Abstract
Coenzyme Q10 (CoQ10) is a key component of the mitochondrial electron transfer chain and is one of the most important cellular antioxidants. We previously reported that glycoprotein saposin B (SapB) binds CoQ10 in human cells. To elucidate the physiological role of SapB and its precursor, prosaposin (Psap), we prepared stable transfectants of HepG2 that overexpress wild-type human Psap (Wt-Tf). We also established a SapB domain mutated Psap (Mt-Tf) in which cysteine198 was replaced with serine to disrupt three dimensional protein structure by the loss of S-S bridging. Psap knockdown (KD) strains were also examined. Western blotting analysis confirmed overexpression or knockdown of Psap in these HepG2 cells. The cellular ratios of CoQ10 to free cholesterol (FC) significantly decreased in the order of Wt-Tf>parental>Mt-Tf>KD. Additionally, the ratios of CoQ10/FC in mitochondrial fractions decreased in the order of Wt-Tf>parental>KD. These data indicate that Psap and/or SapB regulate CoQ10 levels in HepG2 cells, especially in their mitochondria.
Keywords: coenzyme Q10, prosaposin, saposin B, mitochondria, HepG2
Introduction
Coenzyme Q10 (CoQ10) is essential for ATP production in the mitochondria and is an important antioxidant in every cellular membrane and lipoprotein.(1) Their CoQ10 concentrations are variable in quantity,(1–3) although CoQ10 is synthesized only in the mitochondria, endoplasmic reticulum-Golgi system, and peroxisomes.(1) This raises two key questions: what delivers CoQ10 to a desired membrane or lipoprotein, and how are their CoQ10 concentrations controlled?
In regard to the first question, we found that a cytosolic protein, saposin B (SapB), binds and transfers CoQ10.(4) SapB is a housekeeping protein necessary for lysosomal sphingolipid hydrolysis and is present in every tissue.(5,6) Its precursor protein, prosaposin (Psap), is also ubiquitously present in tissues and extracellular spaces such as blood and saliva.(6) Mature Psap exists in two forms, an intracellular form of 68 kDa and an extracellular form of 73 kDa. The 68 kDa Psap is hydrolyzed to 12–15 kDa saposins A, B, C, and D in lysosomes.(5,6) Recently, we found that the 73 kDa Psap isolated from human seminal fluid also binds CoQ10 (unpublished data). These data suggest that Psap and SapB are likely to be extracellular and intracellular CoQ10 transfer proteins, respectively, although this needs to be confirmed by further experimental evidence.
In general, the steady-state level of CoQ10 is regulated by rates of its synthesis and catabolism. For example, genetic impairment of CoQ10 synthesis results in significantly lower cellular levels.(7) However, the rates of CoQ10 synthesis in human organs have not been extensively studied. The half-life of coenzyme Q9 in rats is relatively short, ranging between 49 and 125 h in various tissues.(8) Yet, the rate of CoQ10 catabolism is not known for humans. Obviously, such data is lacking to better understand the steady-state dynamics of cellular CoQ10 metabolism.
Instead, we have focused attention on the possible role of Psap and SapB to regulate CoQ10 concentrations in cells since these proteins control the rate of CoQ10 transfer. In this study, we established two Psap transfected human HepG2 cell strains, a SapB domain mutated Psap, as well as two Psap knockdown cell lines. We have measured the cellular concentrations of Psap, SapB, and CoQ10 and found that cellular CoQ10 levels were dependent on Psap and SapB concentrations. Moreover, mitochondrial levels of CoQ10 were also Psap- and SapB-dependent. These results indicate that the Psap family is an important regulator of CoQ10 concentration in tissues.
