SUMMARY
The regulatory events guiding progenitor activation and differentiation in adult white adipose tissue are largely unknown. We report that induction of brown adipogenesis by β3-adrenergic receptor (ADRB3) activation involves the death of white adipocytes and their removal by M2-polarized macrophages. Recruited macrophages express high levels of osteopontin (OPN), which attracts a subpopulation of PDGFRα+ progenitors expressing CD44, a receptor for OPN. Preadipocyte proliferation is highly targeted to sites of adipocyte clearance and occurs almost exclusively in the PDGFRα+ CD44+ subpopulation. Knockout of OPN prevents formation of crown-like structures by ADRB3 activation and the recruitment, proliferation, and differentiation of preadipocytes. The recruitment and differentiation of PDGFRα+ progenitors are also observed following physical injury, during matrix-induced neogenesis, and in response to high-fat feeding. Each of these conditions recruits macrophages having a unique polarization signature, which may explain the timing of progenitor activation and the fate of these cells in vivo.
INTRODUCTION
Substantial evidence indicates that adipose tissue dysfunction contributes importantly to the adverse outcomes associated with obesity (Rosen and Spiegelman, 2006). Adipose tissues can exhibit pronounced metabolic and cellular plasticity, and modulation of cellular phenotypes within adipose tissue offers a potential means for therapeutic intervention (Sethi and Vidal-Puig, 2007). For example, peroxisome proliferator-activated receptor gamma (PPARγ) agonists improve metabolic function in part by promoting fatty acid sequestration, upregulating adiponectin secretion, and suppressing macrophage inflammation (Maeda et al., 2001; Odegaard et al., 2007; Tontonoz and Spiegelman, 2008). Conversely, β3-adrenergic receptor (ADRB3) agonists also improve metabolic profiles in rodent models of type 2 diabetes in part by promoting a catabolic phenotype in existing white adipocytes (WAs) (Granneman et al., 2003; Himms-Hagen et al., 2000) and by recruiting mitochondrial uncoupling protein 1 (UCP1)+ multilocular adipocytes from progenitors (Granneman et al., 2005; Lee et al., 2012).
Utilizing chemical and genetic tracing techniques, we recently reported that cells expressing the cell-surface markers platelet-derived growth factor receptor alpha (PDGFRα), CD34, and Sca1 (PDGFRα+ cells) proliferate and differentiate into multilocular UCP1+ brown adipocytes (BAs) in response to ADRB3 agonist treatment (Lee et al., 2012). Lineage tracing demonstrated that PDGFRα+ cells also give rise to unilocular white adipocytes (WAs) in gonadal and inguinal WAT following high-fat feeding (Lee and Granneman, 2012; Lee et al., 2012). These results indicate that PDGFRα+ cells have the potential to become BAs or WAs, depending on the nature of inductive signals.
WAT PDGFRα+ cells have a unique morphology in which dendritic processes contact numerous cell types in the micro-environment (Lee et al., 2012). PDGFRα+ cells with similar morphology are involved in cellular restoration and repair in other tissues (Joe et al., 2010; Uezumi et al., 2010; Zawadzka et al., 2010), and we previously proposed that PDGFRα+ cells might serve a similar function in WAT. To explore mechanisms involved in adipocyte progenitor recruitment, we used a model of WAT remodeling induced by treatment with CL 316,243, a highly selective ADRB3 agonist. ADRB3 are almost exclusively expressed in WAs and BAs (Granneman et al., 1991), and activation of these receptors offers a highly selective means of triggering tissue remodeling. In addition, remodeling proceeds rapidly along a well-defined time course of proliferation and differentiation (Lee et al., 2012), which allows for detailed temporal and spatial analysis of remodeling events.
Tissue macrophages play a critical role in normal development and during tissue remodeling and repair (Pollard, 2009). Macrophages are now recognized as a crucial cellular component of adipose tissue involved in physiologic and pathologic remodeling (Chawla et al., 2011; Lumeng and Saltiel, 2011; Sun et al., 2011), and these diverse effects have been linked to variations in phenotypic polarization. Classically activated (M1) macrophages are recruited to adipose tissue during obese states and appear to promote insulin resistance by triggering local and systemic proinflammatory signaling (Lumeng et al., 2007; Strissel et al., 2007; Weisberg et al., 2003). In contrast, alternatively activated macrophages (M2) enhance insulin sensitivity by PPARγ-dependent mechanisms (Odegaard and Chawla, 2011; Odegaard et al., 2007). Nonetheless, it is largely unknown whether and how macrophages participate in adipogenesis during remodeling and repair.
We report that brown adipogenesis induced by ADRB3 agonist treatment is triggered by the recruitment of macrophages to vulnerable white adipocytes undergoing agonist-mediated cell death. The recruited macrophages express markers that are characteristic of alternatively activated macrophages (M2) and lack expression of classically activated macrophage (M1) markers. PDGFRα+ progenitors concentrate at sites of dead adipocyte clearance, the so-called crown-like structure (CLS), where they proliferate and differentiate into adipocytes. M2 macrophages within CLS express high levels of osteopontin (OPN), which is chemotactic for PDGFRα+ progenitors in vitro. Knockout of OPN prevents ADRB3-induced macrophage recruitment and PDGFRα+ progenitor migration, proliferation, and differentiation. The interaction of PDGFRα+ progenitors and macrophages in CLS is also observed during high-fat feeding and following localized tissue damage, and these structures appear to constitute an adipogenic cell niche for tissue repair and remodeling in adult WAT.
RESULTS
Depot-Specific Adipogenesis Identifies Crown-like Structure as an Adipogenic Niche
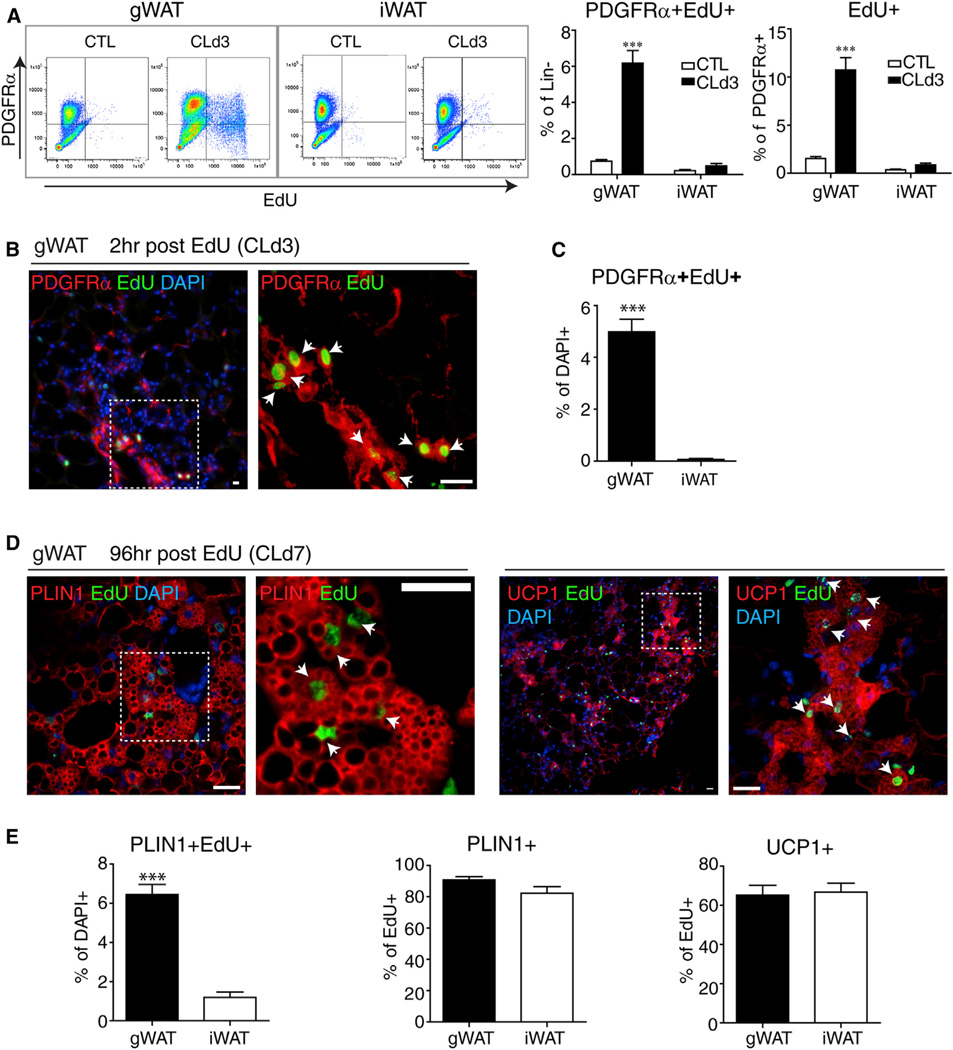
We previously observed that mitogenic responses of PDGFRα+ progenitors varied greatly among adipose tissue depots during ADRB3-mediated remodeling, yet the in vitro proliferation and differentiation of cells isolated from these depots were remarkably similar (Lee et al., 2012). To gain further insight into depot-specific progenitor activation, we characterized proliferating progenitors in gonadal WAT (gWAT) and inguinal WAT (iWAT) by flow cytometry and immunohistochemistry during the course of ADRB3-mediated remodeling. 129S1 mice were continuously infused with CL. On the third day of infusion, mice were injected with EdU to tag dividing cells, and the proliferation and differentiation of labeled cells were assessed 2 or 96 hr later, respectively. Flow cytometric analysis of stromovascular cells (SVCs) obtained from dissociated gWAT confirmed that majority of proliferating cells expressed PDGFRα (Figure 1A and Figures S1A – S1C) and that the proliferation of these cells was markedly higher in gWAT compared to iWAT (Figure 1A and see Figure S1D online). Immunohistochemical analysis confirmed proliferation of PDGFRα+ cells in gWAT on the third day of CL treatment (Figures 1B and 1C) and the differentiation of these cells into multilocular perilipin-1(PLIN1)+ and UCP1+ adipocytes(Figure 1D). Lineage tracing using Pdgfra-CreER confirmed the presence of numerous multilocular adipocytes that were derived from PDGFRα+ progenitors (Figure S1E). Quantification of double-label immunofluorescence demonstrated that CL induced greater proliferation (2 hr post-EdU) (Figure 1C) and differentiation (96 hr post-EdU) of PDGFRα+ cells in gWAT than in iWAT (Figure 1E).Although agonist treatment produced far less proliferation in iWAT, the vast majority of cells that divided on the third day of CL treatment became PLIN1+ adipocytes that expressed UCP1 (Figure 1E). These data indicate that CL produces a depot-specific induction of progenitor proliferation; however, once recruited to divide, progenitors from both depots differentiate into BAs at similarly high frequency.
