Abstract
Indigenous opportunistic bacteria on the interior of the Peyer’s patches play a key role in the development of the mucosal immune, but their population composition has been ignored. The present study was conducted to test the hypothesis that the changes in the composition of indigenous opportunistic bacteria in the Peyer’s patches are associated with obesity. C57BL/6J-male mice had been fed either a control diet or a high-fat diet. After 25 weeks, mice in high-fat diet exhibit either an obesity-prone (OP) or an obesity-resistant (OR) phenotype. Control diet group (CT) and OR group had a significant larger bacteria diversity than that in the OP group. Allobaculum and Lactobacillus were significantly decreased in high-fat diet induced OP mice compared with CT and OR mice, whereas Rhizobium and Lactococcus was significantly increased. The result of quantitative real-time PCR was consistent with that of 454 pyrosequencing. Significant correlations between mRNA expression of inflammation marks and the top 5 abundance genera bacteria on the interior of Peyer’s patches were observed by Pearson’s correlation analysis. Taken together, the indigenous opportunistic bacteria on the interior of Peyer’s patches plays a major role in the development of inflammation for an occurrence of obesity.
Keywords: indigenous opportunistic bacteria, inflammation, obesity, Peyer’s patches, 454 pyrosequencing
Introduction
Obesity is a devastating epidemic worldwide and is associated with severe disorders including diabetes, hypertension and cardiovascular diseases. It has been recently determined that obesity is associated with low grade chronic systemic inflammation.(1,2) Disruption in the intestinal mucosal immunity could increase passage of lipopolysaccharide (LPS) from the lumen to the lamina propria, resulting in an increase in plasma levels of LPS, “metabolic endotoxemia”, which plays a major role in the development of chronic systemic inflammation in response to ingestion of high-fat foods.(3–5)
Peyer’s patches (PPs) are discrete areas of organized lymphoid tissue in the small intestine, with specialized membranous cells (M cells),(6) and lacks an overlying brush border, mucoid glycocalyx, and hydrolytic enzymes characteristic of absorptive epithelium. Due to its specialized structure, adaptation for antigen sampling, and the induction of immune responses, the jejuna Peyer’s patch-containing mucosa is more susceptible to probiotics invasion.(7–9) In addition, mice that lacked PPs and mesenteric lymph nodes, failed to induce antigen-specific IgA responses, suggesting that PPs play a key role in the induction of specific IgA responses to orally administered antigens.(10) Chassaing et al.(11) showed that the earliest lesions of recurrent inflammatory bowel disease were erosions of PPs. Together, PPs are a major inductive and regulatory site for mucosal immunity.
PPs continuously take up gut luminal antigens through M cells, including both beneficial and antigens. Subsequent studies revealed that commensal bacteria persist in PPs dendritic cells (DCs), contributing to induction of local specific immune responses that limit dissemination no farther than the mesenteric lymph nodes, ultimately preventing systemic spread.(12) However, in PPs exposed to pathogenic bacteria (e.g., Shigella and Alcaligenes),(13,14) high expression of proinflammator genes were measured, including tumor necrosis factor-α (TNF-α), interleukin 6 (IL-6), cyclooxygenase-2 (Cox-2), and interferon-γ (IFN-γ). Inflammatory bowel disease exhibited a higher level of pathogenic bacteria interaction with PPs and a higher level of translocation through M cell monolayers.(11) There are positive correlation based between inflammatory bowel disease and obesity.(15) Thus, it is compelling to investigate the relative contributions of indigenous opportunistic bacteria on the interior of the PPs to obesity and metabolic syndrome. However, most studies were focused on the impact of dietary on the luminal microbiota, and few have examined indigenous bacteria structures on the interior of the PPs.
On an high-fat diet (HFD), the C57BL/6J-male mice can be divided into obesity-prone (OP) and obesity-resistant (OR) phenotypes.(16,17) This two-phenotype model can be useful for elucidating mechanisms that drive obesity, and to identify physiological and biochemical differences between the two groups.
In this study, an animal model of a high-fat diet-induced OP or OR phenotype was used to test the hypothesis that the changes of indigenous opportunistic bacteria on the interior of the PPs are associated with obesity induced by high-fat diet.
Materials and Methods
Animal experiments
The animal experiment was performed in compliance with the fundamental guidelines for proper conduct of animal experiments and related activities in academic research institutions under the jurisdiction of the Ministry of Education of China and approved by the Jiangnan University Institutional Animal Care and Use Committee. Thirty-eight, 4-week-old, C57BL/6J-male mice were obtained from Shanghai Slac Laboratory Animal, Co., Ltd. (Shanghai, China); had been fed either a control diet (total calories 3.6 kcal/g, 10% calories in fat) or a HFD (total calories 4.7 kcal/g, 50% calories in fat). Mice were kept in an environmentally controlled breeding room (temperature: 23 ± 2°C, humidity: 60 ± 5%, 12 h light-dark cycle) and had free access to food and water throughout the study. After 25 weeks, 30 mice in HFD were classified by body weight gain (range from 28.1 to 42.3 g) into OP and OR, according to the method used previously.(5,16,17) The 8 mice in the upper tertile of body weight gain (40.8 ± 1.5 g) were designated as OP mice and eight mice chosen at random from the lower tertile of body weight gain (29.3 ± 1.2 g, p<0.05) as OR group. The food intake and body weight of each animal was measured every 2 weeks. Groups of eight mice were killed after a 4 h fast during deep anesthesia. The PPs were aseptically removed, flash frozen, and stored at –80°C until processing. Blood samples were collected and were isolated by centrifugation at 1,500 × g at 4°C for 10 min. Blood glucose was assayed using commercial kits (Jingmei BioTech Co. Ltd, Shenzhen, China) and estimated by the glucose oxidase method.
