Abstract
Background
For the detection of respiratory pathogens, the sampling strategy may influence the diagnostic yield. Ideally, samples from the lower respiratory tract are collected, but they are difficult to obtain.
Objectives
In this study, we compared the diagnostic yield in sputum and oropharyngeal samples (OPS) for the detection of respiratory pathogens in patients with community-acquired pneumonia (CAP), with the objective to optimize our diagnostic testing algorithm.
Methods
Matched sputum samples, OPS, blood cultures, serum, and urine samples were taken from patients (>18 years) with CAP and tested for the presence of possible respiratory pathogens using bacterial cultures, PCR for 17 viruses and five bacteria and urinary antigen testing.
Results
When using only conventional methods, that is, blood cultures, sputum culture, urinary antigen tests, a pathogen was detected in 49·6% of patients (n = 57). Adding molecular detection assays increased the yield to 80%. A pathogen was detected in 77 of the 115 patients in OPS or sputum samples by PCR. The sensitivity of the OPS was lower than that of the sputum samples (57% versus 74%). In particular, bacterial pathogens were more often detected in sputum samples. The sensitivity of OPS for the detection of most viruses was higher than in sputum samples (72% versus 66%), except for human rhinovirus and respiratory syncytial virus.
Conclusion
Addition of PCR on both OPS and sputum samples significantly increased the diagnostic yield. For molecular detection of bacterial pathogens, a sputum sample is imperative, but for detection of most viral pathogens, an OPS is sufficient.
Keywords: Community-acquired pneumonia, oropharyngeal swabs, real-time PCR, respiratory virus, sputum samples, yield
Introduction
Community-acquired pneumonia (CAP) remains a major cause of morbidity and mortality worldwide.1 Streptococcus pneumoniae, Mycoplasma pneumoniae, influenza A virus (InfA), respiratory syncytial virus (RSV), and adenoviruses (AdV) are recognized as important causes of CAP.2,3 Despite efforts to find evidence for bacterial and viral pathogens as etiological agents in patients with CAP, etiology remains elusive in up to 50% of the patients.1,3–5 Reasons reported for this low yield are use of antibiotics before collecting samples, sample type tested, and the diagnostic panel used for patient evaluation.6–8 Diagnostic methods used range from culture (sputum, blood, throat swabs), antigen testing (e.g. urinary antigen testing), and molecular tests. Some studies question the value of bacterial sputum culture findings.8–10 Furthermore, serologic testing requires convalescent-phase samples, and therefore, it is not useful in the initial phase of determining the etiology. Blood cultures provide a microbiological diagnosis in 0–17%, and the addition of urinary antigen detection assays for S. pneumoniae and Legionella pneumophila has improved the yield substantially.3,11 During the past years, PCR has been developed for many viral and bacterial pathogens, resulting again in higher diagnostic yields.12 Knowledge of the probable etiological agent(s) may inform treatment, thereby potentially reducing the use of antibiotics and eventually that of antimicrobial resistance.10
In this study, we aimed to assess the added value of viral and bacterial molecular diagnostics on oropharyngeal swabs (OPS) in comparison with sputum samples for the diagnosis of CAP.
Materials and methods
Study design
This study was embedded within a larger prospective, observational cohort study performed between April 2008 and March 2009. All patients with CAP aged 18 years and older attending the emergency ward of two teaching hospitals in Tilburg, the Netherlands, with the suspicion of CAP were included. CAP was defined as the presence of a new or progressive infiltrate on a chest radiograph with clinical symptoms suggestive of a lower respiratory tract infection. Exclusion criteria were (i) recent hospitalization (<2 weeks) or residence in long-term care facilities, (ii) known bronchial obstruction or a history of post-obstructive pneumonia (with exception of chronic obstructive pulmonary disease), (iii) primary lung cancer or another malignancy metastatic to the lungs, (iv) AIDS, known or suspected Pneumocystis jirovecii pneumonia, and (v) known or suspected active tuberculosis.
A case report form was obtained from all patients to collect data on age, gender, current smoking, comorbidity, clinical symptoms, prior antimicrobial treatment at admission, and blood analysis.
