Abstract
For construction of the bacterial flagellum, FliI ATPase forms the FliH2-FliI complex in the cytoplasm and localizes to the flagellar basal body (FBB) through the interaction of FliH with a C ring protein, FliN. FliI also assembles into a homo-hexamer to promote initial entry of export substrates into the export gate. The interaction of FliH with an export gate protein, FlhA, is required for stable anchoring of the FliI6 ring to the gate. Here we report the stoichiometry and assembly dynamics of FliI-YFP by fluorescence microscopy with single molecule precision. More than six FliI-YFP molecules were associated with the FBB through interactions of FliH with FliN and FlhA. Single FliI-YFP molecule exchanges between the FBB-localized and free-diffusing ones were observed several times per minute. Neither the number of FliI-YFP associated with the FBB nor FliI-YFP turnover rate were affected by catalytic mutations in FliI, indicating that ATP hydrolysis by FliI does not drive the assembly-disassembly cycle of FliI during flagellar assembly. We propose that the FliH2FliI complex and FliI6 ring function as a dynamic substrate carrier and a static substrate loader, respectively.
The bacterial flagellum, which is responsible for motility in liquid media, is a macromolecular assembly made of about 30 different proteins with their copy numbers ranging from a few to a few tens of thousands. The flagellar export apparatus transports flagellar component proteins from the cytoplasm to the distal end of the growing flagellar structure for self-assembly. The export apparatus can coordinate protein export with assembly by ordered export of substrates to parallel with their order of assembly. Thus, the bacterial flagellar export system is a remarkable example of how bacterial cell coordinates protein export with assembly in a highly organized and well-controlled manner1.
The export apparatus consists of an export gate complex made of six membrane proteins, FlhA, FlhB, FliO, FliP, FliQ, and FliR, and a cytoplasmic ATPase complex consisting of three soluble proteins, FliH, FliI, and FliJ2,3. In addition, the C ring, which is formed by FliG, FliM and FliN on the cytoplasmic face of the MS ring of the FBB4, acts as a platform for efficient assembly of the ATPase complex to the export gate5. The whole flagellar protein export system is highly homologous to the type III secretion system of pathogenic bacteria, through which bacteria directly inject virulence factors into their host cells6.
The export gate is located within the central pore of the MS ring. The C-terminal cytoplasmic domains of FlhA (FlhAC) and FlhB (FlhBC) provide binding sites for the ATPase complex, export substrate and chaperone-substrate complexes7,8,9,10,11. A nonameric ring structure of FlhAC has been visualized to project from the gate into the large central cavity of the C ring through a linker region of FlhA (FlhAL)12,13. Consistently, about nine molecules of FlhA-YFP are estimated to be associated with the FBB14. The export gate utilizes proton motive force (PMF) across the cytoplasmic membrane to drive protein export15,16.
FliI is the ATPase of the export apparatus17 and self-assembles into a homo-hexamer to fully exert its ATPase activity18. FliJ binds to the center of the FliI6 ring to form the FliI6FliJ ring complex19. The FliI6FliJ ring complex looks similar to F- and V-type ATPases, suggesting that the flagellar protein export system and F- and V-type ATPases share an evolutionary relationship19,20,21. FliI also forms a hetero-trimer with a homo-dimer of FliH22, whose primary sequence is highly homologous to the components of the peripheral stalk of the FOF1-ATPsynthase23. Because flagellar chaperone-substrate complexes bind to the FliH2FliI complex through cooperative interactions among chaperone, substrate and FliI24,25, the FliH2FliI complex is believed to deliver export substrates and chaperone-substrate complexes from the cytoplasm to the export gate. The FliH2FliI complex binds to the C ring through an interaction between the extreme N-terminal region of FliH (FliHEN) and FliN26. Photo-crosslinking experiments have shown that FliHEN is also in very close proximity to FlhA27. Given that the interaction between FliHEN and FlhA allows FliI to efficiently exert its function for efficient export27 and that FliJ requires the support of FliH and FliI for the interaction with FlhAL to facilitate PMF-driven protein export28,29, FliH is proposed to anchor the FliI6FliJ ring complex to its docking platform formed by the FlhAC nonameric ring and FlhBC. Since FliH and FliI are required for efficient entry of export substrates into the export gate15, the FliH12FliI6FliJ complex is proposed to act as a substrate loader to couple energy transduction with protein transport into the central channel of the growing flagellar structure. In vitro reconstitution experiments of the FliI6 ring have shown that the binding of Mg2+-ATP to FliI induces FliI6 ring formation and that ATP hydrolysis and the following release of ADP and Pi destabilizes the ring structure30. This suggests that FliI couples ATP-binding and hydrolysis to its assembly-disassembly cycle. However, it remains unknown whether the assembly-disassembly cycle of the FliH12FliI6FliJ ring occurs in vivo.
Previous fluorescent imaging experiments have shown that formation of punctate localization patterns of FliI-YFP molecules within the cell requires both FliH and FliN but none of the export gate components26,31. The FliH defect can be significantly overcome by over-expression of FliI or extragenic suppressor mutations in FlhA and FlhB32. The over-expression of FliI also enhances the export activity of a fliN null mutant considerably33. Purified FliH2FliI complex binds to the FliMFliN4 complex to form a stable FliH2FliI–FliMFliN4 complex5. These observations suggest that FliI requires both FliH and FliN for efficient formation of the FliI6 ring to be docked to the FlhAC9–FlhBC platform. However, in situ structural analyses by electron cryotomography have shown that neither deletion of FlhAC nor FlhBC affects the density of the FliI6 ring, which is observed just outside of the C ring at the bottom of the FBB12, raising the question of how FliI6 ring formation occurs in vivo.
To clarify the stoichiometry and assembly dynamics of FliI ATPase in vivo, we used single-molecule fluorescence techniques in this study. We show that the number of FliI-YFP molecules localized to the flagellar base is more than six but deletion of the flhA gene reduces the number to around six. Continuous total internal reflection fluorescence (TIRF) illumination and fluorescence recovery after photobleaching (FRAP) experiments show exchanges of FliI-YFP molecules between localized and diffusing ones.
