Abstract
The outbreak of a novel H7N9 influenza virus in 2013 has raised serious concerns for the potential of another avian-source pandemic influenza. Effective vaccines against H7N9 virus are important in the prevention and control of any major outbreak. Novel vaccination technologies are useful additions to existing approaches. In the current report, DNA vaccine studies were conducted to identify the optimal design of an H7 HA antigen using the HA gene from a previously reported H7N7 virus that is lethal in humans as the model antigen. New Zealand White rabbits were immunized with DNA vaccines expressing 1 of 3 forms of H7 HA antigen inserts encoding the HA gene from the same H7N7 virus. High-level H7 HA-specific IgG was detected by ELISA, and functional antibodies were confirmed by hemagglutination inhibition assay and pseudotyped virus-based neutralization assay against viruses expressing HA antigens from either the previous H7N7 virus or the novel H7N9 virus. HA antigen design under the tissue plasminogen activator leader (tPA) was the most immunogenic. The data presented in the current report confirm the immunogenicity of the H7 HA antigen and provide useful guidance to prepare for an optimized H7 HA DNA vaccine to help to control the emerging H7N9 virus if and when it is needed.
Keywords: influenza infection, H7N7, H7N9, antibody, neutralizing antibody, cross protection, vaccine
Introduction
Influenza virus infections continue to pose a major threat to public health. Seasonal influenza viruses cause high mortality in the human population each year. Emerging influenza viruses, such as the highly pathogenic avian influenza virus, H5N1, and less reported but equally concerning, H9N2 and H7N7 viruses,1-4 remind the world of a potential influenza pandemic threat. The recent outbreak of a novel H7N9 subtype influenza in humans in China further highlights the need to develop fast response measures when the world is facing such emerging threats.
Currently, multiple groups are working to develop vaccines against new H7N9 viruses.5 However, previous reports showed low immunogenicity of H7 subtype vaccines when traditional vaccine approaches were used.6,7 Influenza viruses isolated from humans often do not replicate well in embryonated chicken eggs, the substrate for production of traditional influenza vaccines. Replication of influenza viruses in eggs often results in the generation of mutations at or near the HA receptor binding region. Therefore, alternative vaccine approaches, such as DNA vaccines, offer a unique advantage in studying the immunogenicity of HA antigens from emerging influenza viruses.
Since its discovery in the early 1990s, it is now well established that DNA is a highly useful immunization approach that can elicit immune responses against a very specific antigen.8 Influenza DNA vaccines were among the first DNA vaccines tested in proof-of-concept, small animal studies, and early phase human trials.9-13 Protection has been shown in multiple species, and direct comparisons of HA DNA vaccines and conventional vaccines have demonstrated high immunogenicity for the DNA vaccine approach against both homologous and heterologous challenge in animal models.9,10,14 While influenza H1 HA DNA vaccines have shown good immunogenicity in humans when delivered by a gene gun device,13 the influenza DNA vaccine was most promising when it was used as a prime immunization to significantly enhance the immunogenicity of a follow-up boost by a conventional, inactivated influenza vaccine in humans.15
Most of these studies, so far, have been limited to influenza virus subtypes H1, H3, and H5, due to their continuous threat to the human population. In the current report, we conducted a pilot study to investigate the immunogenicity of H7 HA DNA vaccines just before the outbreak of an H7N9 infection. In this study, we designed 3 forms of HA DNA vaccines using a codon-optimized HA gene from a pathogenic strain of H7 subtype influenza virus that originated in the Netherlands (A/Netherlands/219/03 H7N7) and tested their relative immunogenicity in inducing H7 HA-specific binding antibody and functional antibodies in a rabbit model. Our results showed that different designs of H7 HA DNA vaccines elicited different levels of antibody responses. Furthermore, antibodies elicited by HA from a historical H7 subtype virus could cross-react against the novel 2013 H7N9 virus. Our study provides useful information for the design of DNA vaccines using HA antigens from emerging H7N9 viruses. Our results also confirm the overall high immunogenicity of an H7 HA antigen and this finding is useful for researchers working to develop traditional influenza vaccines against emerging H7N9 viruses.
Results
Construction and in vitro expression of 3 forms of H7 HA DNA vaccine plasmids
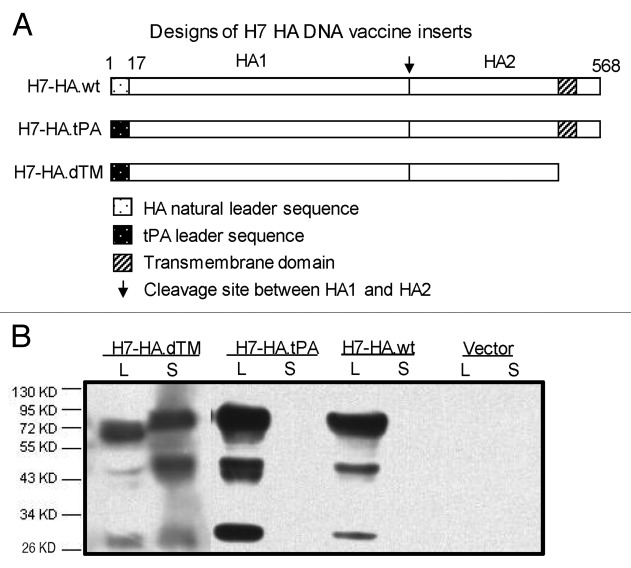
In our previous studies with seasonal (H1 and H3 subtypes) and avian influenza (H5 subtype) HA antigens, differences in immunogenicity were observed with different designs of HA inserts in a DNA vaccine.16,17 In the current study, various forms of DNA plasmids were constructed to express 3 different H7 HA antigen designs using the same HA sequences (H7 HA.NL) from the influenza virus strain, A/Netherland/219/03(H7N7) (Fig. 1A). One design (H7 HA.wt) expresses the full-length H7 HA-NL with the wild type HA leader sequence, the second one (H7 HA.tPA) replaces the original HA leader with a human tissue plasminogen activator (tPA) leader sequence, and the third one (H7 HA.dTM) codes for a truncated HA (without transmembrane and intracellular domains) under the tPA leader sequence. All 3 HA antigen gene inserts were subcloned into the DNA vaccine vector, pJW4303, as previously described.16
Figure 1. Design and expression of H7 HA DNA vaccine inserts. (A) Schematic diagram of influenza A H7 HA gene inserts used in codon optimized DNA vaccines (A/Netherlands/219/03, H7N7), including the full-length HA antigens with natural leader sequence (H7-HA.wt) or a human tissue plasminogen activator (tPA) leader substituting the natural HA leader sequence (H7-HA.tPA) and the transmembrane/cytoplasmic region truncated HA antigens (H7-HA.dTM) with tPA leader sequence. The cleavage site between HA1 and HA2 subunits are marked. The numbers above the HA inserts denote the relevant amino acid positions in natural HA proteins. (B) western blot analysis of the expression of differently designed H7-HA DNA vaccines, including HA protein expression, secretion and susceptibility to cleavage expressed in transiently transfected 293T cell supernatant (S) and cell lysate (L).
