Abstract
Cytochrome P450 (CYP) 3A4 is a membrane protein that catalyzes hydroxylation of endogenous and exogenous substrates. Protein–protein interaction is a crucial factor that regulates the function of enzymes. However, protein–protein interactions involving human CYPs have not been fully understood. In this study, extensive protein–protein interactions involving CYP3A4 were determined by a shotgun analysis of immunoprecipitate utilizing anti-CYP3A4 antibody. Our shotgun analysis revealed that 149 proteins were immunoprecipitated with anti-CYP3A4 antibody in human liver microsomes. We further determined that 51 proteins of 149 proteins were specifically immunoprecipitated with the anti-CYP3A4 antibody. Our analysis demonstrated that other CYP isoforms are interacting with CYP3A4, which is in agreement with previous findings. Based on our current and previous findings, we propose that drug-metabolizing enzymes such as CYP3A4 and UDP-glucuronosyltransferase 2B7 form a metabolosome, which is a functional unit of metabolism consisting of multiple metabolism-related proteins.
Keywords: CYP3A4, cytochrome P450, metabolosome, nano-LC-MS/MS, protein–protein interactions
Introduction
Cytochrome P450s (CYPs; EC 1.14.x.x) are important membrane-bound enzymes that metabolize a number of endogenous and exogenous substrates. Human CYP is a superfamily of enzymes that are divided into 18 families with a total of 57 isoforms (Lewis 2004). The CYP3A4 gene is located on chromosome 7q22.1 and is the main CYP isoform responsible for the metabolism of more than 50% of clinically used drugs (Williams et al. 2004). As CYPs are called as phase I drug-metabolizing enzymes, there is also a group of phase II drug-metabolizing enzymes that catalyze the conjugation of their substrates. UDP-Glucuronosyltransferases (UGTs; EC 2.4.1.17), which are the major phase II drug-metabolizing enzymes, metabolize drugs by transferring the glucuronic acid moiety of UDP-glucuronic acid to the substrates (Dutton 1980; Mackenzie et al. 2005).
While the active site of CYPs locates in the cytoplasmic side of endoplasmic reticulum (ER), the active site of UGT proteins locates in the luminal side of ER (Shepherd et al. 1989). Whereas it has been reported that CYPs and UGTs functionally interact with each other, as CYPs catalyze hydroxylation of substrates so that UGT can transfer glucuronic acid to the hydroxyl group of the substrates (Nakajima and Yokoi 2005; Zheng et al. 2007). Physical interactions between CYPs and UGTs have been demonstrated by immunoprecipitation assays utilizing anti-CYP3A4 and anti-UGT2B7 antibodies (Fremont et al. 2005; Ishii et al. 2007; Takeda et al. 2009), supporting that CYPs and UGTs are interacting with each other to cooperatively metabolize the substrates. Not only UGTs but also other drug-metabolizing enzymes, such as epoxide hydrolase 1, have been reported to interact with CYPs (Taura et al. 2000). However, the physiological role of these protein interactions is still largely unknown.
Metabolosome, which is a functional unit of metabolism, is the multimolecular assembly composed of metabolizing enzymes, transport-related proteins (transporters, channels or pumps), regulatory proteins, scaffold proteins, and other functional cellular components, which are assembled by means of multiple protein–protein interactions and/or protein–lipid interactions (Mori et al. 2011). The fact that human CYPs have been reported to interact with other proteins such as UGTs and epoxide hydrolase led us to investigate whether CYPs form a metabolosome assembled by extensive protein–protein interactions. In this study, large-scale analysis of protein–protein interaction involving CYP3A4 was carried out by shotgun liquid chromatography–mass spectrometry (LC-MS/MS) proteomic analysis of immunoprecipitated proteins to identify proteins interacting with human CYP3A4 in human liver microsomes.
Materials and Methods
Chemicals and reagents
Human liver microsomes were obtained from BD Gentest (Woburn, MA). Rabbit anti-human CYP3A4 antibody (H00001576-D01) was purchased from Abnova (Taipei, Taiwan). Control antibodies (IgG from rabbit serum) were purchased from Sigma–Aldrich (St Louis, MO) and Abcam (Cambridge, MA). Immunoprecipitation Kit Dynabeads ProteinG was purchased from Invitrogen (Carlsbad, CA). All other chemicals and solvents were of analytical grade or the highest grade commercially available.