Figure 1. ADRB3 Stimulation Induces Proliferation and Differentiation of PDGFRα+ Cells in gWAT.
(A) Flow cytometric analysis of depot-specific proliferation of PDGFRα+ cells from gWAT and iWAT of control mice and mice treated with CL for 3 days and injected with EdU 2 hr before analysis. Lineage-negative cells were gated and analyzed for EdU incorporation and PDGFRα expression. Representative density plots and quantification of three independent experiments are shown (values are mean ± SEM; ***p < 0.001).
(B–E) Immunohistochemical analysis of proliferation and differentiation of PDGFRα+ cells in paraffin sections of gWAT and iWAT. Proliferating cells were flashed labeled on the third day of CL treatment, and cellular phenotypes were examined 2 or 96 hr later.
(B) Representative low (left)- and high-magnification (right) images of gWAT paraffin sections stained for EdU and PDGFRα. Most actively dividing cells expressed PGDFRα (arrows).
(C) The level of proliferation of PDGFRα+ cells was significantly greater in gWAT versus iWAT. Data are mean ± SEM, n = 4 (***p < 0.001).
(D) Ninety-six hours after flash labeling, most cells that divided on day 3 have differentiated into multilocular adipocytes with a central nucleus surrounded by PLIN+ lipid droplets and expressed UCP1 (arrows).
(E) Although the rate of proliferation was far greater in gWAT versus iWAT (left), the differentiation of EdU+ cells into PLIN1+ (middle) and UCP1+ (right) multilocular adipocytes did not differ between adipocyte depots. Data are mean ± SEM, n =4 (***p < 0.001). Sections were counterstained with DAPI (blue). Scale bars, 20 µm. See also Figure S1.
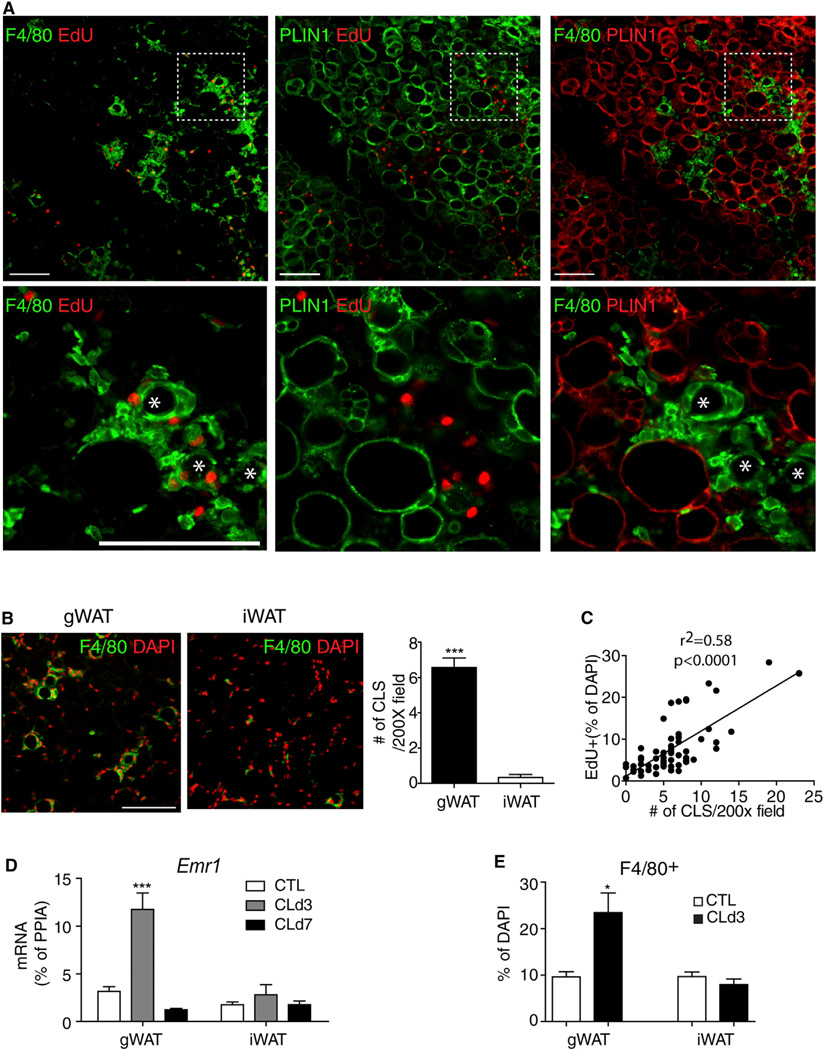
Detailed histological examination at the time of peak proliferation (day 3) demonstrated that dividing cells in gWAT were found near cell clusters that surrounded PLIN1-negative vacancies. These structures were reminiscent of the so-called crown-like structures (CLS) that are observed during efferocytosis of adipocytes (Cinti et al., 2005). Triple staining for F4/80, PLIN1, and EdU confirmed that proliferating cells were near F4/80+ macrophages that surrounded the remains of dead/dying (PLIN1-negative) adipocytes (Figure 2A). Because adipocyte triglycerides are extracted in the process of paraffin embedding, we confirmed the presence of lipid cores surrounded by macrophages in cryosections (Figure S2A) and in whole-mount staining (see below). CLS were rarely found in iWAT (Figure 2B), and the appearance of these structures in gWAT was closely correlated with CL-induced proliferation (Figure 2C). As expected, agonist treatment increased expression of Emr1 (the gene encoding F4/80) and the number of F4/80+ macrophages in gWAT, but not in iWAT, as determined by immunohistochemical and flow cytometric analyses (Figures 2D and 2E and Figures S2B and S2C).
Figure 2. Depot-Specific ADRB3-Mediated Proliferation in WAT Correlates with CLS Formation and Macrophage Recruitment.
(A) Representative images of paraffin sections of gWAT stained for EdU, F4/80, and PLIN1on D3 of CL treatment along with a magnified view of the boxed regions (the bottom row). CLSs are indicated by asterisks in high-magnification fields.
(B) Representative low-magnification images of paraffin sections of gWAT and iWAT stained for F4/80 (left). Quantification of CLS in 200× microscopic fields of paraffin sections of gWAT and iWAT from mice treated with CL for 3 days (mean ± SEM; n = 6, ***p < 0.001). CLS were defined as circular regions with continuous rim of F4/80+ cells.
(C) A positive correlation between CLS number and mitotic index. Symbols represent data from individual microscopic fields, with a minimum 5 of 200× fields of gWAT paraffin sections from individual mice treated with CL for 3 days (n = 6).
(D) Quantitative PCR analysis of Emr1 expression of gWAT and iWAT on D3 and D7 of CL treatment (mean ± SEM; n = 4; ***p < 0.001).
(E) Quantification of F4/80+ cells in paraffin sections of gWAT and iWAT (mean ± SEM; n = 3; *p < 0.05). Scale bars, 100 µm. See also Figure S2.