Expression of the above HA DNA vaccine plasmids was verified by western blot using culture supernatant and cell lysate from 293T cells transiently transfected with different H7 HA DNA plasmids (Fig. 1B). The empty vector, pJW4303, was included as the negative control. All 3 H7 HA DNA vaccine plasmids expressed HA in cell lysate, and only cells transfected with the truncated H7 HA design (H7 HA.dTM) showed HA antigens in supernatant. The full-length HA precursor (HA0, 72 KDa) can be cleaved into HA1 (55 KDa) and HA2 (26 KDa) due to the polybasic cleavage site for all 3 forms of the H7 HA DNA vaccine. No HA-specific bands were recognized in cell samples transfected with the empty pJW4303 vector.
HA-specific antibody responses in NZW rabbits
Four groups of New Zealand White (NZW) rabbits (3 per group) were immunized with each of the 3 H7 HA DNA vaccines or the empty DNA vaccine vector by electroporation. After 4 DNA immunizations at weeks 0, 2, 4, and 8 without adjuvant, temporal HA-specific antibody responses were measured by ELISA using a fixed 1:500 serum dilution for all groups. Positive HA-specific antibodies were detected after 2 immunizations for all 3 HA DNA vaccine groups and response levels were boosted further following 2 additional immunizations (Fig. 2A). Peak anti-HA IgG responses at 2 wk after the fourth DNA immunization were determined by endpoint ELISA titers. All 3 constructs were highly immunogenic and the final binding antibody titers were close to 1:100 000. The H7 HA used to coat the ELISA plates was supernatant from a 293T cell transfection without purification; however, the same batch of supernatant was used for this study to ensure consistency. We also tested control rabbit sera immunized with other non-H7 HA antigens (H1, H3, and H5) and did not see any cross reactivity (data not shown).
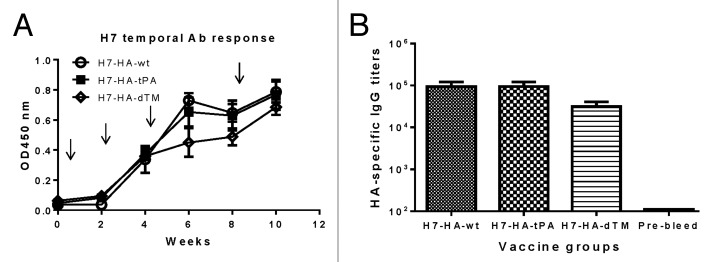
Figure 2. HA-specific antibody responses induced by differently designed H7-HA DNA vaccines (H7-HA-wt, H7-HA-tPA, and H7-HA-dTM) in rabbits. (A) Temporal serum anti-HA IgG responses is measured by ELISA at 1:1000 serum dilution against H7-HA-dTM as the coating antigen. The arrows indicate the time points of DNA immunizations by electroporation. Each curve represents the mean OD values with standard deviations of each group of 3 rabbits. (B) End titration titers of serum anti-H7-HA-dTM IgG responses at 2 wk after the fourth DNA immunization.
Protective antibody responses as measured by the hemagglutinin inhibition (HAI) assay
An HAI assay using horse erythrocytes demonstrated the presence of HAI antibodies in rabbit sera immunized with both full-length H7 HA DNA vaccines (HA.wt and HA.tPA), whereas sera from rabbits that received the truncated H7 HA DNA vaccine (HA.dTM) did not show a meaningful HAI response against the wild type A/African starling/England-Q/938/79(H7N1) virus. Furthermore, levels of HAI antibody responses were higher in rabbits that received HA.tPA than those immunized with HA.wt (Fig. 3A). The difference between HA.wt and HA.tPA was not significant due to a small sample size but the difference between HA.tPA and HA.dTM groups was indeed significant (P < 0.05).
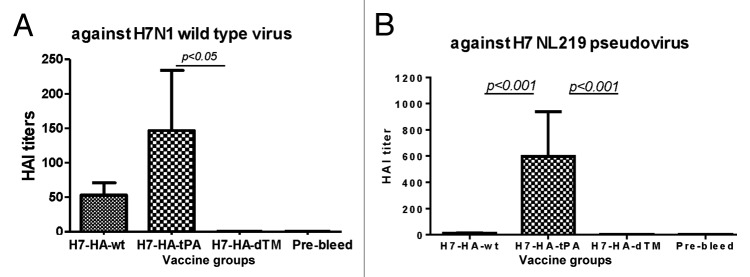
Figure 3. Hemagglutination inhibition (HAI) antibody responses in rabbit sera immunized with different designed H7-HA DNA vaccines (H7-HA-wt, H7-HA-tPA, and H7-HA-dTM) in rabbits. HAI antibody responses were measured against a wild type H7N1 virus (A/African Starling/England-Q/938/79) (A) or H7N1 pseudovirus expressing H7-HA from A/Netherlands/219/03 (H7N7) and H1 N1-NA from A/VietNam/1203/04 (H5N1) (B). The rabbit sera used in the HAI assay were collected at 2 wk after the fourth DNA immunization or relevant pre-bleed sera, as indicated. HAI antibody titers are shown as the geometric mean for each group (3 rabbits per group) with standard deviations. Statistical significance (P < 0.05) between vaccine groups is indicated.
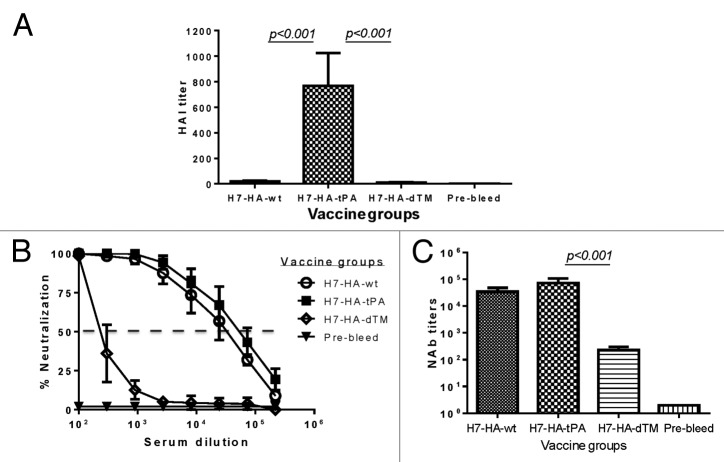
In order to rule out the possibility that the difference in HAI antibodies observed in the above assay was due to the use of a heterologous H7 influenza viral isolate, an additional HAI assay was conducted using a pseudotyped virus (H7-NL219) expressing the autologous HA antigen from A/Netherlands/219/03 (H7N7). Chicken erythrocytes were used in this assay. Similar to the above HAI assay using a wild type heterologous H7 virus, the H7 HA.tPA DNA vaccine elicited the highest HAI titer against the pseudotyped virus expressing autologous HA antigen from H7-NL219, when compared with either HA.wt or HA.dTM DNA vaccine groups, and the difference was statistically significant (both P < 0.001).
