Abstract
The sinoatrial node (SAN) is essential for rhythmic beating of the heart; however, our understanding of what controls proper functioning of the SAN remains primitive. To explore molecular control of SAN function, we specifically deleted Baf250a, a key regulatory component of the ATP-dependent chromatin remodeling complex SWI/SNF, in the SAN. Deletion of Baf250a in the SAN led to sinus bradycardia. Time series analysis of dysregulated genes after deletion of Baf250a reveals a transcriptional hierarchy maintaining pacemaker cell identity, i.e., Baf250a activates the expression of Tbx3, and Baf250a, Tbx3 and histone deacetylase 3 coordinately repress the expression of Nkx2.5. Disruption of this repressive pathway switches on expression of Nkx2.5, which stimulates expression of Gata4 and Tbx5. These three cardiac transcription factors further turn on a contractile cardiomyocyte program in the SAN, which eventually leads to sick sinus disease (SSD). Our study suggests that disruption of key genetic pathways regulating cardiac lineage segregation may cause SSD and cardiac arrhythmias in general.
Keywords: chromatin, epigenetics, transcription
Introduction
Rhythmic contraction of cardiac chambers is essential for efficient blood circulation in the body. Throughout life, the heart contracts 2.5 billion times. This restless activity originates from a tiny pool of specialized cardiomyocytes within the sinoatrial node (SAN), a structure in the right atrium at the junction with the superior vena cava1,2,3. A variety of etiological factors can lead to sick sinus disease (SSD), a collection of disorders defined by abnormal electrical impulse formation and propagation from the SAN4. This disease can affect people of all ages, but mostly occurs in the elderly. The older population is projected to increase to 20% within next two decades in Western countries, thus elucidating the mechanisms underlying SSD is urgent5.
The automaticity of the SAN and its molecular control mechanism have been topics of intense interest. Decades of cellular electrophysiological studies and Ca2+ signaling revealed critical roles of ion channels and Ca2+ signaling in setting the automaticity6,7,8. The critical roles of ion channels in the SAN have been demonstrated by human genetic studies in which mutations altering ion channels and their trafficking can cause SSD in patients9,10,11. Even though the ionic basis of automaticity is relatively well understood, the molecular control of this automaticity remains exclusively unknown. So far, only a few transcription factors including Tbx3, Tbx18, and Shox2 have been demonstrated to play important roles in the SAN12,13,14,15,16.
We are interested in the transcriptional regulatory circuits controlling function of the SAN. Transcription is closely associated with accessibility of DNA. In eukaryotic cells, DNA is hierarchically packaged into nucleosomes and higher-order chromatin, which create barriers, as DNA must be accessible for critical nuclear events such as transcription17. Chromatin remodeling factors, histone acetylases, and histone deacetylases (HDACs) play key roles in regulating gene expression via regulating DNA accessibility18,19,20. The SWI/SNF (switch/sucrose nonfermentable) is an evolutionarily conserved, ATP-dependent chromatin remodeling complex. It acts with transcription factors such as nuclear receptors and myogenic transcription factors to regulate specific transcriptional programs21,22. The critical roles of SWI/SNF chromatin remodeling complex have been exemplified by Baf250a, a key component of SWI/SNF family. Genetic deletions have demonstrated that Baf250a plays key roles in maintaining embryonic stem cell pluripotency and in cardiac development and function23,24. To explore the role of Baf250a in the SAN, we specifically deleted Baf250a using Hcn4-CreERT2 BAC transgenic mice25. Our data revealed that Baf250a orchestrates a critical hierarchy involving Tbx3 and HDAC3 to specifically repress expression of Nkx2.5 in the SAN.
Results
SAN-specific deletion of Baf250a led to sinus bradycardia
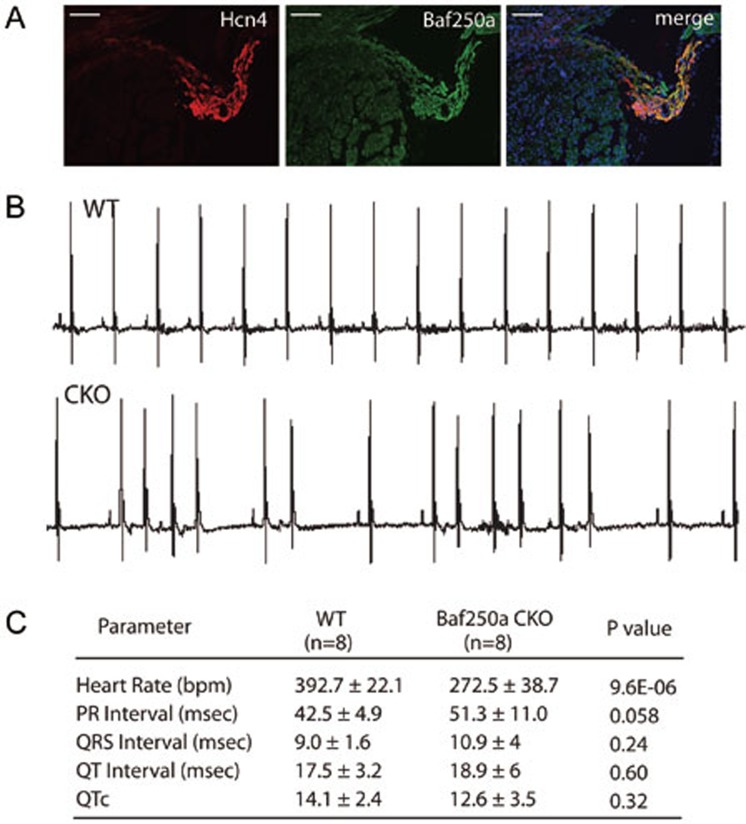
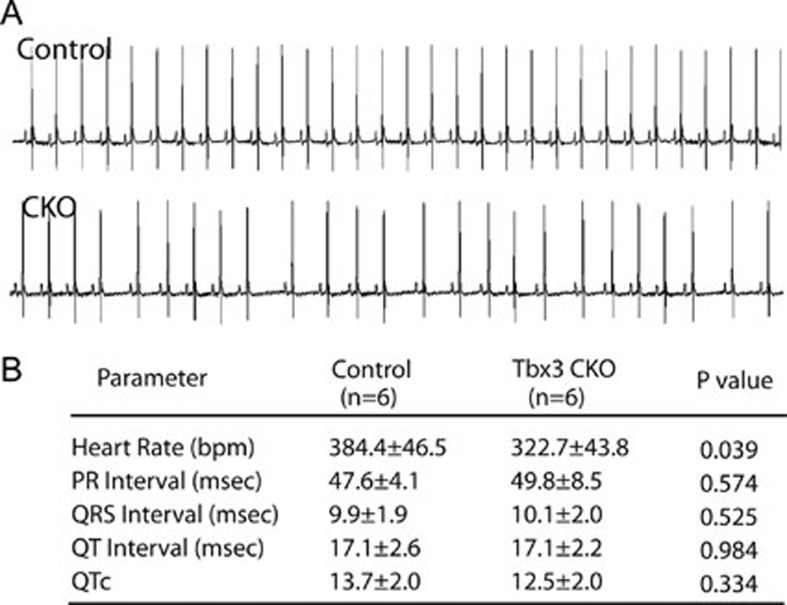
Baf250a is highly expressed in the SAN as demonstrated by double immunohistochemistry (IHC; Figure 1A). To ask whether Baf250a is required for the function of the SAN, we deleted Baf250a in the SAN by using our recently developed Hcn4-CreERT2 BAC transgenic mice. Adult Hcn4-CreERT2;Baf250af/f mice at the age of 3 weeks (henceforth referred to as Baf250a CKO) were injected with tamoxifen and then, surface EKGs were performed to examine the function of the SAN. EKG examination revealed normal PR intervals and QRS morphology in the Baf250a CKO mice; however, PP intervals were irregular, suggesting that the SAN was not discharging regularly (Figure 1B and 1C).
