Abstract
This study describes for the first time the expression levels of genes encoding membrane transporters and drug-metabolizing enzymes in the lungs of ex-smoking patients with chronic obstructive pulmonary disease (COPD). Membrane transporters and drug-metabolizing enzymes are key determinants of drug uptake, metabolism, and elimination for systemically administered as well as inhaled drugs, with consequent influence on clinical efficacy and patient safety. In this study, while no difference in gene expression was found between healthy and COPD subjects, we identified a significant regional difference in mRNA expression of both membrane transporters and drug-metabolizing enzymes between central and peripheral tissue in both healthy and COPD subjects. The majority of the differentially expressed genes were higher expressed in the central airways such as the transporters SLC2A1 (GLUT1), SLC28A3 (CNT3), and SLC22A4 (OCTN1) and the drug-metabolizing enzymes GSTZ1, GSTO2, and CYP2F1. Together, this increased knowledge of local pharmacokinetics in diseased and normal lung may improve modeling of clinical outcomes of new chemical entities intended for inhalation therapy delivered to COPD patients. In addition, based on the similarities between COPD and healthy subjects regarding gene expression of membrane transporters and drug-metabolizing enzymes, our results suggest that clinical pharmacological studies in healthy volunteers could be a valid model of COPD patients regarding drug disposition of inhaled drugs in terms of drug metabolism and drug transporters.
Keywords: central airways, expression pattern, ex-smoker, human lung tissue, mRNA, PCA, PCR, peripheral tissue, pharmacokinetics, TLDA
Introduction
Inhaled drugs are commonly used in the treatment of patients with respiratory diseases, for example, chronic obstructive pulmonary disease (COPD) (Somers et al. 2007). The inhalation route offers efficient delivery of drugs resulting in a high local concentration while keeping the systemic levels low (Patton and Byron 2007). The deposition of inhaled drugs is largely determined by the anatomical structure of the lung together with the physical and chemical characteristics of the inhaled formulation. Smaller hydrophilic particles are likely to reach the periphery and alveolar region and the drug is commonly rapidly absorbed into the systemic circulation after its dissolution. Larger particles are on the other hand more likely to deposit in the central airways and may be partly eliminated by mucociliary clearance (Patton and Byron 2007). In contrast to deposition and dissolution, the mechanisms of drug uptake and distribution into the lung tissue, metabolism, and absorption into the systemic circulation are far from understanding (Bosquillon 2010).
Membrane transporters and metabolizing enzymes have emerged as key determinants of orally administered drugs regarding drug disposition, intracellular concentration of the drugs, and bioactivity with influence on clinical efficacy and patient safety (Giacomini et al. 2010). Several membrane transporters are also present in the lung where some mainly are involved in uptake like members of the solute carrier family (SLCs), while others like the ATP-binding cassette (ABC) transporters are actively secreting compounds out of the cell. Drug-metabolizing enzymes like the cytochrome P450 (CYPs), glutathione -S-transferases (GST), sulfo-transferases (SULT), epoxide hydrolases (EPX), and glucuronyl-transferases (UGT) which are highly expressed metabolizing enzymes primarily in the liver are also present in extrahepatic tissues such as the lung. These enzymes serve in detoxification of inhaled xenobiotics. However, the detoxification process can also lead to activation of compounds and thereby increase their reactivity and promoting cellular damage and disease. Thus, the expression of drug-metabolizing enzymes and membrane transporters in COPD lungs is of great importance not only for drug disposition and treatment efficacy of COPD but their presence could also contribute to the development of this disease.
Cigarette smoking is the predominant cause of COPD, which initiates an inflammatory process. This process induces irreversible and progressive loss of lung function with tissue destruction and structural changes with time, such as chronic bronchitis, small airway disease, and emphysema. The consequences of these structural changes on drug uptake are not understood, but it is established that the inflammatory process in itself affects the uptake, distribution, and elimination of specific drugs, in, for instance, other inflammatory lung diseases such as asthma (Harrison and Tattersfield 2003). In addition, lipopolysaccharide (LPS) or inflammatory cytokines like interleukin (IL)-1, -6, interferon (INF)γ, and tumor necrosis factor (TNF) have all been shown to suppress the expression of several drug-metabolizing enzymes and transporters partly through downregulation of transcriptional activators of membrane transporters and drug-metabolizing enzymes such as the androstane receptor (CAR), the pregnane X receptor (PXR), the retinoid X receptor (RXR), and CCAAT Enhancer binding protein β (C/EBPβ) (Aitken et al. 2006). It is also known that the expression pattern of various CYPs is affected in lung epithelial cells in smokers with or without COPD compared to healthy nonsmokers. For example, components in cigarette smoke like polycyclic aromatic hydrocarbons (PAH) are known to, for instance, induce the expression of CYP1A, CYP1B1, and CYP2E1 with possible consequences of drug efficacy (Kim et al. 2004; Pierrou et al. 2007).
However, to our knowledge, no studies have been published regarding the gene expression of drug transporters in ex-smokers with COPD. As COPD progress even after smoke cessation, it is important to unravel the effect on drug uptake and metabolism after smoke cessation.
Thus, this study aims at examining the gene expression profile of drug transporters and metabolizing enzymes, as well as some common regulatory pathways in peripheral lung and central airways from ex-smokers with COPD and healthy subjects. Increased knowledge in this area would aid establishing models of drug disposition and predictions of local pharmacokinetics in the lung, and facilitate the development of new chemical entities with improved compound retention and activity in the lung.
Experimental Section
Patients
Patients (n = 7) suffering from very severe COPD (GOLD stage IV) who were undergoing lung transplantation at Lund University Hospital were included in the study. The patients had stopped smoking at least 6 months before the lung transplantation. Written consent was obtained from all subjects. Lung explants from healthy donors (n = 3) with no history of lung disease were also included. Lungs which were supposed to be used for transplantation but no matched recipients were available at that moment and could therefore be included in this study. In these cases, written consents were obtained from their closest relatives. All subjects had been treated with the glucocorticoid Solu-medrol, a broad-spectrum antibiotic, propofol, fentanyl, benzodiazepine, and methyl-prednisolone in connection to the transplant surgery. Subject information is summarized in Table 1. Those two studies were approved by the Swedish Research Ethical Committee in Lund (FEK 91/2006 and FEK 413/2008).
Table 1.
Description of subjects
| Diagnosis | |||||
|---|---|---|---|---|---|
| COPD | Age (years) | Sex | Pack years | FEV1 It | FEV1/FVC (%) |
| 1 | 64 | F | 60 | 0.50 | 38 |
| 2 | 62 | F | 50 | 0.52 | 26 |
| 3 | 61 | M | 40 | 0.77 | 39 |
| 4 | 61 | M | 50 | 0.60 | 24 |
| 5 | 62 | M | 50 | 0.90 | 32 |
| 6 | 65 | F | 30 | 0.49 | 32 |
| 7 | 63 | M | 45 | 1.00 | 37 |
| Healthy controls | |||||
| 1 | 46–65 | F | Nonsmoker | ||
| 2 | 46–65 | F | Nonsmoker | ||
| 3 | 46–65 | M | Nonsmoker | ||
FEV1, forced expiratory volume in one second; FVC, forced vital capacity; pack years, number of cigarettes smoked per day × number of years smoked)/20.
Tissue acquisition
Lung explants were dissected directly after removal from the COPD patients as described previously (Hallgren et al. 2010, 2012). Lung tissue from donors were perfused with Stig Steen solution and kept cool on ice until dissection. Bronchial specimens were taken from the fourth or fifth bifurcation and alveolar parenchymal specimens from explants were collected 2–3 cm from the pleura in the lower lobes. All specimens were immediately frozen in liquid nitrogen to prevent RNA degradation.
