Abstract
A novel avian-origin H7N9 influenza strain emerged in China in April 2013. Since its re-emergence in October–November 2013, the number of reported cases has accelerated; more than 220 laboratory-confirmed cases and 112 deaths (case fatality rate of 20–30%) have been reported. The resurgence of H7N9 has re-emphasized the importance of making faster and more effective influenza vaccines than those that are currently available. Recombinant H7 hemagglutinin (H7-HA) vaccines have been produced, addressing the first problem. Unfortunately, these recombinant subunit vaccine products appear to have failed to address the second problem, influenza vaccine efficacy. Reported unadjuvanted H7N9 vaccine seroconversion rates were between 6% and 16%, nearly 10-fold lower than rates for unadjuvanted vaccine seroconversion to standard H1N1 monovalent (recombinant) vaccine (89% to pandemic H1N1). Could this state of affairs have been predicted? As it turns out, yes, and it was.1 In that previous analysis of available H7-HA sequences, we found fewer T-cell epitopes per protein than expected, and predicted that H7-HA-based vaccines would be much less antigenic than recent seasonal vaccines. Novel approaches to developing a more immunogenic HA were offered for consideration at the time, and now, as the low immunogenicity of H7N9 vaccines appears to indicate, they appear to be even more relevant. More effective H7N9 influenza vaccines can be produced, provided that the role of T-cell epitopes is carefully considered, and accumulated knowledge about the importance of cross-conserved epitopes between viral subtypes is applied to the design of those vaccines.
Keywords: CD4+ T cell, H1N1, H7N9, T cell, T helper, T-cell epitope, avian-origin, cell-mediated immunity, genome, hemagglutinin, influenza, recombinant protein
Introduction
A new avian-origin influenza, known initially as H7N9 A/Anhui/1/2013 (H7N9), emerged near Shanghai beginning in February, peaking in May–June and losing momentum in July 2013.2 After a period of quiescence, case reports accelerated in October 2013: more than 220 cases of H7N9 have occurred as of February 2014.3 The high mortality rate (case fatality rate or CFR of 20–30%) and rapid spread of the new strain of H7N9 throughout China4 is renewing concern about the availability of effective anti-H7N9 vaccines. A number of H7N9 vaccines are in clinical trials; however, the efficacy of these vaccines is reported to be extremely low compared with other subunit and seasonal influenza vaccines, as shown in Figure 1. While the addition of adjuvant to the vaccines did increase the antigenicity of the vaccines in young, healthy adults, adjuvant is not currently used in standard seasonal influenza vaccines. Experts have expressed some concern about the use of adjuvant in influenza vaccination due to the recently reported association between adjuvanted influenza vaccines and narcolepsy in Finnish (and Swedish) children.5

Figure 1. Comparative effectiveness of unadjuvanted monovalent recombinant vaccines (IAV H7N9 and H1N1) and seasonal trivalent vaccine. Percent effectiveness is shown, as percent of study cohort achieving “significant” seroconversion (see text). The Novartis vaccine company reported that H7N9 cell culture-produced vaccine without adjuvant produced seroconversion rates of 6%.20 In contrast, the rate of seroconversion to unadjuvanted monovalent pandemic H1N1 was 89% in one study.23 Rates of seroconversion to trivalent seasonal vaccine are 84% on average to the three strains of virus in the vaccine.25
The H7N9 genome was first made available on the GISAID website on April 2, 2013; it was rapidly analyzed and compared with other circulating H7-type viruses6-8 and examined for potential antigenicity by our group.1 By early 2013, European, American, and Asian influenza vaccine companies were entering clinical trials with recombinant subunit vaccines produced in protein-production cell lines.9,10
Our group viewed this approach to the production of H7N9 vaccines with some concern, because our previous analysis of the H7-HA sequence indicated that it had fewer than expected T-cell epitopes and would be poorly antigenic.1 This analysis appears to be even more relevant in light of the new H7N9 vaccine efficacy data. Briefly, we estimated the immunogenic potential of the new influenza using well-established immunoinformatics tools.11-13 We then normalized the H7-HA T-cell epitope content to protein length, and compared it to other circulating influenza strains. As shown in Figure 2, H7-HA proteins and HA tested in H7N7 and H7N1 vaccines were found to contain 14–24% fewer T-cell epitopes (per length) than other circulating strains of IAV (A/California/07/2009, A/Victoria/361/2011, and A/Texas/50/2012).