Protective antibody responses as measured by the neutralization assay
Rabbit immune sera were also analyzed for neutralizing antibody levels. All 3 H7 HA DNA vaccine groups developed neutralizing antibody responses against the pseudotyped virus expressing H7 HA from the autologous virus. Neutralizing activities were determined as the serum dilution that can inhibit 50% of the viral infection in this assay (Fig. 4A). Rabbits that received the H7 HA.tPA DNA vaccine developed the highest neutralizing antibodies against its autologous influenza virus followed by rabbits immunized with HA.wt (P < 0.05) (Fig. 4B). Positive neutralizing antibodies were also observed in rabbits immunized with HA.dTM but titers were much lower than observed in the HA.tPA group (P < 0.001).
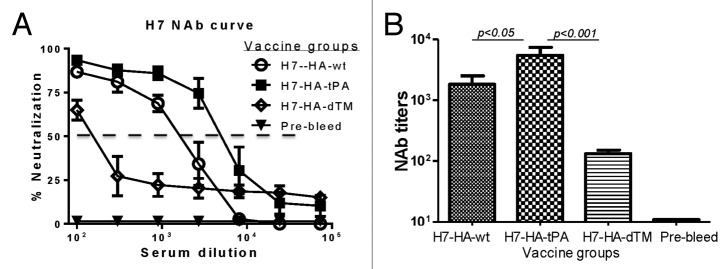
Figure 4. Neutralizing antibody (NAb) responses in rabbit sera immunized with differently designed H7-HA DNA vaccines. Neutralization assays were measured against H7N1 pseudovirus expressing H7-HA from A/Netherlands/219/03 (H7N7) and H1 N1-NA from A/VietNam/1203/04 (H5N1), using rabbit sera collected at 2 wk after the fourth DNA immunization or relevant pre-bleed sera, as indicated. (A) Percentage neutralization titration curves. Each curve represents the mean percentage of neutralization with standard deviation of each group of 3 rabbits. The dashed line indicates the 50% neutralization cut-off. (B) Neutralizing antibody titers were measured at 50% inhibition (IC50) of virus infection to target cells, shown as the geometric mean with standard deviation from each group of 3 rabbits. Statistical significance (P < 0.05) between vaccine groups is indicated
Levels of cross reactivity against the 2013 H7N9 virus
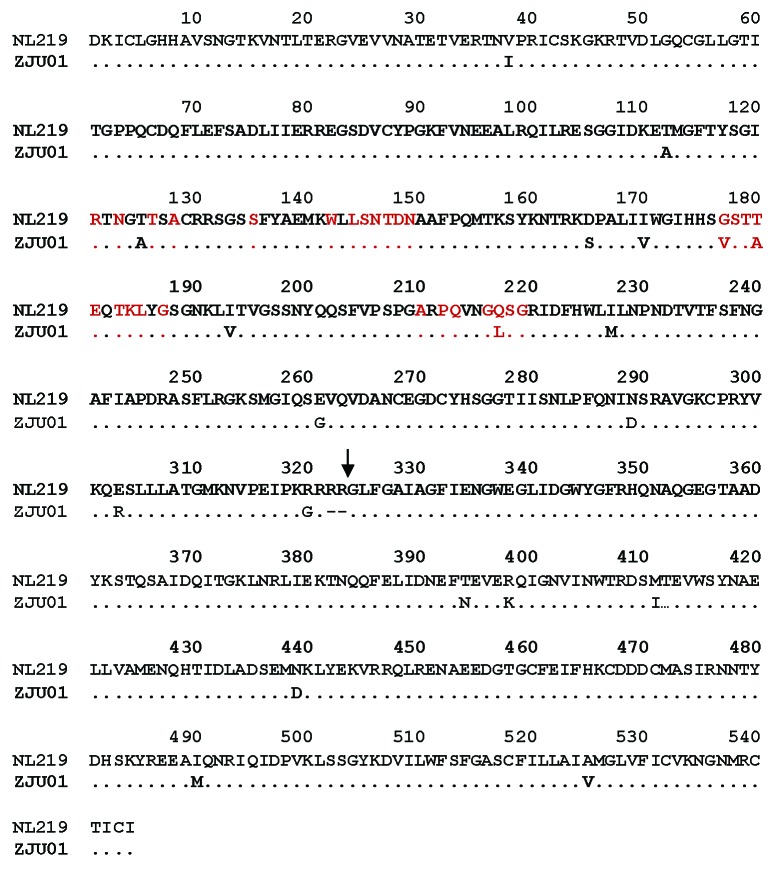
We then tested the cross reactivity of rabbit sera elicited by HA from the previously reported H7N7 virus against the new H7N9 virus that emerged in China in 2013, which was responsible for an influenza outbreak with ~30% mortality in humans. Based on amino acid sequence alignment, there is a ~97% homology between HA from A/Netherlands/219/03 (H7N7) and HA from A/Zhejiang/U01/2013 (H7N9) (Fig. 5), but it was not known whether antibodies elicited by DNA vaccines expressing HA from the 2003 H7N7 virus can recognize or neutralize the 2013 H7N9 virus. In the current report, a pseudotyped virus was constructed expressing HA from A/Zhejiang/U01/2013 (H7N9) and NA from A/Shanghai02/2013 (H7N9). Rabbit sera immunized with the H7 HA.NL DNA vaccine were tested for both HAI and neutralizing antibody activities against this H7N9 pseudotyped virus.
Figure 5. Amino acid sequence alignment of H7 HA proteins from H7N7 A/Netherlands/219/03 (NL219) and H7N9 A/Zhejiang/U01/2013 (ZJU01). The receptor binding site amino acids18 are in red font. Symbol “.” in the sequence alignment indicates the same amino acid sequence as HA in A/Netherlands/219/03 and symbol “-“ represents an amino acid deletion.
Sera from the HA.tPA DNA vaccine group was very effective in eliciting high HAI titer against heterologous H7N9 pseudotyped virus while rabbit sera immunized with either the HA.wt or the HA.dTM vaccines were less effective in eliciting significant HAI antibody responses, and the difference is statistically significant (both P < 0.01) (Fig. 6A). Analysis on neutralizing antibody using the same heterologous H7N9 pseudotyped virus showed again that HA.tPA DNA vaccine was the most effective design to induce the highest level of neutralizing antibody, followed closely by the HA.wt DNA vaccine (Fig. 6B and 6C) although due to small group sizes, this difference fell short of statistical significance (P = 0.13). Rabbit sera immunized with the HA.dTM DNA vaccine elicited the lowest neutralizing antibody response, about 320 times lower than the HA.tPA group (P < 0.001) (Fig. 6C).