RNA extraction
RNA was extracted from ∼3 × 3 × 7 mm sized tissue specimens. The tissue was homogenized in a Tissue Lyser II (Qiagen Nordic, Sollentuna, Sweden) with one prechilled steel ball for 30 sec at 2000 rpm. Thereafter, 1 mL TRIZOL reagent (Life Technologies, Carlsbad, CA) was added to the pulverized tissue. The homogenate was then shaken a few seconds in the tissue lyser to thaw and completely dissolve the tissue powder in TRIZOL reagent. The RNA was extracted according to the TRIZOL protocol according to the manufacturer’s instructions for RNA isolation. The RNA concentration was measured using NanoDrop, Thermo Fisher Scientific, Waltham, MA.
cDNA and quantitative real-time PCR
One μg of RNA was converted into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s recommendations. cDNA was then diluted 1:20. Gene expression was quantified using custom TaqMan low-density arrays (TLDA) (Applied Biosystems). Each array consisted of a 384-well microfluidic card preloaded with primer sets and 6-FAM-labeled TaqMan probes. Each card consisted of 96 gene assays including five control assays. Two samples were run on each card in duplicates. The TLDA cards were loaded with each cDNA template mixed with 2x TaqMan Gene Expression Master Mix (Applied Biosystems) according to the manufacturer’s instructions. After centrifugation (two times 1 min at 1200 rpm) each well contained 1-μL reaction mixture, which corresponded to 1.25 ng of total RNA. The wells were sealed with a TLDA Sealer (Applied Biosystems). The Applied Biosystems 7900HT Real-Time PCR System was used to perform the real-time PCR amplification according to instructions from the manufacturer.
TLDA design
Genes were selected based on their relevance in pharmacokinetics regarding metabolism and transport of small molecular drugs. The two sets of 96 genes contained 91 genes encoding membrane transporters (set 1), 68 genes encoding drug-metabolizing enzymes (set 2), 11 transcriptional regulators (set 2), and 11 cell-type-specific genes (set 2) to account for cell-specific variations among the tissue specimens (Table S1 and S2). Each set also included five potential control housekeeping genes (HKGs): 18S rRNA (included in the TLDA array), beta Actin (ACTB), ubiquitin C (UBC), ribosomal protein, large, (RPLP), and beta-2-microglobulin (B2M) which all had been previously tested on lung tissue samples (data not shown). Expression levels of these genes were relatively constant between samples. Three of them, ACTB-Hs99999903_m1, B2M-Hs00984230_m1, and RPLP0-Hs99999902_m1, showed consistently high correlation for both gene data sets and regardless of central airways or peripheral tissue. The sample means of these three HKGs were therefore used to normalize the target genes. The probes of the target genes all spanned an gene, an exon–exon junction, except for CYP2A6- Hs00868409_s1*, CEBPA- Hs00269972_s1*, CEBPB- Hs00942496_s1*, CEBPD- Hs00270931_s1*, UGT1A1- Hs02511055_s1*, UGT1A4- Hs01655285_s1*, UGT1A7- Hs02517015_s1*, UGT1A9- Hs02516855_sH*, UGT1A10-Hs02516990_s1*, UGT2B4- Hs02383831_s1*, UGT2B10- Hs02556282_s1*, UGT2B15- Hs00870076_s1*, and UGT1A3- Hs04194492_g1* where primers and probes mapped within a single exon, and for the following assays a minor SNP allele frequency is located within the probe sequences: GSTM2- Hs00265266_g1* and SULT1A2-Hs02340929_g1*. All assays and their assay ID numbers are listed in Tables S1 and S2.
TLDA procedure
Analysis of gene expression
The detection threshold was arbitrary determined for all genes. The threshold cycle (Ct) values were determined with the RQ Manager 1.2 software (Applied Biosystems). Median Ct-value >35 was used as the cutoff for nondetected genes (Table S1 (set I) for gene encoding membrane transporters, and Table S2 (set 2) for gene encoding drug-metabolizing enzymes, transcription factors, and cell-specific markers). Ct-values for target genes were normalized to the mean Ct-value of the HKGs ACTB, B2M, and RPLP0 (average HKGs). Gene expression was calculated as (%average HKGs) = 2−(Ct(Gene of interest)−Ct(avg HKGs)) × 100 (Tables S3, S5, S7, and S11). Genes with a higher Ct-value than 35 was denoted not detectable. Expression levels correspond to average% of the expression in each group of the average of the three HKGs. Expression levels were classified according to: nondetectable (−)% < 0.05%; very low (+/−) 0.05% ≤ % < 0.1%; low (+) 0.1 ≤ % < 1%; moderate (++) 1% ≤ % < 10%; high (+++) 10% ≤ % < 100%; very high (++++) 100% ≤ % (Tables 4, S8 and S12). Variability between individuals in gene expression was evaluated by calculating the coefficient of variation (CV = ΔCt standard deviation/mean ΔCt). Fold change in% were calculated using the formula% = 2(−ΔCT sample−ΔCt control (Ct(Gene of interest)−Ct(avg HKGs)) × 100 (Table S4, S6, S10, and S13).
Table 4.
Expression levels of mRNA encoding drug-metabolizing enzymes in upper and lower airways
| Healthy | COPD | ||||
|---|---|---|---|---|---|
| Gene (protein) | Central | Peripheral | Central | Peripheral | |
| Carboxyl esterase | |||||
| High/moderate | CES4A (CES4A) | ++ | ++ | + | + |
| Low | CES2 (CES2) | + | + | + | + |
| CES3 (CES3) | + | + | +/− | + | |
| Not detected | CES5A (CES5A) | − | − | − | − |
| Cytochrome P450 enzymes | |||||
| High/moderate | CYP1B1 (CYP1B1) | ++ | + | ++ | ++ |
| CYP2B6 (CYP2B6) | ++ | ++ | +++ | + | |
| CYP2F1 (CYP2F1) | ++ | ++ | + | + | |
| CYP2S1 (CYP2S1) | ++ | ++ | + | + | |
| CYP4B1 (CYP4B1) | +++ | +++ | +++ | +++ | |
| Low | CYP2A6 (CYP2A61) | + | + | + | +/− |
| CYP2A13 (CYP2A131) | + | + | − | − | |
| CYP2C9 (CYP2C91) | + | − | − | − | |
| CYP2J2 (CYP2J2) | + | + | + | +/− | |
| CYP2E1 (CYP2E1) | + | + | + | + | |
| Very low/not detected | CYP1A1 (CYP1A1) | − | + | − | − |
| CYP1A2 (CYP1A2) | − | + | − | − | |