Figure 2. EpiMatrix Immunogenicity scale comparing the potential antigenicity of H7-HA to recent seasonal influenza A strain HA proteins. Using EpiMatrix and assessing each protein for overall T-cell epitope content, the protein scores are plotted on an immunogenicity scale that ranks the immunogenic potential of HA protein derived from recently-circulating strains of seasonal IAV (H1N1, H3N2) as compared with strains of avian-origin influenza. The set of avian-origin IAV strains have a low immunogenicity potential (below zero), EpiMatrix scores that are commonly associated with low immunogenicity proteins that do not effectively trigger IgG antibody responses. Scores are shown for H7-HA from several different sources, including the vaccines described in the text (refs. 22 and 23 [Anhui, scoring -6.49], 28 [mallard, –7.12], and 29 [chicken, –6.43]). In contrast, H1N1 California, H3N2 Victoria, and H2N3 Texas, rank +19.22, +16.23, and +14.25 respectively). The EpiMatrix Immunogenicity Scale is derived by analyzing more than 10 000 random protein sequences for T cell epitope content and producing a score that reflects the normal distribution of T cell epitope content per protein length. Proteins scoring higher than 10 on this scale are usually antigenic; proteins scoring lower than 10 are generally poorly antigenic. Details on the methodology are available in reference 13 and other publications by the group.
H7-HA also appeared to be “de-immunized” and “tolerized” with respect the quantity and quality of T-cell epitopes expected in proteins of similar size (Fig. 2). De-immunization, or removal of T-cell epitopes, is a method employed by some viruses to evade human immune response. Tolerization relates to the introduction of T-cell epitopes that are highly conserved, on the T-cell receptor face, with a large number of other similar epitopes contained in the human genome. We recently observed de-immunization and tolerization of T-cell epitopes to be prevalent among viruses that are known to be commensal in human beings, such as Herpes simplex virus, Cytomegalovirus, and Epstein Barr virus.14 This type of epitope analysis is especially relevant to influenza vaccine design, since the principles of protein engineering could be applied to correct these features and improve the antigenicity of recombinant subunit HA-based vaccines. Using a similar approach, we correctly predicted the antigenicity of influenza A/California/04/2009 (H1N1), which is due in part to its substantial ration of T helper epitopes with the potential to cross-react with other seasonal H1N1 strains,15,16 a finding that was later validated in studies performed by other groups.17-19
H7-HA stands out as a uniquely non-immunogenic protein, which may be due to the paucity of T-cell epitopes that we described previously and to cross-conserved epitopes (with the human genome) that may further reduce its antigenicity. Here, we reiterate our hypothesis that the emerging avian-origin H7N9 2013 might behave like a “stealth” virus, using the T-cell epitope deimmunization and tolerization methods to evade human cellular and humoral immune response, and review these findings in the context of recently published vaccine efficacy and human serology reports.
H7N9 Vaccines: Current Status
Two vaccine studies20,21 and one clinical serology study of H7N9-infected and exposed subjects22 have been published or announced in the last few months. Novartis produced a monovalent inactivated vaccine in MDCK cells,9 and Novavax produced a virus-like particle vaccine in insect cells.10 The reported seroconversion rates to unadjuvanted H7N9 HA vaccines were extremely low (6–16%) as compared with the 89% seroconversion rate observed in response to subunit influenza vaccines in comparable studies of pandemic H1N1 HA.23-25 According to the Novartis media release, the vaccine’s protective immune response was only achieved after two doses of 15 µg of the MF59-adjuvanted vaccine (protective HAI titers in 85% of healthy subjects, 400 healthy volunteers, 18–64 y of age). In contrast, “…only 6% of subjects achieved a protective response when given two doses of the 15 µg unadjuvanted vaccine.” Similarly low hemagglutination-inhibition (HAI) titers were achieved in the Novavax study: seroconversion and reciprocal antibody titers of 40 or more (…the value cited by regulatory authorities as having a potential association with clinical benefit) were only detected in 5.7% and 15.6% of participants receiving 15 μg and 45 μg of HA, respectively, without adjuvant. Addition of adjuvant to the Novavax vaccine significantly increased the level of HAI titers; more than 80% of participants receiving 5 μg of the HA vaccine with 60 units of adjuvant saponin-based ISCOMATRIX adjuvant seroconverted.
The low immunogenicity of H7-HA has been previously reported in pre-clinical (mouse) and clinical (human) studies. In one such previous study, a low-pathogenicity recombinant H7N7 virus was generated by reverse genetics and shown to protect mice against lethal infection with a high-pathogenicity H7N7 virus. Importantly, protection was only achieved with co-administration of adjuvant.26 This subunit vaccine was developed by cloning the HA of influenza virus A/Mallard/Netherlands/12/00 (H7N3) and the NA of influenza virus A/Netherlands/33/03 (H7N7) into the cell culture-adapted strain A/PR/8/34.
In a second study, a split H7N1 vaccine was prepared by reverse genetics and produced in a specialized cell line (PER.C6). The vaccine contained the internal genes of A/PR/8/34 with surface antigens of the highly pathogenic avian strain (A/Chicken/Italy/13474/l99(H7N1). This vaccine was also observed to be poorly immunogenic in a Phase I clinical trial27 even when given in two doses containing either 12 μg or 24 μg of HA alone or with aluminum hydroxide (Alum) adjuvant. This vaccine was found to induce only low antibody responses in humans, and again, only when formulated with the adjuvant.