Sample collection, processing, and storage
According to protocol, at the emergency ward an OPS, a sputum sample, urine sample, and a serum sample were taken and two sets of blood samples were obtained. For this comparative evaluation, only patients for whom a complete sample set was available were included. Blood and urine specimens were processed immediately. Sputum samples were divided into two equal aliquots: one for bacterial culture and another was stored at −20°C for real-time reverse transcriptase PCR ([RT]-qPCR) testing. The OPS was used to sample the posterior oropharyngeal mucosal membrane using a commercial rigid cotton-tipped swab (MWE, Wiltshire, UK). After swabbing, the OPS specimens were placed in 1·5 ml virus transport medium (Gly medium) and stored at −20°C before performing qPCR assays.
Diagnostics
The sputum samples and blood samples were cultured according to standard microbiological procedures. All sputum samples were examined by microscopy, and sputum samples with the presence of >25 polymorphonuclear leukocytes and <10 squamous cells per field were considered to be acceptable for culture. Significant bacterial growth of the sputum sample was defined as growth of a predominant organism on the culture plates and compatible results from Gram stain.
Urine sample were tested by urinary antigen detection tests for S. pneumoniae and L. pneumophila (BinaxNOW® pneumococcal urinary antigen test and the BinaxNOW® Legionella urinary antigen test, Alere, Portland, ME, USA). All oropharyngeal and sputum samples were tested by (RT)-qPCR for the presence of respiratory viruses and bacteria including AdV, human bocavirus (HBoV), KI polyomaviruses and WU polyomaviruses (KIPyV and WUPyV), human metapneumovirus (hMPV), human rhinovirus (HRV), human coronaviruses (HCoV) (OC43, NL63, HKU1, and 229E), parainfluenza viruses (PIV)1−4, influenza viruses A and B (InfA, InfB), RSV, L. pneumophila, M. pneumoniae, Chlamydophila psittaci, Chlamydophila pneumoniae, and Coxiella burnetii. Serum samples were tested for the presence of C. burnetii. (RT)-qPCR procedures were performed as described.13–19 (RT)-qPCR results were expressed in cycle threshold-values.
Statistical analysis
A consensus standard was used to assess the sensitivity of the OPS or sputum sample: A positive result in either the OPS or sputum sample was considered as the gold standard for the presence of a pathogen and was used to calculate the sensitivity of the OPS or sputum sample for the detection of the respiratory pathogens. McNemar's test was used to assess the significance of the difference between two correlated proportions. Analyses were conducted using pasw Statistics 18 (IBM Company, Chicago, VS, USA).
Results
Characteristics
Of the 408 patients with CAP that were evaluated during the study period, a subset of 115 (28·2%) met the inclusion criteria for completeness of sampling and was included in the study. Patients ranged in age from 20 to 90 years (mean 66 years), 62% of the patients were male. Thirty-two (27·8%) patients had had antibiotic treatment prior to admission.
Microbiological yield
Using conventional methods, that is, blood cultures, sputum culture, urinary antigen tests, 57 patients (49·6%) tested positive for at least one pathogen. Adding the full molecular diagnostic package increased the diagnostic yield to 80%. The most frequently detected bacterial pathogens were S. pneumoniae (n = 27) and C. burnetii (n = 13). In 14 patients, S. pneumoniae was the only detected pathogen, and in six patients, C. burnetii was the only detected pathogen. The most frequently isolated viral pathogens were HRV (n = 13) and PIV1 (n = 8). In the majority of patients, HRV and PIV1 were detected in combination with other pathogens. In 58 patients (50·4%), only one pathogen was detected. Mixed infections were common, with up to three possible pathogens listed (Table 1).
Table 1.