| CYP2C19 (CYP2C19)1 | +/− | + | − | − | |
| CYP2D6 (CYP2D6)1 | − | − | − | − | |
| CYP3A4(CYP3A4) | − | − | − | − | |
| CYP3A5 (CYP3A5) | +/− | − | + | +/− | |
| CYP4F2 (CYP4F2) | − | − | − | − | |
| Epoxide hydrolases | |||||
| High/moderate | EPHX1 (EPHX1) | +++ | ++ | ++ | ++ |
| Low | EPHX2 (EPHX2) | + | + | + | + |
| EPHX3 (EPHX3) | + | +/− | +/− | +/− | |
| Very low/not detected | EPHX4 (EPHX4) | +/− | +/− | − | − |
| Flavin monooxygenases | |||||
| High/moderate | FMO2 (FMO2) | ++ | ++ | ++ | ++ |
| FMO5 (FMO5) | +++ | ++ | +++ | ++ | |
| Low | FMO3 (FMO3) | + | + | + | + |
| FMO4 (FMO4) | + | + | + | + | |
| Very low/not detected | FMO1 (FMO1) | +/− | +/− | − | − |
| Glutathione S-transferases | |||||
| High/moderate | GSTA1 (GSTA1) | +++ | ++ | + | ++ |
| GSTA2 (GSTA2) | ++ | ++ | + | + | |
| GSTA4 (GSTA4) | ++ | ++ | ++ | ++ | |
| GSTK1 (GSTK1) | +++ | +++ | ++ | ++ | |
| GSTM2 (GSTM2)1 | ++ | ++ | ++ | ++ | |
| GSTM3 (GSTM3)1 | ++ | + | ++ | + | |
| GSTO1 (GSTO1) | ++ | ++ | ++ | ++ | |
| GSTP1 (GSTP1) | ++++ | ++++ | ++++ | +++ | |
| Low | GSTA3 (GSTA3) | + | + | +/− | +/− |
| GSTM4 (GSTM4)1 | + | + | + | + | |
| GSTM5 (GSTM5)1 | + | + | + | + | |
| GSTO2 (GSTO2) | + | + | +/− | +/− | |
| GSTT1 (GSTT1)1 | + | − | + | +/− | |
| GSTZ1 (GSTZ1) | + | + | + | + | |
| Very low/not detected | GSTA5 (GSTA5) | − | − | − | − |
| GSTM1 (GSTM1)1 | − | − | − | − | |
| Arylamine N-acetyltransferase | |||||
| Not detected | NAT1 (NAT1)1 | − | − | − | − |
| NAT2 (NAT2)1 | − | − | − | − | |
| SULFO-transferases | |||||
| High/moderate | SULT2B1 (SULT2B1) | ++ | ++ | ++ | ++ |
| Low | SULT1A2 (SULT1A2) | + | +/− | + | + |
| SULT1C4 (SULT1C4) | + | + | + | + | |
| Very low/not detected | SULT1A1 (SULT1A1) | − | − | − | − |
| SULT1C2 (SULT1C2) | − | +/− | − | +/− | |
| SULT4A1 (SULT4A1) | − | − | − | − | |
| SULT6B1 (SULT6B1) | − | − | − | − | |
| UDP-glycosyl-transferases | |||||
| Low | UGT1A1 (UGT1A1) | + | + | + | − |
| UGT1A7 (UGT1A7) | + | + | + | − | |
| UGT2A1 (UGT2A1) | + | +/− | − | − | |
| Very low/not detected | UGT1A3 (UGT1A3) | − | + | − | − |
| UGT1A4 (UGT1A4) | − | + | − | − | |
| UGT1A6 (UGT1A6) | − | +/− | − | − | |
| UGT1A8 (UGT1A8) | − | − | − | − | |
| UGT1A9 (UGT1A9) | +/− | + | +/− | − | |
| UGT1A10 (UGT1A10) | +/− | + | +/− | − | |
| UGT2B4 (UGT2B4) | − | − | − | − | |
| UGT2B7 (UGT2B7) | − | − | − | − | |
| UGT2B10 (UGT2B10) | − | − | − | − | |
| UGT2B15 (UGT2B15) | − | − | − | − | |
Average expression levels within each group expressed in% of the expression of the housekeeping genes ACTB, B2M, and RPLP0 (avg HKGs). Expression was calculated as (% avg HKGs) = 2−(Ct(Gene of interest)−Ct(avg HKGs)) × 100. Nondetectable (−)% < 0.05; very low (+/−) 0.05 ≤ % < 0.1; low (+) 0.1 ≤ % t < 1; moderate (++) 1 ≤ % < 10, high (+++) 10 ≤ % t < 100; very high (++++) 100 ≤ %. The genes were sorted by their expression in the central region of healthy individuals.
Null allele, expression level calculated only on subjects harboring this allele.
Statistical analysis
Principal component analysis (PCA) was applied separately to each of the two gene data sets (to check for outliers, to evaluate sources of variation (within subject duplicates and between subjects), and to look for indications of separation of specific groups). Missing data in the PCA were handled by removing those genes that had more than 50% missing values and setting the remaining missing Ct-values to 35. Difference in gene expression between groups (COPD versus healthy and central airways versus peripheral tissue) was tested using a linear mixed effect model with random effects for subject and for biopsy within subject. The genes that had at least one nonmissing Ct-value for two or more COPD patients and for two or more healthy donors were selected for testing. In the two data sets, 71% and 75%, respectively, of the genes fulfilled the criteria. For each group comparison, the empirical distribution of the P-values of the tested genes was calculated. If the distribution shows a peak for the lower, “significant”, P-values, this is an indication that there are more findings of differences that can be explained by chance alone. Using the method of Storey (Storey and Tibshirani 2003) a cutoff for “significant” P-value was set, and the genes with P-values below the cutoff were defined as the genes of interest. The false discovery rate (FDR) was then calculated, providing an estimate of how many of the significant genes that are likely to be false positives. All analyses were performed in R, version 2.10.1.
Results
Expression of genes encoding membrane transporters in human lung
Of the 91 analyzed genes encoding membrane transporters, 67 genes (74%) were expressed in either central or peripheral region in either healthy or COPD subjects, while 24 genes were not detected at all. Of the 67 genes being expressed, 32 were classified as expressed at high or moderate levels, defined as expressed above 1% of the average HKGs (HKGs) (+++/++) (Table 2).
Table 2.
Expression levels of mRNA encoding membrane transporters in human lung
| Healthy | COPD | ||||
|---|---|---|---|---|---|
| Gene (protein) | Central | Peripheral | Central | Peripheral | |
| ABC transporters | |||||
| High/moderate | ABCA1 (ABCA1) | ++ | ++ | ++ | ++ |
| ABCA3 (ABCA3) | ++ | +++ | ++ | +++ | |
| ABCA5 (ABCA5) | ++ | ++ | ++ | + | |
| ABCA6 (ABCA6) | ++ | ++ | ++ | ++ | |
| ABCA9 (ABCA9) | ++ | ++ | ++ | ++ | |
| ABCB10 (ABCB10) | ++ | + | + | + | |
| ABCB6 (ABCB6) | ++ | ++ | ++ | + | |
| ABCB7 (ABCB7) | ++ | ++ | ++ | ++ | |
| ABCB8 (ABCB8) | ++ | ++ | ++ | + | |
| ABCC10 (MRP7) | ++ | ++ | ++ | + | |
| ABCC1 (MRP1) | ++ | ++ | ++ | + | |
| ABCC6 (MRP6) | ++ | ++ | + | + | |
| ABCC9 (ABCC9) | ++ | ++ | + | ++ | |
| ABCG1 (ABCG1) | ++ | ++ | ++ | ++ | |