Figure 1.
Specific deletion of Baf250a in the SAN caused sinus bradycardia. (A) Expression of Baf250a in the SAN. Double-label fluorescent immnohistochemistry at postnatal day 21 (P21) to show expression of Hcn4 (red) and Baf250a (green). Scale bar, 100 μm. (B) Representative surface EKG tracing in lead II. (C) EKG parameters of littermate control and Baf250a CKO at P21. Bpm, beats per minute; msec, milliseconds; QTc, corrected QT interval.
To further confirm whether Baf250a CKO mice suffered irregular sinus rhythm, we performed ambulatory EKG recording from conscious unstressed mice for 3 consecutive days (three control and three Baf250a CKO mice). Similarly to what we observed using surface EKG, the freely moving Baf250a CKO mice exhibited irregular sinus rhythm as demonstrated by telemetry EKG (Supplementary information, Figure S1). These data confirmed that Baf250a is required for maintaining proper function of the SAN.
Transcriptional regulatory cascades revealed by deletion of Baf250a
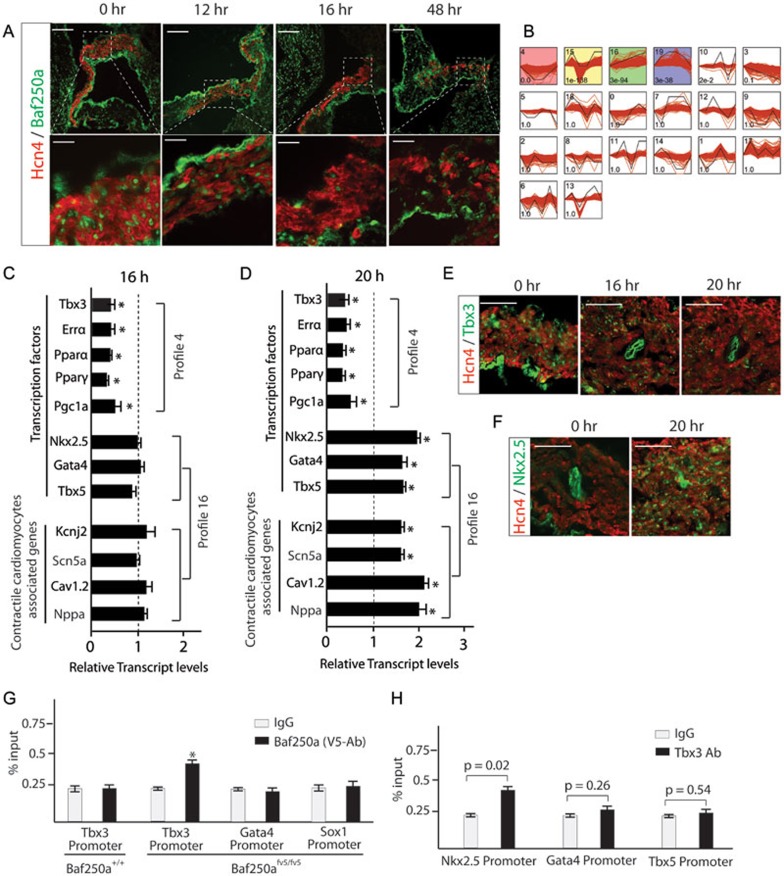
To identify direct transcriptional targets of Baf250a, we first examined protein level of Baf250a in the SAN after tamoxifen induction. Tamoxifen concentration peaks in blood at 6 h and tamoxifen-induced gene deletion are completed within 12 h26. About 80% of Baf250a protein remained in the SAN at 12 h after tamoxifen treatment. However, the protein was decreased by more than 80% at 16 h post administration (Figure 2A). Therefore, dysregulated genes at 16 h would reflect the direct consequence of decreased Baf250a protein, whereas those after 16 h may be regulated by direct transcriptional targets of Baf250a.
Figure 2.
Transcriptional regulatory cascades revealed by deletion of Baf250a in SAN. (A) Protein level of Baf250a in the SAN after tamoxifen induction in Baf250af/f;Hcn4CreERT2 mice. Scale bar, 100 μm (top panel), and 25 μm (bottom panel). (B) Unbiased clustering of time course expression data. (C, D) qPCR validated relative transcript levels of representative genes in profiles 4 and 16 (n = 3 in each group). *P < 0.05. (E) Protein levels of Tbx3 after deletion of Baf250a in the SAN. Scale bar, 50 μm. (F) Enhanced expression of Nkx2.5 after deletion of Baf250a in SAN. Scale bar, 50 μm. (G) Direct binding of Baf250a to the promoter of Tbx3 as examined by ChIP-qPCR (n = 3 in each group). *P < 0.05. (H) Binding of Tbx3 to the promoters of Nkx2.5, Gata4, and Tbx5 as examined by ChIP-qPCR (n = 3 in each group).
Taking turnover of Baf250a protein into account, we performed massively parallel sequencing (mRNA-seq) analysis using SAN samples collected at 0, 16, 20, 24, 48, and 144 h (6 days) time points. mRNA-seq revealed that numerous genes were dysregulated in a sequential manner (Supplementary information, Tables S1,S2,S3,S4,S5). Unsupervised clustering analysis uncovered the sequential transcriptional events27. Genes clustered in a same profile are likely co-regulated, and thus are under the control of the same transcription modules. Clustering analysis across all six time points revealed 20 profiles among which four profiles were significant (Figure 2B). Gene ontology (GO) enrichment analysis revealed that energy metabolism and contractile cardiomyocyte-related terms were most enriched in profiles 4 and 16, respectively. The significantly enriched GO terms and their profiles were listed in Supplementary information, Tables S6,S7,S8.