Results
Stoichiometry of FliI-YFP associated with the FBB
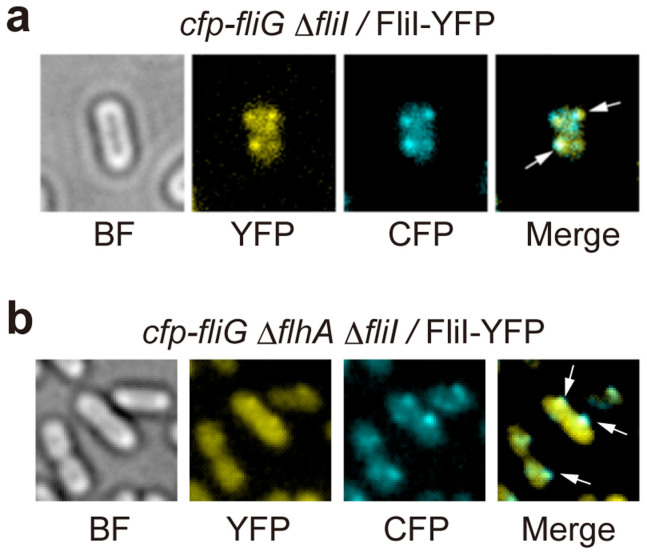
It has been shown that FliI-YFP is functional and that its subcellular localization to the FBB requires FliH and FliN but none of the export gate components26. To clarify how many FliI-YFP molecules associate with the C ring, we first analyzed the subcellular localization of FliI-YFP by high-resolution single-molecule imaging techniques. We used the cfp-fliG ΔfliH fliI-yfp as a negative control. Under Epi-fluorescence illumination, we observed clear fluorescent spots of FliI-YFP molecules on top of a uniform cytoplasmic intensity corresponding to free-diffusing FliI-YFP molecules in the cfp-fliG fliI-yfp cells (Fig. 1a) but not in the cfp-fliG ΔfliH fliI-yfp cells (data not shown), in agreement with previous reports26,31. We tracked the movement of each spot and found that they are not mobile, indicating that each FliI-YFP spot is firmly attached to the flagellar base. The FliI-YFP spots were observed to co-localize with CFP-FliG spots (Fig. 1a). Because the punctate localization pattern of FliI-YFP requires only FliH and FliN26, these results indicate that many of the FliI-YFP molecules are localized to the FBB through a specific interaction between FliH and FliN in Salmonella cells.
Figure 1. Localization of FliI-YFP to the FBB.
Bright field (BF) and epi-fluorescence images of FliI-YFP (YFP) and CFP-FliG (CFP) in (a) Salmonella YVM029 (cfp-fliG ΔfliI) strain transformed with pJSV203 (FliI-YFP) and (b) YVMA013 (ΔflhA ΔfliI cfp-fliG) strain carrying pJSV203. The fluorescence images of CFP-FliG (cyan) and FliI-YFP (yellow) are merged in the right panel. Arrows point to the FliI-YFP spots that co-localize with the CFP-FliG spots.
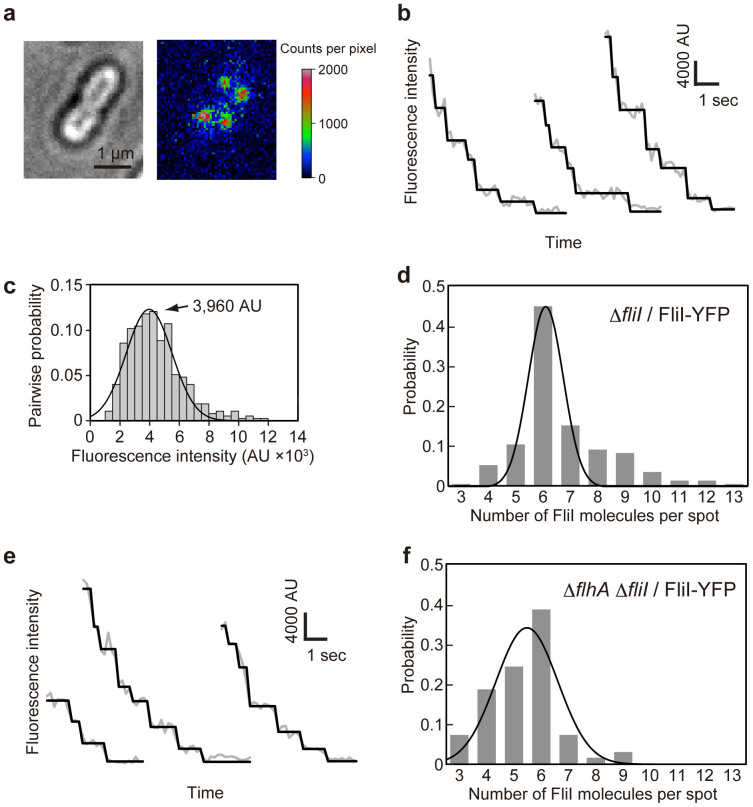
To determine how many FliI-YFP molecules are associated with the FBB, we carried out stepwise photobleaching experiments as described previously by Leake and co-workers34,35,36. As the positions of FliI molecules associated with the FBB are likely at different depth within the height of the cells, we used the epi-fluorescence photobleaching method34 instead of the well-established TIRF method35,36. Bright-field images of cells allowed us to define the boundary of the cell under study (Fig. 2a). The epi-fluorescence images were recorded by an EMCCD camera, and the intensity of each FliI-YFP spot within this boundary was processed by our custom-built software. We modeled the fluorescence intensity distribution (I) within a square region of interest (ROI) centered on each FliI-YFP spot with a side length of 268 nm (67 nm/pixel × 4 pixels) as the sum of a 2D Gaussian function representing the FliI-YFP spot and a uniform local background representing FliI-YFP diffusing in the cytoplasm, non-specific cellular auto-fluorescence and instrumental background (Fig. 2a). Our software automatically separated the spot intensity by subtracting the background intensity (average of the fluorescent intensity in a region within the cell boundary but not containing punctate spot) from the total intensity within the ROI.