Figure 6. Hemagglutination inhibition (HAI) and neutralizing (NAb) antibody responses against H7N9 pseudovirus in rabbit sera immunized with differently designed H7-HA DNA vaccines. H7N9 pseudovirus expressed H7 HA from H7N9 A/Zhejiang/U01/2013 and N9 NA from A/Shanghai/02/2013. Rabbit sera were collected at 2 wk after the fourth DNA immunization or relevant pre-bleed sera, as indicated. (A) HAI antibody titers are shown as the geometric mean with standard deviation for each group of 3 rabbits. (B) Percentage neutralization titration curves. Each curve represents the mean percentage of neutralization with standard deviations of each group of 3 rabbits. The dashed line indicates the 50% neutralization cut-off. (C) Neutralizing antibody titers were measured at 50% inhibition (IC50) of virus infection to target cells, shown as the geometric mean with standard devations from each group of 3 rabbits. Statistical significance (P < 0.05) between vaccine groups is indicated
Evaluation of HA antibody in 2013 H7N9-infected patients using HA antigen derived from A/Netherlands/219/03 strain
Next, we tried to complete the analysis on the levels of cross reactivity among H7 influenza viruses. Since rabbit sera immunized with the HA antigen from the 2013 H7N9 virus was not produced, reciprocal antibody analysis was conducted using sera from 3 patients infected with the 2013 H7N9 influenza virus to study reactivity against the previous H7N7 strain used in the current study. Serum samples were collected from these patients at different days (day 11–day 16) after symptom onset. Binding antibody analysis was conducted against the HA antigen from the early H7N7 NL219 strain. All 3 patient sera showed high levels of binding antibody for the 3 isotypes tested, IgG, IgA, and IgM, with titers of 1 × 103−1 × 106, 1 × 103−1 × 105, and 1 × 103−1 × 105, respectively (Fig. 7).
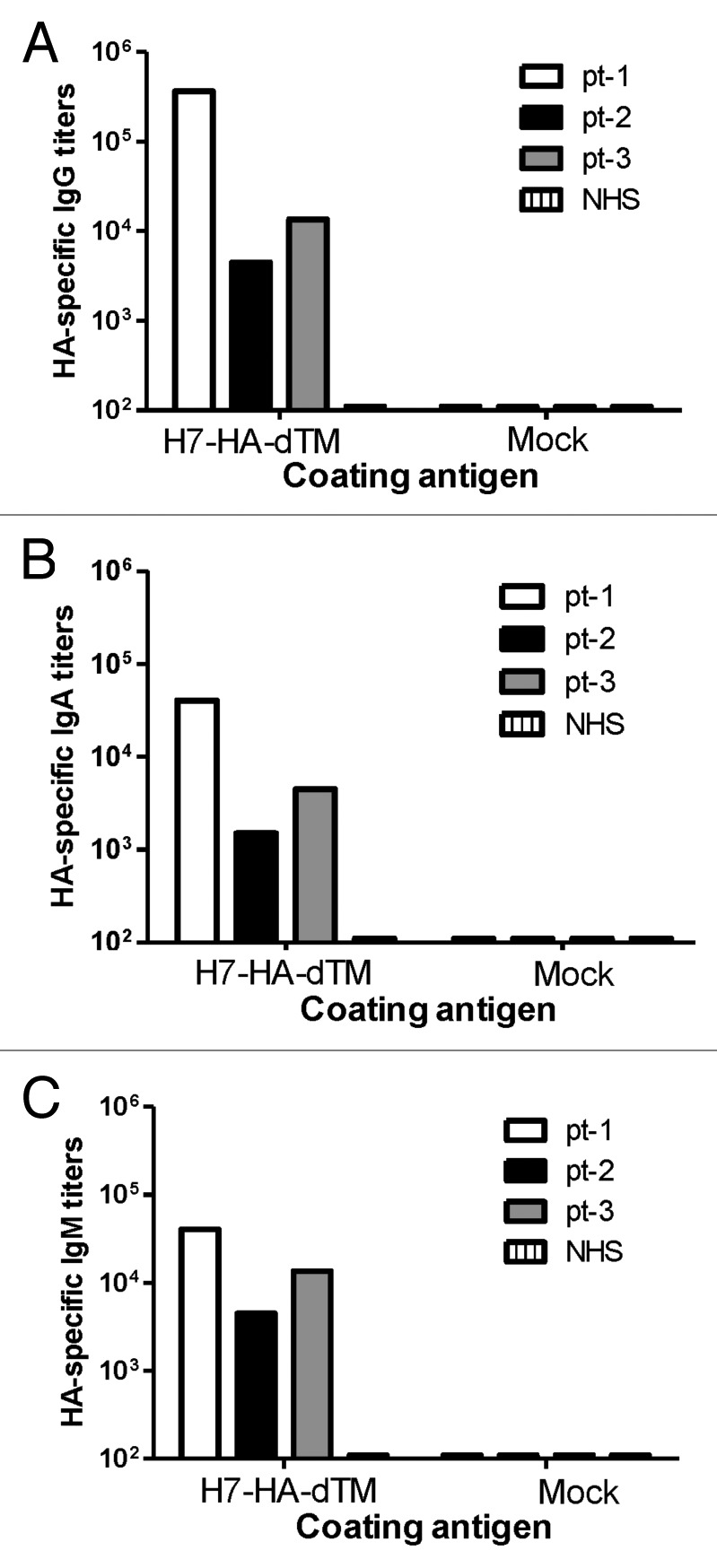
Figure 7. Detection of H7 HA-specific IgG (A), IgA (B), and IgM (C) antibody responses in H7N9 patients (pt-1, pt-2 and pt-3) by ELISA. Patient sera were collected from 3 H7N7 patients: patient 1 (pt-1) at day 11, patient 2 (pt-2) at day 13 or patient 3 (pt-3) at day 16 after onset of fever; normal human sera (NHS) served as negative control. Antibody responses in each patient serum sample were measured against 293T cells supernatant transfected with H7-HA-dTM (A/Netherlands/219/03) DNA vaccine or empty vector (Mock), as indicated.
Functional antibody responses further demonstrated that all 3 patients developed neutralizing antibodies against the pseudotyped virus expressing H7 HA antigen from the NL219 strain at titers between 1:100–1:700 (Fig. 8A). As a control, patient sera had much higher neutralizing antibody titers against the autologous pseudotyped virus expressing HA antigen from the 2013 H7N9 virus (Fig. 8B). However, this difference may only reflect a difference of sensitivity between these 2 pseudotyped viruses as the assays using the rabbit immune sera against these 2 pseudotyped viruses also showed a large difference in neutralizing antibody titers (Figs. 4 and 6). Nevertheless, our data indicates that there is a high level of cross reactivity between the early H7N7 NL219 strain and the newly emerged 2013 H7N9 strain.
