Summary
Enhancer RNAs (eRNAs) are a class of long noncoding RNAs (lncRNA) expressed from active enhancers, whose function and action mechanism are yet to be firmly established. Here we show that eRNAs facilitate the transition of paused RNA Polymerase II (RNAPII) into productive elongation by acting as a decoy for the negative elongation factor (NELF) complex upon induction of immediate early genes (IEGs) in neurons. eRNAs are synthesized prior to the culmination of target gene transcription and interact with the NELF complex. Knockdown of eRNAs expressed at neuronal enhancers impairs transient release of NELF from the specific target promoters during transcriptional activation, coinciding with a decrease in target mRNA induction. The enhancer-promoter interaction was unaffected by eRNA knockdown. Instead chromatin looping might enable eRNAs to act locally at a specific promoter. Our findings highlight the spatiotemporally regulated action mechanism of eRNAs during early transcriptional elongation.
Introduction
Stimulus-induced gene expression in the nucleus is a critical mechanism for cell-wide adaptive responses to environmental cues. In neurons, sensory experience-evoked synaptic activity triggers various calcium-dependent signaling events, which then induce the expression of a group of genes involved in distinct aspects of neuronal function. We have previously shown that the enhancers of these activity-regulated genes rapidly induce eRNA synthesis when cortical neurons are depolarized by 55 mM KCl (Kim et al., 2010). The majority of eRNAs are transcribed bi-directionally with a strong positive correlation with the expression of nearby protein-coding genes, suggesting a possible “activating” function of eRNA as part of a genome-wide activity-dependent epigenetic mechanism (Kim et al., 2010).
eRNAs have also been identified in many non-neuronal cell types and recognized as a reliable marker for active enhancers (Andersson et al., 2014; Creyghton et al., 2010; De Santa et al., 2010; Djebali et al., 2012; Hah et al., 2011; Hsieh et al., 2014; IIott et al., 2014; Rada-Iglesias et al., 2011; Wang et al., 2011). Moreover, eRNAs appear to be functionally important for gene activation as knockdown of eRNAs expressed in different cell types invariably resulted in a reduction of transcription of specific target genes (Hsieh et al., 2014; IIott et al., 2014; Lam et al., 2013; Li et al., 2013; Melo et al., 2013; Mousavi et al., 2013). Despite these exciting findings, precise action mechanisms of eRNAs during the transcriptional induction process have not been well established. A study of eRNAs in human breast cancer cells showed that eRNAs contribute to 17β-oestradiol (E2)-dependent gene activation by stabilizing enhancer–promoter looping through an interaction with cohesin, which forms a complex with Mediator to facilitate chromosomal looping (Kagey et al., 2010; Li et al., 2013). This effect of eRNAs in chromatin looping is reminiscent of the function of activating-ncRNAs (ncRNA-a) that activate gene transcription by facilitating looping through an interaction with Mediator (Lai et al., 2013). In contrast, the eRNA expressed from the distal regulatory region near Myod1 in C2C12 cells does not regulate chromatin looping when judged by the binding levels of the cohesin subunit, RAD21 and a cohesin-loading factor, NIPBL. Instead it was shown to promote transcription of the Myod1 gene by establishing chromatin accessibility through an unknown mechanism (Mousavi et al., 2013). These recent findings suggest that eRNAs might play a regulatory role in various aspects of the transcription process and that further mechanistic study of eRNA function would be imperative for understanding the regulatory capacity of noncoding RNAs in gene expression.
RNAPII pausing is a genome-wide regulatory mechanism in higher eukaryotes, especially enriched at genes in developmentally and environmentally responsive pathways (Adelman and Lis, 2012; Gilchrist et al., 2012). NELF and DRB sensitivity-inducing factor (DSIF) cooperatively induce RNAPII pausing by binding directly to RNAPII and nascent RNA (Adelman and Lis, 2012; Cheng and Price, 2008; Missra and Gilmour, 2010; Yamaguchi et al., 1999). One of the NELF subunits, NELF-E mediates the binding of the NELF complex to nascent RNAs through its RNA recognition motif (RRM), which has been shown to be critical for the transcriptional repression activity of NELF in an in vitro transcription assay (Yamaguchi et al., 2002). Pause release and subsequent elongation are mediated by the positive transcription elongation factor b (P-TEFb), which phosphorylates the RNAPII C-terminal domain (CTD), DSIF, and likely NELF (Adelman and Lis, 2012; Fujinaga et al., 2004; Marshall et al., 1996; Wada et al., 1998a; Wada et al., 1998b; Yamaguchi et al., 1999). Here we not only show that eRNAs are functionally important for proper induction of neuronal immediate early genes (IEGs) in response to an increase in neuronal activity, but also reveal a novel action mechanism of eRNAs during the transition of paused RNAPII to productive elongation. Knockdown of eRNAs caused a reduction in the expression of specific target genes, while the chromosomal looping between the promoter and enhancer was unaffected. However when eRNA levels are reduced, the NELF complex could not be efficiently released from the promoter of the specific target gene during transcriptional induction, and this is accompanied by a reduction in elongating RNAPII and target mRNA. Both ultra-violet RNA immunoprecipitation (UV-RIP) and in vitro RNA pull-down assays demonstrated that eRNAs expressed upon stimulation of neurons are able to directly bind to the RNA recognition motif (RRM) of the NELF-E subunit. Replacement of endogenous NELF-E with the RRM-deletion mutant in neurons significantly reduces the levels of NELF complex binding at the IEG promoters as well as mRNA induction, further illustrating that the interaction with RNA molecules (e.g., eRNA and nascent RNAs) is a critical mechanism for NELF to regulate IEG induction in neurons. Taken together, neuronal eRNAs may facilitate the release of NELF by acting as a decoy for nascent transcripts to allow for the efficient transition of paused RNAPII to productive elongation.