Genes including Tbx3 in the profile 4 were downregulated at 16 h as confirmed by quantitative real-time PCR (qPCR; Figure 2C), suggesting that they are direct transcriptional targets of Baf250a. Different from profile 4, genes in profile 16 did not show expression level changes until 20 h and their expression continued to be upregulated at least until 6 days (Figure 2C and 2D and Supplementary information, Tables S1,S2,S3,S4,S5). Consistent with GO term analysis, genes in this profile are associated with the contractile cardiomyocyte program. For example, Nkx2.5, Gata4, and Tbx5 are well known to play important roles in myocardium development28. Kcnj2, Cav1.2, Scn5a, and Nppa are highly expressed in the atrial myocardium but normally are not expressed or expressed at low levels in the SAN2. The time course-dependent decrease of Tbx3 and increase of Nkx2.5 were further validated by IHC (Figure 2E and 2F). Tbx3 is believed to be a transcriptional repressor18. The sequential expression changes of these genes after deletion of Baf250a suggested a model in which Baf250a activates the expression of Tbx3 and, when Baf250a levels fall, the resulting decreased levels of Tbx3 fails to repress the expression of Nkx2.5, and probably the expression of Gata4 and Tbx5 as well.
To test this model, we first determined whether Tbx3 is a direct transcriptional target of Baf250a using chromatin immunoprecipitation coupled with qPCR (ChIP-qPCR). We found that the currently available commercial Baf250a antibodies were not suitable for ChIP-qPCR assay. In order to overcome this hurdle, we generated Baf250afV5/+ mice in which a DNA fragment encoding V5 tag was inserted in-frame with the endogenous coding region of Baf250a (Supplementary information, Figure S2A and S2B). Homozygous Baf250afV5/fV5 mice were viable and fertile, suggesting that Baf250a-V5 fusion protein functions as normal Baf250a. The specificity of V5 tag was validated by IHC using V5 antibodies and Baf250a antibodies (Supplementary information, Figure S2C). Baf250a contains an ARID domain, which binds to DNA without sequence preference29. The specificity of V5 antibodies was tested in wild-type mice (Figure 2G). As expected, ChIP-qPCR using V5 antibodies failed to identify binding of Baf250a to the Tbx3 promoter in wild-type mice due to lack of V5-tagged fusion protein. However, significant binding of Baf250a to the Tbx3 promoter was seen in Baf250afv5/fv5 mice. This binding specificity was confirmed as no binding was seen in the promoters of Gata4 and Sox1 in the same mice (Figure 2G). Given that mRNA levels of Tbx3 were immediately decreased upon decreased protein levels of Baf250a, these data established that Tbx3 was a direct transcriptional target of Baf250a.
Next, we asked whether Nkx2.5, Gata4, and Tbx5 were direct transcriptional targets of Tbx3. ChIP-qPCR confirmed that Tbx3 directly bound to the promoter of Nkx2.5; however, ChIP-qPCR failed to detect binding of Tbx3 to the proximal promoters of Gata4 and Tbx5 (Figure 2H).
Baf250a orchestrated a genetic program that repressed the expression of Nkx2.5 in the SAN through Tbx3 and HDAC3
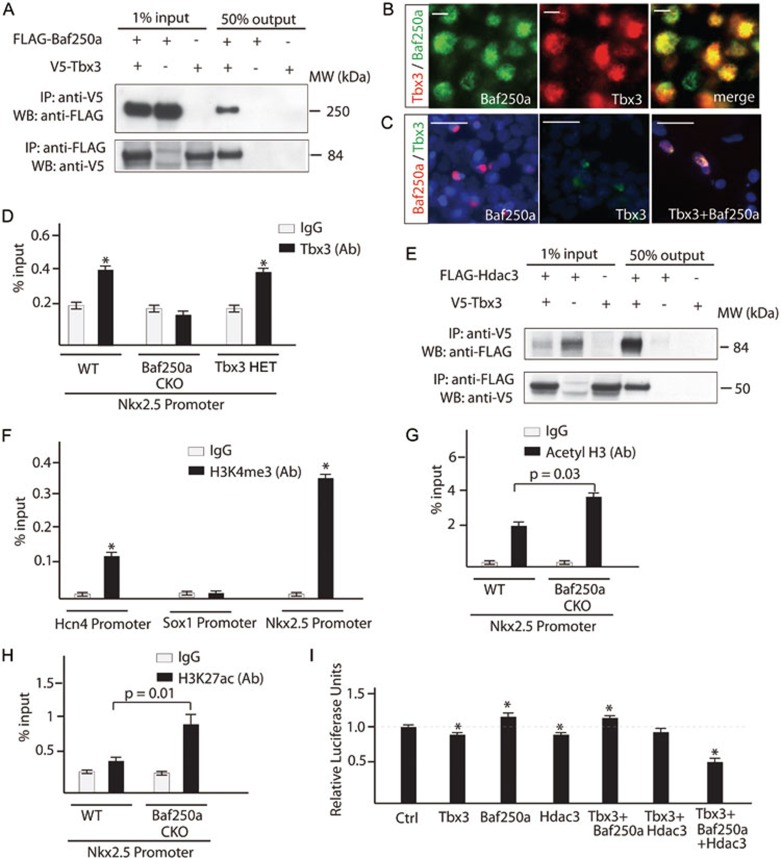
We next attempted to gain further insights into how this genetic pathway operates in the SAN. ATP-utilizing SWI/SNF family remodelers are believed to slide and eject nucleosomes to expose DNA for transcription factor binding30. Reciprocal co-immunoprecipitation (Co-IP) revealed that Tbx3 directly interacted with Baf250a (Figure 3A), suggesting that Baf250a and Tbx3 coordinately regulated gene expression. Double IHC using antibodies against Baf250a and Tbx3 revealed their co-localization in the pacemaker cells of SAN (Figure 3B). The specificity of this direct interaction was further demonstrated in 293T cells. Tbx3 was primarily localized in the cytosol when expressed alone; however, co-expression of Tbx3 and Baf250a in the same cell promoted nuclear localization of Tbx3 in 293T cells (Figure 3C).
Figure 3.
Baf250a, Tbx3, and HDAC3 coordinately repressed the expression of Nkx2.5 in the SAN. (A) Baf250a directly interacts with Tbx3. Baf250a or Tbx3 was immunoprecipitated from nuclear extracts of HEK 293 cells expressing FLAG-tagged Baf250a, V5-tagged Tbx3, or both proteins. Association with Tbx3 or with Baf250a was detected by western blotting with V5- or FLAG-specific antibody. (B) Co-localization of Baf250a and Tbx3 in the pacemaker cells of SAN. Scale bar, 5 μm. Magnification: 400×. (C) Either Baf250a or Tbx3 alone primarily localized in the cytosol of 293T cells. Co-expression of Baf250a and Tbx3 led to exclusive nuclear localization of both proteins. Blue: DAPI. Scale bar, 25μm. (D) Tbx3 binding to promoter of Nkx2.5 required Baf250a. Binding of Tbx3 to the promoter of Nkx2.5 in the SAN of wild-type (WT), Baf250a CKO mice and Tbx3 HET mice. WT: wild type as determined by ChIP-qPCR (n = 3 in each group). (E) Tbx3 directly interacts with HDAC3. Reciprocal Co-IP was similarly performed as described in A. (F) ChIP-qPCR showed the distribution of H3K4me3 in the proximal promoters of Hcn4, Sox1, and Nkx2.5 (n = 3 in each group). *P < 0.05. (G) Increased acetylation of histone 3 in the promoter region of Nkx2.5 at 20 h after tamoxifen induction in the SAN of Baf250a CKO mice as determined by ChIP-qPCR (n = 3 in each group). (H) ChIP-qPCR evaluated the distribution of H3K27ac in the proximal promoter of Nkx2.5 in Baf250 CKO mice. (I) Baf250a, Tbx3, and HDAC3 coordinately repressed the promoter activity of Nkx2.5 in a luciferase reporter assay (n = 3 in each group). *P < 0.05.