Immunoprecipitation and Nano-LC-MS/MS analysis of the immunoprecipitate
Immunoprecipitation assay and nano-LC-MS/MS analysis were conducted as described before (Fujiwara and Itoh 2014).
Results
Identification of proteins immunoprecipitated with human CYP3A4 antibody
CYP3A4 is one of the major CYP isoforms involved in metabolism of a wide variety of drugs in the human liver. To identify proteins that interact with CYP3A4, human liver microsomes were subjected to an immunoprecipitation assay with rabbit anti-human CYP3A4 antibody, followed by a shotgun analysis of the immunoprecipitate by an LC-MC/MC analysis. Our shotgun analysis revealed that 149 proteins including human CYP3A4 were coimmunoprecipitated with anti-human CYP3A4 antibody (Fig. 1). Human CYP3A4 has been reported to interact with human UGTs and epoxide hydrolases. In this study, peptide sequences of these proteins were observed in the LC-MC/MC analysis of the immunoprecipitate, showing the reproducibility of the previous findings. Several peptide sequences of other proteins such as protein transport protein Sec61 were also obtained in the shotgun analysis. These data indicate that CYP3A4 might interact with multiple proteins to form a metabolosome.
Figure 1.
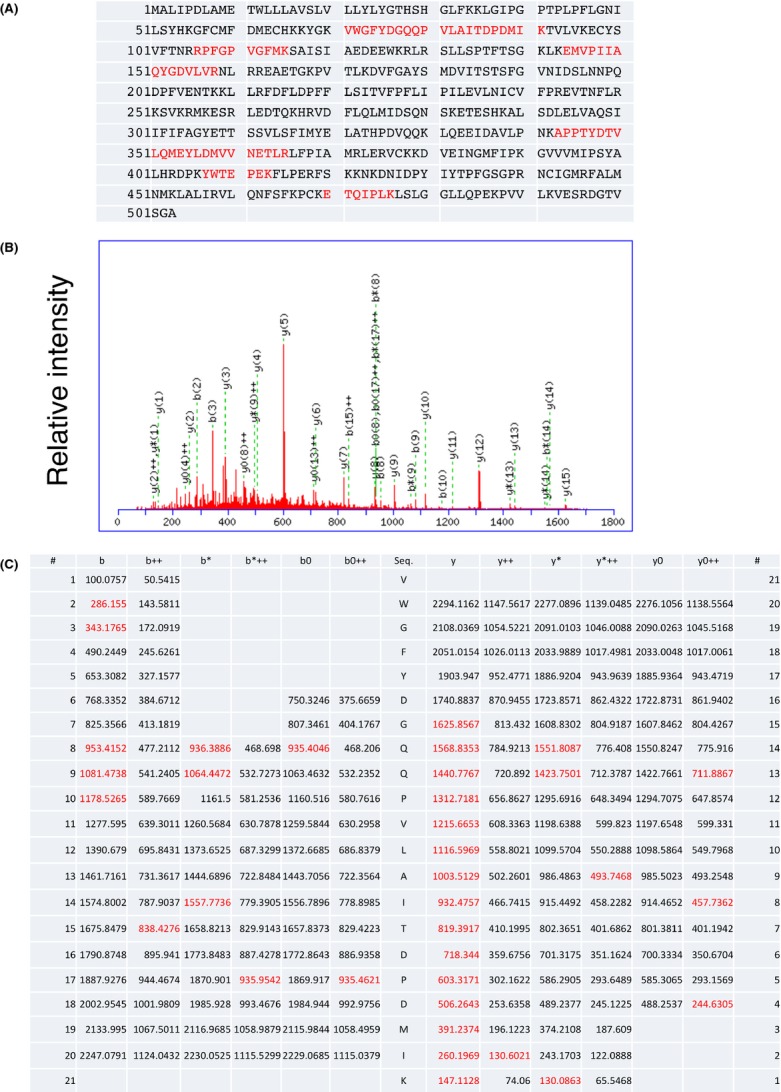
Peptide sequence analysis of human CYP3A4. Six peptide sequences of human CYP3A4 were obtained with the shotgun analysis of the immunoprecipitate with anti-CYP3A4 antibody (A). The middle panel indicates the MS/MS spectra of the peptide (B), whereas the bottom panel indicates ion tables (C).