Results
eRNAs Are Necessary for Target Gene Induction
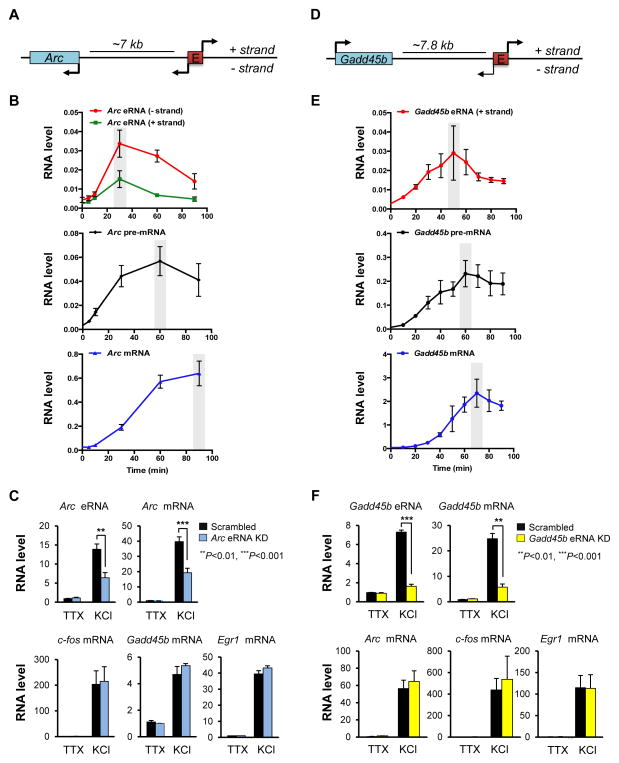
In our initial effort to characterize the function of eRNAs, we performed a time course measurement of the eRNAs that are expressed from the enhancer for Activity-regulated cytoskeletal protein (Arc), an IEG important for brain development and function (Korb and Finkbeiner, 2011) (Figures 1A, 1B, and S1A). Neuronal activity was first suppressed by tetrodotoxin (TTX), a sodium channel blocker that prevents neuronal action potentials, and then expression levels of Arc eRNA, pre-mRNA, and mRNA were monitored for various times following KCl-mediated membrane depolarization. We observed that the bi-directional synthesis of Arc eRNA was induced by membrane depolarization but peaked earlier than Arc pre-mRNA and mRNA (Figure 1B). The difference in peak times between eRNA and pre-mRNA is not due to a significant difference in decay times between the transcripts as both transcripts show a similar decay rate (approximate half-life ≤7.5 min) upon addition of a transcription inhibitor, Actinomycin D following 30 min of KCl-mediated depolarization (Figure S1C). This result could suggest that eRNA synthesis is not merely a byproduct of promoter-driven transcription activity, but instead an independently regulated process.
Figure 1. Characterization of Arc and Gadd45b eRNA.
(A) Schematic diagram of the Arc genomic locus (see also Figure S1A). (B) Cortical neurons were depolarized at DIV 6 with 55 mM KCl for various time points, and expression levels of Arc eRNAs, pre-mRNA, and mRNA were measured using qRT-PCR and normalized to the level of TBP mRNA (n = 4 biological replicates). (C) qRT-PCR analysis of Arc eRNA and mRNA expression after knockdown of Arc eRNA (− strand) or infection with a scrambled control in cortical neurons. Levels of indicated RNAs were measured after 30 min KCl or TTX treatment in cortical neurons and normalized to the level of TBP mRNA (n = 3 biological replicates). (D) Schematic diagram of the Gadd45b genomic locus (see also Figure S1B). (E) Cortical neurons were depolarized at DIV 6 with 55 mM KCl for various time points, and expression levels of Gadd45b eRNA, pre-mRNA, and mRNA were measured using qRT-PCR and normalized to the level of TBP mRNA (n = 4 biological replicates, Gadd45b pre-mRNA: n=3 biological replicates) (F) qRT-PCR analysis of Gadd45b eRNA and mRNA expression after knockdown of Gadd45b eRNA (+ strand) in cortical neurons. Levels of indicated RNAs were measured after 60 min KCl or TTX treatment in cortical neurons and normalized to the level of TBP mRNA (n = 3 biological replicates). Error bar indicates s.e.m.; P-value from two-tailed t-test.
Arc eRNA was also induced by the GABAA receptor antagonist bicuculline, which more closely resembles the physiological activation of synapses (Figure S2A) (Hardingham et al., 2001). By blocking the major inhibitory input in neurons, bicuculline triggers a synchronous burst of action potentials and induces both Arc eRNA and mRNA. Notably, the minus strand of Arc eRNA was predominantly induced in response to bicuculline, suggesting that synaptic activity-driven eRNA induction may occur in a strand-specific manner. When the minus strand of Arc eRNA was sequenced using an RNA circularization method, we found that while there was a distinct 5′ end of the transcript, the 3′ ends were degenerate without noticeable polyadenylation (Figure S2B), although we cannot rule out the possibility that a minor population of the eRNAs could be polyadenylated. We also found that Arc eRNA can be induced by serum stimulation in NIH3T3 cells, but remain localized in the nucleus after their synthesis whereas Arc mRNA is present in both the nucleus and the cytoplasm (Figure S2C). These properties of Arc eRNAs are in good agreement with the latest ENCODE consortium analysis of eRNAs in human cell lines showing that eRNAs are prevalent in the nuclear non-polyadenylated RNA fraction (Djebali et al., 2012).
To test the functionality of eRNAs in activity-induced neuronal gene expression more directly, lentiviral constructs containing short-hairpin RNAs (shRNAs) against the minus strand of Arc eRNA were designed to knockdown the eRNAs and assess the effect on Arc mRNA induction in response to membrane depolarization. Knockdown of Arc eRNAs reproducibly led to a decrease in the level of Arc mRNA induction when compared to a scrambled control shRNA, suggesting that Arc eRNAs are functionally important for neuronal activity-dependent transcriptional induction of the Arc gene (Figure 1C). Arc eRNA appears to specifically regulate Arc gene expression since the expression levels of other neuronal IEGs (e.g., c-fos, Egr-1, Gadd45b) were not affected by the Arc eRNA knockdown. To further evaluate the specificity of eRNA action, we examined the knockdown effect of the eRNAs expressed from an enhancer located nearby the Growth arrest and DNA-damage-inducible, beta (Gadd45b) gene (Figures 1D and S1B). Although expressed from a well-defined enhancer region, the plus strand of Gadd45b eRNAs is predominantly transcribed upon membrane depolarization and peaks ~50 min after KCl treatment (Figures 1E and S1B), which is later than the Arc eRNA peak time. Knockdown of the Gadd45b eRNA plus strand specifically reduces the induction level of Gadd45b mRNA but not other IEGs upon KCl-mediated membrane depolarization of neurons (Figure 1F). We also observed that the impairment in the activity-dependent induction of Arc and Gadd45b transcription caused by the knockdown of corresponding eRNAs leads to a decrease in the levels of ARC and GADD45B proteins (Figure S2D). Taken together, these results suggest that eRNAs can act locally at their specific target genes, which is consistent with recent functional analyses of eRNAs in non-neuronal cells (Hsieh et al., 2014; IIott et al., 2014; Lam et al., 2013; Li et al., 2013; Melo et al., 2013; Mousavi et al., 2013) (see also Figure S5).