Diagnostic yield in 92 patients with CAP
| Single pathogen (n = 58) | 2 pathogens (n = 25) | 3 pathogens (n = 9) | |||
|---|---|---|---|---|---|
| 14 | Streptococcus pneumoniae | 5 | S. pneumoniae+HRV | 1 | S. pneumoniae+InfA+GNB |
| 6 | Coxiella burnetii | 1 | S. pneumoniae+GNB | 1 | S. pneumoniae+Haemophilus influenzae+RSV |
| 6 | GNB | 1 | S. pneumoniae+C. burnetii | 1 | H. influenzae+C. burnetii+HCoV OC43 |
| 5 | H. influenzae | 1 | S. pneumoniae+InfA | 1 | H. influenzae+RSV+KIPyV |
| 3 | Staphylococcus aureus | 1 | S. pneumoniae+PIV1 | 1 | Legionella pneumophila+PIV1+GNR |
| 3 | HRV | 1 | S. pneumoniae+HCoV OC43 | 1 | PIV1+HRV+HCoV NL63 |
| 3 | InfA | 1 | S. pneumoniae+HCoV NL63 | 1 | InfB+WU+HCoV NL63 |
| 3 | RSV | 1 | L. pneumophila+InfB | 1 | S. aureus+P. aeruginosa+HCoV 229E |
| 2 | Chlamydophila psittaci | 1 | L. pneumophila+HRV | 1 | C. burnetii+HCoV 229E+HCoV OC43 |
| 2 | L. pneumophila | 1 | L. pneumophila+C. burnetii | ||
| 2 | HCoV OC43 | 1 | L. pneumophila+M. pneumoniae | ||
| 2 | PIV1 | 1 | C. burnetii+HRV | ||
| 1 | E. coli | 1 | C. burnetii+S. milleri | ||
| 1 | Moraxella catarrhalis | 1 | C. burnetii+E. coli | ||
| 1 | P. luteola | 1 | C. psittaci+S. aureus | ||
| 1 | AdV | 1 | E. coli+HRV | ||
| 1 | InfB | 1 | InfA+H. influenzae | ||
| 1 | HCoV 229E | 1 | PIV1+H. influenzae | ||
| 1 | hMPv | 1 | PIV1+HCoV HKU | ||
| 1 | PIV1+PIV3 | ||||
| 1 | PIV3+HRV | ||||
AdV, adenovirus; KIPyV, KI polyomavirus; WUPyV, WU polyomavirus; hMPV, human metapneumovirus; HRV, human rhinovirus; HCoV OC43, NL63, HKU1 and 229E, human coronaviruses; PIV1–4, parainfluenza viruses 1–4; InfA, influenza A virus; InfB, influenza B virus; RSV, respiratory syncytial virus; GNB, Gram-negative bacteria; CAP, community-acquired pneumonia.
The majority of patients with mixed infections had S. pneumoniae identified. S. pneumoniae was detected in 14 blood cultures, 20 urinary antigen tests, and five sputum samples.
Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus were only detected in sputum cultures. Escherichia coli and other Gram-negative bacteria were isolated from blood cultures and sputum cultures. Pseudomonas aeruginosa was only isolated from a blood culture. Beside qPCR on OPS and sputum samples, L. pneumophila was also detected with the urinary antigen test in three patients. These patients had also a positive qPCR on OPS and/or sputum samples. Coxiella burnetii was detected in four serum samples; these patients had also qPCR-positive sputum samples (Figure 1).
Figure 1.
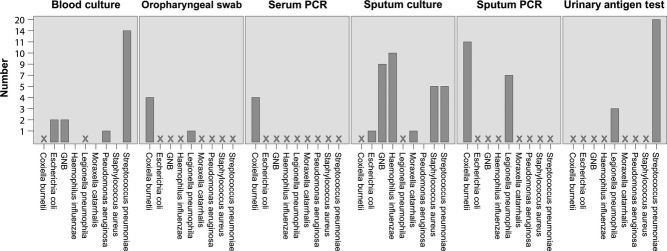
Detection of pathogens in patients with CAP by material. GNB, Gram-negative bacteria; CAP, community-acquired pneumonia; X: method not used or suitable for detection of specific pathogen.
Sensitivity of molecular diagnostics on OPS and sputum samples
A positive qPCR in OPS and/or sputum samples was found in 77 of the 115 patients. For 33 (42·9%) of the 77 patients, the pathogens were only detected in the sputum sample, while for 20 (26·0%) of them, the pathogens were only detected in the OPS. KIPyV, WUPyV, HCoV HKU1, and HCoV 229E were only detected in the OPS, whereas C. psittaci was uniquely found in sputum (Table 2). The sensitivity for detecting any pathogen was 57% (95%CI: 45–68) using OPS and 74% using sputum (95%CI: 63–83). Bacterial pathogens were more often detected in sputum samples than in OPS (92%, 95%CI: 72–99 versus 25%, 95%CI: 11–47, P < 0·001). Except for HRV and RSV, the sensitivity for the detection of viruses using OPS was higher compared with the use of sputum samples (72%, 95%CI: 57–83 versus 66%, 95%CI: 52–78, P = 0·69).
