 10 53BP1 foci as in (D) (asterisks compare to nonT siRNA). (F) Suggested model for HR-dependent replication fork slowing and DSB formation. Forks affected by Gemcitabine treatment are recognised by BRCA2 and RAD51 and remodelled into joint molecule HR intermediates such as D-loops. These intermediates are preferentially cleaved by MUS81 and XPF. Error bars: SEM; Asterisks: * p<0.05, ** p<0.01, Student’s t-test.
Discussion
We report that after release from Gemcitabine treatment, BRCA2 and RAD51 inhibit replication fork progression, promote MUS81/XPF-dependent DSB formation and exacerbate cell death. This supports the idea that initiation of HR is required for DNA damage formation at Gemcitabine-stalled replication forks. HR, normally a pathway that prevents accumulation of DNA damage, can thus promote the formation of DNA damage after Gemcitabine treatment.
We speculate that in response to Gemcitabine, BRCA2-assisted loading of RAD51 onto replication forks promotes the generation of HR intermediates, which inhibit further fork progression. This is likely the same mechanism as the RAD51-mediated fork slowing previously observed in cisplatin- or camptothecin-treated cells (22, 23), although the cellular consequences of this phenomenon have not been described. We speculate that these HR intermediates, likely D-loops and Holliday junctions, present substrates for endonucleolytic processing by MUS81 and XPF to generate DSBs (Fig. 6F). Gemcitabine-induced DSBs are not efficiently repaired, which could explain why HR does not protect from cell death. We speculate that the processes described here also occur at forks that have been stalled by other types of replication inhibitors, but this may not be obvious if the inhibitor also induces DSBs by other mechanisms and does not prevent HR-mediated DSB repair. Our data suggest that for transient treatments, DNA damage response factors that promote rearrangements and nuclease processing of stalled forks can be expected to cause sensitivity to Gemcitabine (Fanconi Anaemia proteins, BRCA2, XRCC3, RAD51), while factors involved in later steps of DSB repair should promote survival or have little effect (DNA Ligase IV, RAD54).
Our data also suggest that the very persistent effects of even short exposures to Gemcitabine are important for its cytotoxic action. Gemcitabine inactivates RNR irreversibly and Gemcitabine nucleotides accumulate in cells after treatment (12), which likely underlies the prolonged replication inhibition observed. After release from Gemcitabine, stalled replication forks and DNA damage signalling therefore persist, but cannot prevent the eventual resumption of cell cycle progression. A similar phenomenon has been observed during prolonged HU treatments and could be common to all situations of prolonged replication fork stalling (4, 28). This cell cycle progression in presence of unresolved DNA lesions contributes to cell death by mitotic catastrophe and likely also to DSB formation and apoptosis, as mitotic CDK1 activity has been suggested to promote MUS81-dependent DSB formation at perturbed forks (29).
These peculiarities of transient Gemcitabine treatments could explain why reports on the impact of HR status on Gemcitabine sensitivity are still conflicting. Previous studies have variously used continuous or transient treatments. For example, RAD51 depletion sensitises cells to continuous treatment with low doses of Gemcitabine (30, 31), but both RAD51 depletion and mutations in BRCA2 decrease sensitivity when combined with transient treatments at higher doses, which seem more relevant for clinical applications (15 and this study). Indeed, a recent study showed that ATR and CHK1 inhibitors could sensitise ovarian cancer cells to transient but not to continuous Gemcitabine treatments (32). As ATR and CHK1 protect stalled forks from DSB formation this suggests that the danger of DSB is higher after release from Gemcitabine than during continuous treatment, possibly due to increased cell cycle progression. In agreement with this, our preliminary data suggest that more DSBs can be detected after release from transient Gemcitabine treatment compared to continuous treatment for the same time (Figure S7). On the other hand, mutations in the HR genes XRCC3 and FANCC can promote resistance even to continuous Gemcitabine treatments (13, 14). This suggests that more research into the time course of Gemcitabine action is needed, especially as this information could be crucial for optimal scheduling in combination treatments such as Gemcitabine/Carboplatin.
There are three published case studies of pancreatic cancer patients carrying BRCA2 or PALB2 mutations that did not respond or responded poorly to Gemcitabine, but responded well to a subsequent treatment with the crosslinking agents cisplatin or Mitomycin C (33-35). While we do not consider these data evidence that HR-deficient pancreatic cancers are more resistant to Gemcitabine than other pancreatic cancers, they do show that cancers that have proven resistant to one DNA damaging agent (Gemcitabine) can be hypersensitive to a different DNA damaging agent (e.g. carboplatin). This also suggests that in the case of Gemcitabine/platinum combination therapies, the hypersensitivity of HR-deficient tumours to platinum compounds could compensate for any Gemcitabine resistance in these tumours.
Taken together, our data have potential implications for the scheduling of Gemcitabine combination treatments in general and pose the question as to whether HR-deficient tumours would respond well to single-agent Gemcitabine treatments.
Supplementary Material
Acknowledgements
We thank Dr Angelo Agathanggelou for ML216 and Dr Agnieszka Gambus for helpful discussions of this manuscript.