The association of Baf250a and Tbx3 in vivo raised a scenario in which Baf250a-associated remodeling complexes open chromatin structure and Tbx3 recognizes specific sequences of target promoters. If this association is important, decreased protein level of Baf250a would affect binding of Tbx3 to the promoter of Nkx2.5. ChIP-qPCR using SAN tissues showed that Tbx3 did not bind to the promoter of Nkx2.5 after deletion of Baf250a (Figure 3D). Since the expression of Tbx3 decreased by about 2-fold in Baf250a CKO mice, to rule out the possibility that the decreased occupancy of Tbx3 was due to a decrease of Tbx3 protein per se, we examined the binding of Tbx3 to the promoter of Nkx2.5 in the Tbx3 heterozygous mice (Tbx3 HET). Compared with wild-type control, the binding of Tbx3 did not show significant change in Tbx3 HET mice, indicating that decreased binding of Tbx3 was attributed to a decrease of Baf250a protein (Figure 3D).
ATP-dependent chromatin remodeling is generally believed to remove nucleosome barriers, thus activating gene expression30. To elucidate how Tbx3 acts with Baf250a to repress the expression of Nkx2.5, we carried out an interaction screen using our expression library and identified HDAC3 as an interaction partner of Tbx3 (Figure 3E). HDAC removes the acetyl groups from lysines of histone tails. As histone deacetylation is correlated with gene silencing, HDACs generally act as transcription repressors. The direct interaction between Tbx3 and HDAC3 is noteworthy as HDAC3 is known to play essential roles in cardiac development and energy metabolism31,32. HDAC3 is highly expressed in the SAN based on mRNA-Seq analysis. These data raised the possibility that Tbx3 acts through HDAC3 to repress the expression of Nkx2.5.
The promoters of silent genes are generally associated with repressive marks but are believed to be devoid of active histone marks. To explore how histone modifications in the promoter of Nkx2.5 are associated with its expression, we examined histone H3 Lys4 trimethylation (H3K4me3), a prominent active histone mark associated with active genes33. The specificity of H3K4me3 signal was confirmed in the promoters of Sox1 and Hcn4 as Sox1 is not expressed in the SAN but Hcn4 is (Figure 3F). To our surprise, H3K4me3 modifications were highly enriched in the Nkx2.5 promoters in the SAN (Figure 3F). The enrichment of an active histone mark, H3K4me3, in the promoter of Nkx2.5 seems paradoxical as Nkx2.5 is silent in the SAN. Histone acetyltransferases (HATs) and HDACs dynamically bind to the promoters of inactive genes with active histone marks, and these dynamic acetylation and deacetylation processes prevent phosphorylated RNA Pol II from binding to the promoters of inactive genes34. The sequence-specific transcription factor Tbx3 interacts directly with HDAC3, suggesting that Tbx3 recruits HDAC3 to the promoter of Nkx2.5 where HDAC3 and an as-yet-unidentified HAT dynamically removes and adds acetyl groups, respectively. Through this dynamic cycle, Tbx3 and HDAC3 may act together to repress the expression of Nkx2.5. To gain support for this model, we examined the acetylation status of the promoter of Nkx2.5. ChIP-qPCR using acetyl H3 antibody revealed that acetylation of histone H3 was significantly increased in the promoter of Nkx2.5 in Baf250a CKO mice compared with littermate controls (Figure 3G). Consistent with this, H3K27ac was increased in the promoter of Nkx2.5 in SAN of Baf250a CKO mice (Figure 3H). These data supported that deacetylation was involved in repression of Nkx2.5 expression.
To provide direct evidence of repression of the promoter of Nkx2.5 by Baf250a, Tbx3, and HDAC3, we adopted a previously established luciferase reporter assay based on pREP4 episomal vector35. A 5.0-kb sequence immediately upstream of Nkx2.5 transcription start site was placed upstream of the luciferase reporter in a way that the luciferase activity was dependent on the histone modifications and binding of transcription factors to this promoter. While Tbx3 and HDAC3 alone slightly repressed luciferase activity, co-expression of Baf250a, Tbx3, and HDAC3 significantly repressed the luciferase activity, documenting a coordinated action of these transcription regulators on the promoter of Nkx2.5 (Figure 3I).
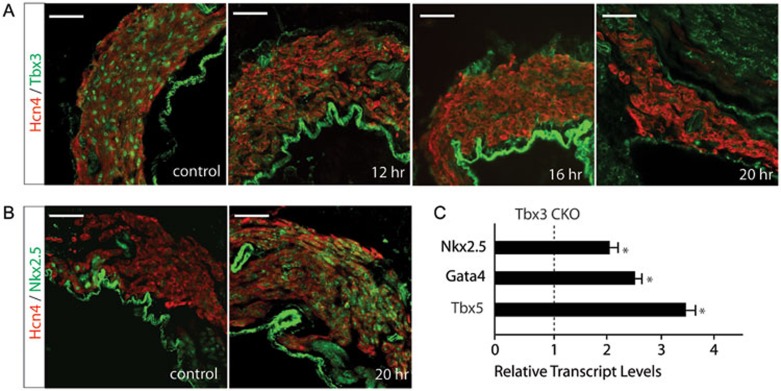
To further confirm that this repressive epigenetic pathway is mediated by Tbx3, we specifically deleted Tbx3 in the SAN using Hcn4-CreERT2 BAC transgenic mice. Comparable to turnover of Baf250a protein, Tbx3 protein began to decrease at 12 h after tamoxifen induction (Figure 4A). The expression of Nkx2.5 was dramatically increased at 20 h (Figure 4B and 4C). Expressions of Gata4 and Tbx5 were also increased as demonstrated by qPCR (Figure 4C). Surface EKG examination revealed that the Tbx3 CKO mice exhibited sinus bradycardia, phenocopying Baf250a CKO mice (Figure 5A and 5B).
Figure 4.
Deletion of Tbx3 in the SAN led to increased expression of Nkx2.5. (A) Protein levels of Tbx3 in the SAN after tamoxifen induction in Tbx3f/f;Hcn4CreERT2 mice. Scale bar, 50 μm. (B) Enhanced expression of Nkx2.5 in the SAN after tamoxifen induction in the Tbx3 CKO mice. Scale bar, 50 μm. (C) qPCR to show relative transcript levels of Nkx2.5, Tbx5, and Gata4 at 20 h after tamoxifen induction in Tbx3 CKO mice (n = 3 in each group). *P < 0.05.