DNA extraction
PPs tissue specimens were placed in Betadine antiseptic solution (Seton Healthcare Group plc., Oldham, UK) for 3 min to disinfect the surface of each, as described by Hooper et al.(18) Subsequently, tissues were vortexed in multiple 500 µl aliquots of phosphate-buffered saline (PBS) to encourage the removal of any bacteria on the tissue surface. Final washes were retained and analyzed by culture-dependent techniques to sure that surface decontamination was successful. Genomic DNA was extracted from each sample individually using a bead beating method followed by phenol/chloroform/isoamyl alcohol extraction as described previously.(19) Briefly, a 100 mg aliquot of each homogenized sample was suspended while frozen in a solution containing 500 µl of DNA extraction buffer {[200 mM Tris (pH 8.0), 200 mM NaCl, 20 mM EDTA], 210 µl of 20% SDS, 500 µl of a mixture of phenol:chloroform:isoamyl alcohol (25:24:1)}, and 500 µl of a slurry of 0.1-mm-diameter zirconia/silica beads. Cells were then lysed by mechanical disruption with a bead beater set on high for 2 min (22°C), followed by extraction with phenol: chloroform: isoamyl alcohol, and precipitation with isopropanol. The quantity and quality of purified DNA was assessed using the E.Z.N.A DNA Assay Kit (Omega Biotech, Doraville, GA).
An Analysis of the indigenous opportunistic bacteria on the interior of PPs by pyrosequencing of 16S rRNA tags
Before pyrosequencing, the above DNA of each sample was amplified with a set of primers targeting the hypervariable V1–V3 region of the 16S rRNA gene (RDP’s Pyrosequencing Pipeline: http://pyro.cme.msu.edu/pyro/help.jsp). The forward primer is 533R of 5'-TTA CCG CGG CTG CTG GCA C-3', and the reverse primer is 27 F of 5'-AGA GTT TGA TCC TGG CTC AG-3' (Shanghai Majorbio Bio-Pharm Tech, Shanghai, China). Barcodes that allow sample multiplexing during pyrosequencing were incorporated between the 454 adaptor and the forward primer. Prior to sequencing, amplicons from the individual PCR samples were quantified using the Quant-iT Pico Green double stranded DNA assay (Invitrogen, Carlsbad, CA) and quality controlled on a GeneAmp 9700 thermal cycler (Applied Biosystems, Foster City, CA).
The pooled DNA samples were adapter ligated with beads and amplified by emulsion PCR using a Roche GS FLX Titanium SV emPCR kit. Beads were counted after emulsion PCR, and the same amount of beads was put in a PicoTiterPlate. Pyrosequencing was performed using the genome sequencer FLX system (Roche, Nutley, NJ). Sequences obtained from pyrosequencing were analyzed with 454 Base Caller 2.3 in genome sequencer FLX system software (Roche). The results are deposited into the NCBI short reads the archive database (accession number: SRP031866).
Taxonomy-based analysis at the phylum and genus levels were performed by assigning taxonomic status to each sequence using BLAST and Ribosomal Database Project (RDP) Classifier.(20,21)
Specific quantification of bacteria by quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) as described by Fierer et al.(22) Primers were published in Table 1. qPCR assays were performed in each 10 µl reaction using AccuPower 2x Greenstar qPCR Mastermix (Bioneer, Seoul, Korea) with the following parameters: 1 cycle of 95°C for 5 min; 45 cycles of 95°C for 20 s, annealing temperature and time given in Table 1 and 72°C for 20 s; and 1 cycle of 72°C for 5 min.(23) A melting curve analysis was performed after amplification. All samples were analysed in duplicate.
Table 1.
Sequences of primers and protocols used for qPCR
| Assay | Primer sequences (5'-3') | Annealing T and time | References |
|---|---|---|---|
| Universal bacteria | |||
| EUB338 | ACTCCTACGGGAGGCAGCAG | 53°C for 10 s | Fierer et al.(22) |
| EUB518 | ATTACCGCGGCTGCTGG | ||
| Phylum Firmicutes | |||
| Lgc353 | GCAGTAGGGAATCTTCCG | 58°C for 10 s | Fierer et al.(22) |
| EUB518 | ATTACCGCGGCTGCTGG | ||
| Phylum Bacteroidetes | |||
| Cfb319 | GTACTGAGACACGGACCA | 60°C for 30 s | Fierer et al.(22) |
| Eub518 | ATTACCGCGGCTGCTGG | ||
| Lactobacillusgroup | |||
| LacRT F | AGCAGTAGGGAATCTTCCA | 58°C for 15 s | Walter et al.(23) |
| LacRT R | CACCGCTACACATGGAG | ||
| Pseudomonasgroup | |||
| Ps-F | GGTCTGAGAGGATGATCAGT | 58°C for 15 s | Widmer et al.(24) |
| Ps-R | TTAGCTCCACCTCGCGGC | ||
| α-Proteobacteria | |||
| Alf685 | TCTACGRATTTCACCYCTAC | 60°C for 10 s | Fierer et al.(22) |
| EUB338 | ACTCCTACGGGAGGCAGCAG | ||
| β-Proteobacteria | |||
| Bet680 | TCACTGCTACACGYG | 60°C for 10 s | Fierer et al.(22) |
| EUB338 | ACTCCTACGGGAGGCAGCAG |
Quantitative real-time PCR for IL-10, IL-6, TNF-α
RT-PCR was performed using an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). The primers used for the amplification are as follows: Zo-1, forward primer: 5'-ACC CGA AAC TGA TGC TGT GGA TAG-3' and reverse primer: 5'-AAA TGG CCG GGC AGA ACT TGT GTA; Occludin, forward primer: 5'-ATG TCC GGC CGA TGC TCT C-3' and reverse primer: 5'-TTT GGC TGC TCT TGG GTC TGT AT-3'; claudin-1, forward primer: 5'-AGC CAG GAG CCT CGC CCC GCA GCT GCA-3' and reverse primer: 5'-CGG GTT GCC TGC AAA GT-3'; JAM (junctional adhesion molecule), forward primer: 5'-AGT CCG TGA CAC GGG AAG AC-3' and reverse primer: 5'-CAC AAG CAC GAT GAG CTT GAC-3'; TNF-α, forward primer: 5'-CAG GGG CCA CCA CGC TCT TC-3' and reverse primer: 5'-CTT GGG GCA GGG GCT CTT GAC-3'; IL-6, forward primer: 5'-CTG CAA GAG ACT TCC ATC CAG TT-3' and reverse primer: 5'-GAA GTA GGG AAG GCC TGG-3'; IL-10, forward primer: 5-GCT CTT ACT GAC TGG CAT GAG-3', reverse primer: 5'-CGC AGC TCT AGG AGC ATG TG-3'. β-Actin was amplified for use as an internal control. Data was normalized to L32 RNA (ΔΔCT analysis).