Figure 2. Measurement of the number of FliI-YFP molecules associated with the FBB using step photobleaching.
(a) Bright field image (left) and 2D intensity plot (right) of an epi-fluorescence image (false-color) of Salmonella MKM30 (ΔfliI) strain harboring pJSV203 (FliI-YFP). (b) Typical examples show continuous photobleaching intensity trace (gray line connecting dots) for a single FliI-YFP spot in MKM30 harboring pJSV203. Filtered intensity (black line) is overlaid on the raw intensity (gray). (c) Histogram of the pairwise difference distribution for photobleaching traces. The highest peak was fitted by a Gaussian function (black line). In total 73 FliI–YFP spots were analyzed. (d) Histogram of the estimated number of FliI-YFP molecules per spot in MKM30 carrying pJSV203. In total 230 FliI-YFP spots were analyzed. (e) and (f) Effect of flhA deletion on the number distribution of FliI-YFP molecules per spot observed in Salmonella NH0027 (ΔflhA ΔfliI) strain harboring pJSV203. (e) Three typical examples show continuous photobleaching intensity trace for a single FliI-YFP spot. (f) Histogram of the estimated number of FliI-YFP molecules per spot in NH0027 carrying pJSV203. In total 70 FliI-YFP spots were analyzed. The number distributions were fitted by Gaussian functions (black line) in (d) and (f).
Fig. 2b shows typical examples of the fluorescence intensity decay of a single FliI-YFP spot (Fig. 2a), under continuous epi-fluorescence illumination (imaged at 5 or 20 Hz frame rate in frame transfer mode). Intensity decay was found to be stepwise with roughly integer multiples of a unitary value, in agreement with irreversible photobleaching of individual YFP molecules. Pairwise difference analysis showed that the single YFP fluorescence intensity (IYFP) was about 3,960 AU in the population of 73 spots (Fig. 2c). Using this number, we quantified the number of FliI-YFP associated with each spot by dividing the initial intensity I0 at the beginning of the photobleaching by IYFP. The number distribution of FliI-YFP molecules in each spot is summarized in Fig. 2d. The number ranged from 3 to 13 with an average of 6.7 ± 1.7 (Fig. 2d).
Considering that imperfect localization of some fluorescent foci to the focal imaging plane would reduce their fluorescence intensity and that premature FliI-YFP may have failed to be activated, this photobleaching measurement would lead to underestimation of the stoichiometry of FliI-YFP associated with the FBB. It is necessary to use another method to confirm our result. Because FliF-YFP forms the MS ring with a known stoichiometry of 26 copies37, we used the fliF-yfp strain as a reference to compare the fluorescence intensity of each FliI-YFP spot with the average intensity of the FliF-YFP spot under the same experimental condition. The average intensity ratio of FliI-YFP and FliF-YFP was 0.28, and this ratio is close to 7/26. The number of FliI-YFP molecules associated with the FBB was thus estimated to be 7 ± 3 (n = 105). Summarizing the results obtained by the above two different methods, we conclude that a dominant number of FliI-YFP associated with the FBB is six, which is consistent with the stoichiometry of the FliI6 homo-hexamer18,30.
Effect of deletion of the flhA gene on the stoichiometry of FliI-YFP
Two conserved Trp residues in FliHEN, Trp-7 and Trp-10, are involved not only in the FliH-FliN interaction but also in the FliH-FlhA interaction26,27, raising the question of whether removal of FlhA affects the number of FliI-YFP molecules associated with the FBB. We therefore measured the number of FliI-YFP molecules localized to the FBB in a Salmonella flhA null mutant strain. When FliI-YFP was expressed in the cfp-fliG ΔflhA mutant, the FliI-YFP spots were still co-localized with the CFP-FliG spots (Fig. 1b), indicating that FliI-YFP associates with the FBB even in the absence of FlhA, in agreement with previous reports12,26. However, careful measurements of the number of bleaching steps indicated that the number distribution of FliI-YFP shifted to smaller values (Fig. 2e and f). The average number of FliI-YFP molecules per spot was 5.4 ± 1.3 (n = 70) for the ΔflhA fliI-yfp strain and showed a statistically significant difference compared to 6.7 ± 1.7 for the fliI-yfp strain (p < 0.001) using two-tailed t-test. The results indicate that the FliI spots are less bright in the absence of FlhA than in its presence. Therefore, we suggest that FlhA may contribute to some additional localization of FliI-YFP to the FBB.
Effect of catalytic mutations of FliI on the stoichiometry of FliI-YFP
It has been shown that catalytic mutations of FliI, such as K188I and Y363S, considerably reduce the ATPase activity by reducing the binding affinity of FliI for ATP17 but do not show much effect on subcellular localization of FliI to the FBB26. In vitro reconstitution experiments of FliI, however, have shown that ATP hydrolysis by FliI destabilizes its hexameric ring structure whereas the E211A mutation, not affecting ATP-binding but resulting in the complete loss of the ATPase activity, does not destabilize the ring18,30,38, raising the possibility that a change in the FliI ATPase activity may affect the stoichiometry of FliI-YFP associated with the FBB. To test this possibility, we analyzed the stoichiometry of FliI(K188I)-YFP and FliI(E211A)-YFP by epi-fluorescence photobleaching techniques (Fig. 3). The average numbers of FliI(K188I)-YFP and FliI(E211A)-YFP molecules per spot were estimated to be 6.4 ± 1.9 (n = 58) and 6.1 ± 1.7 (n = 50), respectively. Two-tailed t-test indicated that these two average values show no statistically significant difference compared to the fliI-yfp strain (p = 0.167 and 0.07, respectively). These results indicate that these two mutations do not affect the localization of FliI-YFP to the FBB.