These findings may have contributed to the decision by vaccine developers to compare unadjuvanted and adjuvanted H7N9 vaccines in side-by-side cohorts in the recent clinical studies described above. Adjuvanting H7N9 influenza vaccines is one approach to improving the antigenicity of H7-HA, and it may become necessary, but emerging associations between adjuvanted influenza vaccination and narcolepsy in Finland and Sweden5,28,29 raise concern about the potential for adverse immune responses to occur.
Human Infection and Seroconversion
In the recent noteworthy seroepidemiology study performed by Guo et al., the protective antibody response against influenza A (H7N9) virus was investigated in 48 acute-phase and convalescent serum samples from 21 subjects with laboratory-confirmed infection.24 Neutralizing anti-H7-HA antibodies were undetectable in samples collected up to 28 d after symptom onset but were observed at 29–37 d. This differs significantly from responses in pandemic H1N1 and H5N1 infections, which are observed at days 14–21. The avidity of serum IgG antibodies for H7-HA was also lower than that observed for H1-HA and H3-HA. Thus, the H7N9 virus infection stimulated delayed and weak antibody responses in the cases investigated by Guo et al., suggesting that vaccination using conventional approaches may also be inefficient.
According to the authors, weak antibody response and low avidity of the resulting anti-HA antibodies might play a role in the severity and duration of infections and the pathogenesis of H7N9 disease. The low titer of antibodies to HA resulting from infection is also noteworthy since serology is also used to detect infection in large population studies. If antibody titers are low among infected individuals, it is possible that the prevalence of exposure to H7N9 might be underreported. One consequence of under-diagnosis is known as the tip of the iceberg phenomenon (prevalence is underestimated due to under-reporting). Indeed, a number of H7N9 cases in China were only identified following routine serosurveillance.30 Should human-to-human transmission of H7N9 begin, other means of identifying infected individuals in large surveys (such as T-cell assays) may be needed, as they may prove to be more accurate than serology.
Epidemiology, Morbidity, and Mortality
The first cases of human infection with H7N9 were detected in residents of the city of Shanghai and in residents of Anhui province.2 The number of cases peaked in May–June and decreased during the summer, but the virus re-emerged in October 2013.6 As of February 1st 2014, more than 280 laboratory-confirmed cases of human infection have already been reported. That is nearly half as many as have occurred for H5N1 (566, as of December 2014), and they are accumulating at a pace that is five times faster than H5N1. Even more worrisome, the number of cases reported in a single province (Zhejiang) over the last two-month period (December 2013–January 2014) already exceeds the peak number of cases in the same region, per two month period, last year.31
H7N9 is “non-pathogenic” in birds but pathogenic in humans. In humans, the CFR is 22%;3 some experts believe that there are quite a few cases that are not laboratory-confirmed, thus the CFR may be lower. Most patients initially developed an influenza-like illness that progressed to respiratory distress syndrome.2 The median age of laboratory-confirmed cases is 56 y, but there is a wide age range (from infant to 91). Infections in men are more frequently reported than in women. In a recent study of 82 confirmed cases with information on possible exposures, 63 (77%) of the cases had contact with live animals at or near the time of infection.4,5 Poultry and other live birds (including pet birds) have been identified as the likely source of infection, although the prevalence of H7N9 influenza in poultry flocks remains extremely low, and a direct link has not yet been proven. The lack of any evidence of circulating H7N9 in poultry has lead some to postulate that another, potentially mammalian, vector is transmitting H7N9 to humans. Of note, the virus appears to have a “mammalian signature,” according to experts at the National Institute for Infectious Disease.9
Public Health Concerns
There are four important reasons why public health officials are on high alert regarding H7N9: First, individual cases of nearly identical H7N9 viral infections have been identified in a wide range of locations within China in a very short period of time. Cases have been reported from 11 provinces in China and two municipalities (Beijing and Shanghai). Taiwan reported two cases imported from Jiangsu, and Hong Kong reported three cases imported from Guangdong.4 The wide distribution of cases strongly suggests that the virus is already widespread in an as-yet-unidentified reservoir population, although human-to-human transmission is rare.32 Second, H7N9 subtype influenza has never before infected humans to a significant extent, meaning the human population has little or no pre-existing immunity. This clearly contributes to the extremely high mortality rate, which ranges between 20 and 30% of individuals.33,34 Third, higher rates of human infection by the emerging H7N9, as reported to occur over the most recent time period, are worrisome because each transmission may be associated with mutations in the viral sequence that increase the likelihood of human-to-human transmission. The pace of human infection appears to have accelerated in recent months.3 According to a recently-published report, the virus can be transmitted between mammals (guinea pigs and ferrets).35,36 More infections may lead to the chance occurrence of the one or two mutations that would enable either drug resistance to occur,37 or enable the virus to achieve full adaptation for human-to-human transmission.30 And fourth, there is no reason to believe that the H7N9 virus will remain geographically confined to China. Not only do wild bird flyways provide a convenient route of spread to Africa, Europe, and North America,38,39 but also human flyways40 (see the recent report of the arrival of H5N1 in North America, ref. 41) provide an easy means to transmit H7N9, should mutations to H7N9 that enable human-to-human transmission occur.