Statistical analysis
Statistical analysis for the animal experiment was performed using SPSS 11.5 (SPSS, Inc., Chicago, IL). One-way analysis of variance (ANOVA) was used to assess the effects of treatments. Significant differences were accepted at p values of <0.05. To assess the diversity of the indigenous opportunistic bacteria on the interior of PPs, the Shannon diversity index and evenness were calculated from the number of OTUs assigned at the phyla and the genus level. Principal component analysis plots (PcoA) were constructed based on the unweighted UniFrac distance metric. Results of qPCR are presented as the ratio of the copy number of the bacteria group to the copy number obtained in the universal bacteria assay. The relative abundances of the bacteria groups on various phylogenetic levels and diversity indices were compared between pet mice samples using various statistical procedures. Pearson’s correlation coefficient was used to determine the relation of inflammation markers in PPs to gut microbiota.
Result
Long-term effects of HFD intake on health phenotypes
After 25 weeks on a high fat diet, two different phenotypes emerged within the HFD group; animals with the highest body weight were assigned to the OP group, the other mice were designated OR. OP mice had a significantly higher body weight compared with CT group (40.8 ± 1.5 g vs 27.6 ± 1.3 g, p<0.05; Fig. 1A) and with the OR group (40.8 ± 1.5 g vs 29.3 ± 1.2 g, p<0.05; Fig. 1A). There was a significant difference in the quantity of energy intake between the groups was observed (Fig. 1B). Furthermore, fasting blood glucose levels in the OP group were significantly increased compared with CT and OR animals (Fig. 1C). However, no significant differences were observed in body weight and fasting blood glucose levels between CT and OR phenotypes.
Fig. 1.
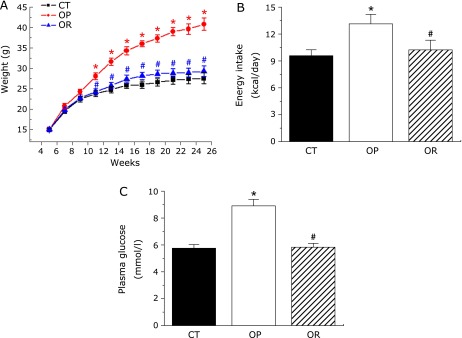
Phenotypes of different mice groups. (A) Growth curve established using the average body weight of three groups every 2 weeks. (B) Average energy intake per day of the three groups of mice. (C) Fasting plasma glucose level (n = 8 in each group). Mean values ± SE are shown. Mean values were significantly different compared with that of CT group: *p<0.05. Mean values were significantly different compared with that of the OP group: #p<0.05. CT, normal chow diet-fed mice; OP, high-fat diet-induced obesity-prone mice; OR, high-fat diet-induced obesity-resistant mice.
Effectiveness check of the raw reads
In this study, we obtained 6,654, 6,871, and 7,345 raw reads for the CT, OP and OR samples, respectively. After initial quality check mentioned above, the chimera and Achaea sequences were also checked and filtered out. As shown in Table 2, 48–53% of the raw reads met the standards of quality and length. The flagged reads were considered as chimeras by using Chimera Slayer in Mothur package. Some of these chimeras may represent naturally generated sequences that do not represent PCR artifacts and are not recommended to be randomly discarded. All the chimeras picked by Chimera Slayer were submitted to RDP classifier for further checking. In this way, only those sequences that cannot be assigned into a genus were considered as real chimeras and excluded in the following analysis. Some sequences were assigned to Archaea by the RDP classifier, although the utilized primers were bacteria specific primers. At last, 4,791 effective bacteria sequences were extracted from each sample to do the downstream analysis.
Table 2.
Sequences from the three samples
| Sample name | Reads |
Sequences |
3% distance |
5% distance |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw read | Valid | Valid | Normalization | Chao 1 richness estimation | Shannon diversity | OTU | Good’s coverage (%) | Chao 1 richness estimation | Shannon diversity | OUT | Good’s coverage (%) | |
| CT | 6,554 | 3,172 | 7,548 | 4,791 | 989 | 4.15 | 604 | 95.2 | 586 | 3.65 | 400 | 97.3 |
| OP | 6,871 | 3,645 | 7,265 | 4,791 | 926* | 3.81* | 568* | 95.1 | 533* | 3.34* | 366* | 97.2 |
| OR | 7,345 | 3,873 | 8,030 | 4,791 | 1,023# | 4.09# | 610# | 95.2 | 601# | 3.59# | 408# | 97.2 |
*p<0.05 compared with CT group, #p<0.05 compared with OP group. CT, normal chow diet-fed mice; OP, high-fat diet-induced obesity-prone mice; OR, high-fat diet-induced obesity-resistant mice.