Materials and Methods
Preparation of HepG2 cells
Cultures of HepG2 cells (Dainippon Sumitomo Pharma Co., Ltd., Osaka, Japan) were maintained in Dulbecco’s MEM (Sigma, St. Louis, MO) supplemented with 10% fetal calf serum (HyClone, Thermo scientific, MA), 100 units/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Preparation of plasmid DNA and establishment of stable transfectants
Full-length human Psap (PubMed Accession No. BC004275) was amplified by PCR and cloned into the pcDNA3.1 plasmid (Invitrogen, Carlsbad, CA). Plasmids were purified with the Qiagen Plasmid Midi Kit (QIAGEN, Venlo, The Netherlands) and the DNA sequence was verified. To obtain SapB domain mutated Psap plasmid DNA (C198S), a QuikChange II Site-Directed Mutagenesis Kit (Agilent Technology, CA) was used. In brief, the human Psap cDNA in the pcDNA3.1 vector was used as a template to mutate cysteine198 to serine; the thiol residue of the cysteine is necessary to maintain S-S binding and thus to maintain protein structure, and its replacement with serine interferes with this S-S binding, causing loss of the protein structure. The C198S mutant of the human Psap vector was synthesized and amplified with primers 5'-CCAAAGGATAATGGGGACGT TAGCCAGGACTGC-3' and 5'-GCAGTCCTGGCTAACGTCC CCATTATCCTTTGG-3' by PCR. Two micrograms of pcDNA3.1 plasmid were transfected into HepG2 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Two days later, cells were plated on 15-cm dishes. After G418 selection, clones were subjected to western blot analysis using an anti-SapB monoclonal antibody.(4)
Design and synthesis of Psap miRNA, construction of vectors, and establishment of stable transfectants
Pre-miRNA sequences for Psap were designed by Invitrogen’s RNAi Designer. Sequences are as follows: Psap miRNA1:
5'- TGCTGTACACTTCTCAGTTCCCAACAGTTTTGGCCACT GACTGACTGTTGGGATGAGAAGTGTA-3';
5'- CCTGTACACTTCTCATCCCAACAGTCAGTCAGTGGCCA AAACTGTTGGGAACTGAGAAGTGTAC-3';
Psap miRNA2:
5'- TGCTGTTGTCCTTCAGCATATCACCAGTTTTGGCCACT GACTGACTGGTGATACTGAAGGACAA-3';
5'- CCTGTTGTCCTTCAGTATCACCAGTCAGTCAGTGGCC AAAACTGGTGATATGCTGAAGGACAAC-3';
Psap miRNA3:
5'- TGCTGTCTCCAAGTAAACAAGGATCTGTTTTGGCCAC TGACTGACAGATCCTTTTACTTGGAGA-3';
5'- CCTGTCTCCAAGTAAAAGGATCTGTCAGTCAGTGGCC AAAACAGATCCTTGTTTACTTGGAGAC-3';
Psap miRNA4:
5'- TGCTGATGACAGGGAGGTAGGAGTCCGTTTTGGCCA CTGACTGACGGACTCCTCTCCCTGTCAT-3';
5'- CCTGATGACAGGGAGAGGAGTCCGTCAGTCAGTGGC CAAAACGGACTCCTACCTCCCTGTCATC-3'.
The synthesized complementary DNA oligos were annealed to generate double-stranded oligos and were cloned into the linearized pcDNATM 6.2-GW/EmGFP-miR vector (Invitrogen) using T4 DNA ligase. All of the vectors were transformed into One Shot TOP10 Chemically Competent E. coli (Invitrogen), and colonies containing spectinomycin-resistant transformants were analyzed for the desired expression clones. The recombinant vectors were purified with a Qiagen Plasmid Midi Kit (Qiagen).