Transport of H7N9 along poultry trade routes or wild bird migration routes appears to be highly likely (Fig. 3). Should H7N9 spill over the borders of China to nearby countries such as Vietnam (through local, unregulated poultry trade), or more distantly by bird flyways to Africa, it is likely to cause an even greater number of human cases. Poultry stock is ubiquitous and contact between humans and food animals is extensive in most developing world countries. In addition, influenza vaccination campaigns are unheard of, although infrastructure does exist for door-to-door vaccination programs; these have been implemented in the context of polio outbreaks. Little is known about the epidemic burden of IAV among African communities, and information about IAV circulation in the population has only emerged recently (with H1N1 in 2009).42 From those studies, it is clear that IAV can be introduced by two routes: first by travelers, (e.g., Malian workers returning home from Europe42); and second by birds. One important location where avian transmission might occur is the inner Niger Delta in Mali, a wetland of 3 000 000 hectares with flood plains, lakes, river branches, and small pockets of flood forest that plays an extremely important role in bird migration throughout Africa. Endogenous circulation of H1N1 has been observed on the Inner Niger Delta, where the human seroconversion rate has been reported to be about 13%.43 A third factor that may also contribute to the spread of H7N9 to developing world countries, should human-to-human transmission become possible, is the increased mobility of Chinese workers worldwide.44 Taken together, the potential for H7N9 is of great concern for developing world countries in Asia and Africa. Scientists, infectious disease experts, and public health officials are watching the re-emergence of H7N9 and its spread within China with growing concern.
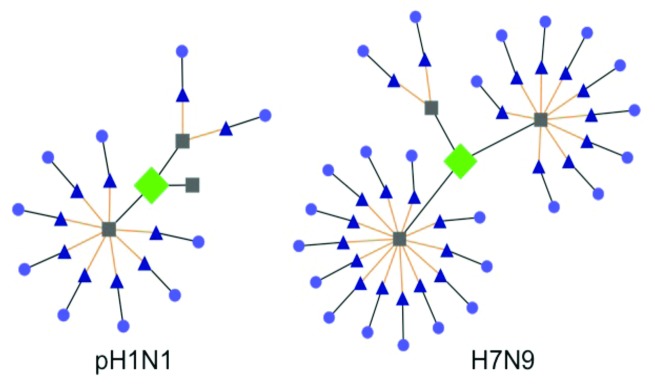
Figure 3. Contrasting TCR-epitope networks of “classic” T-cell epitopes from pH1N1 and H7N9 HA. Human genome T-cell epitope conservation defined by JanusMatrix53 is illustrated using Cytoscape. Here, the more limited human conservation of TCR-facing residues for the T-cell epitope in Influenza A/H1N1 2009/CA (pH1N1; three 9-mers, with 0, 2, or 8 human conserved epitopes), contrasts with a similar epitope in H7N9 (three 9-mers, with 9, 13, or 2 human conserved epitopes). A detailed description of the JanusMatrix tool is available in reference 53.
Novel Vaccines: Strategies That Will Work
Should H7N9 develop pandemic potential, novel strategies for improving the immunogenicity of vaccines for this unique low-immunogenicity strain of avian-origin influenza will be needed. Several approaches to improving the next generation of H7N9 or other IAV vaccines are reviewed here.
Introduction of cross-conserved T-cell epitopes
In 2009, we anticipated the importance of cross-conserved epitopes in the context of pandemic H1N1 influenza in a comprehensive analysis that was published shortly after the emergence of the new strain.15 There was considerable support for this hypothesis in published studies involving exposure or vaccination and heterotypic challenge in animal models.21,45-47 Thus, we argued, the reason for the unusual age distribution of H1N1 2009 cases was that older individuals might have established a cross-reactive cell-mediated immune response to novel H1N1 due to vaccination or exposure to previously-circulating influenza H1N1 (from strains circulating after the 1918 epidemic and again in 197748). While cross-reactive memory T-cell responses (in the absence of a cross-reactive humoral immune response) may not have provided complete protection against infection, it is possible that the severity of the illness was reduced, leading to a lower hospitalization rate and lower reports of H1N1 in this age group, as was observed in a case-control study from Mexico.49 Since then, we have carried this work further by confirming the predictions and developing novel vaccines that incorporate highly cross-strain-conserved CD4+ T-cell epitopes across a broad range of H1N1 influenza sequences.50-52 Similarly, H7N9 epitopes that are conserved in circulating seasonal influenza strains may contribute to recovery from infection. Serum IgG samples from H7N9-infected patients were shown to recognize H1 and H3 hemagglutinins,24 suggesting that a portion of their influenza memory CD4+ T-cell repertoire may cross-react with H7N9 sequences. Thus, introduction of T-cell epitopes that are highly cross-conserved (with other seasonal influenza A strains), or attaching an epitope string to HA, might boost the humoral immune response to H7-HA. We are currently engaged in the production of an epitope-boosted H7 protein.