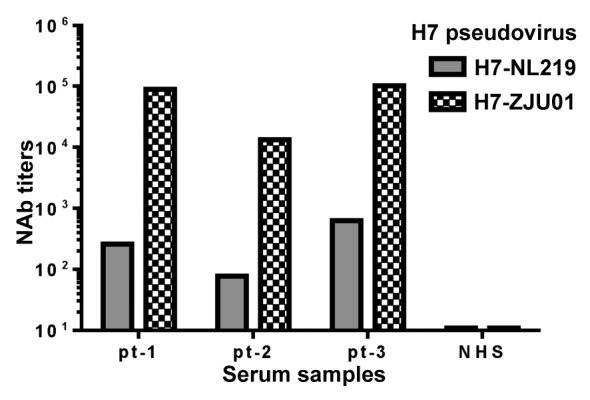
Figure 8. Neutralizing antibody (NAb) responses in H7N9 patients (pt-1, pt-2 and pt-3) against H7N1 and H7N9 pseudoviruses. Patient sera were collected from 3 H7N9 patients: patient 1 (pt-1) at day 11, patient 2 (pt-2) at day 13 or patient 3 (pt-3) at day 16 after onset of fever; normal human sera (NHS) served as negative control. H7N1 pseudovirus (H7-NL219) expressed H7-HA from A/Netherlands/219/03 (H7N7) and N1-NA from A/VietNam/1203/04 (H5N1) while H7N9 pseudovirus expressed H7 HA from H7N9 A/Zhejiang/U01/2013 and N9 NA from A/Shanghai/02/2013. Neutralizing antibody titers in each patient serum sample were measured at 50% inhibition (IC50) of virus infection to target cells.
Discussion
Influenza is a major global infectious disease affecting humans and animals. Influenza viruses continue to undergo antigenic changes and genomic reassortment resulting in new and potentially pathogenic strains for human transmission. New subtypes discovered in the human population can lead to severe pandemics with high morbidity and mortality due to the absence of pre-existing protective immunity.13 A novel H7N9 influenza A virus, first detected in March 2013, has caused more than 130 human infections in China, resulting in 40 deaths19-21 before summer, and new cases have been reported since the Fall of 2013 in this region although it is less publicized. Genetic analyses identified that the new virus is a reassortant with HA from H7N3, NA from H7N9, and 6 internal genes from H9N2 avian influenza viruses and it carries amino acids associated with mammalian receptor binding.18,20,22,23 These new viruses have acquired an ability to break the species barrier to establish infection in humans. Although the strain cannot easily transmit from person to person, it still raises concerns of a potential pandemic in the human population.
While there is an active effort to quickly produce H7N9 vaccines using traditional influenza vaccine approaches, there have been reports showing low immunogenicity with some of these inactivated H7 vaccines.6,7 At the same time, the lack of global capacity to produce sufficient doses of vaccine against any sudden outbreak of pandemic influenza has always been and continues to be a concern. We believe that the DNA vaccine approach offers unique advantages to be considered as part of the overall strategy to control a pandemic influenza. The production process of DNA vaccines is easy. It does not have the issues of low virus yield and mutations that are associated with inactivated or live attenuated vaccine production. With DNA vaccines, HA antigens do not need to be mutated while mutations may be needed to improve the safety profile with the production of virus stocks. More importantly, recent human studies have demonstrated that DNA vaccines are immunogenic when used as a priming step.11,15,24-26 Furthermore, DNA vaccines have been studied extensively in animals and in clinical trials to optimize a wide range of key parameters including antigen design, dosing, adjuvants, optimal routes of delivery (needle injection, gene gun, and electroporation), and novel immunization regimens, including heterologous prime-boost vaccination.16,27-30
Although ferrets are often used as a challenge model to test the protective efficacy of the influenza vaccine, in the current study, our goal is to optimize the DNA vaccine design to enhance its immunogenicity using a less costly animal model. Rabbit was used as the animal model to examine immunogenicity of H7 DNA vaccines because rabbit is highly immunogenic in producing high titer antibody responses and has been widely used for evaluation of vaccine immunogenicity. Rabbits also provide a large volume of immune sera, which allows for a wide range of and repeated antibody assays with limited background, while the other common experimental animal species, such as mouse or guinea pig, can provide only a limited volume of immune sera. Moreover, rabbit antibodies, compared with those from mouse, are able to generate a long CDR3 region, which is important for broad neutralizing antibodies as shown in HIV-1 antibody research.
Our previous studies indicate that the immunogenicity of HA-based DNA vaccines can vary dramatically with different designs of the HA antigen insert, and HA from different subtypes may also have their own preferred design.16,17 Therefore, while the current study did not propose a novel HA design, it is important to identify the optimal design of an HA antigen to be included in a H7 DNA vaccine from the various designs we previously described.
Our study started before the report of the emergence of 2013H7N9 virus and used an HA antigen from a lethal strain of H7 subtype discovered in the Netherlands in 2003. Similar to our report with H5 HA DNA vaccines,16 the same 3 designs of HA antigen inserts were tested with the H7 HA antigen subcloned into the DNA vaccine vector. The results are very similar to our report on H5 HA antigen designs. The DNA vaccine expressing H7 HA with a tPA leader was the most immunogenic in producing the highest H7 HA-specific binding antibody, HAI, and neutralizing antibodies against both the wild type and pseudotyped viruses expressing different H7 HA antigens. A DNA vaccine expressing the wild type HA leader was less immunogenic but still able to elicit higher binding, HAI, and neutralizing antibodies than the DNA vaccine expressing a truncated HA antigen.
While both H5 and H7 HA antigens showed that the HA.tPA design is the most immunogenic, the exact mechanism behind this remains unclear at this point. A leader sequence plays an important role in efficient translation of encoded sequences from mRNA templates31 and subsequent intracellular processing of newly synthesized proteins. Both wild type and tPA leaders, in theory, are highly functional leaders and thus, it is not clear what sequence difference between 2 leaders or any critical intracellular steps may have contributed to a difference in immunogenicity when used to drive the expression of the same HA antigen. One clue is that a significant difference between these 2 groups was observed for functional antibodies, but less for binding antibodies. This would imply that these 2 HA antigen designs elicited different profiles of conformational antibodies, which is important for functional activity, while the binding antibody can easily recognize non-conformational antibodies. Subtle changes in glycosylation patterns may be responsible for this difference but it will require a detailed glycan profile analysis to investigate further.
Our study provides important information on the cross reactivity for immune sera elicited by an early H7N7 virus to recognize an emerging H7N9 virus. Clearly, both HAI antibodies and neutralizing antibodies were present in rabbit immune sera against the heterologous 2013 H7N9 virus. Furthermore, sera from patients infected with the 2013 H7N9 virus were able to neutralize the H7N7 virus included in our study. Our data would support the hypothesis that there is limited mutation within the H7 subtype viruses, while it is known that there are many clades of viruses within the H5 subtype. This is good news for current efforts in developing an H7 influenza vaccine that can be more effective in covering a wide range of H7 viruses as long as ongoing monitoring does not identify new H7 strains with any new critical mutations. Furthermore, the DNA vaccine platform is a quick and simple approach to reliably investigate the degree of cross reactivity with any new emerging viral isolates.
The current report has the following limitations: (1) Due to a lack of access to the live H7N7 and H7N9 virus, pseudotyped virus was used to evaluate functional antibody responses instead of the live H7 virus. (2) A truncated form of HA was used to detect binding antibody responses, which carries a certain bias toward this form of the HA antigen. However, the focus of our study was on functional antibodies so such a bias in binding antibody detection should not impact the final conclusion based on functional antibody results. (3) Human study will be required to confirm the immunogenicity of H7 HA DNA vaccines in humans. However, given the high level of similarity among various DNA vaccines, it is very unlikely that an H7 HA DNA vaccine will behave much differently from other HA DNA vaccines when tested in humans.