Figure 5.
Deletion of Tbx3 in the SAN led to sinus bradycardia. (A) Representative diagrams of EKG in lead II. (B) EKG parameters of littermate control and Tbx3 CKO at 3 months.
Taken together, these data established a Baf250a-orchestrated epigenetic pathway in the SAN, i.e., Baf250a directly activates the expression of Tbx3, and Baf250a, Tbx3, and HDAC3 coordinately repressed the expression of Nkx2.5.
Nkx2.5, Gata4, and Tbx5 coordinately activated contractile cardiomyocyte program
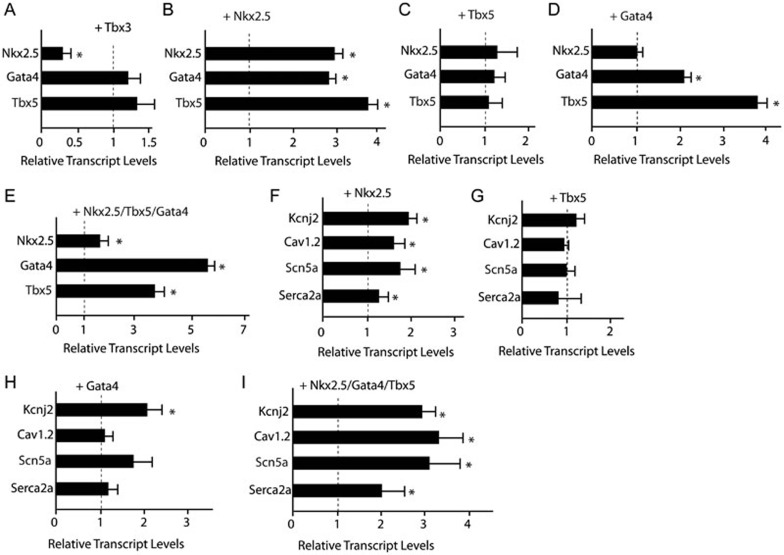
Nkx2.5 is at the top of the cardiac transcription cascade during contractile cardiomyocyte differentiation. It acts with other cardiac transcription factors such as Srf, Gata4, and Tbx5 to regulate myocardial development28,36,37,38,39,40,41,42. To further explore transcriptional regulatory circuits among Tbx3, Nkx2.5, Gata4, and Tbx5, we examined how overexpression of these transcription factors actively regulated expression of other transcription factors. Cultivation of pacemaker cells in vitro has not been established and early embryonic cardiomyocytes resemble pacemaker cells43. For these reasons, we decided to use ex vivo culture of hearts from embryonic day (E) 10 to explore genetic interactions among these transcription factors. Expression of Tbx3 in the embryonic heart at E10 by lentiviral transduction specifically inhibited the expression of Nkx2.5 but not Gata4 and Tbx5 (Figure 6A and Supplementary information, Figure S3). Overexpression of Nkx2.5 did significantly activate the expression of Nkx2.5, Gata4, and Tbx5 (Figure 6B). Overexpression of Gata4 could also activate the expression of Gata4 and Tbx5, while Tbx5 overexpression alone did not have any effect (Figure 6C and 6D). Similar activations of Gata4 and Tbx5 were observed upon overexpression of all three transcription factors (Figure 6E).
Figure 6.
Nkx2.5, Gata4, and Tbx5 activated contractile cardiomyocyte program. (A) Overexpression of Tbx3 specifically repressed the expression of Nkx2.5 (n = 3 in each group). *P < 0.05. (B) Overexpression of Nkx2.5 activated the expression of Nkx2.5, Gata4, and Tbx5 (n = 3 in each group). *P < 0.05. (C) Overexpression of Tbx5 did not significantly affect the expression of Nkx2.5, Gata4, and Tbx5 (n = 3 in each group). (D) Overexpression of Gata4 did not affect the expression of Nkx2.5, but activated the expression of Gata4 and Tbx5 (n = 3 in each group). *P < 0.05. (E) Co-expression of Nkx2.5, Gata4, and Tbx5 significantly activated expression of Gata4 and Tbx5 (n = 3 in each group). (F-I) Activation of ion channel genes by overexpression of Nkx2.5 (F), Tbx5 (G), Gata4 (H), and Nkx2.5/Tbx5/Gata4 triplet (I) (n = 3 in each group). *P < 0.05.
Transcription factors Nkx2.5, Gata4, Tbx5 together with ion channel genes Kcnj2, Scn5a, and Cav1.2 were clustered into the same profile 16, suggesting that these genes are under the control of the same transcriptional module and that these cardiac transcription factors could directly activate the expression of these ion channel genes. Lentiviral vector-mediated transduction of Nkx2.5, Tbx5, and Gata4 did synergistically activate the expression of Kcnj2, Cav1.2, Scn5a, and Serc2a (Figure 6F-6I). Collectively, these data suggested that enhanced expression of Nkx2.5 and subsequent increased expression of Gata4 and Tbx5 in the SAN are responsible for activation of the contractile cardiomyocyte program in the SAN of the Baf250a CKO mice.
Discussion
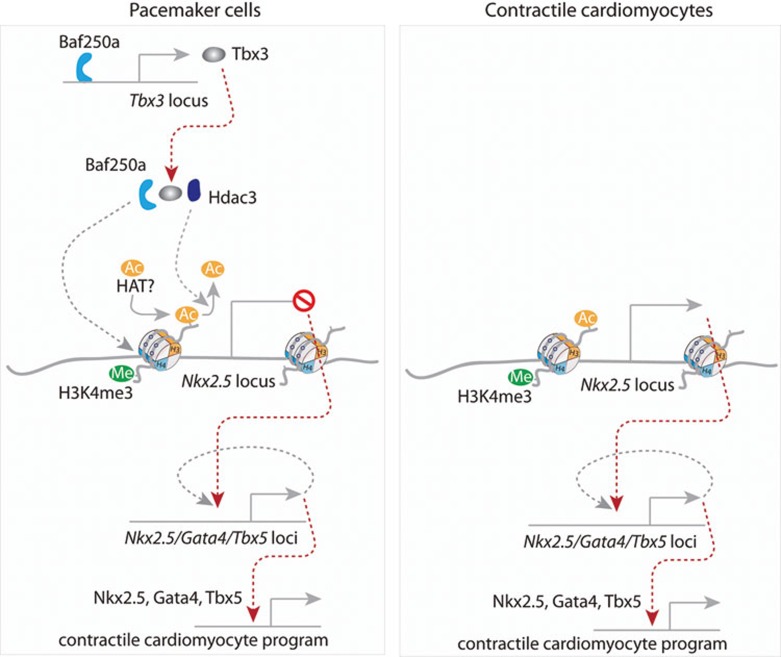
In this study, we combined conditional gene deletion in vivo with time course gene expression analysis to dissect the transcriptional hierarchies. For the first time, we elucidate a repressive epigenetic pathway maintaining proper function of the SAN (Figure 7). Our study provides a paradigm for studying transcriptional hierarchies in vivo. The molecular mechanism that we revealed in this study has important implications and points to new directions.