The community composition of bacteria on the interior of PPs by pyrosequencing
To compare the bacteria species richness on the interior of PPs, operational taxonomic units (OTUs) were analyzed for each sample at distance levels of 3 and 5% (Table 2). The OTU number in the CT and OR group was significantly larger than that in the OP group at distance cutoff levels of 3 and 5%, respectively. The bacteria phylotype richness levels can also be reflected by using Chao 1 estimator and Shannon index, which also showed that CT and OR group had a significantly larger bacteria diversity than that in the OP group (Table 2). The rarefaction curves of the three groups at distance cutoff levels of 3, 5 and 10%, demonstrated that the bacteria phylotype richness of the CT and OR sample was much higher than that of the OP samples (Fig. 2).
Fig. 2.
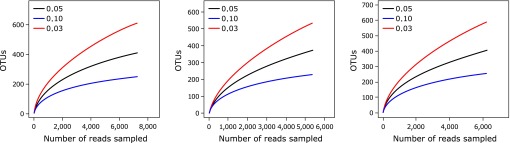
Rarefaction curves of the three groups at cutoff levels of 3, 5 and 10% created by using RDP’s pyrosequencing pipeline. CT, normal chow diet-fed mice; OP, high-fat diet-induced obesity-prone mice; OR, high-fat diet-induced obesity-resistant mice.
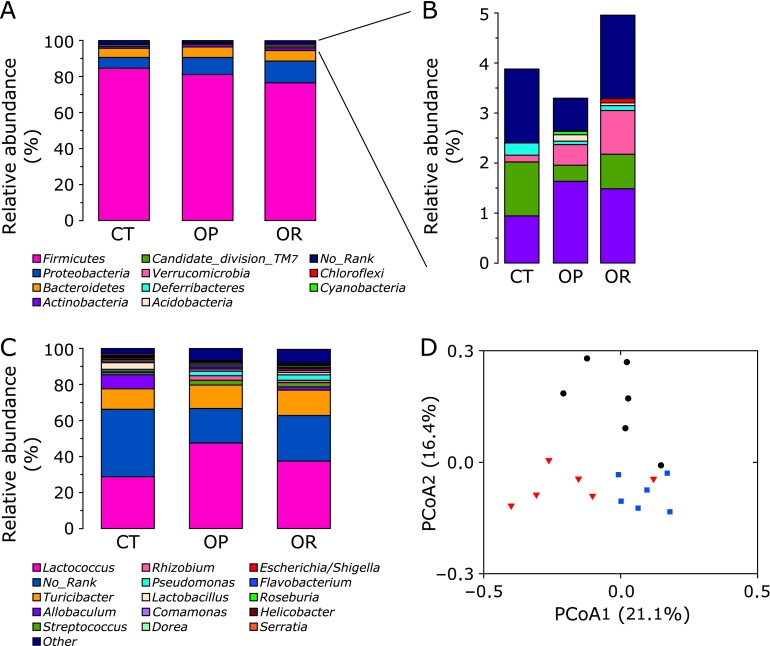
Abundances of different bacteria communities at the phylum and genus level were compared among CT, OP, and OR mice (Fig. 3). The most represented phylum in all subjects was Firmicutes, accounting for about 77–85% of sequences. The second and third most represented phylum was Proteobacteria (about 6–12%) and Bacteroidetes (about 5%), respectively. No significant differences were observed among CT, OP, and OR groups at the phylum level of Firmicutes and Bacteroidetes. At phylum level, Proteobacteria were significantly increased by 1.6-fold in OP mice and by 2.2-fold in OR mice (Fig. 3A). In addition, Actinobacteria and Verrucomicrobia were significantly increased (p<0.05), whereas Candidate_division_TM7 and Deferribacteres were reduced in OP and OR mice compared with CT group. Except for the significant increases in Candidate_division_TM7 and Verrucomicrobia in OR mice, no significant differences were observed in Actinobacteria and Deferribacteres between OP and OR phenotypes (Fig. 3B).
Fig. 3.
Relative abundances of distinct major phyla are shown in panel (A). Other minor phyla with trace abundance are shown in panel (B). Relative abundances of distinct genus are shown in panel (C) (the results were obtained by using BLASTN combined with MEGAN). (D) Principal component analysis plots (PcoA) were constructed based on the unweighted UniFrac distance metric. CT: black circle, OP: red triangle, OR: blue square. CT, normal chow diet-fed mice; OP, high-fat diet-induced obesity-prone mice; OR, high-fat diet-induced obesity-resistant mice.