Enhancer-Promoter Interactions Are Not Dependent on eRNA
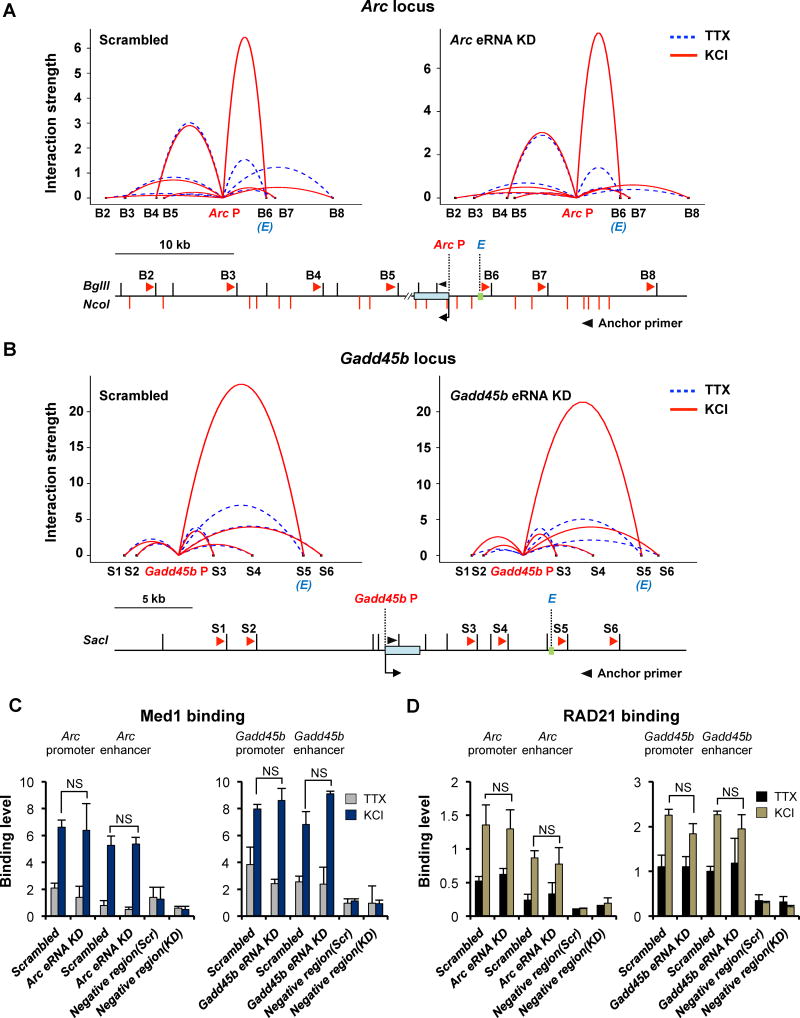
Promoters and distal enhancers can be physically juxtaposed with each other through chromosomal looping as part of a gene regulatory mechanism (Smallwood and Ren, 2013). We reasoned that the target specificity of eRNAs could be mediated by enhancer-promoter looping. In order to see if the interaction between the Arc promoter and enhancer occurs constitutively or in a stimulus-dependent manner (i.e., membrane depolarization of neurons), we performed Chromosome Conformation Capture (3C) to quantitatively measure the chromosomal interactions in the regions surrounding the Arc gene (Figures 2A and S3A). As expected, the eRNA-producing Arc enhancer was the most prominent genomic locus that interacts with the Arc promoter in an activity-dependent manner. The 3C analysis also found another interaction site (B4) that had not been previously identified, but its interaction with the Arc gene locus was constitutive. Some lncRNAs and eRNAs have been shown to promote target gene expression by facilitating chromosomal looping between the enhancer and promoter (Lai et al., 2013; Li et al., 2013). Therefore we tested if Arc eRNA can also mediate the interaction between the Arc promoter and enhancer (Figures 2A and S3A). Despite the efficient knockdown of the Arc eRNA observed in the same experiment (Figure S3B), we found no significant change in the chromosomal interaction between the Arc enhancer and promoter. This was also the case with the Gadd45b locus, as we identified a strong activity-dependent interaction between the Gadd45b promoter and enhancer that was unaffected by Gadd45b eRNA knockdown (Figures 2B, S3C and S3D).
Figure 2. Activity-induced interactions between enhancers and promoters.
(A and B) 3C analysis to examine the effect of eRNA knockdown in enhancer-promoter looping. Chromosomal interactions between the Arc or Gadd45b promoter and surrounding genomic loci were measured by q-PCR using the primers indicated in the schematic diagram. Arc P or Gadd45b P indicates the promoter and E indicates the enhancer. The black arrowhead near the Arc P and Gadd45b P indicates the anchor primer. The restriction enzyme sites (vertical lines) and primers used for q-PCR together with the anchor primer (arrowheads) are also shown (n = 3 biological replicates). (C and D) Binding levels of Med1 and RAD21 at the Arc and Gadd45b promoters, and corresponding enhancers determined by ChIP-qPCR in neurons infected with a scrambled control or eRNA knockdown lentivirus in quiescent (TTX) and KCl stimulated conditions (n = 2 biological replicates). Error bar indicates s.e.m.; P-value from two-tailed t-test.
To corroborate our findings from 3C analysis, we next examined the effect of eRNA knockdown on the binding levels of the Mediator and cohesin complexes at both the Arc and Gadd45b enhancers as well as promoters (Figures 2C and 2D). The Mediator-cohesin complex co-occupies enhancers and promoters to facilitate enhancer–promoter DNA looping, and recent studies have implicated ncRNA-a and eRNAs in chromosomal looping through interactions with Mediator and the cohesion complex, respectively (Lai et al., 2013; Li et al., 2013). Chromatin immunoprecipitation (ChIP) analysis of a common Mediator complex subunit Med1 and a cohesin subunit RAD21 shows that both the promoters and enhancers of Arc and Gadd45b genes are inducibly occupied by the Mediator/cohesin complex upon membrane depolarization, but eRNA knockdown has no effect on their occupancy. Together with the 3C analysis, these results collectively argue that at least for neuronal IEG expression, eRNAs are not required for the enhancer-promoter interaction. Our finding is consistent with other reports showing that eRNA transcription is not necessary for enhancer-promoter looping in human breast cancer cells and that eRNA knockdown has no effect on cohesin complex loading in mouse skeletal muscle (Hah et al., 2013; Mousavi et al., 2013).