Financial Support: Medical Research Council (MR/J007595/1, E. Petermann), Association for International Cancer Research (13-1048, E. Petermann), Wellcome Trust (ISSFPP12, E. Petermann); Cancer Research UK (C17183/A13030, G.S. Stewart)
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 2.Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J. 2012;443:13–26. doi: 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- 3.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 5.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, et al. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol Cell Biol. 2002;22:5869–78. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-Strand Break Repair-Independent Role for BRCA2 in Blocking Stalled Replication Fork Degradation by MRE11. Cell. 2011;145:529–42. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–11. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Groep P, van der Wall E, van Diest PJ. Pathology of hereditary breast cancer. Cell Oncol (Dordr) 2011;34:71–88. doi: 10.1007/s13402-011-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hruban RH, Canto MI, Goggins M, Schulick R, Klein AP. Update on familial pancreatic cancer. Adv Surg. 2010;44:293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinemann V, Boeck S, Hinke A, Labianca R, Louvet C. Meta-analysis of randomized trials: evaluation of benefit from gemcitabine-based combination chemotherapy applied in advanced pancreatic cancer. BMC Cancer. 2008;8:82. doi: 10.1186/1471-2407-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verrill M. Chemotherapy for early-stage breast cancer: a brief history. Br J Cancer. 2009;101(Suppl 1):S2–5. doi: 10.1038/sj.bjc.6605268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewald B, Sampath D, Plunkett W. Nucleoside analogs: molecular mechanisms signaling cell death. Oncogene. 2008;27:6522–37. doi: 10.1038/onc.2008.316. [DOI] [PubMed] [Google Scholar]
- 13.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, Swartz MJ, et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11:7508–15. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 14.Crul M, van Waardenburg RC, Bocxe S, van Eijndhoven MA, Pluim D, Beijnen JH, et al. DNA repair mechanisms involved in gemcitabine cytotoxicity and in the interaction between gemcitabine and cisplatin. Biochem Pharmacol. 2003;65:275–82. doi: 10.1016/s0006-2952(02)01508-3. [DOI] [PubMed] [Google Scholar]
- 15.Issaeva N, Thomas HD, Djureinovic T, Jaspers JE, Stoimenov I, Kyle S, et al. 6-thioguanine selectively kills BRCA2-defective tumors and overcomes PARP inhibitor resistance. Cancer Res. 2010;70:6268–76. doi: 10.1158/0008-5472.CAN-09-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Molecular and cellular biology. 2002;22:669–79. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenthal AS, Dexheimer TS, Gileadi O, Nguyen GH, Chu WK, Hickson ID, et al. Synthesis and SAR studies of 5-(pyridin-4-yl)-1,3,4-thiadiazol-2-amine derivatives as potent inhibitors of Bloom helicase. Bioorg Med Chem Lett. 2013;23:5660–6. doi: 10.1016/j.bmcl.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 19.Veltkamp S, Beijnen J, Schellens J. Prolonged versus standard gemcitabine infusion: translation of molecular pharmacology to new treatment strategy. The Oncologist. 2008;13:261–337. doi: 10.1634/theoncologist.2007-0215. [DOI] [PubMed] [Google Scholar]
- 20.Eisbruch A, Shewach DS, Bradford CR, Littles JF, Teknos TN, Chepeha DB, et al. Radiation concurrent with gemcitabine for locally advanced head and neck cancer: a phase I trial and intracellular drug incorporation study. J Clin Oncol. 2001;19:792–9. doi: 10.1200/JCO.2001.19.3.792. [DOI] [PubMed] [Google Scholar]
- 21.Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12:385–92. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 22.Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol. 2008;183:1203–12. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, et al. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell. 2003;11:1109–17. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 24.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–90. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic acids research. 2006;34:2269–79. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuchi K, Narita T, Pham VT, Iijima J, Hirota K, Keka IS, et al. Structure-specific endonucleases xpf and mus81 play overlapping but essential roles in DNA repair by homologous recombination. Cancer Res. 2013;73:4362–71. doi: 10.1158/0008-5472.CAN-12-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agostinho A, Meier B, Sonneville R, Jagut M, Woglar A, Blow J, et al. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS genetics. 2013;9:e1003591. doi: 10.1371/journal.pgen.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–7. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neelsen KJ, Zanini IM, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. The Journal of cell biology. 2013 doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choudhury A, Zhao H, Jalali F, Al Rashid S, Ran J, Supiot S, et al. Targeting homologous recombination using imatinib results in enhanced tumor cell chemosensitivity and radiosensitivity. Mol Cancer Ther. 2009;8:203–13. doi: 10.1158/1535-7163.MCT-08-0959. [DOI] [PubMed] [Google Scholar]
- 31.Tsai MS, Kuo YH, Chiu YF, Su YC, Lin YW. Down-regulation of Rad51 expression overcomes drug resistance to gemcitabine in human non-small-cell lung cancer cells. J Pharmacol Exp Ther. 2010;335:830–40. doi: 10.1124/jpet.110.173146. [DOI] [PubMed] [Google Scholar]
- 32.Huntoon CJ, Flatten KS, Wahner Hendrickson AE, Huehls AM, Sutor SL, Kaufmann SH, et al. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013;73:3683–91. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnenblick A, Kadouri L, Appelbaum L, Peretz T, Sagi M, Goldberg Y, et al. Complete remission, in BRCA2 mutation carrier with metastatic pancreatic adenocarcinoma, treated with cisplatin based therapy. Cancer biology & therapy. 2011;12:165–8. doi: 10.4161/cbt.12.3.16292. [DOI] [PubMed] [Google Scholar]
- 34.Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, De Jesus-Acosta A, Jones S, Maitra A, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther. 2011;10:3–8. doi: 10.1158/1535-7163.MCT-10-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalasani P, Kurtin S, Dragovich T. Response to a third-line mitomycin C (MMC)-based chemotherapy in a patient with metastatic pancreatic adenocarcinoma carrying germline BRCA2 mutation. Jop. 2008;9:305–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.