Previous literature has reported efforts in developing various vaccination approaches against H7 subtype viruses using reassortant live viruses,32,33 live attenuated vaccine,34,35 and inactivated vaccines.36,37 Now, our study confirms that the DNA vaccine is also effective in eliciting protective antibodies against H7 viruses. The ultimate utility of H7 DNA vaccines will need to be demonstrated in human studies. However, given the recent success in using HA DNA vaccines as a priming immunization in several human studies,15,38,39 it is feasible to include an H7 DNA vaccine as part of a comprehensive strategy to prevent and control any potential major outbreak of H7N9 virus infections in humans.
Materials and Methods
Construction of 3 forms of codon optimized H7 HA DNA vaccines
The codon usage of the HA gene from influenza A human viruses A/Netherlands/219/03 (H7N7) was analyzed using MacVector software 7.2 against codon preference of Homo sapiens. The less optimal codons in the HA genes were replaced by the preferred codons of mammalian systems in order to produce a higher expression of the HA proteins in mammalian systems. Sequence optimization was also performed to make the mRNA more stable and the gene more favorable for transcriptional and translational processes. During sequence optimization, the following cis-acting sequence motifs were avoided: internal TATA boxes, chi-sites, and ribosomal entry sites; AT-rich or GC-rich sequence stretches; ARE, INS, CRS sequence elements; cryptic splice donor and acceptor sites; and branch points. The final codon optimized HA DNA sequence produced the same HA amino acid sequences as in the original H7N7 virus. The codon optimized HA gene was chemically synthesized by Geneart containing 2 enzyme sites, PstI and BamH I, at the 5′ and 3′ ends, respectively. The HA gene insert was removed from Geneart plasmid, pGA4/HA, and directly subcloned into DNA vaccine vector, pJW4303, at PstI and BamH I sites to generate the pJW4303/H7-HA NL219.wt (H7 HA.wt) DNA vaccine. A modified HA gene insert linked to a tPA leader was created by PCR with the template pGA4/HA using forward primer H7 NL219–1 (5′-GTCACTTCGC TAGCGACAAG ATCTGCCTGG GCCAC-3′) and the reverse primer H7 NL219–2 (5′- GAGCTCGGAT CCTCATCAGA TGCAGATGG -3′). The PCR products were then ligated into pJW4303 downstream of the tPA leader. The resulting DNA vaccine was named pJW4303/H7-HA NL219.tPA (H7 HA.tPA). The third version, a truncated HA gene with the removal of transmembrane domain and intracellular sequences was produced by PCR using the same pGA4/HA template with primers H7 NL219-1 and H7 NL219-4 (5′- ATCATCGGAT CCTCAGTAGC CGCTGGACAG CTTCAC-3′). The PCR product was then subcloned into pJW4303 downstream of the tPA leader. The DNA vaccine was designated as pJW4303/H7-HA NL219.dTM (H7 HA.dTM). All 3 forms of H7-HA NL219 DNA plasmids were confirmed by sequencing and enzymatic digestion. Large, individual DNA vaccine plasmid preps were produced from Escherichia coli (HB101 strain) with a Mega purification kit (Qiagen) for animal immunization and in vitro transfection.
In vitro expression of HA antigens in 293T cells
Expression of HA DNA vaccines was verified by transient transfection in 293T cells. Cells were seeded and grown to 50–70% confluence in 10 cm dishes, and plasmids (15 µg per dish) were mixed with 75 µL Polyethylenimine (PEI) for 15 min at room temperature before being added into 293T cells. Medium was changed with fresh FBS-free medium 8 h after transfection. Supernatants and cell lysates were harvested at 72 h after transfection. HA protein expression was examined by western blot analysis. Briefly, the HA proteins from transiently expressed 293T cell lysate (L) and supernatant (S) were subjected to SDS–PAGE and transferred onto PVDF membrane. HA DNA vaccine rabbit sera were diluted at 1:500 and used as the detecting antibody. Subsequently, membranes were washed with PBST solution 5 times for a total of 1 h and then reacted with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG at 1:10 000 dilution. After a final wash, Western-light substrate was applied to the membranes and Kodak films were exposed to the membrane before being developed.
DNA immunization of New Zealand White (NZW) rabbits
NZW rabbits (~2 kg body weight) were purchased from Shanghai Animal Center, Chinese Academy of Science. Rabbits were housed in the Animal Research Center at the Nanjing Medical University in accordance with approved protocol. DNA vaccines were injected intramuscularly followed by electroporation. A Caliper Electrodes style electroporator (SCIENTZ-2C) from Scientz Co., Ltd. was used with the following parameters: 100 V, 60 ms and 60 Hz; 200 µg of HA DNA vaccine or vector control plasmid was delivered each time for each rabbit at 2 different sites in the quadriceps muscle. All immunizations were given at weeks 0, 2, 4, 8. Serum samples were taken prior to the first immunization and every 2 wk after each immunization for the study of HA-specific antibody responses.
Patient serum sample collection
The new H7N9 influenza virus caused human infections in China in 2013; here we collected serum samples from 3 infected patients from Jiangsu Province 11–16 d after symptom onset. Informed consent forms were provided to and signed by each participant.
Evaluation of binding antibody responses by enzyme-linked immunosorbent assay (ELISA)
An ELISA was conducted to measure H7 HA-specific antibody (IgG) responses in immunized rabbits. The 96-well flat-bottom plates were coated with 100 µL/well of HA antigen transiently expressed in 293T cells transfected with the truncated HA NL219 plasmids at 4 °C overnight. After being washed 5 times, the plates were then blocked with 200 µL/well of blocking buffer (5% non-fat dry milk, 4% Whey, 0.5% Tween-20 in PBS at pH 7.2) for 1 h at 37 °C. For each well, 100 µL of serially diluted rabbit serum in dilution buffer was added in duplicate wells and incubated for 1 h at 37 °C. The plates were incubated for 1 h at 37 °C with 100 µL of biotinylated anti-rabbit (Vector Laboratories) diluted at 1:2000 in dilution buffer. Then, 100 µL of horseradish peroxidase (HRP)-conjugated streptavidin (Vector Laboratories) diluted at 1:5000 in dilution buffer was added to each well and incubated for 1 h at 37 °C. After the final wash, the plates were developed with 3,3_,5,5_ tetramethylbenzidine (TMB) solution at 100 µL per well (Sigma) for 3.5 min. The reaction was stopped by adding 50 µL of 1M H2SO4.
To detect HA-specific antibody in H7N9 influenza virus-infected patients, 100 µL of serially diluted human sera in dilution buffer was added in duplicate wells and incubated for 1 h at 37 °C. The plates were incubated for 1 h at 37 °C with 100 µL of HRP-conjugated anti-human IgG, IgM, or IgA (Southernbiotech) diluted at 1:2000 in dilution buffer. After 5 washes, the plates were read at OD 450 nm. The end titration titer was determined as the highest serum dilution that has an OD reading above twice of that from the negative control serum.