Figure 3. Effects of K188I and E211A catalytic mutations on the number of FliI-YFP molecules per spot.
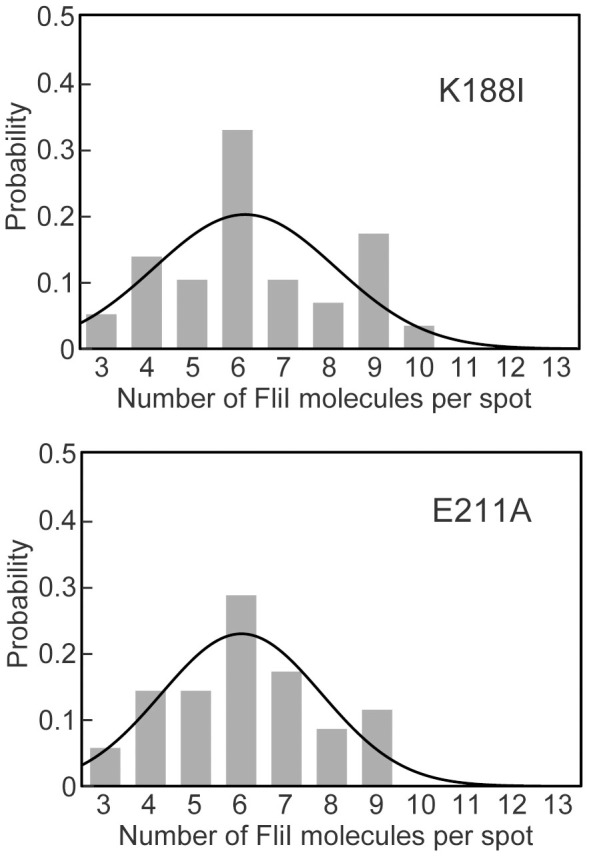
The number of photobleaching steps in each FliI(K188I)-YFP (a) and FliI(E211A)-YFP (b) spot was counted in Salmonella MKM30 (ΔfliI) strain transformed with pSY001 and pSY002, respectively. In total, 58 FliI(K188I)-YFP and 50 FliI(E211A)-YFP spots were analyzed. The number distributions were fitted by Gaussian functions (black line).
FliI turnover investigated by TIRF and FRAP
To investigate whether each FliI-YFP spot shows dynamic exchanges between the FBB and the cytoplasmic pool, we carried out TIRF-FRAP experiments39. In the TIRF experiment, only FliI-YFP molecules that are very close (~100–200 nm) to the glass surface are excited. We first took a single picture with 200 ms exposure of the intact FliI-YFP intensity in living cells under TIRF illumination and then photobleached all the fluorescent proteins within the evanescent field by a strong laser excitation for 400 ms. To estimate the fraction of unbleached FliI-YFP molecules in the cytoplasm after one-shot photobleaching, we compared the cytoplasmic fluorescence intensities of epi-fluorescence images of the fliI-yfp cells before and after photobleaching and found that 45 ± 12% of FliI-YFP molecules are unbleached (n = 64) (Supplementary Fig. 1). Then, we took sequential images of the same area under continuous TIRF illumination at 5 Hz in frame transfer mode to observe changes in the YFP intensity in each FliI-YFP spot for 60 seconds. Of 89 FliI-YFP spots we have analyzed, about 90% of the spots showed turnover between localized and non-localized FliI-YFP (Fig. 4a and Supplementary movie 1). The intensity change of this turnover event is similar to that of the last step in the initial photobleaching event and also approximately the same as IYFP determined from many other photobleaching steps. Turnover of a single FliI-YFP molecule were observed several times per minute (Fig. 4a). The number of times that FliI-YFP showed turnover ranged from 1 to 10 with an average of 2.9 ± 2.1 per minute (Fig. 4b). Because there are about 55% of quenched, free-diffusing FliI-YFP molecules in the cytoplasm (Supplementary Fig. 1), the average rate of FliI-YFP turnover was estimated to be 6.4 molecules per minute.
Figure 4. Observation of FliI-YFP turnover at the flagellar base.
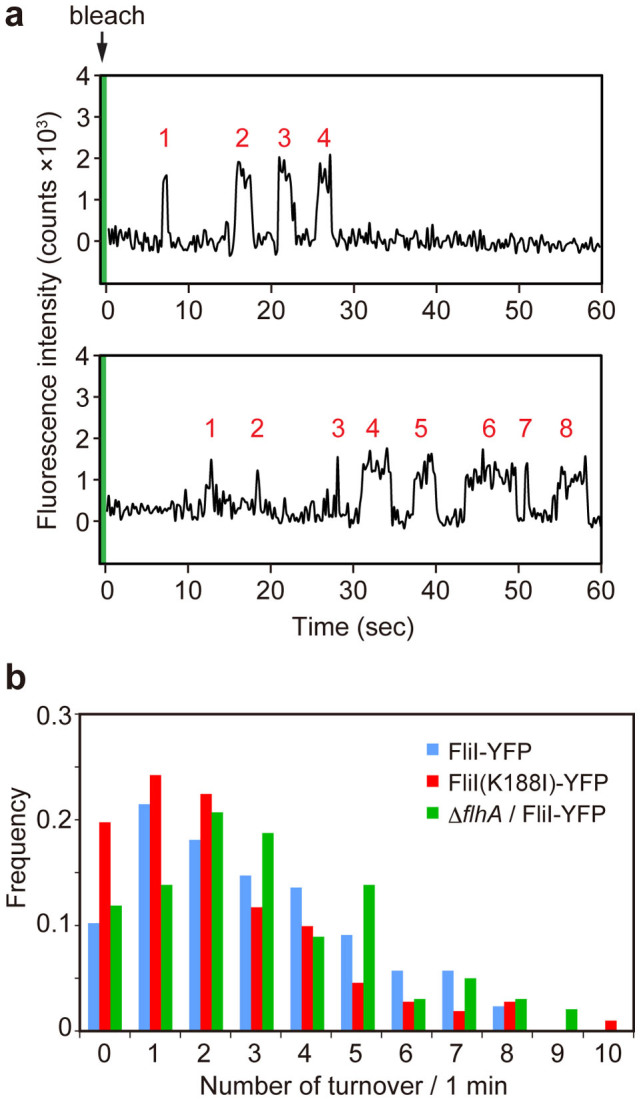
(a) Typical two examples of the fluorescent intensity trace in each FliI-YFP spot in Salmonella MKM30 strain harboring pJSV203 under continuous TIRF illumination after one-shot photobleaching using a strong excitation laser (green band). Peaks labeled with numbers in red show intensity recovery of a single FliI-YFP molecule within a time period of 60 seconds. (b) The number of times of single FliI-YFP molecule turnover per minute. Each YFP spot was counted in Salmonella MKM30 (ΔfliI) strain harboring pJSV203 (as indicated as FliI-YFP, light blue) (n = 89) and pSY001 (as indicated as FliI(K188I)-YFP, red) (n = 112) and NH0027 (ΔflhA ΔfliI) carrying pJSV203 (as indicated as ΔflhA/FliI-YFP, green) (n = 102).