To exclude the possibility that the anti-CYP3A4 antibody used in the immunoprecipitation assay nonspecifically reacted to proteins in human liver microsomes, we incorporated our previous data into the current study. We have previously carried out immunoprecipitation assays with two different control rabbit IgG, followed by shotgun analysis (Fujiwara and Itoh 2014). It was revealed that more than 100 proteins were nonspecifically interacted with IgG in the human liver microsomes. Table S1 shows the list of proteins specifically coimmunoprecipitated with the anti-human CYP3A4 antibody.
While the list of proteins coimmunoprecipitated with the anti-CYP3A4 antibody contains several mitochondrial, outer membrane, and extracellular proteins, it also contains a number of proteins that are present in the ER such as CYP3A5, CYP2A6, CYP4F2, and bile acyl-CoA synthetase (Table 1).
Table 1.
ER Proteins that were coimmunoprecipitated with anti-CYP3A4 antibody
| No. | Accession | Score | Mass | Number of peptide sequences | emPAI | Protein name |
|---|---|---|---|---|---|---|
| 2 | CP3A4_HUMAN | 140 | 57705 | 6 | 0.21 | Cytochrome P450 3A4 |
| 7 | S61A1_HUMAN | 87 | 52687 | 1 | 0.07 | Protein transport protein Sec61 subunit alpha isoform 1 |
| 9 | SSRG_HUMAN | 73 | 21067 | 1 | 0.18 | Translocon-associated protein subunit gamma |
| 10 | NDUB4_HUMAN | 70 | 15256 | 1 | 0.26 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 |
| 12 | ERP29_HUMAN | 64 | 29032 | 1 | 0.13 | Endoplasmic reticulum resident protein 29 |
| 14 | S27A5_HUMAN | 61 | 76420 | 2 | 0.1 | Bile acyl-CoA synthetase |
| 15 | CO4A_HUMAN | 61 | 194247 | 1 | 0.02 | Complement C4-A |
| 16 | CISD2_HUMAN | 59 | 15497 | 2 | 0.56 | CDGSH iron-sulfur domain-containing protein 2 |
| 18 | AT2A3_HUMAN | 52 | 115444 | 1 | 0.03 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 |
| 21 | SSRD_HUMAN | 50 | 19158 | 1 | 0.2 | Translocon-associated protein subunit delta |
| 22 | NDUAC_HUMAN | 49 | 17104 | 1 | 0.23 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 |
| 23 | FAS_HUMAN | 48 | 275877 | 1 | 0.01 | Fatty acid synthase |
| 24 | AT1A1_HUMAN | 48 | 114135 | 1 | 0.03 | Sodium/potassium-transporting ATPase subunit alpha-1 |
| 27 | GLU2B_HUMAN | 43 | 60357 | 1 | 0.06 | Glucosidase 2 subunit beta |
| 28 | GANAB_HUMAN | 43 | 107263 | 1 | 0.03 | Neutral alpha-glucosidase AB |
| 36 | CP3A5_HUMAN | 26 | 57357 | 1 | 0.06 | Cytochrome P450 3A5 |
| 38 | TM109_HUMAN | 25 | 26194 | 1 | 0.15 | Transmembrane protein 109 |
| 39 | CP4F2_HUMAN | 23 | 60442 | 1 | 0.06 | Leukotriene-B(4) omega-hydroxylase 1 |
| 41 | CP2A6_HUMAN | 22 | 56636 | 1 | 0.07 | Cytochrome P450 2A6 |
| 47 | STRUM_HUMAN | 18 | 135113 | 1 | 0.03 | WASH complex subunit strumpellin |
Discussion and Conclusions
CYPs are the predominant proteins that are involved in metabolism of a wide variety of drugs. Due to a greater interest especially in the biomedical field, the insight into the molecular mechanism of CYPs-catalyzed metabolism (i.e., hydroxylation) of the substrate has been thoroughly investigated. Protein–protein interaction is known as a crucial factor that regulates the function of the proteins. CYPs have been reported to interact with a microsomal protein, NADPH-cytochrome P450 reductase (CPR). Indeed, these interactions are required for CYP catalysis (Bernhardt 2006). The physical and functional interactions between CYP and CPR have been widely demonstrated. Recent studies also revealed that CYPs interact with other drug-metabolizing enzymes expressed in ER such as UGTs and epoxide hydrolase (Taura et al. 2000; Ishii et al. 2007). It was further demonstrated that the protein–protein interaction between CYPs and UGTs affected their enzymatic activities (Ishii et al. 2014). Not only with other proteins, but CYPs also interact with CYP enzymes themselves to form homo- and heterodimers (Subramanian et al. 2009, 2010; Davydov 2011; Reed and Backes 2012). The presence of these complexities of the protein–protein interaction in ER suggests that CYP3A4 might be involved in the formation of a metabolosome, a functional unit of metabolism consisting of multiple metabolism-related proteins.