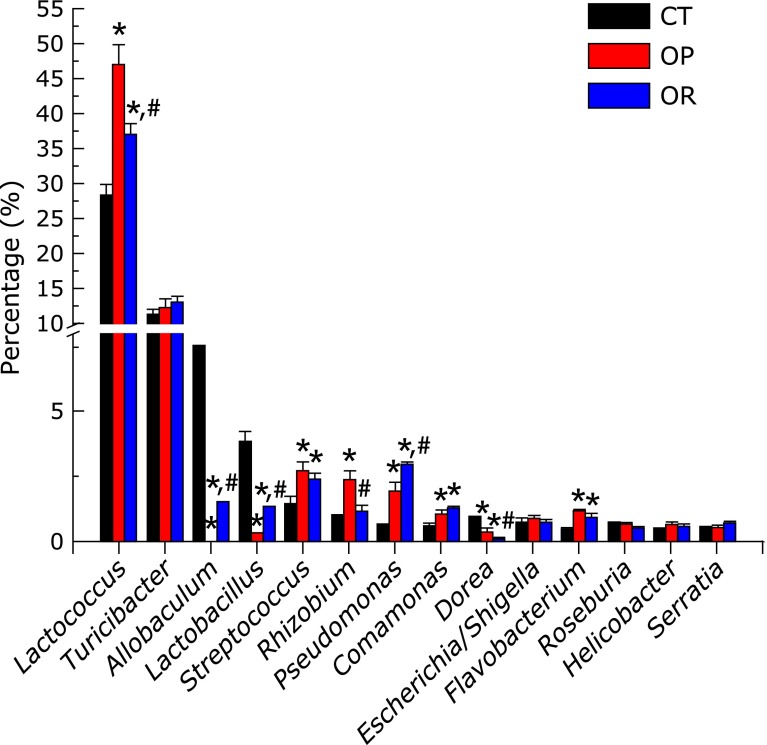
At the genus level (Fig. 3C and 4), the most represented genera in all subjects were Lactococcus and Turicibacter, accounting for about 29–47% and 11–14%, respectively. Allobaculum represented about 7.6% of genes in the CT group, but was 1.5% of genes in the OR group and even 0.1% of genes in the OP group. Significant differences were observed in the composition of bacteria on the interior of PPs at the genera level (Fig. 4). The proportion of Lactococcus and Rhizobium were significantly increased in the OP group compared with the CT and OR group (Lactococcus, OP vs CT, 47.48 ± 2.62% vs 28.72 ± 1.17%, p<0.05; OP vs OR, 47.48 ± 2.62% vs 37.33 ± 1.50%, p<0.05; Rhizobium, OP vs CT, 2.45 ± 0.31% vs 0.94 ± 0.06%, p<0.05; OP vs OR, 2.45 ± 0.31% vs 1.24 ± 0.15%, p<0.05), whereas the proportion of Allobaculum and Lactobacillus were significantly reduced (Allobaculum, OP vs CT, 0.11 ± 0.01% vs 7.56 ± 0.58%, p<0.05; OP vs OR, 0.11 ± 0.01% vs 1.54 ± 0.03%, p<0.05; Lactobacillus, OP vs CT, 0.33 ± 0.06% vs 3.87 ± 0.37%, p<0.05; OP vs OR, 0.33 ± 0.06% vs 1.24 ± 0.09%, p<0.05). Several genera of the microbial community were significantly increased in the OP and OR group compared with the CT mice, including Streptococcus, Pseudomonas, Comamonas and Flavobacterium. Except for the significant increases in Pseudomonas in OR mice, no significant differences were observed in the proportion of Streptococcus, Comamonas, and Flavobacterium between OP and OR phenotypes. Principal component analysis of the indigenous opportunistic bacteria on the interior of PPs among different groups resulted in distinct clusters (Fig. 3D).
Fig. 4.
Abundance of top 14 genera bacteria on the interior of PPs in CT, OP, and OR animals, analyzed by pyrosequencing of 16S rRNA tagsa. Values are expressed as mean ± SE. Mean values were significantly different compared with that of CT group: *p<0.05. Mean values were significantly different compared with that of the OP group: #p<0.05. CT, normal chow diet-fed mice; OP, high-fat diet-induced obesity-prone mice; OR, high-fat diet-induced obesity-resistant mice.
Selected bacteria genera on the interior of PPs was estimated by qPCR
There was no significant difference in abundance of Firmicute and Bacteroidetes among CT, OP, and OR mice (Table 3). α-Proteobacteria and β-Proteobacteria were significantly increased in OP mice compared with CT group. HFD induced OP mice significantly decreased the abundance of Lactobacillus and significantly increased the abundance of Pseudomonas compared with OR and CT groups (Table 3). Together, these differences in bacteria composition were nearly consistent between 454 pyrosequencing and qPCR.
Table 3.
Bacteria groups on the interior of Peyer’s patches from CT, OP, and OR mice measured by qPCR
| Phylum Firmicutes | α-Proteobacteria | β-Proteobacteria | Phylum Bacteroidetes | Lactobacillus group | Pseudomonas group | |
|---|---|---|---|---|---|---|
| CT | 0.43 ± 0.11 | 0.11 ± 0.09 | 0.21 ± 0.06 | 0.11 ± 0.05 | 0.35 ± 0.09 | 0.23 ± 0.06 |
| OP | 0.42 ± 0.10 | 0.22 ± 0.13* | 0.32 ± 0.09* | 0.14 ± 0.09 | 0.10 ± 0.12* | 0.45 ± 0.19* |
| OR | 0.39 ± 0.12 | 0.17 ± 0.10 | 0.24 ± 0.09 | 0.13 ± 0.03 | 0.24 ± 0.06*# | 0.33 ± 0.21*# |
Results are presented as proportion of total bacteria copies. *p<0.05 compared with CT group, #p<0.05 compared with OP group. Values are expressed as mean ± SE (n = 8). CT, normal chow diet-fed mice; OP, high-fat diet-induced obesity-prone mice; OR, high-fat diet-induced obesity-resistant mice.
Intestinal permeability markers in ileal sections and inflammation markers in PPs
We demonstrated that OP mice dramatically increased intestinal permeability by a mechanism associated with a reduced expression of epithelial tight junction proteins such as Zo-1, Occludin, Claudin-1, and JAM (Fig. 5A), although only a tendency was observed for Occludin. There was no significant differences in Zo-1 and Occludin mRNA expression between OR and CT group. qPCR analyses of pro- and anti-inflammatory cytokines in PPs revealed that the expressions of pro-inflammatory cytokines (IL-6 and TNF-α) were significantly increased in the OP mice compared with the CT and OR mice, whereas anti-inflammatory (IL-10) was significantly decreased (Fig. 5B). Except for the significant increases of TNF-α in the OR mice, no significant differences were observed in the relative expression of IL-6 and IL-10 between OR and CT group (Fig. 5B).
Fig. 5.