Vectors with Psap miRNA1, Psap miRNA2, Psap miRNA3, Psap miRNA4 were mixed and transfected into HepG2 cells using Hylimax (Dojindo, Kumamoto, Japan) according to the manufacturer’s protocol. Two days later, cells were plated on 15-cm dishes. After blastcidin selection, clones were subjected to western blot analysis using an anti-SapB monoclonal antibody.(4)
Western blotting analysis
HepG2 cells in each dish were treated with lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 0.1% nonidet P-40 (Nakarai Tesuque, Tokyo, Japan), 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 µg/ml of leupeptin, pepstatin A, N-tosyl-l-phenylalanyl chloromethyl ketone, N-tosyl-l-lysyl chloromethyl ketone) for 1 h. Then the samples were collected and centrifuged at 15,000 × g for 10 min. Protein concentrations in the supernatants were measured with a Pierce BCA Protein Assay Kit (Thermo Scientific, IL). Samples were separated by electrophoresis through a 15% SDS/polyacrylamide gel. After electrophoresis, proteins were transferred to PVDF membranes. The membranes were incubated with mouse anti-SapB IgG for 1 h at room temperature. Proteins were visualized with horseradish peroxidase-conjugated secondary antibodies (Bio-Rad Japan, Tokyo, Japan).
Lipid analysis
Concentrations of CoQ10 and free cholesterol (FC) in cells were determined by HPLC as reported previously.(4) Briefly, cells were seeded in 24-well plates and incubated for various times. The cell media were removed and the cells were washed twice with ice cold PBS. Then, 400 µl of HPLC grade 2-propanol (Fisher Chemicals, Fairlawn, NJ) was added. Samples were collected and centrifuged. Extracts thus obtained were injected into the HPLC-ECD system. Mobile phase: 50 mM NaClO4 in methanol/2-propanol (7/3, v/v); flow rate: 1.0 ml/min; analytical column: KANTO RP-18 (L) GP, 5 µm × 150 mm × 4.6 mm (Kanto Chemical, Tokyo, Japan); post-reduction column: RC-10, 15 mm × 4 mm (IRICA, Kyoto, Japan). CoQ10 and FC concentrations were determined by an electrochemical detector (600 mV; NANOSPACE SI-1, Shiseido, Tokyo, Japan) and a UV detector (210 nm; SPD-10A, Shimadzu, Kyoto, Japan), respectively.
CoQ10 in sucrose gradient fractions was extracted by using a 5-volume quantity of methanol and a 10-volume quantity of hexane. Triolein was used as an internal standard. Hexane fractions thus obtained were dried under a nitrogen gas stream and redissolved in 2-propanol. The samples were directly injected into the HPLC system described above.
Cell fractionation
Cell fractionation was performed as previously described with minor modifications.(9) Briefly, cells were lysed with a glass homogenizer in 0.25 M sucrose buffer containing protease inhibitors. The homogenate was then centrifuged at 600 × g for 10 min, and the pellet containing nuclei and cell debris was isolated as the 600 g pellet fraction. The remaining supernatant was centrifuged at 8,000 × g for 10 min, and the new supernatant was further centrifuged at 100,000 × g for 60 min to obtain the cytosol fraction. The 8,000 g pellets were resuspended in 3 ml of 0.25 M sucrose buffer and centrifuged at 95,000 × g for 90 min on a discontinuous sucrose gradient in bucket tubes by successively layering 3 ml each of 1.6, 1.4, 1.2, 1.0, and 0.8 M sucrose in 2 mM HEPES-KOH buffer (pH 7.4). The 0.25 M/0.8 M sucrose layer (fraction 0), the 0.8 M/1.0 M sucrose layer (fraction 1), the 1.0 M/1.2 M sucrose layer (fraction 2), the 1.2 M/1.4 M (fraction 3), and the 1.4 M/1.6 M contains (fraction 4) contained lysosomes, mitochondria-associated membranes, endoplasmic reticulum and plasma membranes, mitochondria, and mitochondria inner membranes (mitoplast), respectively.(9) The purities of fractions were determined by western blotting by using the following specific antibodies against the cytosol enzyme, lactate dehydrogenase A (LDH-A), and a mitochondrial enzyme, cytochrome c oxidase IV (COX IV). Antibody against LDH-A (N-14) was obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX). Human polyclonal COX IV antibody (#4844) was obtained from Cell Signaling Technology, Inc. (Danvers, MA).