To test whether a catalytic mutation in FliI or deletion of FlhA affects the rate of FliI-YFP turnover, we performed the TIRF-FRAP experiments with the fliI(K188I)-yfp and ΔflhA fliI-yfp strains. FliI(K188I)-YFP showed turnover with an average of 2.2 ± 2.1 times per minute after photobleaching (n = 112) (Fig. 4b). Statistical analysis with two-tailed t-test indicated that this average number of times showed only a weak difference compared to the fliI-yfp strain (p = 0.03), suggesting that ATP hydrolysis by FliI does not influence the turnover rate. In the ΔflhA fliI-yfp strain, the average turnover frequency of FliI-YFP molecules was 3.1 ± 2.2 (n = 102) per minute, which shows no significant difference compared to the fliI-yfp strain (p = 0.55) (Fig. 4b). Because a specific interaction between FliH and FliN is required for the localization of FliI-YFP to the FBB26, we suggest that the FliH-FliN interaction is highly dynamic rather than static.
Discussion
FliI ATPase forms not only the FliH2FliI complex in the cytoplasm but also the FliH12FliI6FliJ ring complex on the FlhAC9-FlhBC docking platform of the export gate. In vitro reconstitution experiments of the FliI6 ring have shown that FliI couples ATP-binding and hydrolysis to its assembly-disassembly cycle3. In this work, we presented in vivo studies of protein stoichiometry and turnover of FliI with single molecule precision in living Salmonella cells and showed that the number of FliI-YFP associated with the FBB ranged from 3 to 13 with an average of 6.7 ± 1.7 per spot (Fig. 2d). The predominant number was six, which is in agreement with the stoichiometry of the FliI6 ring18,30. Such an apparent variation in the number could result from imperfect localization of some fluorescent foci to the focal imaging plane that would reduce their fluorescence intensity, a failure of FliI-YFP to activate, and/or prior bleaching, thereby leading to underestimation of the stoichiometry of FliI-YFP molecules associated with the FBB. However, we actually observed an excess number of FliI molecules up to seven in about 40% of spots (Fig. 2b and d), which raises the possibility that not only the FliI6 ring but also FliH2FliI complexes associate with the FBB. Interestingly, the number of spots with three to five FliI-YFP molecules increased significantly in the ΔflhA mutant, and the populations of spots with more than six FliI-YFP molecules decreased significantly although the majority of FliI-YFP stoichiometry was still six (Fig. 2e and f). This indicates that removal of FlhA reduces the number of FliI-YFP molecules associated with the FBB. Therefore, we suggest that FlhA contributes to some additional localization of FliI-YFP to the FBB. It has been shown that FliH-Trp7 and FliH-Trp10 are responsible not only for the interaction between FliH and FliN but also for the interaction between FliH and FlhA26,27. Because FlhA exists as a homo-nonamer in the export gate12,13,14, we propose that not only the FliI6 ring but also several FliH2FliI complexes may associate with the FBB through both the FliH–FliN and FliH–FlhA interactions (Fig. 5).
Figure 5. Schematic diagrams of the bacterial flagellar export apparatus.
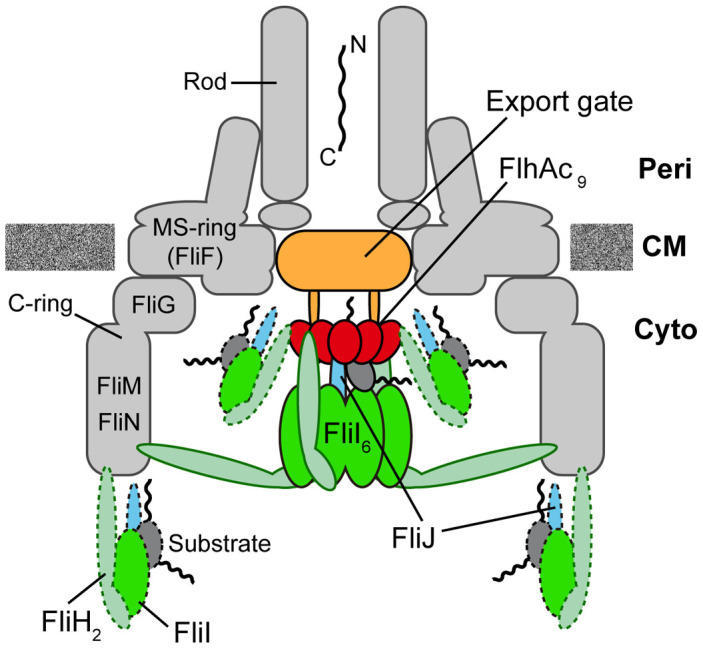
The export gate made of FlhA, FlhB, FliO, FliP, FliQ and FliR are located within the central pore of the MS ring. The C-terminal cytoplasmic domain of FlhA (FlhAC) forms a nonameric ring structure and projects into the cavity of the C ring formed by FliG, FliM, and FliN. FliI forms a homo-hexamer (FliI6). The FliI6 ring firmly associates with the FBB through interactions of FliH with both FliN and FlhA. FliI also forms the FliH2-FliI complex and binds to the FBB through the FliH-FliN and FliH-FlhA interactions. During flagellar assembly, the FliH2FliI complex binds to FliJ and export substrate in the cytoplasm and acts as a dynamic carrier (dashed line) to deliver FliJ and export substrate to the C and FlhAC9 rings. Upon formation of the FliH12FliI6FliJ ring complex (continuous line) on the FlhAC9-FlhBC platform, the FliI6 ring can act as a substrate loader to promote the initial entry of the substrate into the gate. A specific interaction of FliJ located at the center of the FliI6 ring with a flexible linker of FlhA allows the export gate to translocate flagellar protein in a PMF-dependent manner.