Knockdown of eRNAs Causes NELF to Remain Bound to Target Gene Promoters
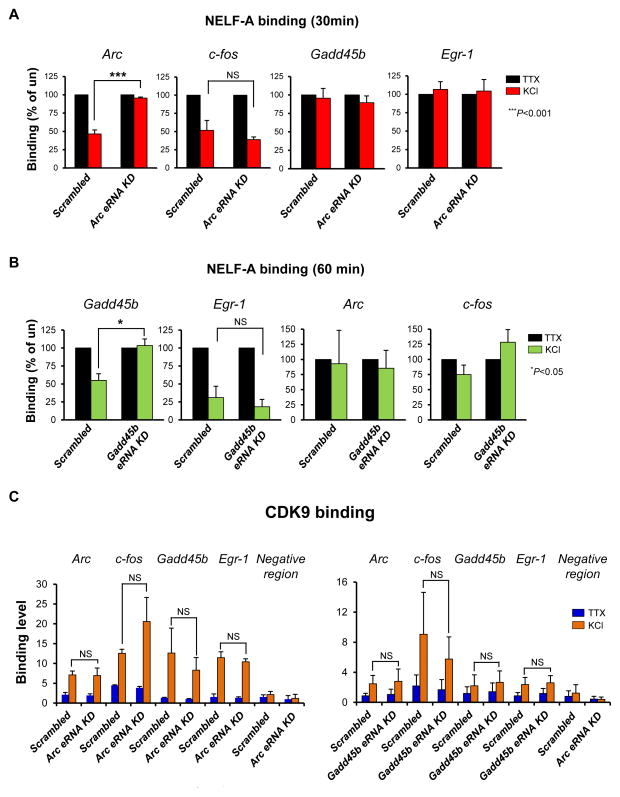
Recent genome-wide studies unambiguously argue that proximal-promoter pausing of RNAPII is a widespread mechanism of transcriptional regulation for controlling expression of stimulus-responsive genes in higher eukaryotes (Adelman and Lis, 2012; Gilchrist et al., 2012). Because of the rapid induction kinetics of eRNAs (Figures 1B and 1E), we investigated whether eRNAs play a role in the early transcription elongation step that involves RNAPII pausing and release. Analyses of RNAPII elongation complexes using a native electrophoretic mobility shift assay have demonstrated that DSIF and NELF complexes are stably associated with paused RNAPII through interactions with both RNAPII and nascent transcripts (Cheng and Price, 2008; Missra and Gilmour, 2010). Therefore, we hypothesized that during target gene activation, eRNAs might destabilize the DSIF/NELF association with RNAPII by mimicking nascent transcripts and thereby facilitate the RNAPII transition from pausing to productive elongation. Expression of neuronal IEGs was also shown to be subject to this RNAPII pausing mechanism (Saha et al., 2011). Consistently we found that NELF binds specifically to the Arc promoter, but not to the Arc enhancer when neuronal gene expression is suppressed by TTX (Figure S4A). NELF is then released from the Arc promoter upon activation of neuronal gene expression by KCl-mediated membrane depolarization. NELF release appears to occur within a narrow time window and to be gene-specific, as NELF occupancy at the promoters of Arc and c-fos was transiently decreased at 30 min after KCl stimulation whereas NELF complexes bound at Gadd45b and Egr1 promoters were released at 1 h (Figures 3A and 3B). Transient release of NELF during transcription activation was also observed in a previous study where NELF occupancy at the TNFα proximal promoter in macrophages was temporarily decreased at 30 min after lipopolysaccharide (LPS) treatment (Adelman et al., 2009). Interestingly, shRNA-mediated knockdown of Arc and Gadd45b eRNAs blocked the NELF release from their corresponding promoters even during membrane depolarization (Figures 3A and 3B). A subunit of P-TEFb, CDK9 was also inducibly recruited to the Arc promoter at 30 min after membrane depolarization (Figure S4B), but its recruitment was unaffected by eRNA knockdown (Figure 3C). This result strongly suggests that eRNAs facilitate transient release of NELF during gene activation.
Figure 3. eRNAs function to facilitate the release of the NELF complex from paused RNAPII.
(A) Effect of Arc eRNA knockdown on NELF-A binding at the Arc, c-fos, Gadd45b and Egr-1 promoters (n = 2 biological replicates). (B) Effect of Gadd45b eRNA knockdown on NELF-A binding at the Arc, c-fos, Gadd45b and Egr-1 promoters (n = 2 biological replicates). (C) Effect of Arc eRNA and Gadd45b eRNA knockdown on CDK9 binding at the Arc, c-fos, Gadd45b and Egr-1 promoters (n = 2 biological replicates). Error bars indicate s.e.m. P-value from two-tailed t-test. NS, not significant.
To further validate our findings, we applied a Locked Nucleic Acid antisense-oligonucleotide (LNA) method to see if we would observe the same result when the level of eRNA is reduced by another independent knockdown method. LNAs can induce degradation of the complementary target RNA by recruiting RNase H without involving the cell’s RNAi machinery (Watts and Corey, 2012). Due to the low efficiency of LNA transfection in neurons, we tested the effect of LNA-mediated knockdown of Arc eRNA in NIH3T3 cells, in which both Arc eRNA and mRNA are induced by serum stimulation (Figure S5A). As seen by shRNA-mediated knockdown of Arc eRNA in neurons, an LNA designed to target Arc eRNA was able to reduce both Arc eRNA and mRNA levels during serum stimulation, without affecting other IEGs induced by serum (compare Figures 1C and S5A). The Gadd45b gene was not induced by serum stimulation in NIH3T3 cells, thus not analyzed in this experiment. Having verified the effect of Arc eRNA by two independent knockdown methods, we then asked if the LNA-mediated knockdown of Arc eRNA would also block NELF release from the Arc promoter during serum stimulation (Figure S5B). In NIH3T3 cells, NELF was transiently released from the promoters of Arc, c-fos, and Egr1 at 30 min after serum stimulation. However, knockdown of Arc eRNA by LNA caused the retention of NELF only at the Arc promoter during serum stimulation, which is consistent with the results from the shRNA-mediated knockdown experiment (compare Figures 3A and S5B). The consistent results obtained by two independent knockdown methods in two different cell types strongly suggest that the effects of Arc eRNA knockdown on NELF release as well as target gene induction are unlikely an artifact or indirect consequence that is associated with a particular knockdown method.
eRNAs facilitate the transition of paused RNAPII to elongation
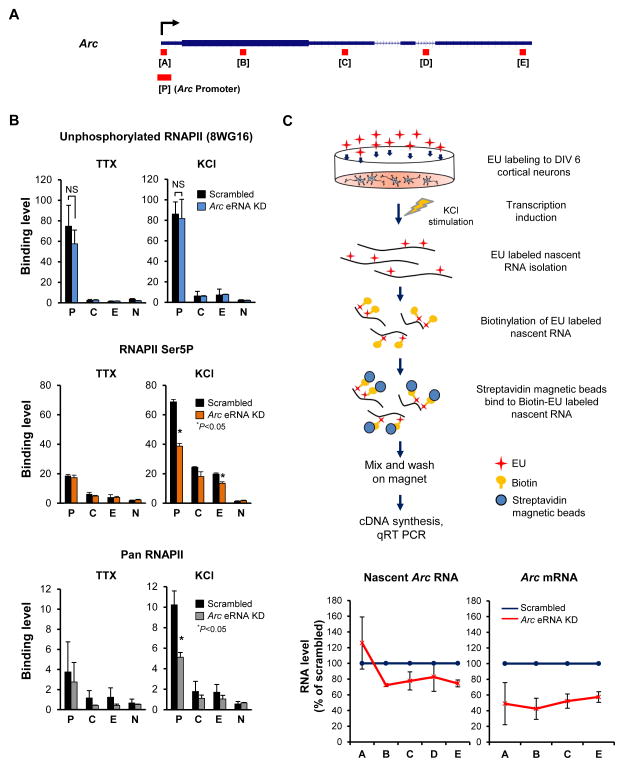
The NELF/DSIF complex pauses RNAPII during the early elongation stage of the transcription cycle (after transcribing 20 – 60 nucleotides of nascent transcript) (Adelman and Lis, 2012; Rasmussen and Lis, 1993; Rougvie and Lis, 1988). If eRNA contributes to the efficient release of NELF from paused RNAPII, thereby facilitating the RNAPII transition to productive elongation, then eRNA knockdown would specifically reduce the level of RNAPII at the elongation stage but not at the initiation and pre-initiation stages. The C-terminal domain (CTD) of RNAPII is subject to sequential phosphorylation events during the transcription cycle (Egloff et al., 2012). Only the unphosphorylated form of RNAPII can be assembled into the preinitiation complex at the promoter. During the promoter escape and early elongation stage, the serine 5 (Ser-5) residue of the CTD is phosphorylated by the CDK7 kinase