Mitochondrial fractions were also obtained by the method of Wallace.(10) In brief, cells were collected in isolation buffer (210 mM mannitol, 70 mM sucrose, 0.1 mM EDTA, 0.5% BSA (fatty acid-free), and 5 mM HEPES, pH 7.2). The suspension was then lysed with a glass homogenizer and centrifuged at 1,000 × g for 10 min. The supernatant was collected and centrifuged at 85,000 × g for 15 min at 4°C, and the pellet was used as the mitochondrial fraction.
Statistical analysis
Values shown are depicted as means with standard deviations. Statistical significance was determined by using the Student’s t test or repeated-measures ANOVA. p<0.05 was considered statistically significant.
Results
Stable transfectant and knockdown of human Psap in HepG2 cells
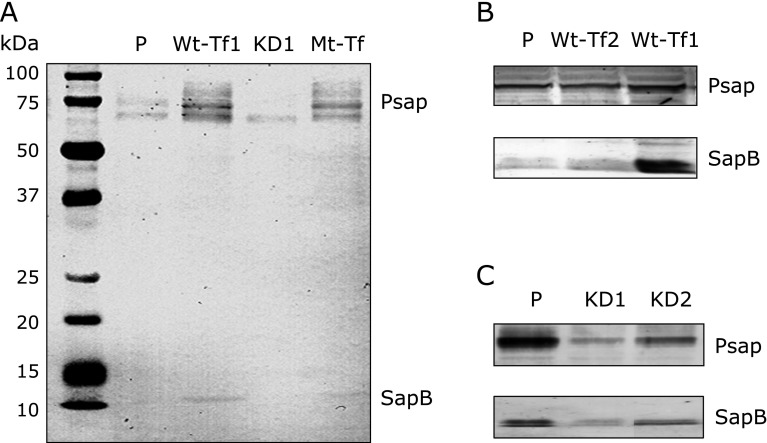
HepG2 cells were transfected with pcDNA3.1 plasmid vector containing the full-length Psap cDNA (Accession No. BC004275). SapB domain mutated Psap cDNA (C198S) was also prepared and transfected. G418-resistant cell populations were subcloned, and subcultures were derived from single cells. Levels of Psap and SapB in wild-type Psap transfected (Wt-Tf) HepG2 cells, Psap knockdown (KD) HepG2 cells, and SapB domain mutated Psap transfected (Mt-Tf) HepG2 cells were compared with parental (P) HepG2 cells. Western blotting analysis using anti-SapB monoclonal antibody revealed that Psap and SapB proteins increased in both Wt-Tf and in Mt-Tf, and decreased in KD HepG2 cells (Fig. 1).
Fig. 1.
Protein measurement by western blot analysis using monoclonal anti-sapB IgG. (A) Prosaposin (Psap) and saposin B (SapB) proteins increase in Psap transfected human (Wt-Tf1) HepG2 cells and SapB domain mutated (Mt-Tf) HepG2 cells, and these proteins decrease in Psap knockdown (KD1) HepG2 cells as compared to those in parental (P) HepG2 cells. (B and C) Psap and SapB increase in Psap transfected (Wt-Tf1 and Wt-Tf2) HepG2 cells and these proteins decrease in Psap knockdown (KD1 and KD2) HepG2 cells as compared to those in parental (P) HepG2 cells.