Recent structural studies by electron cryotomography have identified a spherical density at the bottom of the FBB as the FliI6 ring, but its position is relatively far from the FlhAC9 ring13,40. FliJ binds to the center of the FliI6 ring to form the FliI6FliJ ring complex19 and a specific interaction of FliJ at the center of the FliI6 ring with FlhAL allows the export gate to efficiently utilize the electric potential difference of PMF for flagellar protein export28,29. Therefore, it is proposed that the export apparatus visualized by electron cryotomography is in an export-off state13. The spherical density corresponding to the FliI6 ring is still observed even in the absence of the FlhAC9 ring12. FliHEN containing FliH-Trp7 and FliH-Trp10 binds to a surface-exposed hydrophobic patch formed by three highly conserved Val111, Val112 and Val113 residues of FliN27,41. This suggests that the FliI6 ring associates with the FBB C ring through such strong hydrophobic interactions between FliHEN and FliN. Since removal of FliJ does not affect the subcellular localization of FliI-YFP to the flagellar base26, we suggest that the binding of the FliH2FliI complexes to the C ring through the FliH-FliN interaction increases the local concentration of FliI at the flagellar base, thereby promoting FliH12FliI6 ring formation independently of FliJ as well as the FlhAC9-FlhBC platform in vivo.
In vitro reconstitution experiments have shown that FliI requires ATP or its analogs for FliI ring formation18. In agreement with this, the K188I mutation of FliI, which considerably reduces the affinity of FliI for ATP, remarkably decreases the efficiency of FliI ring formation. On the other hand, the E211Q mutation of FliI, not affecting ATP-binding but resulting in the complete loss of the ATPase activity, does not affect FliI ring formation30. These suggest that in vitro assembly-disassembly cycle of the FliI6 ring occurs in an ATP dependent manner. In contrast, we found that neither the stoichiometry of FliI-YFP associated with the FBB (Fig. 3) nor FliI-YFP exchange between the FBB and free-diffusing cytoplasmic pool were affected. Therefore, we suggest that the chemical energy derived from ATP hydrolysis by FliI is not responsible for the assembly-disassembly cycle of FliI molecules in vivo.
The chaperone–substrate complexes bind to the FliH2FliI complex through co-operative interactions between FliI, chaperone and export substrate, suggesting that the FliH2FliI complex efficiently delivers export substrate and chaperone-substrate complex to the export gate24,25. TIRF-FRAP experiments revealed that about 90% of the FliI-YFP spots show FliI-YFP molecule turnover between the FBB-localized and free-diffusing ones after photobleaching (Fig. 4 and Supplementary Fig. 1). About six FliI-YFP molecules were estimated to exchange within 1 minute (Fig. 4). These observations support the hypothesis that the FliH2FliI complex acts as a dynamic carrier. The maximum rate of flagellar protein export in the very early stage of filament assembly is estimated to be about 20 molecules per sec in Salmonella42, which is much faster than the rate of FliI-YFP turnover. Thus, flagellar protein export is a highly efficient and speedy process. In contrast, when the length of flagellar filaments reaches longer than 10 μm, the rate of filament growth slows down to 0.2–0.3 μm/h, indicating that the export rate of flagellin subunit is about 10 molecules/min43. Because most of fliI-YFP cells that we observed in the present study have quite long filaments, around 10 μm long, the turnover rate of FliI-YFP we observed, about 6 molecules/min, is presumably due to the slow export state of the export apparatus. FlhA forms a nonameric ring in the export apparatus12,13,14 and not only provides the binding sites for chaperone-substrate complexes9,10,11 but also contributes to some additional localization of FliH2FliI molecules to the FBB (Fig. 2e and f). Therefore, we propose that the FliH2FliI complex and the FlhA9 ring act as a dynamic carrier and a sorting platform, respectively, allowing rapid and efficient entry of export substrates into the export gate with the help of the FliI6 ring, which appears to act as a static substrate loader (Fig. 5).
Methods
Bacterial strains, plasmids, and media
Bacterial strains and plasmids used in this study are listed in Table 1. DNA manipulations, site-directed mutagenesis and DNA sequencing were carried out as described previously44. To construct Salmonella cfp-fliG ΔfliI and cfp-fliG ΔfliH-fliI strains, the fliG gene on the chromosome was replaced by the cfp-fliG allele by using the λ Red homologous recombination system45 as described before44. L-broth (LB) and soft tryptone agar plates were used as described previously46. Ampicillin was added to LB at a final concentration of 100 μg/ml.