| Low | ABCA10 (ABCA10) | + | + | + | + |
| ABCA2 (ABCA2) | + | + | + | + | |
| ABCA7 (ABCA7) | + | + | + | + | |
| ABCB1 (MDR1) | + | + | + | + | |
| ABCB9 (ABCB9) | + | + | + | +/− | |
| ABCC3 (MRP3) | + | + | + | + | |
| ABCC4 (MRP4) | + | ++ | + | + | |
| ABCC5 (MRP5) | + | + | + | + | |
| ABCC7 (CFTR) | + | + | + | ++ | |
| ABCC8 (ABCC8) | + | ++ | − | − | |
| ABCG2 (BCRP) | + | +/− | +/− | + | |
| Very low/not detected | ABCA4 (ABCA4) | +/− | − | +/− | − |
| ABCA12(ABCA12) | − | − | + | − | |
| ABCB4 (ABCB4) | − | − | − | − | |
| ABCB5 (ABCB5) | − | − | − | − | |
| ABCC11 (MRP8) | − | − | − | − | |
| ABCC12 (MRP9) | − | − | − | − | |
| ABCC2 (MRP2) | − | +/− | − | − | |
| ABCG4 (ABCG4) | − | − | − | − | |
| ABCG5 (ABCG5) | − | − | − | − | |
| ABCG8 (ABCG8) | − | − | − | − | |
| Solute carrier transporters | |||||
| High/moderate | Slc15A2 (PEPT2) | ++ | ++ | ++ | + |
| SLC19A2 (THT2) | ++ | ++ | + | ++ | |
| SLC22A3 (OCT3) | ++ | ++ | ++ | ++ | |
| SLC22A4 (OCTN1) | ++ | + | ++ | + | |
| SLC29A1 (ENT1) | ++ | ++ | ++ | ++ | |
| SLC2A1 (GLUT1) | ++ | + | ++ | + | |
| SLC31A1 (CTR1) | ++ | + | ++ | ++ | |
| SLC31A2 (CTR2) | ++ | ++ | ++ | ++ | |
| SLC36A4 (PAT4) | ++ | ++ | ++ | ++ | |
| SLC3A2 (4F2HC) | ++ | ++ | ++ | ++ | |
| SLC6A14 (ATB(0+)) | ++ | + | ++ | ++ | |
| SLC6A4 (SERT) | ++ | ++ | + | ++ | |
| SLC7A1 (CAT1) | ++ | ++ | ++ | + | |
| SLC7A6 (LAT-2, LAT3, y+LAT-2) | ++ | ++ | ++ | + | |
| SLC7A8 (LAT2, LPI-PC1) | ++ | ++ | + | + | |
| Low | SLC16A1 (MCT1) | + | + | + | + |
| SLC16A3 (MCT3) | + | + | + | + | |
| SLC16A4 (MCT4) | + | ++ | + | ++ | |
| SLC19A1 (RFP1) | + | ++ | + | + | |
| SLC19A3 (THTR2) | + | ++ | + | ++ | |
| SLC22A5 (OCTN2) | + | + | + | + | |
| SLC28A3 (CNT3) | + | +/− | + | +/− | |
| SLC29A2 (ENT2) | + | + | + | + | |
| SLC29A3 (ENT3) | + | ++ | + | + | |
| SLC29A4 (ENT4) | + | + | + | +/− | |
| SLC47A1 (MATE1) | + | + | + | + | |
| SLC51A (OSTalpha) | + | + | ++ | +/− | |
| SLC51B (OSTbeta) | + | + | + | + | |
| SLC7A11 (SLC7A11) | + | +/− | + | + | |
| SLC7A5 (LAT1) | + | + | + | + | |
| Very low/not detected | SLC15A1 (PEPT1) | − | − | − | − |
| SLC16A7 (MCT2) | − | − | − | − | |
| SLC22A11 (OAT4) | − | − | − | − | |
| SLC22A1 (OCT1) | +/− | + | +/− | +/− | |
| SLC22A2 (OCT2) | − | − | − | − | |
| SLC22A6 (OAT1) | − | − | − | − | |
| SLC22A7 (OAT2) | − | − | − | − | |
| SLC22A8 (OAT3) | − | − | − | − | |
| SLC22A9 (UST3H) | − | − | − | − | |
| SLC28A1 (CNT1) | − | − | − | − | |
| SLC28A2 (CNT2) | − | − | − | − | |
| SLC47A2 (MATE2) | − | − | − | − | |
| SLC6A2 (NAT) | − | − | − | − | |
| SLC6A3 (DAT) | − | − | − | − | |
| Solute carrier organic anion transporters | |||||
| High/moderate | SLCO2B1 (OATP2B1) | ++ | ++ | ++ | ++ |
| SLCO4A1 (OATP4A1) | ++ | + | ++ | + | |
| Low | SLC01A2 (OATP1A2) | + | +/− | +/− | + |
| SLCO3A1 (OATP3A1) | + | + | + | + | |
| SLCO4C1 (OATP4C1) | + | + | + | + | |
| SLCO5A1 (OATP5A1) | + | + | + | +/− | |
| Very low/not detected | SLCO1B1 (OATP1B1) | − | − | − | − |
| SLCO1B3 (OATP1B3) | − | − | − | − | |
| SLCO1C1 (OATP1C1) | − | − | − | − | |
| SLCO6A1 (OATP6A1) | − | − | − | − | |
| Copper-transporting ATPases | |||||
| High/moderate | ATP7B (Copper transporter) | ++ | ++ | + | + |
| Low | ATP7A (Copper transporter) | + | + | + | + |
Average expression levels within each group expressed in% of the expression of the housekeeping genes ACTB, B2M, and RPLP0 (avg HKGs). Expression was calculated as (% avg HKGs) = 2−(Ct(Gene of interest)−Ct(avg HKGs)) × 100. Nondetectable (−)% <0.05; very low (+/−) 0.05 ≤ % < 0.1; low (+) 0.1 ≤ % t < 1; moderate (++) 1 ≤ % < 10; high (+++) 10 ≤ % t < 100; very high (++++) 100 ≤ %. The genes were sorted by their expression in the central region of healthy individuals.
To unravel disease- or region-specific differences in expression levels of genes encoding membrane transporters, an exploratory multivariate analysis using PCA was first performed followed by univariate tests within the linear mixed model. The PCA showed no differences in mRNA expression between biopsies from healthy controls and COPD. However, when ignoring health status, a distinct difference in mRNA expression of genes encoding membrane transporters between central airways and peripheral tissue could be detected (Fig. 1A). The formal statistical tests of differences in gene expression levels confirmed the pattern indicated by the exploratory multivariate analyses. The FDR for the top 16 genes with a P<0.01 was estimated to be 0.36, thus an expected number of false positive within this group is less than 1 (Fig. 1B). Among these genes, 11 mRNAs were higher expressed in the central airways and five mRNAs were higher expressed in the peripheral region independent of health status (Table 3). The individual graphs of four genes, SLC2A1 (GLUT1), SLC28A3 (CNT3), and SLC22A4 (OCTN1) with the highest expression in central airways, and SLC47A1 (MATE1) with the highest difference between peripheral tissue to central airways are shown in Figure 2 and individual graphs of top 5–16 genes with differential expression pattern are shown in Figure S1.
Figure 1.
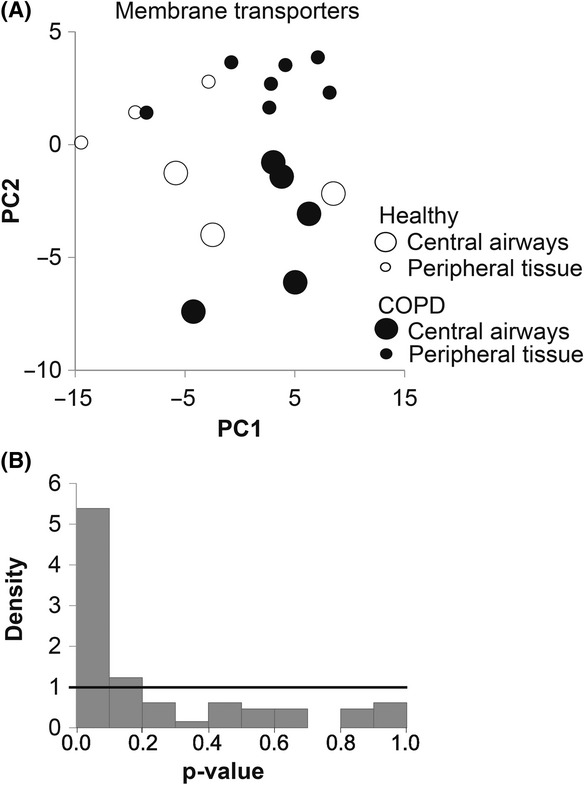
Differential expression of membrane transporters between central and peripheral airways. (A) The relationship between the first two principle components did not indicate any distinct grouping of the data due to differences in expression between healthy lung (unfilled circles) and COPD lung (black circles). However, a separation was seen between central airways (large circles) and peripheral airways (small circles) irrespectively of health status. (B) The distribution of P-values showed a high peak for the lower values, indicating that there are more significances than expected by chance between central and peripheral airways for membrane transporters.
Table 3.