Effect of Psap transfection on CoQ10 content
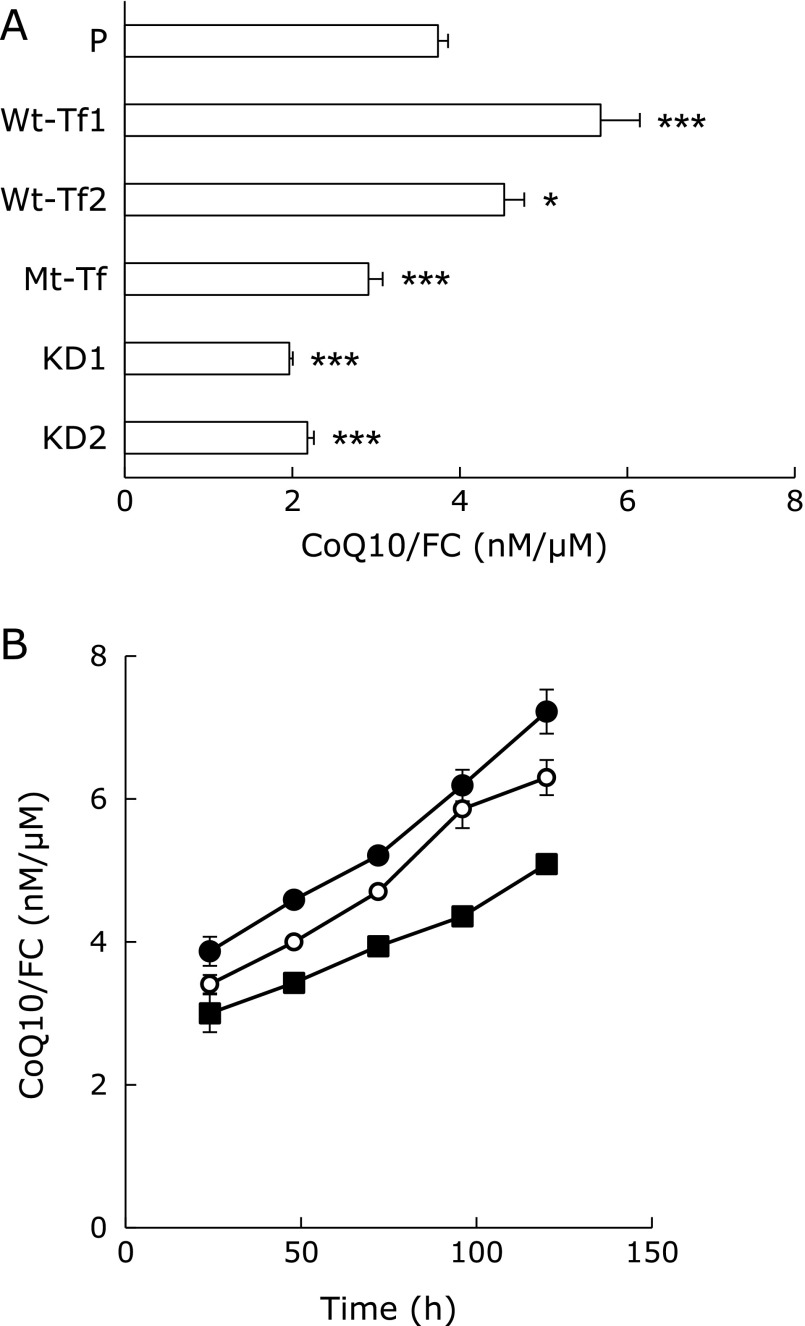
Cells were seeded at a density of 10,000 cells/cm2. After a 96-h incubation in a 24-well plate, 400 µl of 2-propanol was added to the isolated cells. Extracted CoQ10 and FC in each well were then measured by HPLC. The cellular ratios of CoQ10/FC significantly decreased in the order of Wt-Tf>parental>Mt-Tf>KD, as shown in Fig. 2A.
Fig. 2.
Cellular CoQ10/FC ratios. (A) Ratios of CoQ10 to FC (CoQ10/FC) in parental, Psap transfected, SapB domain mutated, and Psap knockdown HepG2 cells. * and *** indicate significant differences (p<0.05 and 0.001, respectively) compared to CoQ10/FC values of parental HepG2 cells. (B) Their time-course changes in parental (P; open circle) and one of each Psap transfected (Wt-Tf1; closed circle) and Psap knockdown (KD1; closed square) HepG2 cell lines. ANOVA indicated that a significant increase in the CoQ10/FC ratio occurs in Wt-Tf1 HepG2 cells (p<0.005) and a significant decrease in the CoQ10/FC ratio occurs in KD1 cells (p<0.001) as compared to that of parental HepG2 cells.