Figure 7.
A working model of Baf250a in maintaining identity of SAN. The transcriptional hierarchy starts with Baf250a, which activates the expression of Tbx3. Baf250a, Tbx3, and HDAC3 act combinatorially to repress the expression of Nkx2.5 through histone deacetylation in the SAN. Increased expression of Nkx2.5 after deletion of Baf250a in the SAN subsequently activates the expression of Gata4 and Tbx5 in a self-reinforcing manner. Nkx2.5, Tbx5, and Gata4 eventually activate a contractile cardiomyocyte program including enhanced expression of Kcnj2 in the SAN, which negatively affects pacemaking activity of the SAN.
Considering the notion that a common pool of cardiac progenitors gives rise to pacemaker cells and contractile cardiomyocytes44,45,46, our work suggests a regulatory framework for how cardiac progenitor cells differentiate. Nkx2.5 is at the top of transcriptional hierarchy and acts as a key node in regulating myocardial development. Nkx2.5, Gata4, and Tbx5 as well as other cardiac transcription factors synergistically regulate the expression of numerous ion channels and connexins in contractile cardiomyocytes. Segregation of cardiac conduction system lineage from cardiac progenitor cells requires repression of the default genetic program that is directed by Nkx2.5. Despite the critical role of Baf250a in repressing the expression of Nkx2.5 in the SAN, the expression of Hcn4 does not change significantly in the Baf250a CKO mice. It appears that two separate mechanisms, the activation of an SAN-specific program and the repression of a contractile cardiomyocyte program, regulate the development and functional organization of the SAN. Identification of genetic pathways responsible for these two mechanisms may provide novel insights into the development of SAN and working myocardium.
It is conceivable that disruption of key transcriptional circuits as we described can lead to dysregulation of ion channels and connexins in the SAN. Compared with contractile cardiomyocytes, the pacemaker cells in the SAN express a unique set of ion channels. The specific expression and organization of those ion channels form the basis for the membrane and Ca2+ clocks, both of which contribute to the spontaneous diastolic depolarization of phase 46,14. Pacemaker cells do not express Kcnj2, which is highly expressed in all contractile cardiomyocytes. The inward-rectifier potassium channel Kir2.1, encoded by Kcnj2, plays an important role in repolarization of the action potential and stabilization of a negative resting potential in contractile cardiomyocytes47. Therefore, Ik1 current, conducted by Kir2.1, is believed to be one of the major mechanisms for suppressing automaticity. In fact, inhibition of this current in the ventricular cardiomyocytes of guinea pigs can induce pacemaker activity in those cells48. SSD is primarily associated with aging, indicating that dysregulation of ion channels and connexins rather than mutations of these genes are its major causes. Dysregulation of ion channels and connexins, which creates electrical remodeling, is a critical feature of SSD and cardiac arrhythmias49. It appears that neither the fully differentiated cardiomyocyte state nor pacemaker cell identity is robust. Ectopic expression of T-box transcription factors, Tbx3 and Tbx18, can turn atrial and ventricular cardiomyocytes, respectively, into pacemaker-like or pacemaker cells50,51. A critical role of Baf250a/Tbx3 genetic pathway in maintaining proper function of pacemaking cells gains further supports from several genome-wide association studies52,53,54. TBX3-TBX5 and NKX2-5 regions have been found to be associated with heart rate and cardiac conduction, highlighting critical interplay among these transcription factors in the cardiac conduction system. In the future, it will be interesting to directly evaluate the clinical relevance of our findings using SAN samples from patients.
In summary, our study reveals that deletion of a single chromatin remodeling factor, Baf250a, can activate the contractile cardiomyocyte program in fully differentiated pacemaker cells in the SAN. Thus, identification of key genetic pathways responsible for regulating ion channels and connexins may provide novel insights into the pathogenesis of SSD and cardiac arrhythmias, laying the foundation for the development of novel treatments.
Materials and Methods
Mouse genetics
BAC recombineering was used to generate V5-tagged Baf250a mice. Targeted ES cells were identified by Southern blot. Two properly targeted ES cells were injected into C57BL6 blastocysts in the Mouse Genetics Shared Resource Facility at the Mount Sinai to generate high-percentage chimeras. Mice with Baf250a-floxed allele were backcrossed six generations to a Sv129 isogenic background. Mice with Baf250a-floxed allele were crossed with Hcn4-CreERT2 BAC transgenic mice. To specifically delete Baf250a in the SAN, tamoxifen was injected intraperitoneally at the amount of 1 mg tamoxifen/25 g body weight for 3 consecutive days as previously described. The surface EKG was performed 3 days after final injection. The Tbx3-floxed allele was generated by inserting two LoxP sites flanking exons 2, 3, and 4. The genotyping primers were provided in Supplementary information, Table S9. Tbx3f/f and Hcn4-CreERT2;Tbx3f/f were on C57BL6 background.
Massively parallel sequencing, clustering analysis, qPCR, and RT-PCR analysis
About 1 μg of total RNAs were extracted from three SAN (control and Baf250a CKO) using TRIzol Reagent. mRNAs were purified using a poly-A selection approach. Sequencing libraries were prepared according to a standard protocol. Samples were sequenced on the Illumina HiSeq 2000 platform in the Genomic Core at the Icahn School of Medicine at Mount Sinai. The single ends (100 bp) were aligned to the mouse reference genome (NCBI Build 37/mm9) using TopHat (Bowtie algorithm). Transcript assembles and identification of differentially expressed genes were achieved using the Cufflinks package. To account for expression bias due to transcript length, each sample transcript expression was normalized by using Cufflinks algorithm with an FDR of 0.05.
The gene expression was measured at six time points, i.e., at 0, 16, 20, 24, 48, and 144 h after tamoxifen induction. These data were processed as follows before clustering analysis. For genes whose expression levels were measured as zero, their expression was replaced by 0.5 times the lowest non-zero expression values among all the genes at each time point. We then transformed the data to obtain log2 (sample/control) for each time point. There were overall 13 803 genes tested at all six time points and they were included for further analysis.
STEM was used to classify genes according to the log2-transformed data at all six time points. Briefly, a set of distinct and representative temporal expression profiles were selected as model profiles, and the rest of the genes were assigned to each of those model profiles based on matching the gene's expression profiles as determined by correlation coefficient. The statistical significance of each module was calculated by comparing to random assignment. The model profiles with P-values < 0.05 were considered as significant and colored in the graph. Differential expression was defined as > 1.5-fold (∼log2(1.5)) in STEM. A total of 8 675 genes passed this criterion and were considered as differentially expressed in at least one time point.