subunit of TFIIH. The serine 2 (Ser-2) residue of the CTD is then gradually phosphorylated by the CDK9 subunit of P-TEFb as RNAPII stably elongates toward the 3′ end of the gene. To test whether eRNAs selectively regulate the RNAPII transition to productive elongation, we examined the effect of Arc eRNA knockdown on RNAPII levels along the Arc gene by ChIP experiments with antibodies recognizing different forms of RNAPII: unphosphorylated RNAPII (8WG16), RNAPII-Ser5P, and Pan RNAPII. We found that when Arc eRNA levels were reduced using shRNAs targeting the Arc minus strand, there was no change in the level of unphosphorylated RNAPII at the Arc promoter detected by the 8WG16 antibody (Jones et al., 2004) (Figure 4B, top). Since only the unphosphorylated form of RNAPII can enter into the pre-initiation complex, this suggests that recruitment of RNAPII to the promoter is unaffected by the eRNA knockdown. However, the level of RNAPII phosphorylated at Ser-5 (Ser-5P) was significantly decreased at the promoter as well as the 3′ end of the Arc gene during KCl stimulated conditions (Figure 4B, middle). The Pan RNAPII antibody also detected a significant reduction in total RNAPII levels at the Arc promoter region during stimulation, and RNAPII levels along the coding region showed a trend toward a decrease, as well, although weak (Figure 4B, bottom). Since the Pan RNAPII antibody cannot distinguish between the different forms of RNAPII, it detects the sum of the levels of RNAPIIs present in different stages of transcription. As the unphosphorylated form of RNAPII is unchanged, the decrease seen in the Pan RNAPII ChIP is likely due to a decrease in the phosphorylated, elongating RNAPII, as is seen with the Ser5P antibody, although we cannot completely rule out a possibility that recruitment of RNAPII is also affected. These results indicate that upon KCl-depolarization, eRNAs play an important role in facilitating the transition of paused RNAPII to elongating RNAPII, as the level of elongating forms of RNAPII is specifically decreased when the Arc eRNA level is lowered.
Figure 4. Arc eRNAs promote efficient transition of RNAPII into productive elongation.
(A) Schematic diagram of primer sets used to measure binding levels (B) or RNA levels (C) at various locations along the Arc gene. (B) Effect of Arc eRNA knockdown on binding of unphosphorylated RNAPII (8WG16), RNAPII phosphorylated at Ser5 (Ser5P), and total levels of RNAPII (Pan RNAPII) (n = 2 biological replicates). Binding was determined at the following locations along the Arc gene: Arc promoter [p], middle [C], 3′ end [E], or a Negative control region [N]. (C) Schematic diagram of nascent RNA detection using a nascent RNA capture kit (top). RNA levels at various points along the transcript (primer sets A–E) are quantified using qRT-PCR (bottom) (n = 2 biological replicates). Error bars indicate s.e.m. P-value from two-tailed t-test. NS, not significant.
To correlate the RNAPII ChIP results with transcription, we then selectively compared the levels of various regions of nascent Arc transcripts induced by KCl treatment with or without shRNA-mediated eRNA knockdown (Figures 4A and 4C) (see methods). The transcript level immediately downstream of the transcription start site (TSS; detected by primer set A) in the nascent RNA sample was not reduced when compared to the scrambled control, whereas all other regions (detected by primer sets B–E) were lower. In contrast, knockdown of Arc eRNA resulted in a uniform decrease in the steady state level of Arc mRNA. Retention of similar or even higher levels of nascent transcription specifically near the 5′ end of the Arc gene after Arc eRNA knockdown is consistent with the RNAPII ChIP results and further supports a role of eRNA in the transition of paused RNAPII to productive elongation.
eRNAs interact with NELF-E in an RRM-dependent manner
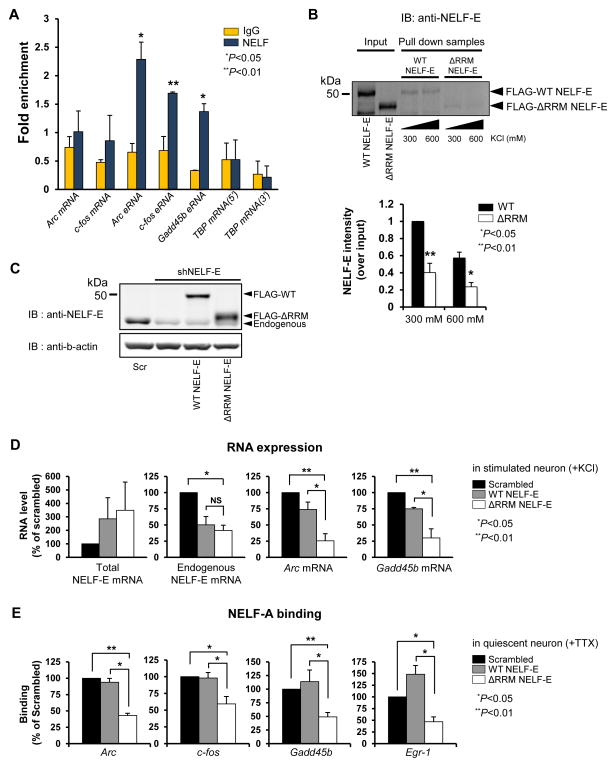
The observed effect of eRNA knockdown on NELF release suggests a possibility that the NELF complex and eRNAs might directly interact with each other. The NELF-E subunit contains an RNA recognition motif (RRM) that mediates direct interactions with various RNA sequences with little or no apparent sequence or structural constraint, which is suitable for binding to nascent RNAs derived from many genes (Rao et al., 2006; Yamaguchi et al., 2002). The RRM of NELF-E was also shown to be critical for the RNAPII pausing activity of NELF in an in vitro transcription assay (Yamaguchi et al., 2002). We postulate that when eRNAs are rapidly induced in neurons by KCl-depolarization, they might compete with the nascent RNA attached to paused RNAPII for binding to NELF, thereby facilitating the release of the NELF complex. To test this idea, we performed UV-RNA Immunopreciptation (UV-RIP) with an antibody directed against the NELF-E subunit that contains the RRM in order to see if eRNAs can directly bind to the NELF-E subunit (Yamaguchi et al., 2002) (Figure 5A). We found that the proportions of Arc, Gadd45b, and c-fos eRNAs brought down with NELF-E at 30 min following membrane depolarization were significantly higher than those with the IgG control, whereas there was no such enrichment of target gene mRNAs or the constitutively expressed TBP mRNA. We also performed eRNA pull-down experiments using biotinylated full-length Arc eRNA transcribed in vitro and lysates from HEK293T cells that overexpress either a FLAG-tagged wild-type (WT) or RRM-deletion mutant (ΔRRM) of the NELF-E protein. When biotinylated Arc eRNAs were pulled down by streptavidin beads under two different salt washing conditions, we reproducibly observed a higher level of WT NELF-E protein co-precipitated than the ΔRRM NELF-E protein (Figure 5B). This data not only provides additional evidence supporting the conclusion that eRNAs are able to interact with NELF-E, but also demonstrates that the interaction is dependent on the RRM.