Table 1. Strains and plasmids used in this study.
| Strains and Plasmids | Relevant characteristics | Source or reference |
|---|---|---|
| Salmonella | ||
| MKM30 | ΔfliI | (48) |
| SJW1684 | ΔfliF | (49) |
| NH0027 | ΔflhA ΔfliI | This study |
| YVM027 | ecfpA206K-fliG | This study |
| YVM028 | ecfpA206K-fliG ΔfliH-fliI | This study |
| YVM029 | ecfpA206K-fliG ΔfliI | This study |
| YVMA013 | ecfpA206K-fliG ΔfliI ΔflhA::tetRA | This study |
| Plasmids | ||
| pET19b | Expression vector | Novagen |
| pJSV203 | pET19b/His-FliI-EYFPA206K | (26) |
| pJSV231 | pET19b/His-FliF-EYFPA206K | (14) |
| pSY001 | pET19b/His-FliI(K188I)-EYFPA206K | (26) |
| pSY002 | pET19b/His-FliI(E211A)-EYFPA206K | This study |
Preparation of Salmonella cells for fluorescence microscopy
Overnight culture of Salmonella cells was inoculated into fresh LB and incubated at 30°C with shaking for 3 hours. The cells were washed twice with motility medium (10 mM potassium phosphate pH 7.0, 0.1 mM EDTA, and 10 mM L-sodium lactate) and resuspended in the motility medium. Then, the cells were put on a coverslip for 10 min at 23°C to attach onto the coverslip surface. Unbound cells were washed out by adding 100 μl of the motility medium.
Fluorescence microscopy
To observe bacterial cell bodies and fluorescence of CFP and YFP, we used a custom-built microscope based on an inverted fluorescence microscope (IX-71, Olympus) with a 150× oil immersion objective lens (UApo150XOTIRFM, NA 1.45, Olympus) and with an Electron-Multiplying Charge-Coupled Device (EMCCD) camera (iXonEM+897-BI, Andor Technology). CFP and YFP were excited by a 29 mW diode laser with a wavelength of 440 nm (LD440, Olympus) with an emission filter of 480AF30 (Omega Optical) and a 150 mW gas laser with a wavelength of 514 nm (35LAS515, Melles Griot) with an emission filter of 535AF26 (Omega Optical), respectively. The primary beam of 514 nm gas laser was split into two independently attenuated paths by a polarizing beam-splitting cube to generate a separate excitation path that was used for continuous fluorescence observation, and a strong laser for photobleaching. 25% transmission ND filter (Olympus) was used for TIRF-FRAP experiments to reduce laser power. The 514 nm laser was gated by a high-speed mechanical shutter (Uniblitz UHS1T2, Vincent Associates). The on/off switching of the mechanical shutter and EMCCD camera were controlled by an electrical stimulator (SEN-8203, Nihon Kohden) and the Andor Solis software (Andor Technology). Fluorescent images were captured by the EMCCD camera with an exposure time of 50 or 200 ms in continuous epi-fluorescence experiments. After photobleaching of FliI-YFP spots by the strong laser excitation for 400 ms under TIRF, the recovery of the FliI-YFP spots were observed with an exposure time of 200 ms under TIRF illumination. To estimate the fraction of unbleached FliI-YFP in the cytoplasm, the fluorescence intensities of epi-fluorescence images of fliI-yfp cells (MKM30/pJSV203) containing no fluorescent spots were compared before and after photobleaching by strong excitation under TIRF.
Image acquisition and data analysis
Fluorescent images were analyzed by an image processing program we developed based on the Igor Pro 6.22j software (WaveMetrics). We applied a square mask for the contribution of each FliI-YFP fluorescent spot of 8 × 8 pixels to the ROI. We defined the spot intensity as the sum of all pixel values within the square mask after subtraction of the total background intensity from each pixel value. We defined the instrumental background intensity as the mean pixel intensity within the same size of ROI in a region contains no cell. The contribution to the background count due to autofluorescence of cells and diffusive FliI-YFP in the cytoplasm was calculated from the area within cell at some pixel distance from fluorescent spots after subtracting the autofluorescence contribution per pixel measured in the non-fluorescent strain (MKM30 harboring pET19b) and the instrumental background.
Estimating stoichiometry of FliI-YFP fluorescent spots
With continuous epi-fluorescence imaging of single fluorescent spot of FliI-YFP, step-wise photobleaching was detected from the fluorescent intensity signal. Detection of photobleaching steps was performed separately for each fluorescent spot. Each photobleaching intensity trace was filtered using a step finding algorithm we have developed based on Igor Pro 6.22j software47. Pairwise differences for all filtered photobleaching traces were calculated to identify the unitary step size of FliI-YFP intensity decay as described before14. To repress the misdetection of noise, a threshold for fluorescence intensity was defined. The number of steps was thus obtained for each fluorescent spot.
Author Contributions
F.B., Y.V.M., K.N. and T.M. conceived and designed research; F.B., Y.V.M., S.D.J.Y., N.H. and T.M. preformed research; F.B., Y.V.M., N.K. and T.M. analysed the data; and F.B., Y.V.M., K.N. and T.M. wrote the paper based on discussion with other authors.
Supplementary Material
Supplementary movie 1
Supplementary Information
Acknowledgments
We acknowledge Masahiro Ueda for continuous support and encouragement. FB and YVM were research fellows of the Japan Society for the Promotion of Science (JSPS). This work was supported in part by JSPS KAKENHI Grant Numbers 21227006 and 25000013 (to K.N.) and 26293097 (to T.M.) and MEXT KAKENHI Grant Numbers 23115008, 24117004 and 25121718 (to T.M.) and 26115720 (to Y.V.M).