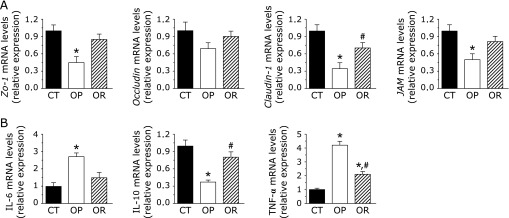
Intestinal permeability markers in ileal sections and inflammation markers in PPs. For qPCR assays of the epithelial tight junction proteins markers (Zo-1 and Occludin) (A) and IL-6, IL-10, and TNF-α mRNAs (B), each relative mRNA expression was determined by the ratio of intensity to β-actin production. Values are expressed as mean ± SE (n = 8). Mean values were significantly different compared with that of CT group: *p<0.05. Mean values were significantly different compared with that of the OP group: #p<0.05. CT, normal chow diet-fed mice; OP, high-fat diet-induced obesity-prone mice; OR, high-fat diet-induced obesity-resistant mice.
Relationship between gut microbiota and inflammation markers in PPs
The relationship between top 5 abundance genera bacteria on the interior of PPs and inflammation markers was analyzed (Table 4). The mRNA expression of IL-6 was positively and significantly correlated with Lactococcus, whereas negatively correlated with Allobaculum and Lactobacillus (p<0.05). A positive correlation was found between IL-10 mRNA and Allobaculum and Lactobacillus (p<0.05), but a negative correlation was found between IL-10 mRNA and Lactococcus. The mRNA expression of TNF-α was positively correlated with Lactococcus and Rhizobium, whereas negatively correlated with Lactobacillus (p<0.05).
Table 4.
Inflammation markers in Peyer’s patches correlated with Lactococcus, Allobaculum, Streptococcus, Lactobacillus and Rhizobium
| Lactococcus (%) | Allobaculum (%) | Lactobacillus (%) | Streptococcus (%) | Rhizobium (%) | |
|---|---|---|---|---|---|
| IL-6 mRNA (Relative expression) | r = 0.865 | r = –0.903 | r = –0.863 | r = 0.804 | r = 0.990 |
| p = 0.084 | p = 0.036* | p = 0.023* | p = 0.091 | p = 0.061 | |
| IL-10 mRNA (Relative expression) | r = –0.774 | r = 0.726 | r = 0.785 | r = –0.824 | r = –0.985 |
| p = 0.021* | p = 0.035* | p = 0.030* | p = 0.074 | p = 0.077 | |
| TNF-α mRNA (Relative expression) | r = 0.982 | r = –0.752 | r = –0.908 | r = 0.846 | r = 0.718 |
| p = 0.032* | p = 0.131 | p = 0.024* | p = 0.156 | p = 0.040* |
Correlations between IL-6 mRNA and the percentage of Lactococcus, Allobaculum, Streptococcus, Lactobacillus and Rhizobium; Correlations between IL-10 mRNA and the percentage of Lactococcus, Allobaculum, Streptococcus, Lactobacillus and Rhizobium; Correlations between TNF-α mRNA and the percentage of Lactococcus, Allobaculum, Streptococcus, Lactobacillus and Rhizobium. *p<0.05; inset corresponds to Pearson’s r correlation and corresponding p value.
Discussion
The current study is the first to investigate possible differences in the composition of the indigenous opportunistic bacteria on the interior of PPs among CT, obesity-prone and obesity-resistant mice induced by HFD. Using both pyrosequencing and qPCR techniques, we did observe clear differences in the interior of PPs microbial populations of CT, OP, and OR mice.
According to pyrosequencing analysis, the OTU number, Chao 1 estimator, and Shannon index in CT and OR groups was higher than that of OP mice, suggesting that CT and OR group had a significantly larger bacteria diversity than that in the OP group. These data are in agreement with that gut luminal bacteria diversity was reduced in obese individuals.(25) This may be because that bacteria on the interior of PPs is mostly sampled by M cells or DCs from gut luminal bacteria, which resides outside the layer of mucus and covers the intestinal epithelial cells.(26) Interesting, Firmicutes was found to be the dominant phylum bacteria in the PPs in all groups, followed by Proteobacteria. This observation is, however, in disagreement with recently published data on Firmicutes and Bacteroidetes were the major luminal microbiota phylum in obesity mice and humans.(24,27,28) Not all bacteria can gain access to M cells, given that size restrictions are set by a glycocalix.(29) Considering the growing knowledge about the complexity of the intestinal microbiome, it becomes obvious that the analysis of the composition on the phylum level can not reveal compositional changes that affect only certain bacteria subgroups, we then analysed the corresponding genera in more detail. Most of the sequences from Firmicutes belonged to the genera Lactococcus, Turicibacter, and Allobaculum, whereas most of the sequences from Proteobacteria belonged to the genera Pseudomonas and Rhizobium.