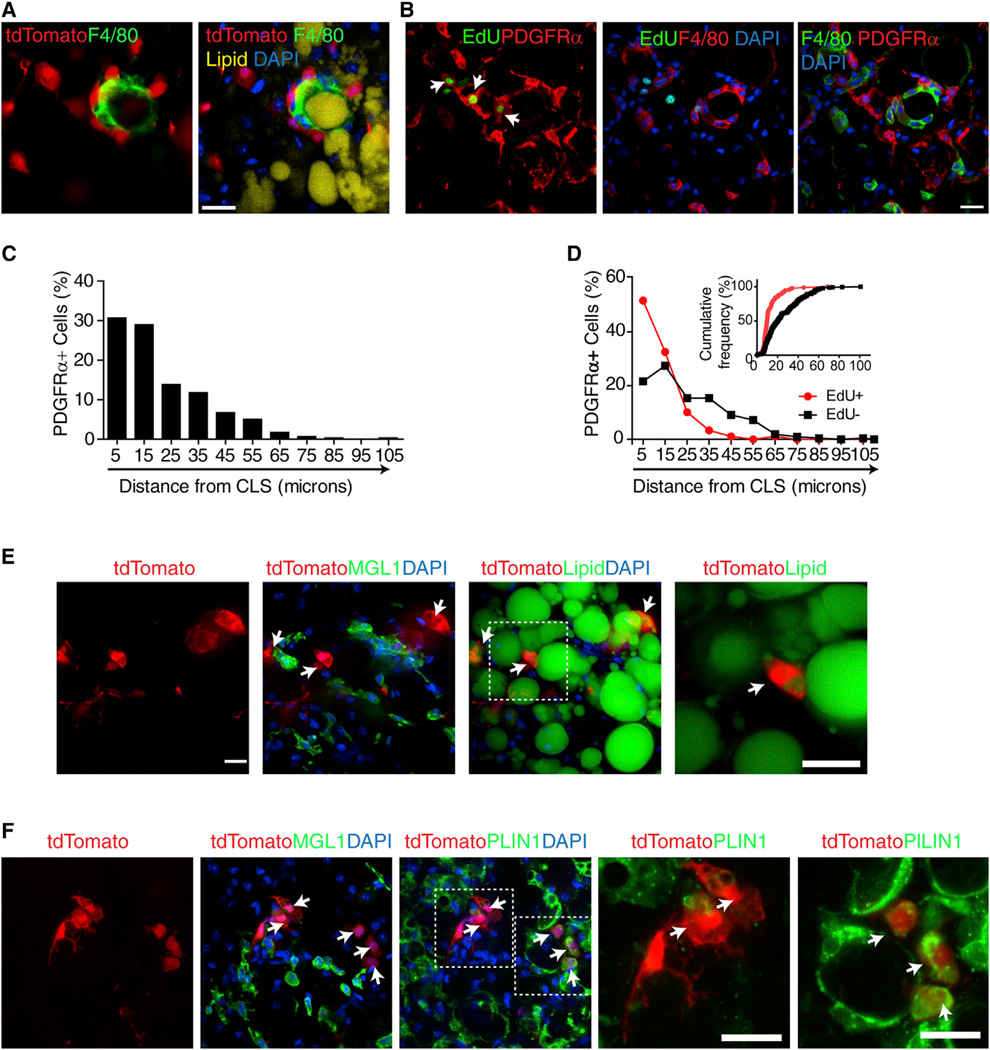
We next characterized the distribution of recruited, proliferating, and newly differentiated PDGFRα+ progenitors by genetic tagging and immunohistochemistry. Genetic tracing was performed by crossing mice harboring a stop codon-floxedtd Tomato reporter gene (R26-LSL-tdTomato) with mice expressing a tamoxifen-sensitive Cre recombinase under the control of the Pdgfra promoter (Pdgfra-CreERT2 mice). Upon treatment with tamoxifen, tdTomato reporter is expressed in PDGFRα+ progenitors, and the migration and fate of these cells can be traced over time. Whole-mount imaging of tdTomato+ cells in gWAT after 3 days of CL treatment showed that PDGFRα+ cells were closely associated with macrophage aggregates as a part of the CLS(Figure 3A). The highest density of PDGFRα+ cells was near CLS, indicating recruitment to these sites. Flash labeling with EdU demonstrated that PDGFRα+ cells proliferate within the CLS (Figure 3B and Figure S3). Indeed, 84% of proliferating PDGFRα+ progenitors were within 20µm of active CLS (Figures 3C and 3D). This distribution differed significantly from nonmitotic PDGFRα+ cells (p < 0.01) and identifies the CLS as a zone of progenitor proliferation. Small newly born (<20 µmdiameter) tdTomato-tagged adipocytes were occasionally observed near CLS (Figures 3E and 3F); however, these adipocyte clusters did not contact active CLS.
Figure 3. PDGFRα+ Progenitors Are Recruited to CLS, where They Form a Zone of Proliferation.
(A) Confocal images of whole-mount gWAT from tamoxifen-induced Pdgfra-CreERT2/tdTomato mice stained for lipid (LipidTox, yellow) and F4/80 (green), showing close association of PDGFRα+ cells with CLS.
(B) F4/80, PDGFRα, and EdU staining in paraffin sections of gWAT from mice treated with CL for 3 days. Left panel merges images of PDGFRα (red) and EdU (green) fluorescence. Arrows indicate proliferating PDGFRα+ cells. Middle panel is the merged image of F4/80 (red) with EdU (green), showing close association between proliferating cells and F480+ macrophages that form CLS.
(C) Frequency distribution of PDGFRα+ cell density near CLS in gWAT from mice treated with CL for 3 days.
(D) Frequency distribution of proliferating and nonproliferating PDGFRα+ cells near CLS. Bins are centered at 10 µm intervals. Insert shows data plotted as distance versus cumulative frequency.
(E and F) Confocal images of cryosectioned gWAT from tamoxifen-induced Pdgfra-CreERT2/tdTomato mice stained for MGL1 and lipid (E) or PLIN1 (F). Magnified views of boxed regions show that tdTomato+ multilocular adipocytes near CLS were labeled with lipid (E) or PLIN1 (F). Double positive cells are indicated by arrows. Scale bars, 20 µm. Nuclei were counterstained with DAPI (blue). See also Figure S3.
Alternatively Activated Macrophages that Express High Levels of Osteoponin Are Recruited to CLS
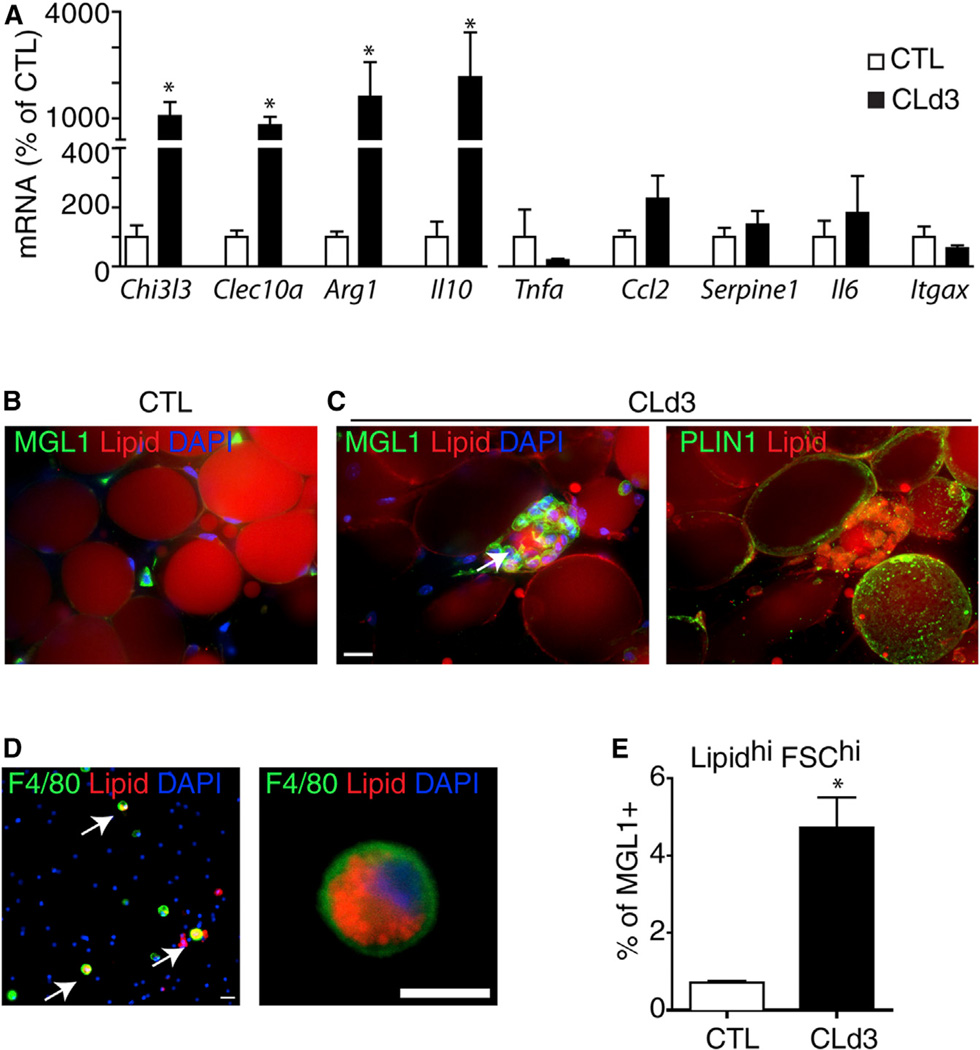
Functionally distinct subtypes of macrophages have been described that range from classically activated proinflammatory macrophages (M1 macrophages) to alternatively activated anti-inflammatory macrophages (M2 macrophages) (Gordon and Taylor, 2005). Expression profiling of gWAT demonstrated that CL treatment strongly upregulated expression of M2 macro-phage markers (Arginase 1, Il10, Chitinase3-like 3, Clec10a), whereas expression levels of M1 markers (Serpine1 [Pai1], Ccl2, Il6, Tnfa, Itgax) were unaffected (Figure 4A). Triple staining for MGL1 (M2 marker, encoded by Clec10a), PLIN1, and lipid in whole-mount gWAT confirmed that M2 macrophages formed CLS and surrounded lipid droplets devoid of PLIN1 (i.e., the remnant adipocyte lipid core) (Figure 4C).