Reduction of T-cell epitopes that might lower immunogenicity
Before moving forward with vaccines containing highly cross-conserved T-cell epitopes, we have become interested in determining whether epitopes conserved with the human genome might also influence the immunogenicity of H7N9 HA. We recently published a description of the differential distribution of epitopes that are highly cross-conserved with the human genome among human viral pathogens. Not surprisingly, commensal viruses such as Herpes simplex virus 1 and 2, Epstein Barr virus, and Cytomegalovirus had more of these types of epitopes that are cross-conserved with the human genome, than human pathogens that are not commensal (Variola and Marburg viruses, for example14). Access to advanced computational TCR-modeling technology such as the JanusMatrix tool53 make it possible to screen for such potential cross-reactivities. In research that is in preparation for publication, we have identified human genome-conserved epitopes derived from H7N9 influenza that may diminish T-cell responsiveness and significantly influence the outcome of vaccination and infection. Shown in Figure 4 are similar HA protein epitopes derived from H1N1 and H7N9. In preliminary studies using PBMCs from H7N9-naïve donors, we found that (1) H7N9-unique epitopes generated limited immune responses in human IFNγ ELISpot assays; (2) H7N9 epitopes that were highly cross-conserved with other circulating strains generated strong T-cell responses, and (3) epitopes that are cross-conserved with the human genome and with other circulating strains generated diminished T-cell responses, by comparison (Rui Liu, unpublished). What is not yet known, and will be critical to define, is how these opposing T-cell responses are integrated in an individual’s protective immune response, as measured by the generation of effective neutralizing (HAI) and binding antibodies.
Figure 4. Wild bird flyways showing the connection between Asia and Africa. From Ian Mackay’s blog “Viruses Down Under,” bird flyways that may contribute to H7N9 spread. Used with permission from http://virologydownunder.blogspot.com/2013/09/h7n9-in-wild-birdsa-review-of-literature.html, accessed 1 February 2014.
Conclusion
H7N9 viruses, along with many other emerging subtypes influenza, challenge our thinking on how to develop faster, more efficacious vaccines. Seasonal influenza viruses have adapted to the human host for many years following the introduction of a novel subtype. Therefore, humans have memory immune responses that can be recalled in a traditional influenza vaccine each flu season. In contrast, people have no memory responses to new, emerging influenza. Following the emergence of novel H1N1, we determined that many cross-reactive T-cell epitopes were present in the viral genome, and a cross-reactive T-cell response was subsequently shown to contribute to significant protection against the novel strain. In contrast, as the analysis of critical H7N9 gene products (HA, NA) demonstrates, there is only limited potential for cross-reactive immune responses to this novel, avian-origin influenza strain.
The challenge of developing an efficacious H7N9 vaccine is further heightened by the presence of T-cell epitopes in the H7N9 HA that may cross-react with the human genome, which may alter or suppress immune response to the emerging H7N9 virus. This landscape forces vaccinologists and vaccine manufacturers to think outside the box and develop novel strategies to enhance the immunogenic potential for vaccines against these viruses. Careful vaccine engineering that enriches for immunogenic T-cell epitopes appears to be an excellent strategy to enhance both the antibody and cellular responses against H7N9 antigens, thereby reducing viral titers in the nasal and lung mucosa, reducing transmission, and protecting against severe morbidity and death.
Disclosure of Potential Conflicts of Interest
Coauthors A.D.G., W.M., L.M., and F.T. are employees of EpiVax, and two (A.D.G., W.M.) are majority stockholders. These authors recognize the presence of a potential conflict of interest and affirm that the information represented in this paper is original and unbiased observations. R.L., A.H.G., O.A.K., and T.M.R. declare no competing interests.
Acknowledgments
The authors each made substantial contributions to conception and design, acquisition of data or analysis, and interpretation of data; they also contributed to drafting the article or reviewing/revising it critically for important intellectual content. They are grateful to AJ Vincelli, Christine Boyle, and Kelsey Confreda of EpiVax for their many contributions to the finalization of this manuscript, and to Matt Ardito for the initial immunoinformatics analysis.