Figure 5. NELF-E directly interacts with eRNAs.
(A) Ultraviolet-crosslinking RNA immunoprecipitation using KCl depolarized cultured cortical neuron lysates. Fold enrichment indicates the amount of RNA normalized to its respective input (n = 3 biological replicates). (B) Pull-down of FLAG-tagged wildtype (WT) or an RRM-deletion mutant (ΔRRM) of NELF-E overexpressed in HEK293T cells by in vitro transcribed biotinylated Arc eRNA. The top panel shows a representative western blot probed with anti-NELF-E. The bottom panel shows the quantification of the results, normalizing each lane to the corresponding input (n = 3 biological replicates). (C) Representative western blot showing the knockdown of NELF-E compared to a scrambled control shRNA and overexpression of either WT or ΔRRM NELF-E in cortical cultures. β-actin was used as a loading control. (D) Effect of replacement of endogenous NELF-E with FLAG-WT or FLAG-ΔRRM NELF-E on RNA levels during KCl depolarized conditions for total Nelf-e mRNA, endogenous Nelf-e mRNA, Arc mRNA, and Gadd45b mRNA. (n = 3 biological replicates). (D) Effect of replacement of endogenous NELF-E with FLAG-WT or FLAG-ΔRRM NELF-E on NELF-A binding during unstimulated conditions (n = 3 biological replicates, except Arc which has n = 2 biological replicates). Error bars indicate s.e.m. P-value from two-tailed t-test.
The RRM of the NELF-E subunit is critical for IEG induction in neurons
Having found an interaction between the eRNA and the NELF-E RRM, we next examined how critical the RRM domain is for NELF function for IEG expression in neurons. To do this, we co-infected neurons with two different lentiviruses that express an shRNA against the 3′ UTR of endogenous NELF-E mRNA, and the shRNA-resistant forms of FLAG-tagged WT or ΔRRM NELF-E protein, which lack both the 5′ and 3′ UTRs. In this replacement experiment, we titrated the amount of NELF-E protein variants to be similar to the level of endogenous NELF-E protein before knockdown, to avoid any complication resulting from excessive expression of exogenous protein in neurons (Figure 5C). We observed in KCl-depolarized neurons that the induction of both Arc and Gadd45b mRNAs was similar to the scrambled control levels when endogenous NELF-E protein was replaced by the FLAG-tagged version of WT NELF-E, but replacement with the FLAG-ΔRRM NELF-E protein led to a significant decrease in the induction levels of Arc and Gadd45b mRNAs compared to the scrambled control or WT NELF-E (Figures 5C and 5D).
We also measured the binding levels of NELF complexes assembled with exogenously expressed NELF-E variants at various IEG promoters. NELF complexes formed with either endogenous NELF-E (scrambled condition) or the WT NELF-E variant showed similar levels of occupancy at the promoters of Arc, Gadd45b, c-fos and Egr1 when neuronal activity was suppressed by TTX. However, the NELF complex assembled with the FLAG-ΔRRM NELF-E protein showed a much lower level of binding at the IEG promoters, suggesting that the RRM interaction with nascent RNA emerging from initiating RNAPII is important for NELF to stably associate with RNAPII to mediate pausing in quiescent neurons (Figure 5E). A previous study in neurons showed that NELF-dependent RNAPII pausing allows rapid induction of neuronal IEG expression (Saha et al., 2011). Reduction of RNAPII pausing by knockdown of Nelf-a or Nelf-e prevented rapid Arc transcription, resulting in a lower level of Arc pre-mRNA induction than a scrambled control upon neuronal activity increase. Our results would further suggest that the interactions with various RNAs via the NELF-E RRM might be a critical mechanism for NELF to regulate IEG induction in neurons. Deletion of the RRM in NELF-E disrupted NELF binding at the promoters of these IEGs (Figure 5E), which we propose causes a reduction of RNAPII pausing, and in turn impairs rapid and synchronous induction of neuronal IEGs such as Arc and Gadd45b (Figure 5D).
Discussion
RNAPII pause and release is a key rate-limiting step in the transcription of many eukaryotic genes, integrating multiple regulatory signaling pathways, and is thought to support the establishment of permissible chromatin architecture, as well as rapid and/or synchronous gene activation (Adelman and Lis, 2012; Min et al., 2011). In neurons, precise coordination and timing of gene induction in response to changes in neural activity is critical for the consolidation of synaptic plasticity, and NELF has been shown to be an important player in the rapid induction of neuronal IEGs by maintaining poised RNAPII at the promoters when neurons are quiescent (Saha et al., 2011; West and Greenberg, 2011). Our study has revealed a novel mechanism of eRNA action during this process to facilitate the rapid induction of IEGs. By acting as a decoy for NELF, eRNAs facilitate the transient release of NELF from paused RNAPII, which then enters into a productive elongation stage.
We did not, however, find any noticeable impact of eRNA knockdown on the chromatin looping between the enhancer and the promoter, which has been found to be commonly targeted by both ncRNA-a and some eRNAs (Lai et al., 2013; Li et al., 2013). In fact our previous study suggested that the Arc eRNA was unlikely to regulate the interaction between the Arc enhancer and promoter (Kim et al., 2010). In neurons in which the coding region, as well as the promoter, for the Arc gene has been deleted, Arc eRNA was not induced at all by membrane depolarization despite the normal level of RNAPII binding at the enhancer. An implication from this result is that the Arc eRNA might be transcribed only after the Arc enhancer is juxtaposed to the Arc promoter by chromosomal looping. Consistent with this idea, our 3C and ChIP analyses argue that, at least for IEGs in neurons, eRNAs do not contribute to enhancer-promoter looping (Figure 2). The eRNA (termed as CERNA) expressed from the enhancer for the Myod1 gene during differentiation of mouse C2C12 muscle cells is another example showing no regulatory activity of eRNA in looping (Mousavi et al., 2013). Instead, the CERNA increases RNAPII occupancy at the promoter as well as transcription of the Myod1 gene by regulating the chromatin accessibility, although the exact mechanism is yet to be determined. These results collectively show that eRNAs can positively regulate gene expression by various mechanisms depending on the context.