Differential expressed genes encoding membrane transporters between central airways and peripheral tissue
| P-value | ||
|---|---|---|
| Central > peripheral | ||
| 1 | SLC2A1 (GLUT 1) | 0.00001 |
| 2 | SLC28A3 (CNT3) | 0.0003 |
| 3 | SLC22A4 (OCTN1) | 0.0006 |
| 4 | SLC51A (OSTalpha) | 0.0009 |
| 5 | ABCA4 (ABCA4) | 0.0017 |
| 6 | SLC15A2 (PEPT2) | 0.0018 |
| 7 | ATP7B (Copper transporter) | 0.0030 |
| 8 | ABCB6 (ABCB6) | 0.0030 |
| 9 | ABCA5 (ABCA5) | 0.0031 |
| 10 | SLC31A1 (CTR1) | 0.0071 |
| 11 | SLC7A1 (CAT1) | 0.0087 |
| Peripheral > central | ||
| 1 | SLC47A1 (MATE1) | 0.0006 |
| 2 | SLCO4C1 (OATP4C1) | 0.0017 |
| 3 | SLCO2B1 (OATP2B1) | 0.0025 |
| 4 | ABCA3 (ABCA3) | 0.0039 |
| 5 | ABCC4 (MRP4) | 0.0092 |
The P-values are unadjusted P-values from two-sided tests in the linear mixed model. The set of genes correspond to a FDR < 0.5.
Figure 2.
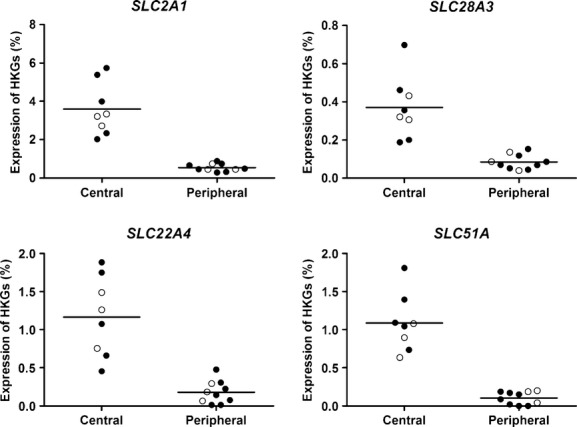
Individual expression pattern of top four differential expressed genes encoding membrane transporters. Graphs show expression levels of individual genes encoding membrane transporters with the lowest P-values (unadjusted and from two-sided tests) regarding differential expression between central airways and peripheral tissue. Black circles denote COPD patients and unfilled circles denote healthy subjects.
To investigate the interindividual variability in expression among subjects, the CV were calculated for the expressed genes. Variability in expression of genes encoding membrane transporters between subjects could determine individual susceptibility for smoking-induced lung diseases as well as differential responses to treatment strategies. The majority of the genes showed a low interindividual variability below 5% (Table S4).
Expression of genes encoding drug-metabolizing enzymes in human lung
Analysis of gene expression encoding drug-metabolizing enzymes showed that 53 of the 69 investigated genes (77%) were expressed in the lung to a varying degree (Table 4). Of these 53 genes 18 genes were classified as expressed to a high or moderate level (>1% of the expression of the average HKGs.
Similar to the results obtained for membrane transporters, no differences in gene expression between healthy and COPD subjects were detected when tissue specimens from central and peripheral regions were included in the analysis (Fig. 3A). Likewise, a separation of mRNA expression between the central and peripheral respiratory regions could be detected regardless of health status (Fig. 3A). In this analysis, genes encoding drug-metabolizing enzymes, as well as cell-specific markers, and transcriptional regulators were investigated. Among the 15 highest expressed genes with a P < 0.007, the FDR was 0.21 (Fig. 3B). The majority of these genes were higher expressed in the central airways compared to that in the peripheral lung tissue (Table S9). Of the genes encoding drug-metabolizing enzymes, all differentially expressed genes were higher expressed in the central airways (Table 5). The individual expression patterns of these genes with a statistically significant difference (P < 0.01) are presented in Figure S2 and the genes with highest significant difference between central and peripheral tissue GSTZ1, GSTO2, CYP2F1, and CYP2J2 are shown in Figure 4.
Figure 3.
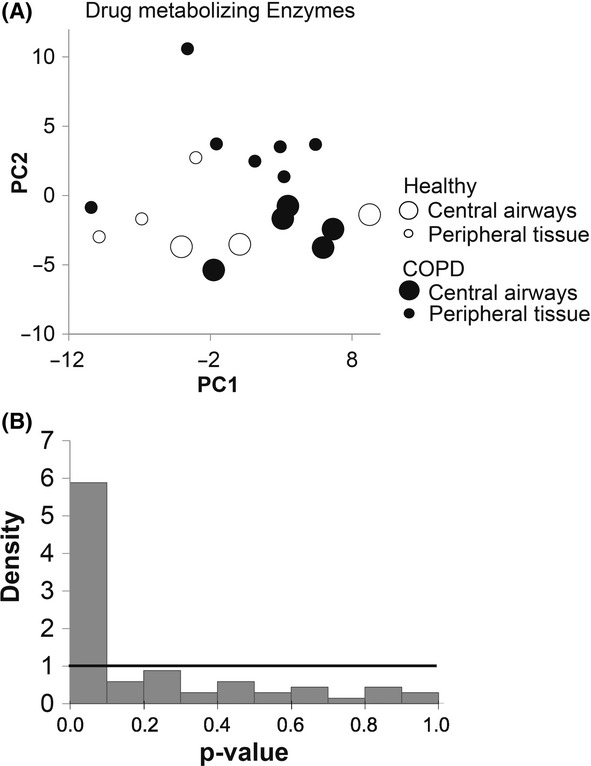
Differential expression of drug-metabolizing enzymes between central and peripheral airways. (A) The relationship between the first two principle components did not indicate any distinct grouping of the data due to differences in expression between healthy lung (unfilled circles) and COPD lung (black circles). However, a separation is seen between central airways (large circles) and peripheral airways (small circles) irrespectively of health status. (B) Distribution of p-values showed a high peak for the lower values, indicating that there are more significances than expected by chance between central and peripheral airways for metabolizing enzymes and transcriptional regulators.
Table 5.
Differential expressed genes encoding metabolizing enzymes between central airways and peripheral tissue
| P-value | ||
|---|---|---|
| Central > peripheral | ||
| 1 | GSTZ1 | 0.0006 |
| 2 | GST02 | 0.0007 |
| 3 | CYP2F1 | 0.0010 |
| 4 | CYP2J2 | 0.0019 |
| 5 | CYP2S1 | 0.0027 |
| 6 | CES4A | 0.0029 |
| 7 | GSTA3 | 0.0038 |
| 8 | FMO4 | 0.0039 |
| 9 | CES3 | 0.0039 |
| 10 | GSTP1 | 0.0047 |
| 11 | GSTA1 | 0.0052 |
| 12 | CYP2C9 | 0.0062 |
| 13 | GSTA2 | 0.0099 |
The P-values are unadjusted P-values from two-sided tests in the linear mixed model. The set of genes correspond to a FDR < 0.5.
Figure 4.
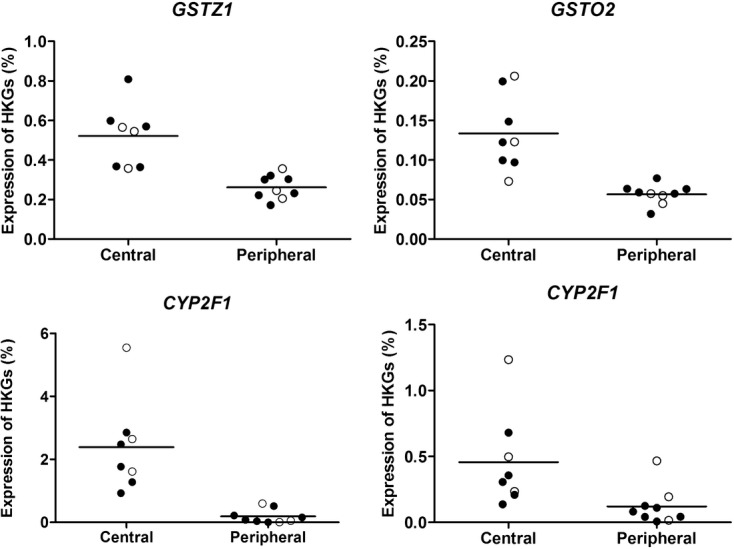
Individual expression pattern of top four genes encoding drug-metabolizing enzymes with higher expression in central airways. Graphs show expression levels of individual genes encoding drug-metabolizing enzymes with the lowest P-values (unadjusted and from two-sided tests) regarding differential expression between central airways and peripheral tissue. Black circles denote COPD patients and unfilled circles denote healthy subjects.