Most of the metabolites of endogenous and exogenous compounds produced by CYPs are subsequently metabolized by phase II drug-metabolizing enzymes such as UGTs to further increase in their hydrophilicity. The fact that the majority of CYPs are localized on the cytoplasmic and UGTs on the luminal side of the ER membrane leads us to believe that their substrates and metabolites need to be efficiently translocated across the ER membrane. However, CYPs and UGTs solely possess a transmembrane helix, which make it impossible to form a pore, suggesting that other proteins might be involved in the transport of the hydrophilic substrates/metabolites across the ER membrane. In this study, it was revealed that a number of microsomal proteins were interacting with CYP3A4 in the microsomal fraction (Table 1). While the functional relationship between CYP3A4 and the interacting proteins is largely unknown, the formation of a metabolosome might contribute to the accelerated translocation of the hydrophilic substrates/metabolites across the ER membrane. This hypothesis is supported by the finding that several translocators such as Sec61 and translocon-associated proteins were coimmunoprecipitated with CYP3A4 in this study (Table 1). While the primary role of the translocators is to translocate polypeptides across the ER membrane, it has been reported that Sec61 was also involved in efflux of a small molecule, calcium, from the ER (Lang et al. 2011). The functional relationship between CYP3A4 and proteins listed in Table 1 needs to be further investigated in the future.
To exclude the proteins nonspecifically interacting with the antibody used in this study, we incorporated our previous data (Fujiwara and Itoh 2014), which showed that more than 100 proteins were nonspecifically interacted with IgG in the human liver microsomes. However, many mitochondrial, outer membrane, and extracellular proteins such as ATP synthase and transhydrogenase were still included in the list of proteins that were found to interact with CYP3A4 (Table S1). While it is possible that those proteins are transiently present in the ER to interact with CYP3A4, it could be a result of indirect/nonspecific interactions.
In conclusion, we performed a shotgun analysis of immunoprecipitate obtained with anti-CYP3A4 antibody by utilizing nano-LC-MS/MS analysis. Twenty ER proteins were newly identified as proteins interacting with CYP3A4 in human liver microsomes. These proteins might be associated with a formation of a metabolosome involving CTP3A4. Functional roles of newly identified proteins interacting with CYP3A4 in metabolism and translocation of substrates or metabolites need to be examined in future studies.
Acknowledgments
This work was supported by the Naito Foundation (R. F.).
Glossary
- CPR
NADPH-cytochrome P450 reductase
- CYP
cytochrome P450
- ER
endoplasmic reticulum
- LC-MS/MS
liquid chromatography–mass spectrometry
- UGTs
UDP-Glucuronosyltransferases
Disclosures
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Proteins that were coimmunoprecipitated with anti-CYP3A4 antibody. Proteins specifically immunoprecipitated with the anti-CYP3A4 antibody are shown. Proteins expressed in the ER are highlighted with a red color.