Fig. 2B shows the time-course changes in CoQ10/FC ratios in parental, Wt-Tf1, and KD1 cells. Cells were seeded at a density of 19,000 cells/cm2 for incubation. The ratios of CoQ10/FC in each cell line increased with incubation time. ANOVA indicated that a significant increase in the CoQ10/FC ratio of Wt-Tf1 cells (p = 0.0019) and a significant decrease in the CoQ10/FC ratio of KD1 cells (p<0.0001) was observed in comparison to those of parental cells.
Distribution of CoQ10 in cells
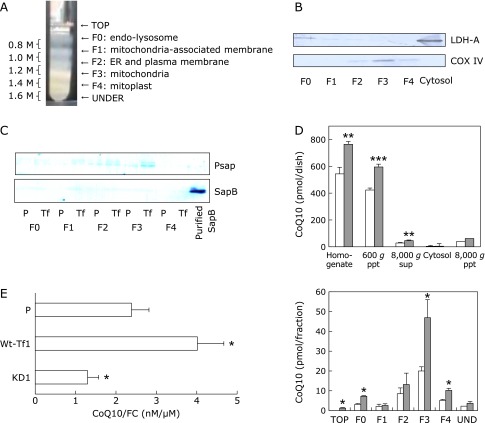
Next, we compared the cellular distribution of CoQ10 in parental HepG2 and Wt-Tf1 cells. The lysosome fraction (F0), mitochondria-associated membrane fraction (F1), endoplasmic reticulum and plasma membrane fraction (F2), mitochondria-enriched fraction (F3), and mitochondria-inner membrane (mitoplast) fraction (F4) were obtained by the sucrose-gradient separation (Fig. 3A), and the mitochondria content was confirmed by western blotting using an antibody against the mitochondrial marker COX IV enzyme (Fig. 3B). No contamination by the cytosol fraction was found by western blotting using an antibody against the cytosolic marker LDH enzyme (Fig. 3B).
Fig. 3.
Preparation of subcellular fractions and their CoQ10 content. (A) Sucrose gradient separation of mitochondria (F3) and mitoplasts (F4). (B) Confirmation of fractionation by western blot analysis using mitochondria-specific COX IV and cytosol-specific LDH-A antibodies. (C) Psap and SapB measurement of each fraction obtained from parental (P) and Psap transfected (Wt-Tf1) HepG2 cells by western blot analysis using monoclonal anti-sapB IgG. (D) CoQ10 contents in homogenates of parental (white bars) and Psap transfected (Wt-Tf1) HepG2 cells (with similar protein concentrations, gray bars) and their separated fractions by step-wise centrifugation as well as sucrose gradient fractions of 8,000 g pellets. *, ** and *** indicate significant differences (p<0.05, 0.01 and 0.001, respectively) between the two cells. (E) Ratios of CoQ10 to FC (CoQ10/FC) in mitochondrial fractions obtained by sucrose-mannitol separation of parental (P) and one of each Psap transfected (Wt-Tf1) and Psap knockdown (KD1) HepG2 cells. * indicates a significant difference (p<0.05) compared to values for parental HepG2 cells.
Fig. 3C shows the Psap and SapB content in each fraction obtained from parental and Wt-Tf1 cells. Psap levels in cellular fractions from parental HepG2 cells decreased in the order of F3 (mitochondria)>F2>F0, F1, F4, while the levels of SapB were much smaller than those of Psap. This was also the case for Wt-Tf1 HepG2 cells. Fig. 3D shows the CoQ10 content in each fraction obtained from parental and Wt-Tf1 cells. Contents of CoQ10 in cell homogenate, nuclei and cell debris (600 g pellet), cytosol and microsomes (8,000 g supernatant), lysosomes (F0), mitochondria (F3), and mitochondria-inner membrane (F4) from Wt-Tf1 cells were significantly greater than those from parental cells.