References
- Chevance F. F. & Hughes K. T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Imada K. & Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. BioSyst. 4, 1105–1115 (2008). [DOI] [PubMed] [Google Scholar]
- Minamino T. Protein export through the bacterial flagellar type III export pathway. Biochim. Biophys. Acta. 1843, 1642–1648 (2014). [DOI] [PubMed] [Google Scholar]
- Francis N. R., Sosinsky G. E., Thomas D. & DeRosier D. J. Isolation, characterization, and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235, 1261–1270 (1994). [DOI] [PubMed] [Google Scholar]
- González-Pedrajo B., Minamino T., Kihara M. & Namba K. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60, 984–998 (2006). [DOI] [PubMed] [Google Scholar]
- Cornelis G. R. The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825 (2006). [DOI] [PubMed] [Google Scholar]
- Minamino T. & Macnab R. M. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35, 1052–1064 (2000). [DOI] [PubMed] [Google Scholar]
- Minamino T. et al. Role of the C-terminal cytoplasmic domain of FlhA in bacterial flagellar type III protein export. J. Bacteriol. 192, 1929–1936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. et al. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 83, 775–788 (2012). [DOI] [PubMed] [Google Scholar]
- Bange G. et al. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. USA 107, 11295–11300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Hara N., Imada K., Namba K. & Minamino T. Interactions of bacterial chaperone-substrate complexes with FlhA contribute to coordinating assembly of the flagellar filament. Mo.l Microbiol. 90, 1249–1261 (2013). [DOI] [PubMed] [Google Scholar]
- Abrusci P. et al. Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto A. et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci. Rep. 3, 3369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto Y. V. et al. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol. Microbiol. 91, 1214–1226 (2014). [DOI] [PubMed] [Google Scholar]
- Minamino T. & Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451, 485–488 (2008). [DOI] [PubMed] [Google Scholar]
- Paul K., Erhardt M., Hirano T., Blair D. F. & Hughes K. T. Energy source of the flagellar type III secretion. Nature 451, 489–492 (2008). [DOI] [PubMed] [Google Scholar]
- Fan F. & Macnab R. M. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271, 31981–31988 (1996). [DOI] [PubMed] [Google Scholar]
- Claret L., Susannah C. R., Higgins M. & Huges C. Oligomerisation and activation of the FliI ATPase central to the bacterial flagellum assembly. Mol. Microbiol. 48, 1349–1355 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibuki T. et al. Common architecture between the flagellar protein export apparatus and F- and V-ATPases. Nat. Struct. Mol. Biol. 18, 277–282 (2011). [DOI] [PubMed] [Google Scholar]
- Imada K., Minamino T., Tahara A. & Namba K. Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc. Natl. Acad. Sci. USA 104, 485–490 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa J. et al. Common evolutionary origin for the rotor domain of rotary ATPases and flagellar protein export apparatus. PLoS One. 8, e64695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. & Macnab R. M. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37, 1494–1503 (2000). [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Bailey C. M. & Beatson S. A. Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases. Protein Sci. 15, 935–941 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J., Stafford G. P. & Hughes C. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. USA 101, 3945–3950 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Kinoshita M., Imada K. & Namba K. Interaction between FliI ATPase and a flagellar chaperone FliT during bacterial flagellar protein export. Mol. Microbiol. 83, 168–178 (2012). [DOI] [PubMed] [Google Scholar]
- Minamino T. et al. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol. Microbiol. 74, 1471–1483 (2009). [DOI] [PubMed] [Google Scholar]
- Hara N., Morimoto Y. V., Kawamoto A., Namba K. & Minamino T. Interaction of the extreme N-terminal region of FliH with FlhA is required for efficient bacterial flagellar protein export. J. Bacteriol. 194, 5353–5360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Morimoto Y. V., Hara N. & Namba K. An energy transduction mechanism used in bacterial type III protein export. Nat. Commun. 2, 475 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibuki T. et al. Interaction between FliJ and FlhA, components of the bacterial flagellar type III export apparatus. J. Bacteriol. 195, 466–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazetani K., Minamino T., Miyata T., Kato T. & Namba K. ATP-induced FliI hexamerization facilitates bacterial flagellar protein export. Biochem. Biophys. Res. Commun. 388, 323–327 (2009). [DOI] [PubMed] [Google Scholar]
- Li H. & Sourjik V. Assembly and stability of flagellar motor in Escherichia coli. Mol. Microbiol. 80, 886–899 (2011). [DOI] [PubMed] [Google Scholar]
- Minamino T., González-Pedrajo B., Kihara M., Namba K. & Macnab R. M. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator FliH. J. Bacteriol. 185, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry J. L., Murphy J. W. & González-Pedrajo B. The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45, 11790–11798 (2006). [DOI] [PubMed] [Google Scholar]
- Leake M. C. et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. USA 105, 15376–15381 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake M. C. et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 (2006). [DOI] [PubMed] [Google Scholar]
- Delalez N. J. et al. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc. Natl. Acad. Sci. USA 107, 11347–11351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Yonekura K. & Namba K. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J. Mol. Biol. 337, 105–113 (2004). [DOI] [PubMed] [Google Scholar]
- Stafford G. P. et al. Sorting of early and late flagellar subunits after docking at the membrane ATPase of the type III export pathway. J. Mol. Biol. 374, 877–882 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka H., Inoue Y., Terasawa S., Takahashi H. & Ishijima A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem. Biophys. Res. Commun. 394, 130–135 (2010). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. Structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K., Harmon J. G. & Blair D. F. Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J. Bacteriol. 188, 5240–5248 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T. Assembly of Salmonella flagellin in vitro and in vivo. J. Supramol. Struct. 2, 372–384 (1974). [DOI] [PubMed] [Google Scholar]
- Aizawa S. & Kubori T. Bacterial flagellation and cell division. Genes Cells 3, 625–634 (1998). [DOI] [PubMed] [Google Scholar]
- Hara N., Namba K. & Minamino T. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS One 6, e22417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A. & Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. & Macnab R. M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181, 1388–1394 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Kami-ike N., Yokota J. P., Minamino T. & Namba K. Evidence for symmetry in the elementary process of bidirectional torque generation by the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 107, 17616–17620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. et al. Oligomerization of the bacterial flagellar ATPase FliI is controlled by its extreme N-terminal region. J. Mol. Biol. 360, 510–519 (2006). [DOI] [PubMed] [Google Scholar]
- Kubori T., Shimamoto N., Yamaguchi S., Namba K. & Aizwa S. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226, 433–446 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary movie 1
Supplementary Information