Figure 4. CLSs Induced by ADRB3 Stimulation Contain M2-Polarized Macrophages.
(A) Quantitative PCR analysis of anti- and proinflammatory gene expression in gWAT from control (CTL) and CL-treated mice (CLd3). CL treatment significantly upregulated anti-inflammatory genes (M2 markers) by 3 days, whereas expression of proinflammatory markers was not affected by CL treatment (mean ± SEM; n = 4; *p < 0.05).
(B and C) 3D confocal projection images of whole-mount gWAT from control mice or mice treated with CL for 3 days, stained for lipid (nile red), MGL1 (B and C, left panel, green) or PLIN1 (C, right panel, green). (C) MGL1+ macrophages surround a lipid droplet devoid of PLIN1, forming CLS.
(D) Dissociated gWAT SVC from CL treated-mice (CLd3), stained for lipid (LipidTox, red) and F4/80 (green). Arrows indicate double positive cells. Nuclei were counterstained with DAPI (blue). Scale bars, 20 µm.
(E) Flow cytometric quantification of large (FSC-A hi) MGL+ cells with intense LipidTox staining (Lipidhi) from SVC of control mice and mice treated with CL for 3 days (mean ± SEM; n = 4;*p = 0.014). See also Figure S4.
Macrophages within the CLS were engorged with neutral lipid, indicating that these cells play an active role in clearing of the lipid remains of dead fat cells. Consistent with immunohistochemical analysis, flow cytometry demonstrated that CL treatment induced the appearance of MGL1+ macrophages that were large (forward scatter area, FSC-A), granular (side scatter area, SSC-A), and intensely positive for neutral lipid (LipidTox) (Figures 4D and 4E and Figure S4). As expected, expression of M2 markers, but not M1, and Pparg were enriched in F4/80+ cells isolated from mice treated with CL (Figures S4D and S4E).
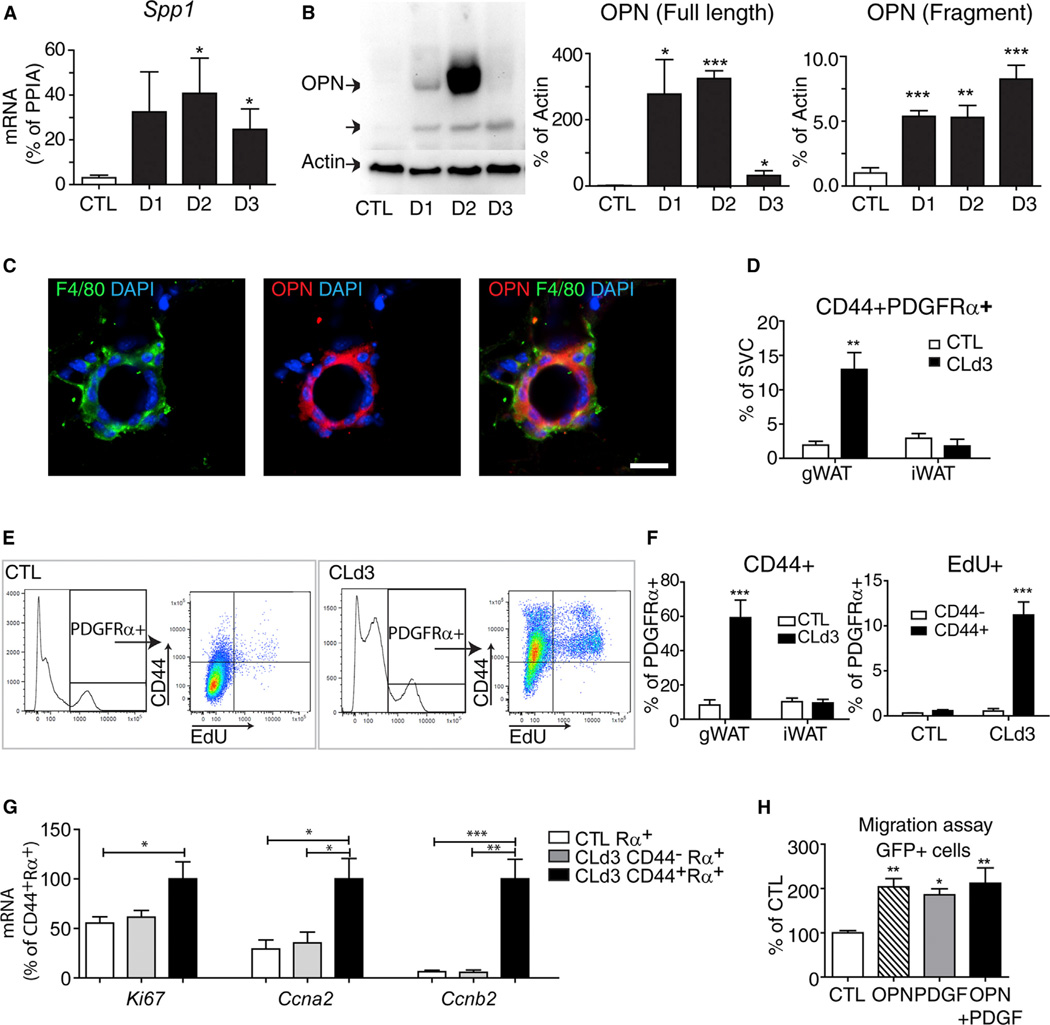
Macrophage trafficking depends on the orchestrated release of chemoattractants, and microarray analysis of gWAT (Granneman et al., 2005; and our unpublished data) indicated that expression of Spp1 (the gene for OPN) was highly upregulated after 3 days of CL treatment. Detailed time course analysis by immunoblot and expression profiling confirmed the dramatic upregulation of mRNA and OPN protein levels (Figures 5A and 5B and Figure S5A). Expression was transient and involved proteolytic processing (Denhardt et al., 2001; Scatena et al., 2007). Upregulation of OPN coincided with macrophage recruitment and preceded maximal activation of PDGFRα+ cell proliferation. High-resolution confocal imaging determined that OPN was highly expressed in macrophages that formed CLS (Figure 5C). Gene expression analysis of fractionated dissociated adipose tissue showed almost exclusive enrichment of Spp1 expression in F4/80+ cells, indicating that macrophages are a major cellular source of OPN (Figure S5B).
Figure 5. ADRB3 Treatment Upregulates OPN Expression in CLS-Associated Macrophages and CD44 Expression in a Subpopulation of PDGFRα-Expressing Cells.
(A) Quantitative PCR analysis of Spp1 expression in gWAT from control mice and mice treated with CL at indicated days after treatment (mean ± SEM; n = 4–6, *p < 0.05).
(B) Immunoblot analysis of gWAT from control mice and mice treated with CL at indicated points. (mean ± SEM; n = 3–5, *p < 0.05, **p < 0.01, ***p < 0.001). CL increased levels of full-length and cleaved OPN.
(C) Immunohistochemical detection of OPN in F4/80+ cells in paraffin sections of gWAT from mice treated with CL for 3 days. Nuclei were counterstained with DAPI. Scale bar, 20 µm.
(D) FACS analysis of PDGFRα+CD44+ cells in SVC obtained from gWAT and iWAT of control mice and CL-treated mice (mean ± SEM; n = 3, **p < 0.01).
(E and F)FACS analysis of CD44 expression and EdU incorporation in PDGFRα+ cells in SVC of gWAT from control mice and mice treated with CL for 3 days. Mice were injected with EdU 2 hr before sacrifice. (E) Representative flow profiles of each condition are shown. (F) Quantification of CD44+PDGFRα+ cells and EdU+CD44+ cells by FACS (mean ± SEM; n = 3; ***p < 0.001).
(G) Quantitative PCR analysis of proliferation maker expression in FACS-isolated cells from gWAT of control mice or mice treated with CL for 3 days (n = 3–4, mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001).
(H) Effect of OPN on in vitro migration of PDGFRα+ cells from Pdgfra-H2BeGFP mice (mean ± SEM; n = 3; *p < 0.05, **p < 0.01). See also Figure S5.