Similar to our results for membrane transporter, about 67% of the expressed genes encoding drug-metabolizing enzymes showed a low interindividual variability (CV < 5%). The highest frequency of mRNAs with an interindividual variation above 5% (CV > 5%) was found within the healthy subjects, where 32% of the genes expressed in the central airways and 26% of the genes expressed in the peripheral tissue genes showed a CV higher than 5% (Table S6).
Expression of genes encoding transcriptional regulators in the human lung
In an attempt to unravel the expression pattern of common regulatory pathways in the lung, we investigated the transcriptional regulators NR1H4 encoding the farnesoid X receptor (FXR), NR1l12 encoding CAR, NR1l2 encoding PXR, NR3C1 encoding the glucocorticoid receptor, several genes encoding proteins involved in the aryl hydrocarbon receptor (AHR) pathway known to be induced by cigarette smoking, and members of the CCAAT enhancer binding protein family. However, no clear correction between these transcriptional regulators and mRNA encoding drug-metabolizing enzymes or membrane transporters were observed. In addition, no differential expression between COPD and healthy subjects could be determined for these transcriptional regulators, but a higher expression of CEBPA, CEBPB, and CEBPD was found in peripheral lung tissue compare to central airways (P = 0.0071, 0.0090, and 0.0016, respectively) (Table S9).
Discussion
The clinical importance of drug transporters in drug disposition has recently been highlighted leading to an extensive update of regulatory guidelines from the European Medicines Agency (EMA) and the US Food and Drug Administration (FDA) (Giacomini et al. 2010). However, compared to the extensive knowledge on membrane transporters and drug-metabolizing enzymes in liver, intestine, kidney, and their role in drug pharmacokinetics of orally administered drugs, their expression in healthy and diseased lung is unexplored. This study describes for the first time the regional expression levels of genes encoding membrane transporters and drug-metabolizing enzymes in lungs from ex-smokers with severe COPD.
While no differential expression of drug transporters and metabolizing enzymes was found between healthy and COPD subjects, significant differences in mRNA expression between central airways and peripheral tissue were found when health status was ignored. The majority of these differentially expressed genes were higher expressed in the central region of the lung. This expression pattern likely reflect the different cell populations and functions represented in the different regions, where the central region represents bronchial epithelium, smooth muscle layers, seromucous glands, and isolated islands of cartilage as well as some alveolar tissue whereas the peripheral region represents alveolar and bronchiolar structures. Knowledge of these regional differences could be valuable for drug design since small particles (1–2 μm) are probably deposited in the alveolar region, whereas larger particles are more likely to deposit in the central regions of the lung (Byron 1986; Martin 2002; Liu et al. 2013). As irreversible airflow obstruction, reduced airway diameters, and emphysema occurring in COPD are suggested to increase the deposition of inhaled drug particles to the central airways compared to that in healthy individuals, it is possible that these regional differences could play an even more important role in drug delivery and treatment efficacy in COPD patients (Darquenne 2012). One of the genes identified to be higher expressed in central airways and with potential importance in COPD was the solute carrier SLC22A4 encoding the organic cation transporter OCTN1, as it transports bronchodilators such as muscarinic antagonists and β2-agonists, which together with corticosteroids are the main treatment of COPD patients (Nakamura et al. 2010). These results were in line with previously published data showing higher expression of OCTN1 in tracheal epithelium then in alveolar tissue in healthy individuals (Horvath et al. 2007), validating our results. Other members of this family, that is, SLCO1B1 (OATP1B1) and SLCO1B3 (OATP1B3), and SLC22A2 (OCT2), SLC22A6 (OAT1), and SLC22A8 (OAT3), were recently denoted particular clinically important in hepatic and renal clearance, respectively (Giacomini et al. 2010). Interestingly, all of these solute carriers were absent from lung and instead SLC15A2 (PEPT2), SLC22A3 (OCT3), SLC22A4 (OCTN1), SLCO2B1 (OATP2B1), and SLCO4A1 (OATP4A1) were dominant, displaying a lung-specific profile that could be important in local pharmacokinetics of inhaled drugs.
Additional genes of potential interest for COPD treatment are genes encoding multidrug resistance (MDR) like proteins. Even here, a different profile of expression was confirmed in lung compared to other organs. ABCB1 (MDR1) and ABCG2 (BCRP) are pointed out as functionally important for drug absorption, clearance, and drug distribution to the brain, and is also highly expressed in intestine, liver, and kidney but known to be rather low in healthy lung. This correlated well with the low expression in detected in the COPD lung (Seree et al. 1998; van der Deen et al. 2005). Still as ABCB1 is known to be repressed by glucocorticoids and all subjects in the study have been treated with glucocorticoids prior to the lung transplant surgery and sampling of tissue specimens, these results might reflect the glucocorticoid therapy rather than the true levels in this study (Seree et al. 1998). ABCC1 is another gene in this family encoding the multidrug resistance-associated protein 1 (MRP1) that is proposed to be protective against development of COPD although it has been described to be decreased in smokers with severe COPD as well as by cigarette smoke (Cole et al. 1992; Bandi and Kompella 2002; van der Deen et al. 2007, 2008; Budulac et al. 2012). However, in this study, it was expressed at high levels in both central and peripheral regions and no disease-specific differences were found, possible reflecting an effect of cigarette smoke rather than disease progression. Additional studies on protein levels will be needed to further investigate the impact of MRP1 and drug resistance in the different subgroups of COPD patients.
Both phase I and phase II drug-metabolizing enzymes are known to be expressed in lung tissue, although the overall metabolic rate is lower compared to that in liver (Somers et al. 2007). However, in the lung, the CYP expression profile is different from that in the liver. In the liver, the phase I enzymes CYP1A2, 2C9, 2C19, 2D6, and 3A4 are considered to be of most importance for drug metabolism based on the relative expression level and broad substrate specificity (Shimada et al. 1994; Raunio et al. 1999; Hukkanen et al. 2002; Ding and Kaminsky 2003; Zhang et al. 2006). In the lung, instead CYP1A1, CYP1B1, CYP2B6, CYP2E1, and CYP2J2 are often is reported to have the highest metabolic rates and to be the most abundant enzymes at both mRNA and protein levels. Our results in both healthy and COPD lung are in line with these previous findings except for CYP1A1 coherent with CYP1A1 being heavily induced by cigarette smoke and that our subjects are ex-smokers and nonsmokers (Kim et al. 2004). Additionally, we found high levels of CYP4B1 in both healthy and COPD subjects in both central and peripheral region as well as high expression levels of CYP2F1 and CYP2S1 specifically in the central region. The regional differences found for CYP2S1 are novel for both healthy and COPD subjects, and the results for CYP4B1 and CYP2F1 are in agreement with previously published mRNA data for healthy subjects, validating our results (Leclerc et al. 2011). Furthermore, the phase II enzymes GSTs were expressed to a higher degree than the phase I enzymes in both healthy and COPD lung. This is in contrast to the liver where phase I and II enzymes are expressed to a similar high degree (Bauer et al. 2006; Somers et al. 2007; Courcot et al. 2012). This is in agreement with previous observations in lung tissue and cell lines (Bauer et al. 2006; Leclerc et al. 2011; Courcot et al. 2012). Phase II enzymes have been proposed to have a protective role against the development of COPD by detoxifying toxic compounds in tobacco smoke. Thus, even though the lung has not been found to play a major role in the systemic metabolism of drugs, it is plausible that the high expression of phase II enzymes could take part in the development of COPD and in pharmacokinetics of inhaled drugs.