We also employed a sucrose-mannitol separation to prepare pure mitochondria fractions. Mitochondrial CoQ10 contents were significantly greater in Wt-Tf1 cells and significantly lower in KD1 cells as compared to those in parental cells, as shown in Fig. 3E.
Discussion
Overexpression of wild-type Psap in HepG2 cells significantly increased the cellular contents of CoQ10, and in contrast, knockdown of Psap had significantly decreased CoQ10 concentrations (Fig. 2A). These changes were especially significant in the mitochondrial fractions (Fig. 3D). We found elsewhere that the knockdown of Psap in Caco-2 cells results in a significant decrease in cellular CoQ10 content, and that Psap knockout mice have significantly lower levels of the CoQ9/FC ratio in the stomach, small and large intestines, and plasma than do wild-type mice (our unpublished data).
The C-S mutation introduced in the SapB domain of Psap resulted in overexpression of Psap in HepG2 cells (Fig. 1), but overexpression of mutated Psap had decreased cellular CoQ10 concentrations. This is likely because mutated SapB does not bind CoQ10 since the cysteine residue is essential for the formation of a S-S bond in the Psap protein, which is necessary for three dimensional protein structure and function.(5,6,11) Our mutated Psap protein is assumed to produce intact saposins A, C, and D, and our observation would suggest that saposins A, C, and D are not important in CoQ10 binding. All saposins have a similar structure characterized by the presence of three internal disulfide linkages and several hydrophobic residues,(5,6,12,13) however, only SapB forms a dimer having a large hydrophobic cavity where CoQ10 is likely to reside, whereas saposins A, C, and D exist only as monomers.(11–13)
Psap levels in cellular fractions from parental HepG2 cells decreased in the order of F3 (mitochondria)>F2>F0, F1, F4 (Fig. 3C) that correlates with a decreasing order of CoQ10 concentrations (Fig. 3D). A similar decreasing order of Psap levels was observed in the cellular fractions of Wt-Tf1 cells, and their Psap levels were markedly increased in comparison to those in parental HepG2 cells. The above results indicate that Psap transport of CoQ10 plays an important role in regulating cellular concentrations of CoQ10, especially in the mitochondria.
Overexpression of Psap resulted in an increase of CoQ10 content not only in the mitochondria but also in plasma, lysosomes, and nuclear membranes (Fig. 3D). However, the precise mechanism by which Psap and SapB determine the CoQ10 cellular destination, and how CoQ10 delivery is controlled, are still unknown. We predict that a kind of receptor-oriented mechanism may be involved, but this awaits further examination.
It is also of interest how human Psap concentrations change with age, given that the CoQ10 contents in most human tissues have been reported to be highest at the age of 20–30 and decrease with age thereafter.(14) It is interesting that Koochekpour et al.(15) observed that serum Psap levels in human males are the lowest before puberty (<age 13 year), peak at the most reproductive age group (age 20–39 year), and then decrease to a range between the two groups for men above 40. Accordingly, the relationship between Psap and CoQ10 should be investigated more extensively. We are currently studying age-dependent changes of Psap and CoQ9 content in rat tissues.
In summary, we prepared Psap-overexpression and knockdown HepG2 cells together with a Psap mutation in its SapB domain. Cellular levels of CoQ10 were well correlated with Psap content and, as excepted, mutated Psap cells had a significantly lower CoQ10 content. These results suggest that Psap and SapB regulate cellular CoQ10 content, especially in the mitochondria.
Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers 22500681 and 24500872. We thank Mr. Wataru Nemoto, Mr. Tatsuya Sakemi, Mr. Yuto Tanaka, and Miss Yukari Yokoo for their technical assistance. We thank Dr. Walter C. Dunlap for his valuable comments on the manuscript.