Several OPN receptors have been reported, including integrin family members and CD44 (Denhardt et al., 2001). We found that mRNA levels of CD44 (Figure S5C) were transiently upregulated in a pattern similar to that of OPN but that expression levels of several integrins were unchanged or slightly decreased (Figure S5D). OPN is a well-known chemoattractant for macrophages (Wang and Denhardt, 2008), and flow cytometric analysis confirmed upregulation of CD44 in recruited macrophages (Figures S5E and Figure S6E). Interestingly, we identified a subpopulation of PDGFRα+ cells that also expressed CD44. Importantly, the abundance of this subpopulation expanded dramatically during remodeling (Figure 5D). FACS analysis of EdU incorporation in PDGFRα+ cells demonstrated that proliferation was largely restricted to the subpopulation of PDGFRα+ cells that expressed CD44 2 and 12 hr after EdU injection (Figures 5E and 5F and Figure S5G). As expected, CL selectively upregulated expression of proliferation markers in this subpopulation (Figure 5G). Consistent with a role of OPN in progenitor recruitment, we found that PDGFRα+ cells migrated to OPN in vitro (Figure 5H). Collectively, these results strongly suggest that OPN could be a major chemokine for establishing CLS and the adipogenic tissue niche.
OPN Is Required for CLS Formation and ADRB3-Mediated Adipogenesis from PDGFRα+ Cells
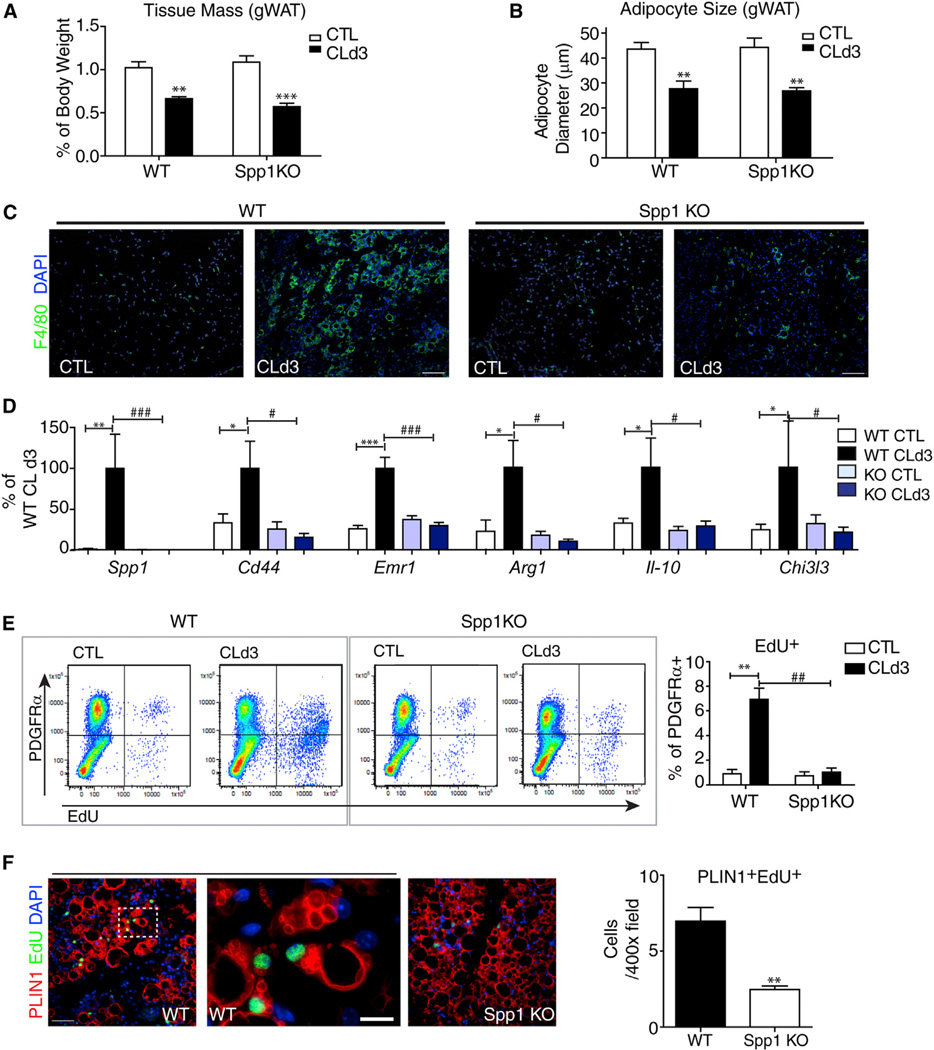
We next used Spp1 knockout mice to investigate the role of OPN in macro-phage recruitment and progenitor activation during CL treatment. Spp1 mutants were maintained on a C57/Bl6 back- ground, so we confirmed that CL induction of OPN expression and progenitor proliferation in wild-type C57/Bl6 mice was similar to that seen in 129S1/SvImJ used above (Figure S6). CL treatment reduced fat tissue mass and adipocyte size equivalently in wild-type and Spp1-KO mice (Figures 6A and 6B). Nonetheless, genetic disruption of OPN expression greatly suppressed recruitment of M2 macrophages and nearly eliminated formation of CLS (Figures 6C and 6D). As expected, CL treatment failed to increase proliferation of PDGFRα+ cells, as assessed by FACS (Figures 6E). As a result of defective macro-phage recruitment during CL treatment, differentiation of adipocytes from proliferating progenitors was greatly reduced in Spp1-KO mice (Figure 6F).
Figure 6. OPN Is Required for CLS Formation and ADRB3-Mediated Adipogenesis from PDGFRα+ Cells.
(A) Reduction in weight of gWAT from WT and Spp1-KO mice after 3 days of CL treatment (mean ± SEM; n = 4–6; **p < 0.01, ***p < 0.001).
(B) Reduction in size of gWAT adipocytes from WT and Spp1-KO mice after 3 days of CL treatment (mean ± SEM; n = 4–6; **p < 0.01).
(C) Immunohistochemical analysis of CLS in paraffin sections of gWAT from wild-type (WT) and Spp1-KO mice under control condition (CTL) or after 3 days of CL treatment (CLd3), stained for F4/80 (green).
(D) Quantitative PCR analysis of macrophage marker expression in gWAT from control and Spp1KO mice (mean ± SEM; n=10–11 from two independent cohorts; *p < 0.05, **p < 0.01, ***p < 0.001).
(E) FACS analysis of EdU incorporation in PDGFRα+ cells from gWAT of WT and Spp1-KO mice treated with CL for 3 days and injected with EdU 2 hr before analysis (mean ± SEM; n = 3–4; **p < 0.01).
(F) Representative images of paraffin sections of gWAT stained for PLIN1 and EdU and quantification of PLIN1+EdU+ cells (mean ± SEM; n = 4; **p < 0.01). Scale bars, 100 µm except high-magnification field in (F), where scale bar, 20 µm. See also Figure S6.
PDGFRα+ Cells Interact with Macrophages and Contribute to Adipogenesis in Response to Environmental, Nutritional, and Physical Stimuli
Adipogenesis can be triggered in adult WAT under numerous conditions, although the cells that contribute to this plasticity have not been defined. To test whether PDGFRα+ cell recruitment is a general feature of WAT tissue restoration, we examined adipogenesis in response to cold stress and high-fat feeding, following sterile tissue damage, and during ECM-induced tissue neogenesis.
Cold-induced upregulation of BA gene expression occurs mainly within fully differentiated adipocytes of subcutaneous white adipose and does not involve recruitment from progenitors (Barbatelli et al., 2010). Nonetheless, we found that cold stress (3 days at 4°C) induced formation of CLS tissues, recruitment of M2 macrophages, and proliferation of PDGFRα+ cells in gWAT (Figures S7A – S7C). The overall pattern of response was qualitatively similar to CL treatment, although the magnitude of macrophage recruitment and proliferation was far less, as would be expected in this sparsely innervated depot.