References
- Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Davydov DR. Microsomal monooxygenase as a multienzyme system: the role of P450-P450 interactions. Expert Opin Drug Metab Toxicol. 2011;7:543–558. doi: 10.1517/17425255.2011.562194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton GJ. Acceptor substrates of UDP glucuronosyltransferase and their assay. In: Dutton GJ, editor. Glucuronidation of drugs and other compounds. Boca Raton, FL: CRC Press; 1980. pp. 69–78. [Google Scholar]
- Fremont JJ, Wang RW, King CD. Coimmunoprecipitation of UDP-glucuronosyltransferase isoforms and cytochrome P450 3A4. Mol Pharmacol. 2005;67:260–262. doi: 10.1124/mol.104.006361. [DOI] [PubMed] [Google Scholar]
- Fujiwara R, Itoh T. Extensive protein-protein interactions involving UDP-glucuronosyltransferase (UGT) 2B7 in human liver microsomes. Drug Metab Pharmacokinet. 2014;29:259–265. doi: 10.2133/dmpk.dmpk-13-rg-096. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Iwanaga M, Nishimura Y, Takeda S, Ikushiro S, Nagata K, et al. Protein-protein interactions between rat hepatic cytochromes P450 (P450s) and UDP-glucuronosyltransferases (UGTs): evidence for the functionally active UGT in P450-UGT complex. Drug Metab Pharmacokinet. 2007;22:367–376. doi: 10.2133/dmpk.22.367. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Koba H, Kinoshita K, Oizaki T, Iwamoto Y, Takeda S, et al. Alteration of the function of the UDP-glucuronosyltransferase 1A subfamily by cytochrome P450 3A4: different susceptibility for UGT isoforms and UGT1A1/7 variants. Drug Metab Dispos. 2014;42:229–238. doi: 10.1124/dmd.113.054833. [DOI] [PubMed] [Google Scholar]
- Lang S, Erdmann F, Jung M, Wagner R, Cavalie A, Zimmermann R. Sec61 complexes form ubiquitous ER Ca2+ leak channels. Channels (Austin) 2011;5:228–235. doi: 10.4161/chan.5.3.15314. [DOI] [PubMed] [Google Scholar]
- Lewis DF. 57 varieties: the human cytochromes P450. Pharmacogenomics. 2004;5:305–318. doi: 10.1517/phgs.5.3.305.29827. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- Mori Y, Kiyonaka S, Kanai Y. Transportsomes and channelsomes: are they functional units for physiological responses? Channels (Austin) 2011;5:387–390. doi: 10.4161/chan.5.5.16466. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Yokoi T. Interindividual variability in nicotine metabolism: C-oxidation and glucuronidation. Drug Metab Pharmacokinet. 2005;20:227–235. doi: 10.2133/dmpk.20.227. [DOI] [PubMed] [Google Scholar]
- Reed JR, Backes WL. Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol Ther. 2012;133:299–310. doi: 10.1016/j.pharmthera.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd SR, Baird SJ, Hallinan T, Burchell B. An investigation of the transverse topology of bilirubin UDP-glucuronosyltransferase in rat hepatic endoplasmic reticulum. Biochem J. 1989;259:617–620. doi: 10.1042/bj2590617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Low M, Locuson CW, Tracy TS. CYP2D6-CYP2C9 protein-protein interactions and isoform-selective effects on substrate binding and catalysis. Drug Metab Dispos. 2009;37:1682–1689. doi: 10.1124/dmd.109.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Tam H, Zheng H, Tracy TS. CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos. 2010;38:1003–1009. doi: 10.1124/dmd.109.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Ishii Y, Iwanaga M, Nurrochmad A, Ito Y, Mackenzie PI, et al. Interaction of cytochrome P450 3A4 and UDP-glucuronosyltransferase 2B7: evidence for protein-protein association and possible involvement of CYP3A4 J-helix in the interaction. Mol Pharmacol. 2009;75:956–964. doi: 10.1124/mol.108.052001. [DOI] [PubMed] [Google Scholar]
- Taura KI, Yamada H, Hagino Y, Ishii Y, Mori MA, Oguri K. Interaction between cytochrome P450 and other drug-metabolizing enzymes: evidence for an association of CYP1A1 with microsomal epoxide hydrolase and UDP-glucuronosyltransferase. Biochem Biophys Res Commun. 2000;273:1048–1052. doi: 10.1006/bbrc.2000.3076. [DOI] [PubMed] [Google Scholar]
- Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, et al. Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Sun D, Sharma AK, Chen G, Amin S, Lazarus P. Elimination of antiestrogenic effects of active tamoxifen metabolites by glucuronidation. Drug Metab Dispos. 2007;35:1942–1948. doi: 10.1124/dmd.107.016279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Proteins that were coimmunoprecipitated with anti-CYP3A4 antibody. Proteins specifically immunoprecipitated with the anti-CYP3A4 antibody are shown. Proteins expressed in the ER are highlighted with a red color.