Abbreviations
- CoQ10
coenzyme Q10
- COX IV
cytochrome c oxidase IV
- KD
Psap knockdown
- LDH-A
lactate dehydrogenase A
- mitoplast
mitochondria inner membrane
- Mt-Tf
SapB domain mutated Psap transfected
- Psap
prosaposin
- SapB
saposin B
- Wt-Tf
wild-type Psap transfected
Conflict of Interest
We have not received any financial support or other benefits from commercial sources for the work reported in the manuscript. None of the authors have financial interests that could create a potential conflict of interest or the appearance of a conflict of interest with regard to this work.
References
- 1.Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Aberg F, Appelkvist EL, Dallner G, Ernster L. Distribution and redox state of ubiquinones in rat and human tissues. Arch Biochem Biophys. 1992;295:230–234. doi: 10.1016/0003-9861(92)90511-t. [DOI] [PubMed] [Google Scholar]
- 3.Runquist M, Parmryd I, Thelin A, Chojnacki T, Dallner G. Distribution of branch point prenyltransferases in regions of bovine brain. J Neurochem. 1995;65:2299–2306. doi: 10.1046/j.1471-4159.1995.65052299.x. [DOI] [PubMed] [Google Scholar]
- 4.Jin G, Horinouchi R, Sagawa T, et al. Coenzyme Q10-binding/transfer protein saposin B also binds gamma-tocopherol. J Clin Biochem Nutr. 2008;43:95–100. doi: 10.3164/jcbn.2008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Brien JS, Kishimoto Y. Saposin proteins: structure, function, and role in human lysosomal storage disorders. FASEB J. 1991;5:301–308. doi: 10.1096/fasebj.5.3.2001789. [DOI] [PubMed] [Google Scholar]
- 6.Kishimoto Y, Hiraiwa M, O'Brien JS. Saposins: structure, function, distribution, and molecular genetics. J Lipid Res. 1992;33:1255–1267. [PubMed] [Google Scholar]
- 7.Multiple-System Atrophy Research Collaboration; . Mitsui J, Matsukawa T, Ishiura H, et al. Mutations in COQ2 in familial and sporadic multiple-system atrophy. New Engl J Med. 2013;369:233–244. doi: 10.1056/NEJMoa1212115. [DOI] [PubMed] [Google Scholar]
- 8.Bentinger M, Dallner G, Chojnacki T, Swiezewska E. Distribution and breakdown of labeled coenzyme Q10 in rat. Free Rad Biol Med. 2003;34:563–575. doi: 10.1016/s0891-5849(02)01357-6. [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Ayala DJ, Brea-Calvo G, López-Lluch G, Navas P. Coenzyme Q distribution in HL-60 human cells depends on the endomembrane system. Biochim Biophys Acta. 2005;1713:129–137. doi: 10.1016/j.bbamem.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 11.Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Privé GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci U S A. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn VE, Leyko P, Alattia JR, Chen L, Privé GG. Crystal structure of saposins A and C. Protein Sci. 2006;15:1849–1857. doi: 10.1110/ps.062256606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popovic K, Privé GG. Structures of the human ceramide activator protein saposin D. Acta Crystallogr D Biol Crystallogr. 2008;64:589–594. doi: 10.1107/S0907444908003120. [DOI] [PubMed] [Google Scholar]
- 14.Kalén A, Appelkvist EL, Dallner G. Age-related changes in the lipid compositions of rat and human tissues. Lipids. 1989;24:579–584. doi: 10.1007/BF02535072. [DOI] [PubMed] [Google Scholar]
- 15.Koochekpour S, Hu S, Vellasco-Gonzalez C, et al. Serum prosaposin levels are increased in patients with advanced prostate cancer. Prostate. 2012;72:253–269. doi: 10.1002/pros.21427. [DOI] [PMC free article] [PubMed] [Google Scholar]