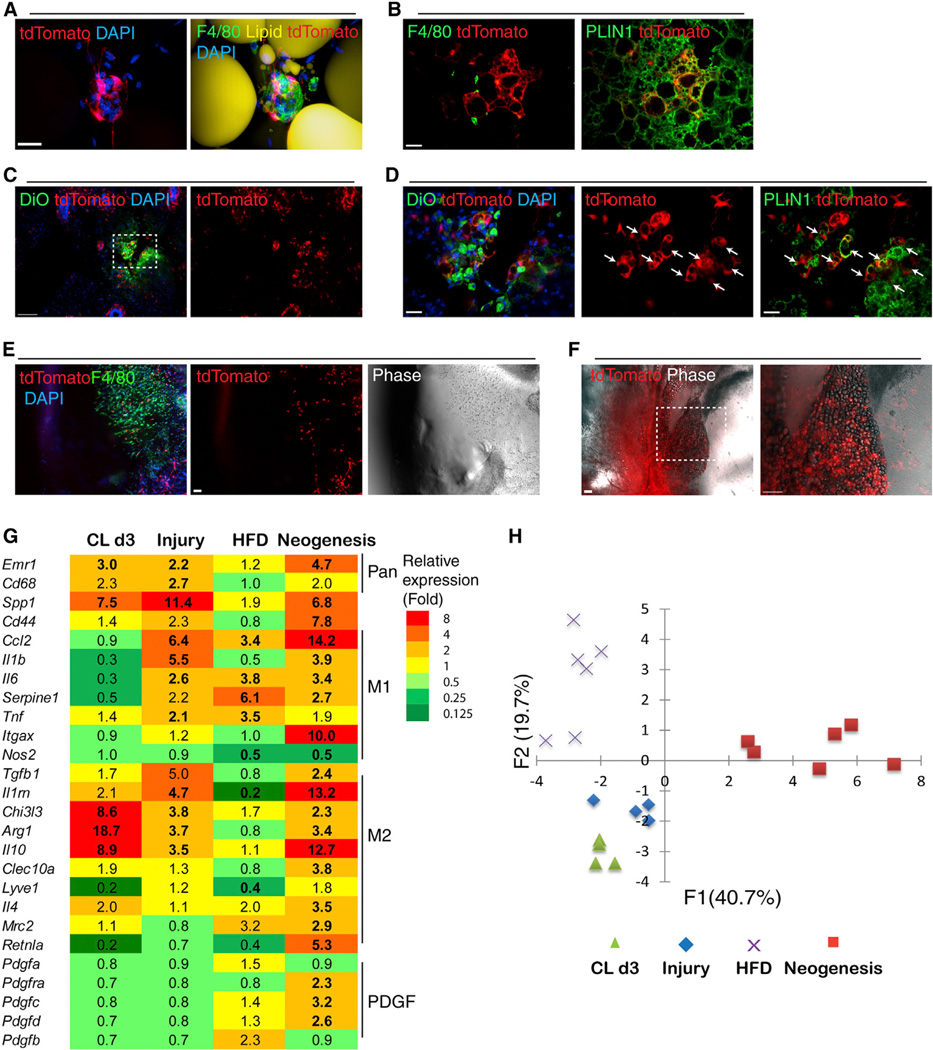
We recently reported that PDGFRα+ cells contribute up to 25% of total adipocytes during 2 months of high-fat feeding (Lee et al., 2012), and that this phenomenon is more pronounced in gWAT versus iWAT. However, it is not known whether adipogenesis from PDGFRα+ cells is related to CLS-mediated inflammation or whether these cells are recruited to active CLS. We found that high-fat feeding induced CLS formation in both gWAT and iWAT; however, the numbers of CLS and the expression of proinflammatory markers were greater in gWAT compared to iWAT (Figure S7D). The frequency of CLS observed after 8 weeks of HFD (Figure S7E) corresponded to the level of adipogenesis from tdTomato+ cells, which is about 2-fold greater in gWAT versus iWAT (Lee and Granneman, 2012). High-resolution imaging clearly demonstrated that PDGFRα+ progenitors were recruited to CLS (Figure 7A and Figure S7F). Newly formed tdTomato+ adipocytes were rarely observed close (100 µm) to active CLS (Figure 7B), suggesting that differentiation might be suppressed by local inflammatory signals.
Figure 7. PDGFRα+ Cells Interact with Macrophages in Adult WAT and Contribute to Adipogenesis during Tissue Repair, Tissue Neogenesis, and Nutritional Hyperplasia.
(A) 3D projection images of CLS in iWAT whole mounts from Pdgfra-CreERT2/tdTomato mice fed HFD for 8 weeks.
(B) tdTomato+ adipocyte clusters that were found in iWAT of HFD-fed mice were not associated with CLS.
(C and D) Needle injury induced the appearance of tdTomato+ adipocyte clusters in iWAT. Shown are cryosections of Pdgfra-CreERT2/tdTomato mice 10 days after needle injury. (D) A magnified view of the boxed region from (C). Injured sites were marked with DiO (green, left). tdTomato+ multilocular adipocytes (arrows) contained PLIN1+ lipid droplets.
(E and F) Representative confocal images of Matrigel plugs 7 days or 4 weeks after injection. (E) tdTomato+ progenitors and F4/80+ macrophages were the major cells infiltrating Matrigel plugs 7 days after injection. (F) Numerous tdTomato+ adipocytes formed 4 weeks after Matrigel injection.
(G) Heatmap showing the unique gene expression profiles of macrophage-associated genes under different adipogenic conditions. Gene expression values are relative to each control condition. (n = 4–6; bold, p < 0.05).
(H) PCA score plot illustrates distinct gene expression profiles across adipogenic conditions (CLd3, gWAT of mice treated with CL for 3 days; injury, iWAT of mice 3 days after needle injury; HFD, gWAT of mice fed HFD for 8 weeks; neogenesis, Matrigel plug 7 days after injection). Scale bars, 20 µm in (A), (B), and (D). Scale bars, 100 µm in (C), (E), and (F). See also Figure S7.
As shown above (Figure 1), iWAT is resistant to CL-induced cell death and de novo adipogenesis; nevertheless, this depot readily responded to localized damage produced by hypodermic needle microinjections with new fat cell formation from PDGFRα+ progenitors. This adipogenesis occurred within 10 days of initial damage and was not observed in untreated pads or in regions of the damaged pad that distal to the microinjection sites (Figures 7C and 7D). We also found that PDGFRα+ cells contribute to ectopic fat pad formation within subcutaneous Matrigel plugs (Figures 7E and 7F). In this model, macrophages and PDGFRα+ cells tagged with tdTomato infiltrated the plug within 1 week (Figure 7E). This migration began from the aspect of the plug that contacted the iWAT pad and was led by tissue macrophages. By 4 weeks, tdTomato+ adipocytes were found throughout the newly formed fat pad (Figure 7F), and based on the efficiency of reporter recombination, we estimate that >40% of adipocytes within the pad were derived from PDGFRα+ cells.
To characterize phenotype and polarization status of macrophages that were recruited during various adipogenic conditions, expression levels of macrophage-associated cytokines and their receptors were analyzed by quantitative PCR (Figure 7G). In comparison to CL-mediated remodeling, injured adipose tissue (3 days after injury) recruited macrophages with mixed M1/M2 phenotypes, which resembled the characteristics of early-phase wound macrophages reported in other tissues (Brancato and Albina, 2011). HFD upregulated expression of M1, but not M2, markers, unlike ADRB3 remodeling (Figure 7G and Figure S7D). Gene expression analysis of Matrigel 7 days after injection confirmed recruitment of macrophages and PDGFRα+ progenitor cells. Principal component analysis (PCA) of macrophage gene expression indicated unique patterns of macrophage polarization under the various adipogenic conditions (Figure 7H). ECM-induced adiponeogenesis was associated with the most distinct pattern of gene expression that was mainly explained by component 1. Several genes (e.g., resistin-like alpha, Retlna; interleukin 1 receptor antagonist, Il1rn) with high loading values to component 1 were identified as potential markers for macrophage subtypes involved in ECM-induced adipose tissue neogenesis. The gene expression profile of macrophages recruited to WAT during CL-mediated remodeling was more similar to injury than HFD. Component 2, which captured this variation, was enriched with M1 markers that were indicative of the proinflammatory characteristics of adipose from HFD-fed mice.
DISCUSSION
Recent work indicates that resident PDGFRα+ cells are committed progenitors of both BA and WA in adult WAT (Berry and Rodeheffer, 2013; Lee et al., 2012); however, the mechanisms involved in activation and recruitment of these progenitors in situ has not been investigated. Under basal conditions, quiescent PDGFRα+ cells are evenly distributed throughout WAT pads, where they extend lengthy dendritic processes that contact numerous cells in the tissue microenvironment. Based on these morphological characteristics, we hypothesized that PDGFRα+ cells sense disturbances in the microenvironment and contribute to tissue repair in adult WAT. In this report, we investigated the mechanisms and location of progenitor recruitment and proliferation, focusing mainly on WAT remodeling induced by ADRB3 activation.
ADRB3 agonists induced greater proliferation and differentiation of PDGFRα+ cells in gWAT versus iWAT. However, fate analysis demonstrated that the main difference between depots involved rates of progenitor proliferation, but not the ability of proliferating cells to differentiate into BAs in the presence of CL. FACS-isolated PDGFRα+ cells from gWAT and iWAT are also similar with respect to clone formation and adipogenicity in vitro (Lee et al., 2012). While intrinsic differences likely exist between PDGFRα+ cells from abdominal and subcutaneous depots (Wu et al., 2012), these observations strongly suggested that differences in tissue microenvironment related to cell activation are most important in determining whether cellular plasticity involves progenitor recruitment, as occurs in gWAT, or metabolic/phenotypic conversion of existing fat cells, as occurs in iWAT.
Detailed histological analysis demonstrated that ADRB3-mediated progenitor proliferation was highly correlated with the appearance CLS. Indeed, individual PDGFRα+ cells accumulated in CLS, where they closely associated with recruited macrophages. Flash labeling with EdU demonstrated that the vast majority of proliferating PDGFRα+ cells were within 20 microns of CLS and formed a zone of proliferation at the site of adipocyte efferocytosis. These observations strongly suggested that PDGFRα+ cells are mobile and sense chemotactic and mitogenic signals emanating from the CLS microenvironment.