Total RNAs for qPCR were extracted from SAN tissues of Hcn4-CreERT2; R26fstopGFP/+ mice (control) and Baf250af/f;Hcn4-CreERT2;R26fstopGFP/+ mice. qPCR was performed using StepOne plus (Applied Biosystems) per the manufacturer's protocol. At least three biological samples in each group were used. Primer sequences are provided in Supplementary information, Table S9. 18S rRNA was used as an internal control.
Co-IP and western blot
For Co-IP, HEK293T cells were co-transfected with plasmids expressing FLAG- or V5-tagged proteins. Cells were lysed with NP-40 lysis buffer (50 mM HEPES, pH 7.6, 250 mM NaCl, 0.1% NP-40, 0.2 mM EDTA, 1.4 mM β-mercaptaethanol, 0.2 mM PMSF, and 1× protease inhibitor cocktail). Total lysates were incubated with anti-M2 FLAG agarose or anti-V5 Agarose Affinity Gel overnight. Unbound proteins were washed away. Protein complexes precipitated by agarose were eluted by boiling in SDS-Laemmli buffer and subjected to western blot analyses. Western blot images were acquired as uncompressed bitmapped digital data (TIFF format). Images shown are representative of at least three independent experiments. The sources of antibodies and their dilutions are provided in Supplementary information, Table S10.
ChIP-qPCR
SAN tissues were dissected out under epifluorescent microscope (Leica MZ16 FA) in Baf250af/f;Hcn4-CreERT2;R26fstopGFP/+ mice. The tissues were cut into small pieces and homogenized using a DUALL tissue grinder in PBS. The homogenized lysates were resuspended in 200 μl of cross-linking solution and incubated at room temperature (RT) for precisely 30 s. Glycine (1 M; 25 μl) was applied for 5 min at RT with gentle shaking to stop fixation. Fetal bovine serum (FBS; 10%) was added and cell lysates were collected by centrifuging at 1 600× g for 5 min at RT. The pellet was washed by PBS with 10% FBS. The pellet was resuspended in 100 μl SDS lysis buffer (100 mM NaCl, 50 mM Tris-Cl, pH 8.1, 5 mM EDTA, and 1% SDS) with freshly added protease inhibitors on ice for 10 min. Ice-cold ChIP dilution buffer (1 000 μl; 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 167 mM NaCl) with protease inhibitors was added to the lysates and sonication was carried out on a Covaris S220 sonicator to achieve chromatin fragments of 150-300 bp. A ∼25-μl aliquot of sonicated chromatin solution was saved as an 'input' whole-cell extract. Appropriate amount of antibody was added to the remaining chromatin solution and incubated at 4 °C in a rotator overnight. Chromatin fragments were pulled down by incubating protein A-sepharose beads for 1 h at 4 °C on a rotating platform. The chromatin-antibody complex was washed twice in ice-cold low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, and 150 mM NaCl), and was further purified using a filter column (Spin-X wash columns, CLS8160-96EA, Sigma). Chromatin was eluted using 250 μl of prewarmed ChIP elution buffer (0.1 M NaHCO3 and 1% SDS) with freshly added 5 nM DTT. The samples were reverse cross-linked by incubating at 65 °C for 5 h. RNAs in the samples were digested by RNase and degradation of proteins was achieved by adding Proteinase K. DNA fragments were extracted by phenol/chloroform extraction. The DNA pellet was dissolved in TE buffer.
Real-time PCR was performed using StepOne plus (Applied Biosystems) as per the manufacturer's protocol. Difference between samples and controls was calculated based on the 2−ΔΔCT method and normalized to 18S RNA. Measurements were performed in triplicate. Primers sequences are provided in Supplementary information, Table S1.
Fluorescence IHC
IHC was performed on cryosections. Hearts were infused in 0.5 M sucrose for 2-3 h, and then in OCT frozen in dry ice. Samples were sectioned at a thickness of 10 μm. Immediately before IHC, the sections were immersed in methanol at −20 °C for 5 min and washed three times with PBS for 5 min. The sections were blocked with 1× Animal-Free Blocker (SP-5030, Vector lab) for 1 h. Hcn4 antibody and anti-Baf250a antibody were applied overnight at 4 °C. The slides were washed in PBS and then were incubated with secondary antibodies for 1 h at RT. The slides were mounted in VECTASHIELD Mounting Medium with DAPI. Fluorescence images were acquired as uncompressed bitmapped digital data (TIFF format). Images shown are representative of at least three independent experiments. The sources of antibodies and their dilutions are provided in Supplementary information, Table S2.
Luciferase reporter assay
The pREP4-Luc vector was from Keji Zhao's laboratory at the National Heart, Lung, and Blood Institute. As the vector did not provide basic promoter activity, which is not ideal for testing transcriptional repressor activity, we replaced the original luciferase cassette with a Thymidine kinase (TK)-Luciferase-pA cassette. A 5.0-kb sequence immediately upstream of Nkx2.5 transcription start site was placed upstream of basic TK promoter. Plasmids were introduced into 293T cells by TurboFect (Thermo Scientific). A renilla reporter was used to normalize transfection efficiency. Luciferase activity was measured with a Dual Luciferase Reporter kit (Promega) 48 h after transfection. All experiments were repeated at least three times.
Lentiviral transduction
Nkx2.5, Tbx3, Gata4, and Tbx5 were introduced into embryonic heart by lentivirus vector-mediated gene transfer. Briefly, cDNAs of these transcription factors were subcloned into a third-generation lentiviral vector construct. Lentivirus vectors were generated in HEK293T cells. Embryonic hearts were maintained by the addition of Dulbecco's modified Eagle media with 10% FBS in organ culture dishes (No. 3037 Falcon, Lincoln Park, NJ; USA) at 37 °C in 95% air/5% CO2 in a water-saturated environment. The embryonic hearts were infected with lentivirus vectors expressing Nkx2.5, Tbx3, Gata4, and Tbx5 for 24 h with 5 × 106 transducing units/ml. Total RNA was isolated 48 h after lentiviral transduction.
Surface EKG and telemetry EKG recording
Cardiac electrophysiological function was assessed using electrocardiograms (lead II) as previously described. Data analysis was performed using the Chart5Pro (v 5.4.2, AD Instruments). For telemetry EKG, ETA-F10 transmitters (Data Sciences International) were implanted in the abdominal cavity with electrodes in lead II configuration. Mice were allowed to recover completely for 10 days after surgery. EKG data were recorded for 3 consecutive days, and EKG data were analyzed using Chart5Pro.
Statistical analysis
Data were presented as mean ± standard deviation. Results were tested for statistical significance using two-tailed t-test. P-values < 0.05 were considered to be statistically significant. The number of biological replicates (n) is indicated in the figure legends or in the main text.
Accession number
Massively parallel sequencing data have been deposited to GEO with accession number GSE55682.
Acknowledgments
We thank B Gelb for critical reading of the manuscript. YZ was supported by a SDG grant (AHA), a Basil O'Connor Starter Scholar Research Award (March of Dimes Foundation), and NIH (1R01HL107376 and 1K02HL103597). ZW was supported by NIH (7R01HL109054). C-LC was supported by NIH (1R01HL095810 and 1K02HL094688), AHA (0855808D), and the March of Dimes Foundation (5-FY07-642).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Loss of Baf250a causes sinus bradycardia as demonstrated by ambulatory ECG recording (three Hcn4CreERT2;Baf250af/f as controls and three Baf250a CKO mice).