Glossary
Abbreviations:
- H7N9
avian origin influenza A/Anhui/1/2013 and other similar avian-origin strains
- HA
hemagglutinin
- H7-HA
HA derived from H7N9
- H1-HA
HA derived from H1N1
- CFR
case fatality rate
- IAV
influenza A virus
- GISAID
Global Initiative on Sharing Avian Influenza Data
References
- 1.De Groot AS, Ardito M, Terry F, Levitz L, Ross T, Moise L, Martin W. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum Vaccin Immunother. 2013;9:950–6. doi: 10.4161/hv.24939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–97. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 3.As 8 new H7N9 influenza infections confirmed in Jiangsu Province, Zhejiang Province and Guangdong Province in China, CECC for H7N9 influenza advises people traveling overseas to take preventive measures to ward off infection [Internet]. Taipei City, Taiwan (R.O.C.): CDC. (Centers for Disease Control), R.O.C. (Taiwan); 2014 29 Jan [cited 2014 29 Jan]. Available from: http://www.cdc.gov.tw/english/info.aspx?treeid=bc2d4e89b154059b&nowtreeid=ee0a2987cfba3222&tid=03ECBB84CD82E4FB
- 4.Rapid risk assessment: Human infection with a novel avian influenza A(H7N9) virus, China - Third update [Internet]. Stockholm, Sweden: European Centre for Disease Prevention and Control; 2014 27 Jan [cited 2014 27 Jan]. Available from: http://ecdc.europa.eu/en/publications/Publications/influenza-AH7N9-China-rapid-risk-assessment-27-January-2014.pdf
- 5.Partinen M, Saarenpää-Heikkilä O, Ilveskoski I, Hublin C, Linna M, Olsén P, Nokelainen P, Alén R, Wallden T, Espo M, et al. Increased incidence and clinical picture of childhood narcolepsy following the 2009 H1N1 pandemic vaccination campaign in Finland. PLoS One. 2012;7:e33723. doi: 10.1371/journal.pone.0033723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GISAID: Global Initiative on Sharing all Influenza Data [Internet]. Munich: GISAID; [cited date: 1 Feb 2014]. Available from: http://platform.gisaid.org
- 7.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–32. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 8.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 9.Dormitzer PR, Suphaphiphat P, Gibson DG, Wentworth DE, Stockwell TB, Algire MA, Alperovich N, Barro M, Brown DM, Craig S, et al. Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci Transl Med. 2013;5:85ra68. doi: 10.1126/scitranslmed.3006368. [DOI] [PubMed] [Google Scholar]
- 10.Smith GE, Flyer DC, Raghunandan R, Liu Y, Wei Z, Wu Y, Kpamegan E, Courbron D, Fries LF, 3rd, Glenn GM. Development of influenza H7N9 virus like particle (VLP) vaccine: homologous A/Anhui/1/2013 (H7N9) protection and heterologous A/chicken/Jalisco/CPA1/2012 (H7N3) cross-protection in vaccinated mice challenged with H7N9 virus. Vaccine. 2013;31:4305–13. doi: 10.1016/j.vaccine.2013.07.043. [DOI] [PubMed] [Google Scholar]
- 11.De Groot AS, Ardito MA, Tassone R, Knopf P, Moise L, Martin W. Development of Vaccines: From Discovery to Clinical Testing. First Edition. John Wiley and Sons; c2011. Chapter 3, Tools for Vaccine Design: Prediction and Validation of Highly Immunogenic and Conserved Class II Epitopes and Development of Epitope-driven Vaccines. pp. 65-94. Available from: http://tinyurl.com/Dvt-of-Vaccines-Chapt-3
- 12.Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27:6471–9. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moise L, Buller RM, Schriewer J, Lee J, Frey SE, Weiner DB, Martin W, De Groot AS. VennVax, a DNA-prime, peptide-boost multi-T-cell epitope poxvirus vaccine, induces protective immunity against vaccinia infection by T-cell response alone. Vaccine. 2011;29:501–11. doi: 10.1016/j.vaccine.2010.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L, De Groot AS, Gutierrez AH, Martin WD, Moise L, Bailey-Kellogg C. Integrated assessment of predicted MHC binding and cross-conservation with self reveals patterns of viral camouflage. BMC Bioinformatics. 2014 doi: 10.1186/1471-2105-15-S4-S1. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008-2009 conventional influenza vaccine. Vaccine. 2009;27:5740–7. doi: 10.1016/j.vaccine.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 16.De Groot AS, McClaine E, Moise L, Martin W. Time for T?: Thoughts about the 2009 novel H1N1 influenza outbreak and the role of T-cell epitopes in the next generation of influenza vaccines. Hum Vaccin. 2010;6:161–3. doi: 10.4161/hv.6.2.11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, Vita R, Ponomarenko J, Scheuermann RH, Sette A, et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A. 2009;106:20365–70. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge X, Tan V, Bollyky PL, Standifer NE, James EA, Kwok WW. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol. 2010;84:3312–9. doi: 10.1128/JVI.02226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellebedy AH, Fabrizio TP, Kayali G, Oguin TH, 3rd, Brown SA, Rehg J, Thomas PG, Webby RJ. Contemporary seasonal influenza A (H1N1) virus infection primes for a more robust response to split inactivated pandemic influenza A (H1N1) Virus vaccination in ferrets. Clin Vaccine Immunol. 2010;17:1998–2006. doi: 10.1128/CVI.00247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novartis announces positive clinical trial results for novel H7N9 vaccine [Internet]. Basel, Switzerland: Novartis, Inc; 2013 Nov 13 [cited 2014 Jan 28]. Available from: http://www.novartis.com/newsroom/media-releases/en/2013/1743124.shtml
- 21.Fries LF, Smith GE, Glenn GM. A recombinant viruslike particle influenza A (H7N9) vaccine. N Engl J Med. 2013;369:2564–6. doi: 10.1056/NEJMc1313186. [DOI] [PubMed] [Google Scholar]
- 22.Guo L, Zhang X, Ren L, Yu X, Chen L, Zhou H, Gao X, Teng Z, Li J, Hu J, et al. Human Antibody Responses to Avian Influenza A(H7N9) Virus, 2013. Emerg Infect Dis. 2014;20:192–200. doi: 10.3201/eid2002.131094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin MR, Monto AS, Belongia EA, Treanor JJ, Chen Q, Chen J, Talbot HK, Ohmit SE, Coleman LA, Lofthus G, et al. U.S. Flu-VE Network Effectiveness of non-adjuvanted pandemic influenza A vaccines for preventing pandemic influenza acute respiratory illness visits in 4 U.S. communities. PLoS One. 2011;6:e23085. doi: 10.1371/journal.pone.0023085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24:1159–69. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 25.Grohskopf LA, Shay DK, Shimabukuro TT, Sokolow LZ, Keitel WA. Bresee, JS, Cox, NJ. Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices – United States, 2013-2014 [Internet]. Atlanta, Georgia: Centers for Disease Control) MMWR; 20 Sep 2013 [cited 01 Feb 2014]. Available from: http://www.cdc.gov/mmwr/preview/mmwrhtml/rr6207a1.htm
- 26.de Wit E, Munster VJ, Spronken MI, Bestebroer TM, Baas C, Beyer WE, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Protection of mice against lethal infection with highly pathogenic H7N7 influenza A virus by using a recombinant low-pathogenicity vaccine strain. J Virol. 2005;79:12401–7. doi: 10.1128/JVI.79.19.12401-12407.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox RJ, Madhun AS, Hauge S, Sjursen H, Major D, Kuhne M, Höschler K, Saville M, Vogel FR, Barclay W, et al. A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine. 2009;27:1889–97. doi: 10.1016/j.vaccine.2009.01.116. [DOI] [PubMed] [Google Scholar]
- 28.CDC statement on narcolepsy following Pandemrix influenza vaccination in Europe [Internet]. Atlanta, Georgia: Centers for Disease Control and Prevention; 2013 Feb 26 [cited 2014 Jan 29] Available from: http://cdc.gov/vaccinesafety/Concerns/h1n1_narcolepsy_pandemrix.html
- 29.Eurosurveillance editorial team Swedish Medical Products Agency publishes report from a case inventory study on Pandemrix vaccination and development of narcolepsy with cataplexy. Euro Surveill. 2011;16:19904. doi: 10.2807/ese.16.26.19904-en. [DOI] [PubMed] [Google Scholar]
- 30.Ip DK, Liao Q, Wu P, Gao Z, Cao B, Feng L, Xu X, Jiang H, Li M, Bao J, et al. Detection of mild to moderate influenza A/H7N9 infection by China’s national sentinel surveillance system for influenza-like illness: case series. BMJ. 2013;346:f3693. doi: 10.1136/bmj.f3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackay IM. H7N9: Zhejiang in 50 cases... Virology Down Under blog; 2014 30 Jan [cited 2014 31 Jan]. Available from: http://virologydownunder.blogspot.com.au/
- 32.Number of confirmed human cases for avian influenza A(H7N9) reported to WHO: Report 3 – data in WHO/HQ as of 24 April 2013, 14:45 GMT+1 [Internet]. Geneva, Switzerland; World Health Organization; 2013 Apr 24 [cited 2014 Jan 29]. Available from from: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/03_ReportWebH7N9Number.pdf
- 33.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med. 2014;370:520–32. doi: 10.1056/NEJMoa1304617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudley JP, Mackay IM. Age-specific and sex-specific morbidity and mortality from avian influenza A(H7N9) J Clin Virol. 2013;58:568–70. doi: 10.1016/j.jcv.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabbard JD, Dlugolenski D, Van Riel D, Marshall N, Galloway SE, Howerth EW, Campbell PJ, Jones C, Johnson S, Byrd-Leotis L, et al. JD Novel H7N9 Influenza Virus Shows Low Infectious Dose, High Growth Rate, and Efficient Contact Transmission in the Guinea Pig Model. J Virol. 2014;88:1502–12. doi: 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 2013;501:560–3. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Ramírez E, Gerrikagoitia X, Barral M, Höfle U. Detection of low pathogenic avian influenza viruses in wild birds in Castilla-La Mancha (south central Spain) Vet Microbiol. 2010;146:200–8. doi: 10.1016/j.vetmic.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Bahl J, Krauss S, Kühnert D, Fourment M, Raven G, Pryor SP, Niles LJ, Danner A, Walker D, Mendenhall IH, et al. Influenza a virus migration and persistence in North American wild birds. PLoS Pathog. 2013;9:e1003570. doi: 10.1371/journal.ppat.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colizza V, Barrat A, Barthelemy M, Valleron AJ, Vespignani A. Modeling the worldwide spread of pandemic influenza: baseline case and containment interventions. PLoS Med. 2007;4:e13. doi: 10.1371/journal.pmed.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.First Human Avian Influenza A. (H5N1) Virus Infection Reported in Americas [Internet]. Atlanta, Georgia: Centers for Disease Control and Prevention; 2014 Jan 8 [cited 2014 Jan 29]. Available from: http://cdc.gov/flu/news/first-human-h5n1-americas.htm
- 42.Pandemic (H1N1) 2009 in the African region: update 66 [Internet]. Geneva, Switzerland: World Health Organization; 2009 [cited 01 Feb 2014]. Available from: http://www.who.int/csr/don/2009_09_18/en/ [Google Scholar]
- 43.Koita OA, Sangare L, Poudiougou B, Aboubacar B, Samake Y, Coulibaly T, Pronyk P, Salez N, Kieffer A, Ninove L, et al. A seroepidemiological study of pandemic A/H1N1(2009) influenza in a rural population of Mali. Clin Microbiol Infect. 2012;18:976–81. doi: 10.1111/j.1469-0691.2011.03725.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuang EMM. The new Chinese migration flows to Africa. Soc Sci Inf (Paris) 2008;47:643–59. doi: 10.1177/0539018408096452. [DOI] [Google Scholar]
- 45.Perrone LA, Ahmad A, Veguilla V, Lu X, Smith G, Katz JM, Pushko P, Tumpey TM. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J Virol. 2009;83:5726–34. doi: 10.1128/JVI.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bright RA, Carter DM, Crevar CJ, Toapanta FR, Steckbeck JD, Cole KS, Kumar NM, Pushko P, Smith G, Tumpey TM, et al. Cross-clade protective immune responses to influenza viruses with H5N1 HA and NA elicited by an influenza virus-like particle. PLoS One. 2008;3:e1501. doi: 10.1371/journal.pone.0001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laurie KL, Carolan LA, Middleton D, Lowther S, Kelso A, Barr IG. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J Infect Dis. 2010;202:1011–20. doi: 10.1086/656188. [DOI] [PubMed] [Google Scholar]
- 48.Hodder RA, Gaydos JC, Allen RG, Top FH, Jr., Nowosiwsky T, Russell PK. Swine influenza A at Fort Dix, New Jersey (January-February 1976). III. Extent of spread and duration of the outbreak. J Infect Dis. 1977;136(Suppl):S369–75. doi: 10.1093/infdis/136.Supplement_3.S369. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Garcia L, Valdespino-Gómez JL, Lazcano-Ponce E, Jimenez-Corona A, Higuera-Iglesias A, Cruz-Hervert P, Cano-Arellano B, Garcia-Anaya A, Ferreira-Guerrero E, Baez-Saldaña R, et al. Partial protection of seasonal trivalent inactivated vaccine against novel pandemic influenza A/H1N1 2009: case-control study in Mexico City. BMJ. 2009;339:b3928. doi: 10.1136/bmj.b3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schanen BC, De Groot AS, Moise L, Ardito M, McClaine E, Martin W, Wittman V, Warren WL, Drake DR., 3rd Coupling sensitive in vitro and in silico techniques to assess cross-reactive CD4(+) T cells against the swine-origin H1N1 influenza virus. Vaccine. 2011;29:3299–309. doi: 10.1016/j.vaccine.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moise L, Terry F, Ardito M, Tassone R, Latimer H, Boyle C, Martin WD, De Groot AS. Universal H1N1 influenza vaccine development: identification of consensus class II hemagglutinin and neuraminidase epitopes derived from strains circulating between 1980 and 2011. Hum Vaccin Immunother. 2013;9:1598–607. doi: 10.4161/hv.25598. [DOI] [PubMed] [Google Scholar]
- 52.Moise L, Tassone R, Latimer H, Terry F, Levitz L, Haran JP, Ross TM, Boyle CM, Martin WD, De Groot AS. Immunization with cross-conserved H1N1 influenza CD4+ T-cell epitopes lowers viral burden in HLA DR3 transgenic mice. Hum Vaccin Immunother. 2013;9:2060–8. doi: 10.4161/hv.26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moise L, Gutierrez AH, Bailey-Kellogg C, Terry F, Leng Q, Abdel Hady KM, VerBerkmoes NC, Sztein MB, Losikoff PT, Martin WD, et al. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix. Hum Vaccin Immunother. 2013;9:1577–86. doi: 10.4161/hv.24615. [DOI] [PMC free article] [PubMed] [Google Scholar]