Macrophages are a major cellular component of CLS generated by HFD (Cinti et al., 2005). The number of F4/80+ macrophages in gWAT and expression of macrophage-specific markers increased after the first day of agonist treatment, before significant proliferation of PDGFRα+ cells (Lee et al., 2012). CL treatment increased the number of EdU-labeled macrophages present in gWAT SVC (by FACS) and in CLS (by immunohistochemistry); however, it is presently unclear whether the increase in WAT macrophages involves mainly recruitment from circulation or proliferation of resident cells. As expected, agonist treatment did not affect macrophage number or gene expression in iWAT, where de novo adipogenesis was low. Confocal imaging of whole-mount gWAT demonstrated that PDGFRα+ progenitors contacted lipid-filled macrophages that surrounded the lipid remains of dead fat cells. Interestingly, macrophage recruitment was associated with strong upregulation of markers of alternative activation (M2), including Arg1, Chi3l3, and Il10, whereas proinflammatory markers were either unchanged or slightly reduced. These observations indicate that ADRB3-mediated adipogenesis involves recruitment of macrophages that mediate noninflammatory tissue repair (Murray and Wynn, 2011).
Tissue-remodeling macrophages express a complex mixture of cytokines, matrix remodeling enzymes, and growth factors that could be involved in progenitor migration, proliferation, and differentiation (Pollard, 2009; Sica and Mantovani, 2012). CL upregulated Spp1 expression and OPN protein on the first day of treatment, before significant progenitor proliferation, and high levels of OPN were restricted to macrophages associated within CLS. OPN is a well-known chemotactic factor for macrophage recruitment through its receptor CD44 (Denhardt et al., 2001; Ponta et al., 2003; Zhu et al., 2004). Interestingly, CD44 was also present in a subpopulation of PDGFRα+ cells in the basal state, and this population nearly tripled in number in gWAT following CL treatment. In contrast, ADRB3 treatment had no effect on the number or distribution of CD44+ PDGFRα+ cells in iWAT, where Spp1 was not induced and de novo adipogenesis was minimal. We found that OPN was chemotactic for PDGFRα+ cells in vitro, and the vast majority of proliferating progenitors in gWAT expressed CD44. Together, these results identify PDGFRα+CD44+ cells as a subpopulation of progenitors that is poised to respond to macrophage-derived signals.
We directly tested involvement of OPN and found that the recruitment of alternatively activated macrophages and the migration, proliferation, and differentiation of PDGFRα+ cells were severely attenuated in Spp1 null mice. CLS number was also greatly reduced, indicating that recruited macrophages promote the death of vulnerable WA. Spp1 deletion did not affect the expected reduction in fat mass and adipocyte size, indicating that OPN did not directly affect ADRB3 signaling. These results indicate that macrophage and progenitor recruitment require OPN; however, progenitor proliferation and differentiation may involve additional signals, such as release of macro-phage-derived and matrix-bound growth factors (Pollard, 2009), and lipid metabolites. In this regard, we have previously shown that PPARα ligands derived from lipolysis promote catabolic remodeling of gWAT, and that in the absence of PPARα, ADRB3 agonists trigger persistent adipose tissue inflammation (Li et al., 2005).
OPN has previously been implicated in recruitment of macrophages to adipose tissue during HFD. This recruitment is associated with systemic inflammation and insulin resistance (Chapman et al., 2010; Nomiyama et al., 2007), which are largely corrected by genetic deletion of Spp1. Although OPN appears to be a critical chemokine for macrophage recruitment in both CL and HFD models, polarization to M1 or M2 status within the CLS clearly differs and is likely influenced by additional factors such as levels of lipid mediators and endotoxin (Cani et al., 2007; Kosteli et al., 2010). Because adipose tissue develops normally in Spp1 null mice, OPN involvement might be restricted to conditions of adipose tissue restoration and repair.
Pharmacological induction of WAT remodeling by ADRB3 agonists has certain experimental advantages for investigating in vivo adipogenesis, including stimulus control and specificity, and rapid time course. We investigated whether recruitment of PDGFRα+ progenitors was a general feature of adipogenesis by genetic tracing under diverse adipogenic conditions. Adipogenesis from PDGFRα+ cells was observed in response to physical and nutritional stimuli, and could be induced by exposure to extracellular matrix. In each case, progenitor recruitment was associated with, or preceded by, macrophage recruitment. Interestingly, the phenotypic character of recruited macrophages varied according to activating stimulus.
CL treatment and high-fat feeding both result in fat cell death and clearance by macrophages (Cinti et al., 2005; Lumeng et al., 2007; Strissel et al., 2007). Interestingly, gWAT is more susceptible than iWAT to CLS formation induced by diet (Strissel et al., 2007) or drug treatment, and this susceptibility strongly correlates to greater adipogenesis from PDGFRα+ progenitors (Lee and Granneman, 2012; Lee et al., 2012). While macrophage-mediated remodeling appears to underlie adult adipogenensis under both conditions, there are interesting differences in these models that may be instructive. One obvious difference is that CLS induced during CL treatment resolves quickly (by 7 days), and newly differentiated UCP1+ adipocytes can be found near active structures. In contrast, CLS formation persists for weeks or months during HFD, and newly differentiated adipocytes, while in clusters, are not found near active CLS. We suggest that the transient versus chronic nature of tissue remodeling involves differences in immune cell recruitment, and specifically the fact that high-fat feeding strongly upregulates proinflammatory cytokines that are indicative of M1 polarization, but CL-induced remodeling does not. Persistent M1 activation is known to delay tissue restoration following damage (Murray and Wynn, 2011), and the high expression of cytokines like TNF-α is known to suppress adipogenesis in vitro (Xu et al., 1999). It thus seems likely that high-fat feeding produces metabolic wounds to WAT that largely fail to heal.
In summary, recruitment of PDGFRα+ progenitors is a common feature of de novo adipogenesis induced by pharmacological, nutritional, and physical provocation to adult WAT. In the case of ADRB3-mediated remodeling, this involves a subpopulation of progenitors that express CD44 and are responsive to macrophage-derived OPN. WAT macrophages assume a range of phenotypes under various adipogenic conditions and play a critical role in establishing microenvironments that are conducive to resolution or persistent inflammation.
EXPERIMENTAL PROCEDURES
Mice
129S1/SvImJ (129S1; stock number 002448), C57BL/6J (C57B/6; stock number 000664), B6.Cg-Gt(ROSA)26Sor_tm9(CAG-tdTomato)Hze/J (Madisen et al., 2010) (R26-LSL-tdTomato; stock number 007909), B6.129S4-Pdgfra_tm11(EGFP)Sor/J (Pdgfra-H2BeGFP; stock number 007669), and B6.129S6(Cg)-Spp1tm1Blh/J (Spp1KO; stock number 004936) were purchased from the Jackson Laboratory. Pdgfra-CreERT2 mice (Rivers et al., 2008) were obtained from William Richardson (University College London). All animal protocols were approved by the Institutional Animal Care and Use Committee at Wayne State University.
For continuous ADRB3 stimulation, mice were infused with CL (0.75 nmol/hr) by osmotic minipumps (ALZET) for up to 7 days. For EdU flash labeling, mice were injected with EdU (Invitrogen, 2 nmol/mouse, i.p.) at times indicated in the text. Cre recombination in double transgenic mice (Pdgfra-CreERT2/R26-LSL-tdTomato) was induced by administering tamoxifen (Sigma, 300 mg/kg, P.O.) on each of 5 consecutive days. CL treatment or HFD was started 1 week after the first dose of tamoxifen. For high-fat diet experiment, 60% fat diet (Research Diet D12492) was introduced at 5–6 weeks of age and continued for 8 weeks. For cold exposure, 129S1 mice (three mice per cage) were kept at 4°C for 3 days or maintained at room temperature as controls. Mice were injected with EdU 2 hr before sacrifice.
Matrix-induced WAT neogenesis was performed as previously described (Kawaguchi et al., 1998). Briefly, 250 µl cold Matrigel (BD Biosciences) was injected subcutaneously into the hind limb regions of tamoxifen-induced Pdgfra-CreERT2/R26-tdTomato mice. After 1 or 4 weeks, the plugs were collected and analyzed for gene expression and histology.
Localized cellular injury in WAT was induced by repeated (∼10 times/pad) hypodermic needle microinjection of PBS containing Vibrant DiO (Invitrogen, 1:100) to identify sites of damage. Tissues were collected 3 or 10 days after injection and analyzed for gene expression and histology.
Tissue Processing and Histology
Tissues were processed for histological sections or whole-mount tissue and subjected to immunohistochemical analysis, as previously described (Lee et al., 2012). Paraffin sections of iWAT and gWAT (5 µm thick) were analyzed for CLS number and adipocyte size. CLS was defined as circular region surrounded by F4/80 cells in paraffin sections stained for F4/80 and DAPI. CLS was counted in at least 5 of 200× fields from each mouse, and the mean of CLS number/200× field obtained from individual mice was used to compare CLS density between iWAT and gWAT. The values of CLS number/200× field and EdU/DAPI (%) from the same field were analyzed to correlate CLS number and mitotic index. Adipocyte size was measured in paraffin sections stained for hematoxylin and eosin, and was calculated as the mean diameter of at least 100 random adipocytes in 5 of 200× fields per each sample. The level of EdU incorporation in PDGFRα+cells or PLIN1 cells were determined by counting at least 400 cells per each mouse. All quantification of histologic samples was carried out as blind analyses.