Upon gene activation, P-TEFb is recruited to the paused RNAPII complex by interacting with various transcription and/or chromatin regulators (Zhou et al., 2012). The CDK9 kinase subunit of P-TEFb then phosphorylates the RNAPII CTD at Ser-2, DSIF, and possibly NELF, which has been shown to be critical for the RNAPII transition into productive elongation (Fujinaga et al., 2004; Marshall et al., 1996; Wada et al., 1998b). P-TEFb-dependent phosphorylation of NELF-E occurs in a region next to the RRM (Fujinaga et al., 2004). A mutant NELF-E protein that mimics its phosphorylated form no longer binds the transactivation response (TAR) element nor represses HIV transcription. However, it is not known whether the P-TEFb-dependent phosphorylation of NELF is necessary and sufficient for the release of NELF from all of its cellular target promoters in vivo. Based on our findings, we envision that in an in vivo chromatin context, efficient transition of RNAPII from pausing to productive elongation is mediated by the coordinated actions of multiple factors, in which eRNAs contribute to the topological rearrangement of NELF in conjunction with P-TEFb action. In this scenario, eRNAs would play a modulatory role in gene induction. eRNAs may not be absolutely required for basic transcription, but their role is instead to enhance the transcriptional response by allowing NELF to be released more efficiently, resulting in a larger and precisely timed response.
Biochemical evidence clearly supports the role of NELF in transcription repression in vitro, as NELF addition to the in vitro transcription system selectively inhibits RNAPII elongation (Yamaguchi et al., 2002; Yamaguchi et al., 1999). However, RNAPII pausing in an in vivo context does not necessarily mean that transcription is completely silenced, but instead may be a tuning mechanism for tailoring transcriptional outputs of subjected genes depending on the cellular context (Adelman and Lis, 2012; Yamaguchi et al., 2013). The magnitude and impact of RNAPII pausing at each gene varies depending on many factors such as the strength and composition of the core promoter elements and nucleosome positioning. Genome-wide analysis pointed out that an important function of RNAPII pausing in metazoans is to maintain the chromatin architecture surrounding the promoter-proximal regions in an open and accessible state for regulatory factors and efficient RNAPII elongation (Gilchrist et al., 2010; Gilchrist et al., 2008; Leibovitch et al., 2002; Yamaguchi et al., 2013). Although the promoter sequences of highly paused genes intrinsically favor nucleosome assembly, RNAPII pausing effectively antagonizes the assembly of nucleosomes immediately downstream of the TSS, thereby indirectly contributing to transcriptional up-regulation. This explains why many NELF target genes are down-regulated upon removal of pausing (Gilchrist et al., 2010; Gilchrist et al., 2008; Leibovitch et al., 2002). Neuronal IEGs appear to be subject to this mode of regulation as well, as their expression levels are also decreased by depletion of NELF-A or E (Saha et al., 2011). We have observed a similar decrease in IEG expression levels by simply deleting the RRM region from NELF-E, which causes a reduced level of NELF binding at the IEG promoters (Figures 5D and 5E). This finding, and the effect of eRNA on NELF release described earlier, highlight the complex role of NELF in vivo during gene expression. Without the NELF complex to induce pausing at the promoter during unstimulated conditions, the proper chromatin architecture is not maintained, and gene expression is impaired. However, if NELF does induce pausing, but is not efficiently released in response to stimulation, as with the case in which eRNAs are knocked down, gene induction is also impaired. Our findings from the replacement experiments collectively emphasize that RNA plays an important regulatory role in both RNAPII pausing and release in neurons.
The ability of RNA to act as a decoy was previously demonstrated by several studies (Hung et al., 2011; Kino et al., 2010; Rinn and Chang, 2012; Sun et al., 2013). The growth arrest–specific 5 (Gas5) noncoding RNA contains a sequence that resembles the glucocorticoid response element (GRE) and blocks the glucocorticoid-mediated induction of several genes in growth-arrested cells by competing with GREs for binding to the glucocorticoid receptor (GR). Another lncRNA, PANDA is induced during the DNA damage response by p53 and inhibits the expression of apoptotic genes by sequestering the transcription factor NF-YA away from chromatin. Another decoy action of RNA can be found during X chromosome inactivation (XCI). The transcription of Xist ncRNA that initiates XCI is controlled by a dynamic balance between CTCF and Jpx RNA. At pre-XCI cells, CTCF is bound to the promoter of the Xist gene to suppress Xist expression, but upon induction of XCI, Jpx RNA is induced and titrates CTCF away from the promoter of the Xist gene to induce Xist transcription. Together with our findings, these examples illustrate that the decoy mechanism is probably a common strategy for ncRNAs to regulate gene expression.
Our study also highlights the importance of the spatiotemporally controlled expression and the stability of regulatory RNAs in the gene expression network (Figure 6). Both the tight control of eRNA synthesis requiring prior communication with the target promoter and the transient nature of eRNA (Figure S1C) (De Santa et al., 2010) are well-suited to ensure the locus-specific action of eRNAs during gene induction. The NELF-E subunit was shown to bind various RNA sequences with little or no apparent sequence or structural constraint, which is suitable for binding to nascent RNAs derived from many genes (Rao et al., 2006; Yamaguchi et al., 2002). In this regard, the spatiotemporally controlled local abundance of eRNAs might be a critical factor for allowing them to effectively compete with nascent RNA for NELF binding at the target promoter.
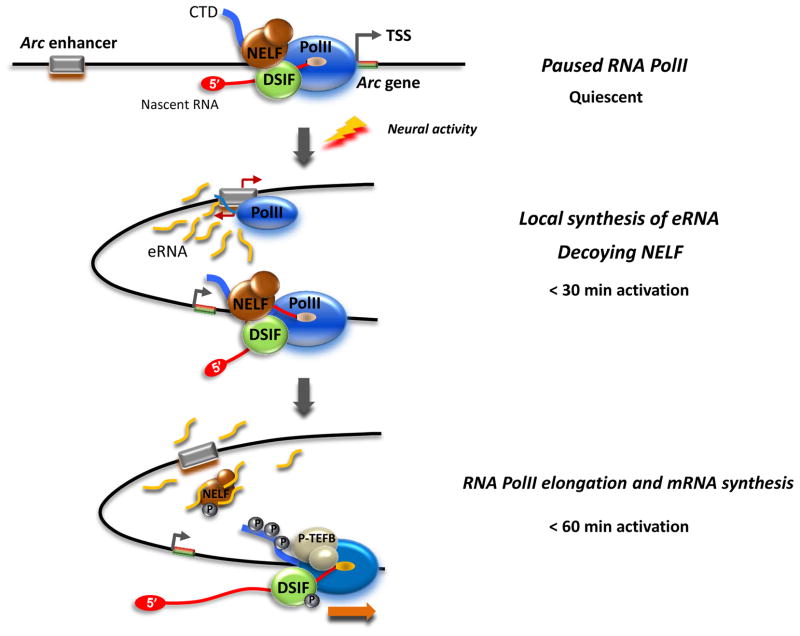
Figure 6. A model for Arc eRNA action during early transcription elongation.
In response to neuronal activity, the enhancer of Arc is brought into close proximity with the promoter. The rapid local rise of Arc eRNA facilitates the dissociation of the NELF complex from paused RNAPII by competing with the nascent Arc mRNA emerging from paused RNAPII for NELF-E binding. P-TEFb is also recruited and phosphorylates RNAPII, DSIF, and NELF. The Arc eRNA is degraded before diffusing out, thus its effect is confined to the Arc gene. RNAPII is able to enter into productive elongation and Arc mRNA induction occurs.