The tissue specimen used for this study was dissected from transplanted lung from COPD patients and healthy donor lungs during highly standardized procedures and from specific regional locations, thus it is a very rare and limited material. Despite this limitation, we did detect significant regional differences between central airways and peripheral tissue. The mRNA expression in the COPD subjects correlated very well with the expression in the healthy subjects and was also in line with previous findings for membrane transporters and drug-metabolizing enzymes in healthy human lung tissue or cells (Bleasby et al. 2006; Leclerc et al. 2011; Courcot et al. 2012). This strengthens the results and suggests that drug pharmacokinetic mechanisms could be maintained in COPD lung and that previous differences found in COPD subjects are related to cigarette smoke and not to the tissue destruction and inflammation (Pierrou et al. 2007). However, we cannot exclude that additional regional or disease-specific differences do exist. Differences in expression might be suppressed by medication taken in conjunction with the transplant surgery prior to the sampling of the tissue specimens, or other components of this complex disease in this very severe stage. The tissue specimen also represents a mixed cell population that could mask any potential cell-specific differences related to COPD. Similar studies on a larger subset of individuals in combination with protein expression analysis in different cell types and functional analysis would thus be useful to further unravel if disease-specific differences exist in individual cell types and their functional importance for pharmacokinetics in the COPD lung. This study also highlights the fact that although some expression patterns are shared between lung and other organs, there are also some distinguishable differences in the expression of transporters and drug-metabolizing enzymes in the pulmonary barrier that may be of relevance for the local pharmacokinetics of inhaled drugs and that possibly could aid the development of more specific drugs to COPD or to avoid organ-specific side effects. Thus, our data could be of potential interest to COPD drug therapy and add value in predictive modeling of disposition, uptake, and metabolism of inhaled drugs in this patient group, as well as for the design of new chemical entities targeting either central or peripheral regions.
Acknowledgments
The authors specifically would like to thank Ann Tronde for the enthusiasm and engagement in the initiation phase of this project. Additionally, Robert Virtala, Martina Kvist Reimer, and Constanze Hilgendorf, Astra Zeneca for valuable advice and Magnus Dahlbäck, AstraZeneca for selection of patients and collection of tissue specimen.
Glossary
- ABC
ATP-binding cassette
- AHR
aryl hydrocarbon receptor
- AHRR
AHR repressor
- AIP
AH receptor-interacting protein
- ARNT
Aryl hydrocarbon receptor nuclear translocator
- AW
airway
- CAR
constitutive androstane receptor
- CES
Carboxylesterase enzymes
- COPD
chronic obstructive pulmonary disease
- Ct
threshold cycle
- CV
coefficient of variation
- CYP
cytochrome P450
- DDI
drug–drug interactions
- EMA
European Medicines Agency
- EPX
epoxide hydrolase
- FDA
Food and Drug Administration
- FDR
false discovery rate
- FEV1
Forced expiratory volume in one second
- FMO
Flavin-containing monooxygenase
- FVC
Forced vital capacity
- FXR
farnesoid X receptor
- GST
glutathione-S-transferase
- HKG
housekeeping gene
- IL
Interleukin
- INF
Interferon
- LPS
lipopolysaccharide
- MDR
multidrug resistance transporter
- MRP
multidrug resistance-related protein
- NAT
N-acetyl-transferases
- OST
organic solute transporter
- PAH
polycyclic aromatic hydrocarbons
- PCA
principal component analysis
- PXR
pregnane X receptor
- qPCR
Quantitative real-time PCR
- RXR
retinoid X receptor
- SLCO
solute organic and steroid transporters
- SLC
solute carrier transporters
- SULT
sulfo-transferase
- TLDA
TaqMan low-density arrays
- TNF
tumor necrosis factor
- UGT
UDP-glucuronosyl transferase
Disclosures
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Individual expression pattern of differential expressed genes encoding membrane transporters. Graphs show expression levels of individual genes encoding membrane transporters regarding differential expression between central airways and peripheral tissue. Black circles denote COPD patients and unfilled circles denote healthy subjects.
Figure S2. Individual expression pattern of differential expressed genes encoding drug-metabolizing enzymes. Graphs show expression levels of individual genes encoding drug-metabolizing enzymes regarding differential expression between central airways and peripheral tissue. Black circles denote COPD patients and unfilled circles denote healthy subjects.
Table S1. TLDA probes and median Ct-values for membrane transporters and housekeeping genes in healthy and COPD lung.
Table S2. TLDA probes and median Ct-values for drug-metabolizing enzymes, transcription factors, and cell-specific markers in healthy and COPD lung.
Table S3. Gene expression of membrane transporter from healthy and COPD lung in % of expression of housekeeping genes.
Table S4. Interindividual variability of expressed genes encoding membrane transporters in healthy or COPD human lung in central or peripheral airways.
Table S5. Gene expression of drug-metabolizing enzymes from healthy and COPD lung in % of expression of housekeeping genes.
Table S6. The interindividual variability among genes encoding metabolizing enzymes with in healthy or COPD human lung from both central airways and peripheral tissue.
Table S7. Expression of genes encoding transcription factors from healthy and COPD lung in % of expression of housekeeping genes.
Table S8. Expression of genes encoding transcription factors from healthy and COPD lung in % of expression of housekeeping genes.
Table S9. Differential expressed genes encoding metabolizing enzymes between central airways and peripheral tissue.
Table S10. Interindividual variability of genes encoding transcription factors in healthy or COPD human lung in both central airways and peripheral tissue.
Table S11. Expression of genes encoding cell-specific markers in % of expression of housekeeping genes.
Table S12. Expression of genes encoding cell-specific markers in % of expression of housekeeping genes.
Table S13. Interindividual variability among genes encoding cell-specific markers within healthy or COPD human lung in both central airways and peripheral tissue.