Nile red (Sigma, 1 µg/ml) or LipidTox (Invitrogen, 1:300) was used for lipid staining. EdU was detected according to the instructions of the manufacturer (Invitrogen).
WAT Stromovascular Cell Fractionation and Flow Cytometry
For FACS analysis, SV fractions from mouse WAT were isolated, as previously described (Lee et al., 2012). Lipid staining (LipidTox, 1:1000) was performed on cells following live-cell-surface marker staining and fixation. For EdU detection, fixed SVCs were processed for Click-it reaction first, followed by cell-surface marker staining. Anti-CD45-PE/Cy7 and CD11b–PE/Cy7 antibodies (Biolegend, rat, 1:300) were used to exclude hematopoietic lineage. Flow cytometry was performed at the Microscopy, Imaging, and Cytometry Resources Core of Wayne State University. Cell sorting and analytic cytometry were using FACS Vantage SE SORP and BD LSR II (BD Biosciences) flow cytometers, respectively. All compensation was performed using single color controls in BD FACS DiVa software at the time of acquisition. Representative cell density plots of negative and positive controls were provided to show that fluorescence signals from single color controls do not overlap with any other color channels (in supplemental information). Raw data were processed using FlowJo software (Tree Star) or CellQuest (BD Biosciences). Alternatively, dissociated adipose tissue was fractionated for gene expression analyses by magnetic sorting with anti-F4/80-FITC/ anti-FITC-microbeads and anti-PDGFRα-PE/anti-PE-microbeads (Miltenyi Biotech).
Statistical Analysis
Statistical analyses were performed with GraphPad Prism 5. Data are presented as mean ± SEM. Statistical significance between two groups was determined by unpaired t test or Mann-Whitney test, as appropriate. Comparison among groups was performed using one-way ANOVA or two-way ANOVA, with Bonferroni posttests to determine the relevant p values. Statistical significance in distance analysis was evaluated by nonlinear regression. PCA was performed with XLStat to detect the common variations between variables and to visualize clusters of correlated observations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. W. Richardson for providing Pdgfra-CreER mice, and Dr. C.M. Giachelli for providing OPN antibody. We thank Drs. T. Leff, R. MacKenzie, J. Wang, and members of CIMER for discussions. We also thank E. Van Buren and J. Back for technical support with FACS analyses. This study was supported by NIH grants (RO1DK62292 and RO1DK76629) to J.G.G. The Microscopy, Imaging, and Cytometry Resources Core was supported, in part, by NIH Center grant P30CA022453 to the Karmanos Cancer Institute, Wayne State University, and the Perinatology Research Branch of the National Institutes of Child Health and Development, Wayne State University.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, one table, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2013.08.003.
REFERENCES
- Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K, Giacobino JP, De Matteis R, Cinti S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte trans differentiation. Am. J. Physiol. Endocrinol. Metab. 2010;298:E1244–E1253. doi: 10.1152/ajpendo.00600.2009. [DOI] [PubMed] [Google Scholar]
- Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancato SK, Albina JE. Wound macrophages as key regulators of repair: origin, phenotype, and function. Am. J. Pathol. 2011;178:19–25. doi: 10.1016/j.ajpath.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Chapman J, Miles PD, Ofrecio JM, Neels JG, Yu JG, Resnik JL, Wilkes J, Talukdar S, Thapar D, Johnson K, Sears DD. Osteopontin is required for the early onset of high fat diet-induced insulin resistance in mice. PLoS ONE. 2010;5:e13959. doi: 10.1371/journal.pone.0013959. http://dx.doi.org/10.1371/journal.pone.0013959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, Goh YPS. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS, Obin MS. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res. 2005;46:2347–2355. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- Denhardt DT, Noda M, O’Regan AW, Pavlin D, Berman JS. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J. Clin. Invest. 2001;107:1055–1061. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Burnazi M, Zhu Z, Schwamb LA. White adipose tissue contributes to UCP1-independent thermogenesis. Am. J. Physiol. Endocrinol. Metab. 2003;285:E1230–E1236. doi: 10.1152/ajpendo.00197.2003. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Lahners KN, Chaudhry A. Molecular cloning and expression of the rat beta 3-adrenergic receptor. Mol. Pharmacol. 1991;40:895–899. [PubMed] [Google Scholar]
- Granneman JG, Li P, Zhu Z, Lu Y. Metabolic and cellular plasticity in white adipose tissue I: effects of b3-adrenergic receptor activation. Am. J. Physiol. Endocrinol. Metab. 2005;289:E608–E616. doi: 10.1152/ajpendo.00009.2005. [DOI] [PubMed] [Google Scholar]
- Himms-Hagen J, Melnyk A, Zingaretti MC, Ceresi E, Barbatelli G, Cinti S. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol. 2000;279:C670–C681. doi: 10.1152/ajpcell.2000.279.3.C670. [DOI] [PubMed] [Google Scholar]
- Joe AWB, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FMV. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi N, Toriyama K, Nicodemou-Lena E, Inou K, Torii S, Kitagawa Y. De novo adipogenesis in mice at the site of injection of basement membrane and basic fibroblast growth factor. Proc. Natl. Acad. Sci. USA. 1998;95:1062–1066. doi: 10.1073/pnas.95.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosteli A, Sugaru E, Haemmerle G, Martin JF, Lei J, Zechner R, Ferrante AW., Jr Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J. Clin. Invest. 2010;120:3466–3479. doi: 10.1172/JCI42845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-H, Granneman JG. Seeking the source of adipocytes in adult white adipose tissues. Adipocyte. 2012;1:230–236. doi: 10.4161/adip.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-H, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zhu Z, Lu Y, Granneman JG. Metabolic and cellular plasticity in white adipose tissue II: role of peroxisome proliferator-activated receptor-α. Am. J. Physiol. Endocrinol. Metab. 2005;289:E617–E626. doi: 10.1152/ajpendo.00010.2005. [DOI] [PubMed] [Google Scholar]
- Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J. Clin. Invest. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Takahashi M, Funahashi T, Kihara S, Nishizawa H, Kishida K, Nagaretani H, Matsuda M, Komuro R, Ouchi N, et al. PPARgamma ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes. 2001;50:2094–2099. doi: 10.2337/diabetes.50.9.2094. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama T, Perez-Tilve D, Ogawa D, Gizard F, Zhao Y, Heywood EB, Jones KL, Kawamori R, Cassis LA, Tschöp MH, Bruemmer D. Osteopontin mediates obesity-induced adipose tissue macrophage infiltration and insulin resistance in mice. J. Clin. Invest. 2007;117:2877–2888. doi: 10.1172/JCI31986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu. Rev. Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard JW. Trophic macrophages in development and disease. Nat. Rev. Immunol. 2009;9:259–270. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat. Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scatena M, Liaw L, Giachelli CM. Osteopontin: a multifunctional molecule regulating chronic inflammation and vascular disease. Arterioscler. Thromb. Vasc. Biol. 2007;27:2302–2309. doi: 10.1161/ATVBAHA.107.144824. [DOI] [PubMed] [Google Scholar]
- Sethi JK, Vidal-Puig AJ. Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J. Lipid Res. 2007;48:1253–1262. doi: 10.1194/jlr.R700005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strissel KJ, Stancheva Z, Miyoshi H, Perfield JW, DeFuria J, II, Jick Z, Greenberg AS, Obin MS. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes. 2007;56:2910–2918. doi: 10.2337/db07-0767. [DOI] [PubMed] [Google Scholar]
- Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J. Clin. Invest. 2011;121:2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu. Rev. Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S-i, Yamamoto N, Takeda Si, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- Wang KX, Denhardt DT. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev. 2008;19:333–345. doi: 10.1016/j.cytogfr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sethi JK, Hotamisligil GS. Transmembrane tumor necrosis factor (TNF)-α inhibits adipocyte differentiation by selectively activating TNF receptor 1. J. Biol. Chem. 1999;274:26287–26295. doi: 10.1074/jbc.274.37.26287. [DOI] [PubMed] [Google Scholar]
- Zawadzka M, Rivers LE, Fancy SP, Zhao C, Tripathi R, Jamen F, Young K, Goncharevich A, Pohl H, Rizzi M, et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell. 2010;6:578–590. doi: 10.1016/j.stem.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Suzuki K, Goldberg HA, Rittling SR, Denhardt DT, McCulloch CAG, Sodek J. Osteopontin modulates CD44-dependent chemotaxis of peritoneal macrophages through G-protein-coupled receptors: evidence of a role for an intracellular form of osteopontin. J. Cell. Physiol. 2004;198:155–167. doi: 10.1002/jcp.10394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.