Differences in certain bacteria subgroups among CT, OP, and OR mice have been investigated. High-fat feeding induced a significant increase in Streptococcus, Comamonas, and Flavobacterium; moreover, these changes were independent of the obesity-prone or obesity-resistant phenotype. This suggests that high-fat feeding induces changes in several genera (i.e., Streptococcus, Comamonas, and Flavobacterium) but that this is not always associated with obesity, which is in agreement with the previous report about gut microbiota.(5) Interesting, Allobaculum and Lactobacillus was significantly decreased in OP mice compared with CT and OR mice, accounting for less than 0.5% of the sequences, whereas Rhizobium and Lactococcus were significantly increased. Several independent studies showed that obese mice have lower abundances of Allobaculum in gut lumen than animals fed the control diet,(4,30) whichwas enriched in the weight reduced mice.(19) Furthermore, Allobaculum relative abundance has been reported to be positively correlated with plasma HDL concentrations in hamsters fed a diet supplemented with grain sorghum lipid extract(31) and negatively correlated with leptin concentration.(19) Lactobacillius was significantly decreased in colonic contents of HFD mice.(32) Moreover, Mantis et al.(33) found that the association of a Lactobacillus with non-specific SigA enhanced probiotic adhesion by a factor of ⩾3.4-fold and transiting through an M cell from the lumen to the PPs. Tsai et al.(34) showed that feeding with Lactobacillus induced stronger CD4+ T cell-DCs interaction, enhanced CD4+ T cell and B cell proliferation, and increased IL-1β, IL-10, IL-12, IFN-γ, and TNF-α mRNA expression in PPs, which could enhance immunosurveillance to prevent intestinal infections or other intestinal pathologies. Poutahidis et al.(35) discovered that oral Lactobacillus reuteri therapy alone was sufficient to change the pro-inflammatory immune cell profile and prevent abdominal fat pathology and age-associated weight gain in mice Westernized ’fast food’ diet. Rhizobium, as symbionts in plant root nodule cells, is contact with the cytoplasm of the host cell by symbiosomes, which are closely related to compounds in the plant’s immune system. Recent studies demonstrated that Rhizobium LPSs induce CD14 expression in bone marrow cells by a TLR4 and TLR2 mechanism.(36)
Pseudomonas genus is an opportunistic, gram-negative pathogen associated with many hospital-acquired infections and disease states,(37) such as P. aeruginosa and P. fluorescens. show an important pathogenic potential and trigger a specific inflammatory response in enterocytes.(38,39) Moreover, Madi et al.(40) found recently that P. fluorescens can be cytotoxic in Caco-2/TC7 intestinal cells, invade the target cell, and translocate from the luminal to the internal compartment. In our current study, the genus Pseudomonas was significantly increased in high-fat diet groups (OP and OR) compared with the control diet. Surprisingly, obesity-resistant mice with the lower body weight had a significant higher Pseudomonas relative abundance than obesity-prone mice.
The reason for the different composition of indigenous opportunistic bacteria on the interior of PPs between obesity-prone and obesity-resisitant is not clear, so it remains to be established how luminal bacteria communities gain access to PPs. One possibility is that commensal bacteria first become opsonized by natural polyreactive IgA antibodies and then undergo IgA-mediated apical-to-basal transepithelial migration across M cells into a PPs.(41) Obata et al.(14) showed that the numbers of Alcaligenes decreased in the absence of B cells and mucosal Abs, suggesting that Abs may play a critical role in the PPs tissue colonization by these bacteria. In addition, these enteroinvasive pathogens (e.g., Salmonella typhimurium, Listeria, and Yersinia) can express the nvasion that permits the invasion of nonphagocytic cells, and then enter the PPs, presumably via a DC-mediated mechanism.(9)
Some bacteria in OP mice can potentially trigger a specific inflammatory response, and in ture, play a key role in the development of obesity. Some bacteria-loaded DCs migrate from epithelial and subepithelial areas to the T cell-rich interfollicular regions of PPs, where they initiate a polarized T helper type-2 (Th2) response characterized by the release of noninflammatory cytokines with B cell-activating functions, including IL-6,(42) TNF-α, retinoic acid, and IL-10, to induce CD4+CD25+Foxp3+ T regulatory (Treg) cells, CD4+CD25−Foxp3− T regulatory type 1 (Tr1) cells, and regulatory Th17 cells. In our study, significant correlations between mRNA expression of IL-6, IL-10, TNF-α and the top 5 abundance genera bacteria on the interior of PPs were also observed (Table 4). Moreover, it has already been demonstrated that Alcaligenes produce antimicrobial substances inhibiting growth of other bacteria, such as E. coli, Streptococcus pyogenes, Pseudomonas aeruginosa, and Staphylococcus aureus, and may be beneficial for the host by eliminating other opportunistic and pathogenic bacteria at their portal of entry.(14) Thus, the composition of the bacteria species that initially colonizes our guts at birth may have a significant impact on our metabolic health later in life. This line of investigation is now being intensively studied in our laboratory to further elucidate the significance of commensal microbiota that inhabits both systemic and mucosal lymphoid tissues.
In summary, indigenous opportunistic bacteria on the interior of PPs was significantly different among control diet mice, high-fat diet induced obesity-prone mice, and obiestiy-resistant mice. By cohabiting within the organized PPs, these bacteria affect the development and maturation of the host mucosal immune system. Further, the PP-inhabiting, commensal microbiota are an additional element that contributes to maintaining immunologic homeostasis in the host.
Acknowledgments
This work was supported by a grant from National Natural Science Foundation of China (No. 31201805) and the Fundamental Research Funds for the Central Universities (No. JUDCF10058), and 12th 5-Year Plan for Science and Technology Development (No. 2012BAD33B05).