Experimental Procedures
Additional methods can be found in the Supplemental Experimental Procedures
Chromatin Immunoprecipitation (ChIP)
Cultured cortical neurons were treated overnight with 1 μM TTX. Next day, they were incubated with 55mM KCl for 30 min and then fixed with 1% formaldehyde for 10 min. ChIP was performed as described (Flavell et al., 2008; Kim et al., 2010). Detailed procedures and antibody information can be found in the Supplemental Experimental Procedures.
Analysis of EU-labeled nascent transcripts
The EU-labeled transcription experiment was performed with the Click-it Nascent RNA Capture kit (Invitrogen) protocol. Briefly, cultured cortical neurons were pulsed with 0.5 mM EU for 1 h at 37 °C and then total RNA was isolated. EU-labeled RNA was biotinylated with azide-modified biotin. Biotin-EU-labeled nascent RNA was captured on streptavidin T1 magnetic beads (Invitrogen) and then cDNA was synthesized using the High-Capacity reverse transcription kit and analyzed by qRT-PCR. Primers employed are listed in the Supplemental Experimental Procedures.
Ultraviolet-crosslinking RNA Immunoprecipitation (UV-RIP)
Cultured cortical neurons (1×108 millions) were harvested and UV-crosslinked at 400 nm (400 mJ/cm2) in 10 ml ice-cold PBS with protease inhibitors. Neurons were incubated with ice-cold Low-salt lysis buffer (50 mM Hepes KOH, pH 7.5, 10 mM NaCl, 1 mM EDTA, pH 8.0, 10% Glycerol, 0.2% NP-40, 1% Triton X-100) containing protease inhibitor and RNase inhibitor (Promega) for 10 min at4 °C using rotating wheel. Nuclei were resuspended in the ice-cold High-salt buffer (1 mM EDTA, pH 8.0, 0.5 mM EGTA, pH 8.0, 10 mM Tris, pH 8.0, 600 mM NaCl, 1% Triton X-100, 0.1% DOC) containing protease inhibitor and RNase inhibitor for 1 h at 4 °C using rotating wheel. After centrifugation, supernatants were diluted with Immunoprecipitation buffer (1 mM EDTA, pH 8.0, 0.5 mM EGTA, pH 8.0, 10 mM Tris, pH 8.0, 1% Triton X-100, 0.1% DOC) containing protease inhibitor and RNase inhibitor and incubated overnight with anti-NELF-E (H-140; Santa Cruz) or anti-normal rabbit IgG (Santa Cruz), and then the lysate was incubated with Protein A/G agarose beads for 2 h at 4 °C. Subsequently, the agarose beads were washed with ChIP washing buffers. Bound proteins were eluted by ChIP elution buffer containing RNase inhibitor for 10 min at 65 °C. Samples were treated with Proteinase K and DNase I (Roche) for post-immunoprecipitation and then the RNA was extracted by phenol/chloroform and ethanol precipitation. The extracted RNA samples were reverse transcribed into cDNA and used as a qPCR template.
Biotinylated RNA Pull-down
The minus Arc eRNA sequence was amplified from a BAC clone containing the Arc gene and cloned into the pBluescript SK(-) vector. The Arc minus strand was transcribed from the T7 promoter using a commercial in vitro transcription kit (MegaScript; Ambion), supplementing the dUTP with 25% bio-16-UTP to produce biotinylated transcripts. The pull-down was performed as described previously (Tsai et al., 2010), with the following modifications. 10 ug of biotinylated Arc eRNA was heated to 85°C for 2 min, and then placed on ice for 2 min and supplemented with RNA structure buffer (10 mM Tris pH 7, 0.1 M KCl, 10 mM MgCl2), and incubated at room temperature for 20 min. HEK293T cells were transfected with constructs containing FLAG-tagged wildtype or ΔRRM NELF-E and harvested using lysis buffer (150 mM NaCl, 1% TX-100, 2mM EDTA, 50 mM Tris pH 7.5). 500 μg of lysate were diluted with Pull-down buffer (100 mM KCl, 20 mM Hepes pH 7.5, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT) for a final concentration of 0.1% TX-100. The biotinylated RNA was incubated with the lysate for 1 h at room temperature with rotation. 100 μl of M-280 Streptavidin Dynabeads (Invitrogen) were added to the lysate-RNA mixture and further incubated for 1h at room temperature. Beads were washed with wash buffer (20 mM Hepes pH 7.5, 1.5 mM MgCl2, 0.5 mM DTT, 0.2 mM EDTA, 0.1% TX-100) containing either 300 mM or 600 mM KCl. Protein was eluted using SDS buffer (120 mM Tric-HCl pH 6.8, 20% glycerol, 4% SDS) at 100°C for 10 min. Lysate was run alongside 1/40 input on a 10% SDS-PAGE gel, transferred using the Trans-blot Turbo (Biorad), and probed with anti-NELF-E (Abcam; 1:1000).
Supplementary Material
Acknowledgments
We would like to thank H.C. Huang for assistance with figure preparation and D. Lazzareschi and B. Gary for assistance with cell culture. We would also like to thank D. Corey for insight into LNA strategies and advice on the manuscript, and Y. Yamaguchi for generously sharing Nelf-e constructs. This work was supported by National Institute of Mental Health Institutional Training Grant, T32-MH76690 (K.S.), The Whitehall foundation, The Welch foundation I-1786, The Klingenstein Fund, and NIH-NINDS R01NS085418 (T.-K.K).
Footnotes
Author Contributions
T.-K.K., K.S., and J.-Y.J. designed the project, and K.S. and J.-Y.J. performed most of the experiments. K.S. contributed to the design and preparation of the Arc eRNA knockdown reagents, expression kinetics analyses of IEGs, transcript mapping and functional characterization of Arc eRNAs in gene expression, as well as NELF in vitro pulldown and replacement assays. K.S. and J.-Y.J. jointly performed UV-RIP experiments. J.-Y.J. performed the characterization of NELF, CDK9, Med1, RAD21 and RNAPII binding, 3C analysis, and nascent transcript analysis. X.L. contributed to the designing and preparation of lentiviral vectors encoding shRNAs against Gadd45b. J. K. Watts contributed to the design and preparation of the LNAs, and C. Martinez produced the LNAs. K.S. and T.-K.K. wrote the manuscript with input from J.-Y.J. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 2008;36:e135. doi: 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Arnold FJ, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci. 2001;4:261–267. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
- Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GS, Chen S, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IIott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Finkbeiner S. Arc in synaptic plasticity: from gene to behavior. Trends Neurosci. 2011;34:591–598. doi: 10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitch BA, Lu Q, Benjamin LR, Liu Y, Gilmour DS, Elgin SC. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol Cell Biol. 2002;22:6148–6157. doi: 10.1128/MCB.22.17.6148-6157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs Promote Transcription by Establishing Chromatin Accessibility at Defined Genomic Loci. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JN, Neumann L, Wenzel S, Schweimer K, Rosch P, Wohrl BM. Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription. Biochem J. 2006;400:449–456. doi: 10.1042/BJ20060421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998a;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998b;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim Biophys Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.