Production and titration of the pseudovirus
Pseudovirus production and titration were performed as previously described with modifications.40 Briefly, 293T cells were cultured in 10cm cell culture dishes with high glucose DMEM growth medium supplemented with 10% fetal bovine serum and were cotransfected with 1.2 μg HA-expressing plasmid, 300 ng NA-expressing plasmid, and 13.5μg Env-deficient HIV-1 genomic backbone plasmid (pNL4–3 LucR-E-) with 75μL 1mg/mL PEI (Polysciences, Eppelheim, Germany) in 750 μL DMEM when reached 80% confluence. At 6–8h after transfection, the medium was changed with freshly-made FBS-free DMEM medium. Supernatants were harvested 72 h after transfection, filtered (0.45 μm), then treated with Trypsin-TPCK (Sigma) at a final concentration 40 µg/mL at RT for 1h, and the reaction was stopped with Tripsin inhibitor (Sigma) at 10 µg/mL, and stored at −80 °C in aliquots until use. For virus titration, serial 5-fold dilutions of pseudovirus were made in quadruplicate wells in 96-well cell culture plates in 100 μL growth medium for a total of 11 dilution steps. 100 μL growth medium was added to the last row as a negative control. Freshly trypsinized 293A cells in 100 μL growth medium were added to each well and the plate was incubated at 37 °C with 5% CO2. After 48 h, the cells were lysed and the relative luminescence unit (RLU) was measured using Luciferase Assay Kit (Promega). Wells producing RLU > 2.5 × background were scored as positive. The 50% tissue culture infectious dose (TCID50) was calculated using Spearman-Karber method.
Wild virus-based hemagglutin inhibition (HAI) assays
The influenza virus used for the HAI assay was A/African Starling/England-Q/938/79(H7N1) provided by the Ha’erbin Veterinarian Institute, Chinese Academy of Agriculture Science. The HAI assay used horse erythrocytes instead of turkey red blood cells, in order to increase assay sensitivity. This assay was performed following WHO influenza laboratory procedures. All sera were treated with receptor destroying enzyme (RDE), at 1:4 dilution for 16 h prior to heat inactivation for 30 min at 56 °C and absorbed with the erythrocytes to remove non-specific hemagglutination. A 2-fold serial dilution of rabbit serum (in 25 μL of PBS) was mixed with 4 HA units of viral antigen (in 25 μL of PBS) and incubated at room temperature for 1 h, after which, 0.75% horse erythrocytes with 0.75% BSA in 50 μL of PBS was added and incubated. HAI titer was read after 1 h and was defined as the reciprocal of the last dilution of serum that completely inhibited hemagglutination.
Pseudovirus-based hemagglutination inhibition (HAI) assays
After treatment with receptor destroying enzyme (RDE) to remove non-specific hemagglutination, sera were 2-fold serially diluted (1:8–1:1024) in 50 μL of PBS in V-shaped well plates, an equal volume of 4 hemagglutinin (HA) units of viral antigen was added and then incubated at room temperature for 1 h. 50 μL of 1% chicken erythrocytes in suspension was then added to the wells, and mixed by shaking the plates on a mechanical vibrator. Agglutination patterns were read after 30 min and the HAI titer was defined as the reciprocal of the last dilution of serum that completely inhibited hemagglutination.
Pseudovirus-based neutralizing antibody assays
A neutralization experiment was conducted, as previously reported.16 Briefly, serial dilutions of serum in duplicate were incubated with pseudovirus (200 TCID50 in 50 μL growth medium per well) at 37 °C for 1 h in 96-well cell culture plate, and 293A cells (1 × 104) in 100 μL growth medium were added to each well. After 48 h incubation at 37 °C, 5% CO2, cells were lyzed and RLU were determined. The 50% inhibitory dose (ID50) was determined.
Statistical analysis
The student t test and One-Way ANOVA was used to analyze the differences in antibody responses between animal immunization groups, as measured by binding antibody titers, HAI titers, and neutralizing antibody titers. A P value of less than 0.05 was considered significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported in part with research grants from the Ministry of Science and Technology and the Ministry of Health (China Mega-Project for Infectious Diseases Grants: 2013ZX10004905 and 2013ZX10002005-002-005), Jiangsu Innovation of Medical Team and Leading Talents (LJ201121), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801), National Institutes of Health, National Institutes of Allergy and Infectious Disease (U01-AI078073). The authors thank Dr Jill M Serrano for her careful reading and editing of the manuscript.
Glossary
Abbreviations:
- TMB
3,3_,5,5_ tetramethylbenzidine
- ID50
50% inhibitory dose
- TCID50
50% tissue culture infectious dose
- BSA
bovine serum albumin
- DMEM
Dulbecco's modified Eagle's medium
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- HA
hemagglutinnin
- HAI
hemagglutinin inhibition
- HRP
horseradish peroxidase
- NZW
New Zealand White
- OD
optical density
- PEI
polyethylenimine
- PCR
polymerase chain reaction
- PVDF
polyvinylidene difluoride
- PBS
phosphate buffered saline
- PBST
phosphate buffer saline tween-20
- RDE
receptor destroying enzyme
- RLU
relative luminescence unit
- RT
room temperature
- tPA
tissue plasminogen activator
- TIV
trivalent inactivated vaccine
References
- 1.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–93. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 2.Palese P. Influenza: old and new threats. Nat Med. 2004;10(Suppl):S82–7. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 3.Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med. 2006;355:2174–7. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 4.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, et al. Writing Committee of the World Health Organization (WHO) Consultation on Human Influenza A/H5 Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–85. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 5.Fries LF, Smith GE, Glenn GM. A recombinant viruslike particle influenza A (H7N9) vaccine. N Engl J Med. 2013;369:2564–6. doi: 10.1056/NEJMc1313186. [DOI] [PubMed] [Google Scholar]
- 6.Couch RB, Patel SM, Wade-Bowers CL, Niño D. A randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One. 2012;7:e49704. doi: 10.1371/journal.pone.0049704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox RJ, Madhun AS, Hauge S, Sjursen H, Major D, Kuhne M, Höschler K, Saville M, Vogel FR, Barclay W, et al. A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine. 2009;27:1889–97. doi: 10.1016/j.vaccine.2009.01.116. [DOI] [PubMed] [Google Scholar]
- 8.Webster RG. Potential advantages of DNA immunization for influenza epidemic and pandemic planning. Clin Infect Dis. 1999;28:225–9. doi: 10.1086/515123. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly JJ, Friedman A, Ulmer JB, Liu MA. Further protection against antigenic drift of influenza virus in a ferret model by DNA vaccination. Vaccine. 1997;15:865–8. doi: 10.1016/S0264-410X(96)00268-X. [DOI] [PubMed] [Google Scholar]