References
- Aitken AE, Richardson TA, Morgan ET. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol. 2006;46:123–149. doi: 10.1146/annurev.pharmtox.46.120604.141059. [DOI] [PubMed] [Google Scholar]
- Bandi N, Kompella UB. Budesonide reduces multidrug resistance-associated protein 1 expression in an airway epithelial cell line (Calu-1) Eur J Pharmacol. 2002;437:9–17. doi: 10.1016/s0014-2999(02)01267-0. [DOI] [PubMed] [Google Scholar]
- Bauer M, Herbarth O, Aust G, Hengstler JG, Dotzauer A, Graebsch C, et al. Expression patterns and novel splicing variants of glutathione-S-transferase isoenzymes of human lung and hepatocyte cell lines. Cell Tissue Res. 2006;324:423–432. doi: 10.1007/s00441-005-0150-8. [DOI] [PubMed] [Google Scholar]
- Bleasby K, Castle JC, Roberts CJ, Cheng C, Bailey WJ, Sina JF, et al. Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: a resource for investigations into drug disposition. Xenobiotica. 2006;36:963–988. doi: 10.1080/00498250600861751. [DOI] [PubMed] [Google Scholar]
- Bosquillon C. Drug transporters in the lung–do they play a role in the biopharmaceutics of inhaled drugs? J Pharm Sci. 2010;99:2240–2255. doi: 10.1002/jps.21995. [DOI] [PubMed] [Google Scholar]
- Budulac SE, Postma DS, Hiemstra PS, Lapperre TS, Kunz LI, Vonk JM, et al. Multidrug resistance-associated protein 1 and lung function decline with or without long-term corticosteroids treatment in COPD. Eur J Pharmacol. 2012;696:136–142. doi: 10.1016/j.ejphar.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Byron PR. Prediction of drug residence times in regions of the human respiratory tract following aerosol inhalation. J Pharm Sci. 1986;75:433–438. doi: 10.1002/jps.2600750502. [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- Courcot E, Leclerc J, Lafitte JJ, Mensier E, Jaillard S, Gosset P, et al. Xenobiotic metabolism and disposition in human lung cell models: comparison with in vivo expression profiles. Drug Metab Dispos. 2012;40:1953–1965. doi: 10.1124/dmd.112.046896. [DOI] [PubMed] [Google Scholar]
- Darquenne C. Aerosol deposition in health and disease. J Aerosol Med Pulm Drug Deliv. 2012;25:140–147. doi: 10.1089/jamp.2011.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Deen M, de Vries EG, Timens W, Scheper RJ, Timmer-Bosscha H, Postma DS. ATP-binding cassette (ABC) transporters in normal and pathological lung. Respir Res. 2005;6:59. doi: 10.1186/1465-9921-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Deen M, Timens W, Timmer-Bosscha H, van der Strate BW, Scheper RJ, Postma DS, et al. Reduced inflammatory response in cigarette smoke exposed Mrp1/Mdr1a/1b deficient mice. Respir Res. 2007;8:49. doi: 10.1186/1465-9921-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Deen M, Homan S, Timmer-Bosscha H, Scheper RJ, Timens W, Postma DS, et al. Effect of COPD treatments on MRP1-mediated transport in bronchial epithelial cells. Int J Chron Obstruct Pulmon Dis. 2008;3:469–475. doi: 10.2147/copd.s2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, et al. Membrane transporters in drug development. Nat Rev Drug Discovery. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgren O, Nihlberg K, Dahlback M, Bjermer L, Eriksson LT, Erjefalt JS, et al. Altered fibroblast proteoglycan production in COPD. Respir Res. 2010;11:55. doi: 10.1186/1465-9921-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgren O, Rolandsson S, Andersson-Sjoland A, Nihlberg K, Wieslander E, Kvist-Reimer M, et al. Enhanced ROCK1 dependent contractility in fibroblast from chronic obstructive pulmonary disease patients. J Transl Med. 2012;10:171. doi: 10.1186/1479-5876-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison TW, Tattersfield AE. Plasma concentrations of fluticasone propionate and budesonide following inhalation from dry powder inhalers by healthy and asthmatic subjects. Thorax. 2003;58:258–260. doi: 10.1136/thorax.58.3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath G, Schmid N, Fragoso MA, Schmid A, Conner GE, Salathe M, et al. Epithelial organic cation transporters ensure pH-dependent drug absorption in the airway. Am J Respir Cell Mol Biol. 2007;36:53–60. doi: 10.1165/rcmb.2006-0230OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukkanen J, Pelkonen O, Hakkola J, Raunio H. Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit Rev Toxicol. 2002;32:391–411. doi: 10.1080/20024091064273. [DOI] [PubMed] [Google Scholar]
- Kim JH, Sherman ME, Curriero FC, Guengerich FP, Strickland PT, Sutter TR. Expression of cytochromes P450 1A1 and 1B1 in human lung from smokers, non-smokers, and ex-smokers. Toxicol Appl Pharmacol. 2004;199:210–219. doi: 10.1016/j.taap.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Leclerc J, Courcot-Ngoubo Ngangue E, Cauffiez C, Allorge D, Pottier N, Lafitte JJ, et al. Xenobiotic metabolism and disposition in human lung: transcript profiling in non-tumoral and tumoral tissues. Biochimie. 2011;93:1012–1027. doi: 10.1016/j.biochi.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Liu X, Jin L, Upham JW, Roberts MS. The development of models for the evaluation of pulmonary drug disposition. Expert Opin Drug Metab Toxicol. 2013;9:487–505. doi: 10.1517/17425255.2013.754009. [DOI] [PubMed] [Google Scholar]
- Martin RJ. Therapeutic significance of distal airway inflammation in asthma. J Allergy Clin Immunol. 2002;109:S447–S460. doi: 10.1067/mai.2002.121409. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Nakanishi T, Haruta T, Shirasaka Y, Keogh JP, Tamai I. Transport of ipratropium, an anti-chronic obstructive pulmonary disease drug, is mediated by organic cation/carnitine transporters in human bronchial epithelial cells: implications for carrier-mediated pulmonary absorption. Mol Pharm. 2010;7:187–195. doi: 10.1021/mp900206j. [DOI] [PubMed] [Google Scholar]
- Patton JS, Byron PR. Inhaling medicines: delivering drugs to the body through the lungs. Nat Rev Drug Discovery. 2007;6:67–74. doi: 10.1038/nrd2153. [DOI] [PubMed] [Google Scholar]
- Pierrou S, Broberg P, O’Donnell RA, Pawlowski K, Virtala R, Lindqvist E, et al. Expression of genes involved in oxidative stress responses in airway epithelial cells of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;175:577–586. doi: 10.1164/rccm.200607-931OC. [DOI] [PubMed] [Google Scholar]
- Raunio H, Hakkola J, Hukkanen J, Lassila A, Paivarinta K, Pelkonen O, et al. Expression of xenobiotic-metabolizing CYPs in human pulmonary tissue. Exp Toxicol Pathol. 1999;51:412–417. doi: 10.1016/S0940-2993(99)80031-1. [DOI] [PubMed] [Google Scholar]
- Seree E, Villard PH, Hever A, Guigal N, Puyoou F, Charvet B, et al. Modulation of MDR1 and CYP3A expression by dexamethasone: evidence for an inverse regulation in adrenals. Biochem Biophys Res Commun. 1998;252:392–395. doi: 10.1006/bbrc.1998.9662. [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- Somers GI, Lindsay N, Lowdon BM, Jones AE, Freathy C, Ho S, et al. A comparison of the expression and metabolizing activities of phase I and II enzymes in freshly isolated human lung parenchymal cells and cryopreserved human hepatocytes. Drug Metab Dispos. 2007;35:1797–1805. doi: 10.1124/dmd.107.015966. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Wang Y, Prakash C. Xenobiotic-metabolizing enzymes in human lung. Curr Drug Metab. 2006;7:939–948. doi: 10.2174/138920006779010575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Individual expression pattern of differential expressed genes encoding membrane transporters. Graphs show expression levels of individual genes encoding membrane transporters regarding differential expression between central airways and peripheral tissue. Black circles denote COPD patients and unfilled circles denote healthy subjects.
Figure S2. Individual expression pattern of differential expressed genes encoding drug-metabolizing enzymes. Graphs show expression levels of individual genes encoding drug-metabolizing enzymes regarding differential expression between central airways and peripheral tissue. Black circles denote COPD patients and unfilled circles denote healthy subjects.
Table S1. TLDA probes and median Ct-values for membrane transporters and housekeeping genes in healthy and COPD lung.
Table S2. TLDA probes and median Ct-values for drug-metabolizing enzymes, transcription factors, and cell-specific markers in healthy and COPD lung.
Table S3. Gene expression of membrane transporter from healthy and COPD lung in % of expression of housekeeping genes.
Table S4. Interindividual variability of expressed genes encoding membrane transporters in healthy or COPD human lung in central or peripheral airways.
Table S5. Gene expression of drug-metabolizing enzymes from healthy and COPD lung in % of expression of housekeeping genes.
Table S6. The interindividual variability among genes encoding metabolizing enzymes with in healthy or COPD human lung from both central airways and peripheral tissue.
Table S7. Expression of genes encoding transcription factors from healthy and COPD lung in % of expression of housekeeping genes.
Table S8. Expression of genes encoding transcription factors from healthy and COPD lung in % of expression of housekeeping genes.
Table S9. Differential expressed genes encoding metabolizing enzymes between central airways and peripheral tissue.
Table S10. Interindividual variability of genes encoding transcription factors in healthy or COPD human lung in both central airways and peripheral tissue.
Table S11. Expression of genes encoding cell-specific markers in % of expression of housekeeping genes.
Table S12. Expression of genes encoding cell-specific markers in % of expression of housekeeping genes.
Table S13. Interindividual variability among genes encoding cell-specific markers within healthy or COPD human lung in both central airways and peripheral tissue.