Table 2.
Detection of respiratory pathogens and the sensitivity by OPS and sputum sample
| Pathogens detected in: | Both sputum and OPS | OPS only | Sputum only | Total | OPS sensitivity 95% CI | Sputum sensitivity 95% CI |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| Legionella pneumophila | 1 | 0 | 6 | 7 | 14 (0–58) | 100 (56–100) |
| Mycoplasma pneumoniae | 1 | 0 | 0 | 1 | 100 (5–100) | 100 (5–100) |
| Coxiella burnetii | 2 | 2 | 9 | 13 | 36 (12–68) | 85 (54–97) |
| Chlamydophila psittaci | 0 | 0 | 3 | 3 | 0 (0–69) | 100 (31–100) |
| Chlamydophila pneumoniae | 0 | 0 | 0 | 0 | – | – |
| Total bacteria | 4 | 2 | 18 | 24 | 25 (11–47) | 92 (72– 99) |
| Viruses | ||||||
| Adenovirus | 1 | 0 | 0 | 1 | 100 (5–100) | 100 (5–100) |
| Human bocavirus | 0 | 0 | 0 | 0 | – | – |
| KI polyomavirus | 0 | 1 | 0 | 1 | 100 (5–100) | 0 (0–95) |
| WU polyomavirus | 0 | 1 | 0 | 1 | 100 (5–100) | 0 (0–95) |
| Human metapneumovirus | 1 | 0 | 0 | 1 | 100 (5–100) | 100 (5–100) |
| Human rhinovirus | 3 | 0 | 10 | 13 | 23 (6–54) | 100 (72–100) |
| Human coronaviruses | ||||||
| OC43 | 5 | 0 | 0 | 5 | 100 (46–100) | 100 (46–100) |
| NL63 | 1 | 2 | 0 | 3 | 100 (31–100) | 33 (2–87) |
| HKU1 | 0 | 1 | 0 | 1 | 100 (5–100) | 0 (0–95) |
| 229E | 0 | 3 | 0 | 3 | 100 (31–100) | 0 (0–69) |
| Parainfluenza viruses | ||||||
| 1 | 0 | 7 | 1 | 8 | 88 (47–99) | 13 (1–53) |
| 2 | 0 | 0 | 0 | 0 | – | – |
| 3 | 1 | 0 | 1 | 2 | 50 (3–97) | 100 (20–100) |
| 4 | 0 | 0 | 0 | 0 | – | – |
| Influenza A virus | 6 | 0 | 0 | 6 | 100 (52–100) | 100 (52–100) |
| Influenza B virus | 1 | 2 | 0 | 3 | 100 (31–100) | 33 (2–87) |
| Respiratory syncytial virus | 1 | 1 | 3 | 5 | 40 (7–83) | 80 (30–99) |
| Total viruses | 20 | 18 | 15 | 53 | 72 (57–83) | 66 (52– 78) |
| Total | 24 | 20 | 33 | 77 | 57 (45–68) | 74 (63–83) |
OPS, oropharyngeal swab, CI, confidence interval.
Discussion
This study demonstrates that a possible etiological diagnosis can be found in a high proportion (80%) of patients with CAP, when optimal sampling and a broad diagnostic package are used.
The reality of clinical practice is that the majority of patients with CAP undergo limited diagnostic tests to demonstrate an etiological agent, other than urine antigen test and, only if available, a bacterial sputum culture. Good quality sputum samples are obtained in 40–60% of patients with CAP, but the diagnostic yield using the classical methods (culture) may be limited. In our study, a bacterial pathogen was cultured from 27% of the sputum samples, slightly higher than was found in published reports (9–14·4%).8,9,20
Isolation of atypical respiratory bacterial pathogens, for example M. pneumoniae, C. pneumoniae is difficult because these pathogens are hardly culturable and cell culture is time-consuming.
In our study, the use of qPCR on sputum samples increased the yield significantly for these pathogens; OPS were not suitable for the detection of them.