Abbreviations
- CT
control diet group
- IL-6
interleukin 6
- IL-10
interleukin 10
- JAM
junctional adhesion molecule
- OP
obesity-prone
- OR
obesity-resistant
- OTUs
operational taxonomic units
- PPs
peyer’s patches
- TNF-α
tumor necrosis factor-α
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hajihashemi P, Azadbakht L, Hashemipor M, Kelishadi R, Esmaillzadeh A. Whole-grain intake favorably affects markers of systemic inflammation in obese children: a randomized controlled crossover clinical trial. Mol Nutr Food Res. 2014;58:1301–1308. doi: 10.1002/mnfr.201300582. [DOI] [PubMed] [Google Scholar]
- 2.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 4.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 5.de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;299:G440–G448. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owen RL. Uptake and transport of intestinal macromolecules and microorganisms by M cells in Peyer’s patches—a personal and historical perspective. Semin Immunol. 1999;11:157–163. doi: 10.1006/smim.1999.0171. [DOI] [PubMed] [Google Scholar]
- 7.Buda A, Sands C, Jepson MA. Use of fluorescence imaging to investigate the structure and function of intestinal M cells. Adv Drug Deliv Rev. 2005;57:123–134. doi: 10.1016/j.addr.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Jang MH, Kweon MN, Iwatani K, et al. Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proc Natl Acad Sci U S A. 2004;101:6110–6115. doi: 10.1073/pnas.0400969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007;27:975–984. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto M, Kweon MN, Rennert PD, et al. Role of gut-associated lymphoreticular tissues in antigen-specific intestinal IgA immunity. J Immunol. 2004;173:762–769. doi: 10.4049/jimmunol.173.2.762. [DOI] [PubMed] [Google Scholar]
- 11.Chassaing B, Etienne-Mesmin L, Bonnet R, Darfeuille-Michaud A. Bile salts induce long polar fimbriae expression favouring Crohn’s disease-associated adherent-invasive Escherichia coli interaction with Peyer’s patches. Environ Microbiol. 2013;15:355–371. doi: 10.1111/j.1462-2920.2012.02824.x. [DOI] [PubMed] [Google Scholar]
- 12.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 13.Boullier S, Tanguy M, Kadaoui KA, et al. Secretory IgA-mediated neutralization of Shigella flexneri prevents intestinal tissue destruction by down-regulating inflammatory circuits. J Immunol. 2009;183:5879–5885. doi: 10.4049/jimmunol.0901838. [DOI] [PubMed] [Google Scholar]
- 14.Obata T, Goto Y, Kunisawa J, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc Natl Acad Sci U S A. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krane MK, Allaix ME, Zoccali M, et al. Does morbid obesity change outcomes after laparoscopic surgery for inflammatory bowel disease? Review of 626 consecutive cases. J Am Coll Surg. 2013;216:986–996. doi: 10.1016/j.jamcollsurg.2013.01.053. [DOI] [PubMed] [Google Scholar]
- 16.Macotela Y, Emanuelli B, Mori MA, et al. Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes. 2012;61:1691–1699. doi: 10.2337/db11-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz TJ, Huang TL, Tran TT, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper SJ, Crean SJ, Lewis MA, Spratt DA, Wade WG, Wilson MJ. Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol. 2006;44:1719–1725. doi: 10.1128/JCM.44.5.1719-1725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravussin Y, Koren O, Spor A, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye L, Zhang T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biotechnol. 2013;97:2681–2690. doi: 10.1007/s00253-012-4082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter J, Hertel C, Tannnock GW, et al. Detection of Lactobacillus, Pediococcus, Leuconostoc and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:2578–2585. doi: 10.1128/AEM.67.6.2578-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widmer F, Seidler RJ, Gillevet PM, et al. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol. 1998;64:2545–2553. doi: 10.1128/aem.64.7.2545-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 27.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 28.Murphy EF, Cotter PD, Healy S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 29.Frey A, Giannasca KT, Weltzin R, et al. Role of the glycocalyx in regulating access of microparticles to apical plasma membranes of intestinal epithelial cells: Implications for microbial attachment and oral vaccine targeting. J Exp Med. 1996;184:1045–1059. doi: 10.1084/jem.184.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S, Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104:919–929. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- 31.Martínez I, Wallace G, Zhang C, et al. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao Y, Sun J, Ding Y, Le G, Shi Y. Alterations of the gut microbiota in high-fat diet mice is strongly linked to oxidative stress. Appl Microbiol Biotechnol. 2013;97:1689–1697. doi: 10.1007/s00253-012-4323-6. [DOI] [PubMed] [Google Scholar]
- 33.Mantis NJ, Cheung MC, Chintalacharuvu KR, Rey J, Corthésy B, Neutra MR. Selective adherence of IgA to murine Peyer’s patch M cells: evidence for a novel IgA receptor. J Immunol. 2002;169:1844–1851. doi: 10.4049/jimmunol.169.4.1844. [DOI] [PubMed] [Google Scholar]
- 34.Tsai YT, Cheng PC, Liao JW, Pan TM. Effect of the administration of Lactobacillus paracasei subsp. paracasei NTU 101 on Peyer’s patch-mediated mucosal immunity. Int Immunopharmacol. 2010;10:791–798. doi: 10.1016/j.intimp.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 35.Poutahidis T, Kleinewietfeld M, Smillie C, et al. Microbial reprogramming inhibits Western diet-associated obesity. PLoS One. 2013;8:e68596. doi: 10.1371/journal.pone.0068596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girard R, Pedron T, Uematsu S, et al. Lipopolysaccharides from Legionella and Rhizobium stimulate mouse bone marrow granulocytes via Toll-like receptor 2. J Cell Sci. 2011;116:293–302. doi: 10.1242/jcs.00212. [DOI] [PubMed] [Google Scholar]
- 37.Nelson RK, Poroyko V, Morowitz MJ, Liu D, Alverdy JC. Effect of dietary monosaccharides on Pseudomonas aeruginosa virulence. Surg Infect (Larchmt) 2013;14:35–42. doi: 10.1089/sur.2011.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperandio D, Rossignol G, Guerillon J, et al. Cell-associated hemolysis activity in the clinical strain of Pseudomonas fluorescens MFN1032. BMC Microbiol. 2010;10:124. doi: 10.1186/1471-2180-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madi A, Lakhdari O, Blottière HM, et al. The clinical Pseudomonas fluorescens MFN1032 strain exerts a cytotoxic effect on epithelial intestinal cells and induces Interleukin-8 via the AP-1 signaling pathway. BMC Microbiol. 2010;10:215. doi: 10.1186/1471-2180-10-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madi A, Svinareff P, Orange N, Feuilloley MG, Connil N. Pseudomonas fluorescens alters epithelial permeability and translocates across Caco-2/TC7 intestinal cells. Gut Pathog. 2010;2:16. doi: 10.1186/1757-4749-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mantis NJ, Rol N, Corthésy Bf. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rimoldi M, Chieppa M, Salucci V, et al. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nat Immunol. 2005;6:507–514. doi: 10.1038/ni1192. [DOI] [PubMed] [Google Scholar]