- 10.Justewicz DM, Morin MJ, Robinson HL, Webster RG. Antibody-forming cell response to virus challenge in mice immunized with DNA encoding the influenza virus hemagglutinin. J Virol. 1995;69:7712–7. doi: 10.1128/jvi.69.12.7712-7717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ledgerwood JE, Hu Z, Gordon IJ, Yamshchikov G, Enama ME, Plummer S, Bailer R, Pearce MB, Tumpey TM, Koup RA, et al. VRC 304 and VRC 305 Study Teams Influenza virus h5 DNA vaccination is immunogenic by intramuscular and intradermal routes in humans. Clin Vaccine Immunol. 2012;19:1792–7. doi: 10.1128/CVI.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bresson JL, Perronne C, Launay O, Gerdil C, Saville M, Wood J, Höschler K, Zambon MC. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet. 2006;367:1657–64. doi: 10.1016/S0140-6736(06)68656-X. [DOI] [PubMed] [Google Scholar]
- 13.Drape RJ, Macklin MD, Barr LJ, Jones S, Haynes JR, Dean HJ. Epidermal DNA vaccine for influenza is immunogenic in humans. Vaccine. 2006;24:4475–81. doi: 10.1016/j.vaccine.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Donnelly JJ, Friedman A, Martinez D, Montgomery DL, Shiver JW, Motzel SL, Ulmer JB, Liu MA. Preclinical efficacy of a prototype DNA vaccine: enhanced protection against antigenic drift in influenza virus. Nat Med. 1995;1:583–7. doi: 10.1038/nm0695-583. [DOI] [PubMed] [Google Scholar]
- 15.Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, et al. VRC 306 Study Team DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis. 2011;11:916–24. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Hackett A, Jia N, Zhang C, Zhang L, Parker C, Zhou A, Li J, Cao WC, Huang Z, et al. Polyvalent DNA vaccines expressing HA antigens of H5N1 influenza viruses with an optimized leader sequence elicit cross-protective antibody responses. PLoS One. 2011;6:e28757. doi: 10.1371/journal.pone.0028757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Taaffe J, Parker C, Solórzano A, Cao H, García-Sastre A, Lu S. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codon-optimized HA DNA vaccines. J Virol. 2006;80:11628–37. doi: 10.1128/JVI.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Shriver Z, Sasisekharan V, Sasisekharan R. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486–93. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–4. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–97. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 21.WHO. Number of confirmed human cases of avian influenza A(H7N9) reported to World Health Organization. World Health Organization, 2013. [Google Scholar]
- 22.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–32. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 23.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Parker C, Taaffe J, Solórzano A, García-Sastre A, Lu S. Heterologous HA DNA vaccine prime--inactivated influenza vaccine boost is more effective than using DNA or inactivated vaccine alone in eliciting antibody responses against H1 or H3 serotype influenza viruses. Vaccine. 2008;26:3626–33. doi: 10.1016/j.vaccine.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suguitan AL, Jr., Cheng X, Wang W, Wang S, Jin H, Lu S. Influenza H5 hemagglutinin DNA primes the antibody response elicited by the live attenuated influenza A/Vietnam/1203/2004 vaccine in ferrets. PLoS One. 2011;6:e21942. doi: 10.1371/journal.pone.0021942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S. Two is better than one. Lancet Infect Dis. 2011;11:889–91. doi: 10.1016/S1473-3099(11)70256-0. [DOI] [PubMed] [Google Scholar]
- 27.Kemble G, Greenberg H. Novel generations of influenza vaccines. Vaccine. 2003;21:1789–95. doi: 10.1016/S0264-410X(03)00074-4. [DOI] [PubMed] [Google Scholar]
- 28.Betts RF, Treanor JJ. Approaches to improved influenza vaccination. Vaccine. 2000;18:1690–5. doi: 10.1016/S0264-410X(99)00508-3. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Parker C, Taaffe J, Solórzano A, García-Sastre A, Lu S. Heterologous HA DNA vaccine prime--inactivated influenza vaccine boost is more effective than using DNA or inactivated vaccine alone in eliciting antibody responses against H1 or H3 serotype influenza viruses. Vaccine. 2008;26:3626–33. doi: 10.1016/j.vaccine.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Zhang C, Zhang L, Li J, Huang Z, Lu S. The relative immunogenicity of DNA vaccines delivered by the intramuscular needle injection, electroporation and gene gun methods. Vaccine. 2008;26:2100–10. doi: 10.1016/j.vaccine.2008.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, Lu S. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine. 2006;24:4531–40. doi: 10.1016/j.vaccine.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Pappas C, Matsuoka Y, Swayne DE, Donis RO. Development and evaluation of an Influenza virus subtype H7N2 vaccine candidate for pandemic preparedness. Clin Vaccine Immunol. 2007;14:1425–32. doi: 10.1128/CVI.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajesh Kumar S, Syed Khader SM, Kiener TK, Szyporta M, Kwang J. Intranasal immunization of baculovirus displayed hemagglutinin confers complete protection against mouse adapted highly pathogenic H7N7 reassortant influenza virus. PLoS One. 2013;8:e63856. doi: 10.1371/journal.pone.0063856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min JY, Vogel L, Matsuoka Y, Lu B, Swayne D, Jin H, Kemble G, Subbarao K. A live attenuated H7N7 candidate vaccine virus induces neutralizing antibody that confers protection from challenge in mice, ferrets, and monkeys. J Virol. 2010;84:11950–60. doi: 10.1128/JVI.01305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Q, Chen Z, Cheng X, Xu L, Jin H. Evaluation of live attenuated H7N3 and H7N7 vaccine viruses for their receptor binding preferences, immunogenicity in ferrets and cross reactivity to the novel H7N9 virus. PLoS One. 2013;8:e76884. doi: 10.1371/journal.pone.0076884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bos ME, Nielen M, Koch G, Stegeman A, De Jong MC. Effect of H7N1 vaccination on highly pathogenic avian influenza H7N7 virus transmission in turkeys. Vaccine. 2008;26:6322–8. doi: 10.1016/j.vaccine.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 37.Jadhao SJ, Achenbach J, Swayne DE, Donis R, Cox N, Matsuoka Y. Development of Eurasian H7N7/PR8 high growth reassortant virus for clinical evaluation as an inactivated pandemic influenza vaccine. Vaccine. 2008;26:1742–50. doi: 10.1016/j.vaccine.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 38.Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol. 2009;21:346–51. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ledgerwood JE, Zephir K, Hu Z, Wei CJ, Chang L, Enama ME, Hendel CS, Sitar S, Bailer RT, Koup RA, et al. VRC 310 Study Team Prime-boost interval matters: a randomized phase 1 study to identify the minimum interval necessary to observe the H5 DNA influenza vaccine priming effect. J Infect Dis. 2013;208:418–22. doi: 10.1093/infdis/jit180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almansour I, Chen H, Wang S, Lu S. Cross reactivity of serum antibody responses elicited by DNA vaccines expressing HA antigens from H1N1 subtype influenza vaccines in the past 30 years. Hum Vaccin Immunother. 2013;9:2049–59. doi: 10.4161/hv.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]