Generation of V5-tagged Baf250a floxed allele.
Overexpression of Tbx3 in embryonic heart by Lentiviral vector-mediated transduction.
A list of differentially expressed genes at 16 h time point
A list of differentially expressed genes at 20 h time point
A list of differentially expressed genes at 24 h time point
A list of differentially expressed genes at 48 h time point
A list of differentially expressed genes at 6 day
Differential Expressed genes and their grouping information
Significantly enriched GO terms at correct P-value < 0.001 for all 4 profiles
A representative list of differentially expressed genes after loss of Baf250a over several time points.
List of primers used in the study
List of Antibodies using in the immunohistochemistry, immunoflurescence, co-IP, ChIP-qPCR, and Western blot
References
- Boyett MR, Honjo H, Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc Res. 2000;47:658–687. doi: 10.1016/s0008-6363(00)00135-8. [DOI] [PubMed] [Google Scholar]
- Mangoni ME, Nargeot J. Genesis and regulation of the heart automaticity. Physiol Rev. 2008;88:919–982. doi: 10.1152/physrev.00018.2007. [DOI] [PubMed] [Google Scholar]
- Park DS, Fishman GI. The cardiac conduction system. Circulation. 2011;123:904–915. doi: 10.1161/CIRCULATIONAHA.110.942284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation. 2007;115:1921–1932. doi: 10.1161/CIRCULATIONAHA.106.616011. [DOI] [PubMed] [Google Scholar]
- Kumar P, Kusumoto FM, Goldschlager N. Bradyarrhythmias in the elderly. Clin Geriatr Med. 2012;28:703–715. doi: 10.1016/j.cger.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart's pacemaker. Circ Res. 2010;106:659–673. doi: 10.1161/CIRCRESAHA.109.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrancesco D. The role of the funny current in pacemaker activity. Circ Res. 2010;106:434–446. doi: 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- Chen B, Wu Y, Mohler PJ, et al. Local control of Ca2+-induced Ca2+ release in mouse sinoatrial node cells. J Mol Cell Cardiol. 2009;47:706–715. doi: 10.1016/j.yjmcc.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesi R, Baruscotti M, Gnecchi-Ruscone T, et al. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- Le Scouarnec S, Bhasin N, Vieyres C, et al. Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease. Proc Natl Acad Sci USA. 2008;105:15617–15622. doi: 10.1073/pnas.0805500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DW, Wang DW, Dyment M, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Dev Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Carter KL, Thomas KR, et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci USA. 2012;109:E154–E163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. Chromatin remodeling in heart development. Curr Opin Genet Dev. 2010;20:505–511. doi: 10.1016/j.gde.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, de la Serna IL, Imbalzano AN. The myogenic basic helix-loop-helix family of transcription factors shows similar requirements for SWI/SNF chromatin remodeling enzymes during muscle differentiation in culture. J Biol Chem. 2002;277:33818–33824. doi: 10.1074/jbc.M205159200. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Lei I, Gao X, Sham MH, et al. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J Biol Chem. 2012;287:24255–24262. doi: 10.1074/jbc.M112.365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Peng S, Zhao Y. Inducible gene deletion in the entire cardiac conduction system using Hcn4-CreERT2 BAC transgenic mice. Genesis. 2014;52:134–140. doi: 10.1002/dvg.22730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Sun X, Nichol P, et al. System for tamoxifen-inducible expression of cre-recombinase from the Foxa2 locus in mice. Dev Dyn. 2008;237:447–453. doi: 10.1002/dvdy.21415. [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsker D, Patsialou A, Zumbrun SD, et al. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 2004;32:1345–1353. doi: 10.1093/nar/gkh277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Trivedi CM, Lu M, et al. Histone deacetylase 3 regulates smooth muscle differentiation in neural crest cells and development of the cardiac outflow tract. Circ Res. 2011;109:1240–1249. doi: 10.1161/CIRCRESAHA.111.255067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Liu H, Chen X, et al. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risebro CA, Riley PR. Formation of the ventricles. ScientificWorldJournal. 2006;6:1862–1880. doi: 10.1100/tsw.2006.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, et al. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, et al. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell. 2012;23:280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Zaffran S, Kuroiwa A, et al. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2-5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc Natl Acad Sci USA. 2012;109:18273–18280. doi: 10.1073/pnas.1215360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ML, Christoffels VM, Moorman AF. The cardiac pacemaker and conduction system develops from embryonic myocardium that retains its primitive phenotype. J Cardiovasc Pharmacol. 2010;56:6–15. doi: 10.1097/FJC.0b013e3181e775d3. [DOI] [PubMed] [Google Scholar]
- Liang X, Wang G, Lin L, et al. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spater D, Abramczuk MK, Buac K, et al. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 2013;15:1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Moreno-Rascon N, Beyer S, et al. Biphasic development of the mammalian ventricular conduction system. Circ Res. 2010;107:153–161. doi: 10.1161/CIRCRESAHA.110.218156. [DOI] [PubMed] [Google Scholar]
- Dhamoon AS, Jalife J. The inward rectifier current (IK1) controls cardiac excitability and is involved in arrhythmogenesis. Heart Rhythm. 2005;2:316–324. doi: 10.1016/j.hrthm.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Miake J, Marban E, Nuss HB. Biological pacemaker created by gene transfer. Nature. 2002;419:132–133. doi: 10.1038/419132b. [DOI] [PubMed] [Google Scholar]
- Jones SA, Lancaster MK, Boyett MR. Ageing-related changes of connexins and conduction within the sinoatrial node. J Physiol. 2004;560(Part 2):429–437. doi: 10.1113/jphysiol.2004.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N, Liang W, Marban E, et al. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ML, Boink GJ, Boukens BJ, et al. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94:439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- den Hoed M, Eijgelsheim M, Esko T, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm H, Gudbjartsson DF, Arnar DO, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loss of Baf250a causes sinus bradycardia as demonstrated by ambulatory ECG recording (three Hcn4CreERT2;Baf250af/f as controls and three Baf250a CKO mice).
Generation of V5-tagged Baf250a floxed allele.
Overexpression of Tbx3 in embryonic heart by Lentiviral vector-mediated transduction.
A list of differentially expressed genes at 16 h time point
A list of differentially expressed genes at 20 h time point
A list of differentially expressed genes at 24 h time point
A list of differentially expressed genes at 48 h time point
A list of differentially expressed genes at 6 day
Differential Expressed genes and their grouping information
Significantly enriched GO terms at correct P-value < 0.001 for all 4 profiles
A representative list of differentially expressed genes after loss of Baf250a over several time points.
List of primers used in the study
List of Antibodies using in the immunohistochemistry, immunoflurescence, co-IP, ChIP-qPCR, and Western blot