Respiratory viruses are poorly detected by conventional techniques.21 Rapid assessment of viruses is now possible with sensitive and highly specific real-time PCR assays, but the utility of swabs versus washes and nose versus oropharyngeal versus nasopharyngeal samples is subject to considerable debate. Lieberman et al.22 found that viral pathogens are better detected by nasopharyngeal washes as they offer a better yield than nasal or OPS, but this procedure is poorly tolerated and rarely used in hospitalized patients. On the other hand, de la Tabla et al.23 reported that a combined nose–throat swab was superior to nasopharyngeal aspirates for the detection of InflA (H1N1) and that the combination of both methods increases the detection rate. In our study, we used a sputum sample and OPS for the detection of viral pathogens and found that OPS was equally or more sensitive for most viruses except HRV and RSV. Falsey et al.24 found in their study that more viruses were detected in sputum samples compared with nose–throat swabs, 44% of the viruses were detected by both methods, 23% were positive by nose–throat swabs alone, and 33% were positive only with sputum samples. Similar to our study, nose–throat swabs and sputum testing yield complementary results. For bacterial pathogens, sputum samples clearly were superior to OPS. Similar to studies elsewhere, we found S. pneumoniae as the most common potential pathogen.1,25 In the literature, S. pneumoniae PCR on sputum samples as a diagnostic tool for pneumococcal CAP has had mixed results because distinguishing colonization from infection is difficult even by quantifying the load.26–29 However, patients with CAP tend to be more frequently colonized with pneumococci than asymptomatic patients and an important hypothesis is that aspiration of oropharyngeal contents the most common route is of developing pneumonia.30–32 Similarly, culturing Streptococci from sputum samples are not conclusive evidence for their etiological role. Therefore, the value of routine detection of S. pneumoniae remains a matter of debate. Similar to others, our study showed that molecular detection of L. pneumophila on sputum could replace urinary antigen testing.33,34 Practically, however, this requires a laboratory setup capable of providing such diagnostics with a rapid turnaround time, and 24/7, a situation that is currently not achievable in many settings.
In our study population, a relatively large number of C. burnetii in patients with CAP were found. This was due to a Q fever outbreak in our area with over 4000 notified cases in the Netherlands between 2007 and 2010.35 For viruses, results comparing sample types were more variable: Overall, based on our data and the convenience of the sampling procedure, OPS would be the preferred sample type, with the exception of HRV and RSV. Our findings are in agreement with published studies focusing on viral pathogens as primary causes of CAP remain an issue of considerable debate.21,36,37 The majority of patients with CAP positive for HRV had a second respiratory virus or a bacterial pathogen, and HCoVs were never found as a single infection. In addition, HRV are highly prevalent, and case–control studies have also found HRV to be common in asymptomatic persons as well.21,36 Our findings do suggest, however, that IF these pathogens are included in the diagnostic package and HRV testing should be integrated in a sputum panel, consisting of bacterial targets in addition to HRV and RSV. This may be more relevant for RSV for which therapeutic options are available, although the efficacy of antivirals in this patient category remains to be determined.38
Limitations in our study include the lack of a control group to determine the prevalence of respiratory pathogens. More than one pathogen was isolated in 34 (2·6%) of the 115 patients and in nine patients three pathogens were found. Real-time PCR significantly improves the sensitivity of detecting pathogens, and often it is not possible to determine the contribution of each pathogen as the detection of viral or bacterial nucleic acids may not always represent causation. In a study by van Gageldonk et al.39, in approximately 20% of the subjects with no respiratory complaints, respiratory viral pathogens were detected. On the other hand, Lieberman et al.22 found a much lower prevalence (7·1%) of respiratory viruses in subjects with no respiratory complaints. In this study, all subjects enrolled were symptomatic, and this would increase the likelihood that isolated pathogens were causative, unfortunately observational cohort studies such as this are not able to directly determine causation. Quantitative (RT)-qPCR data would have been useful to help address the question whether there is active infection in the lower respiratory tract instead of detecting residual DNA/RNA from a prior infection or asymptomatic carriage. Finally, we only included a subset of patients with CAP, but we have no reason to believe that the patients who were not included would be substantially different compared with the study group.
Conclusions
Based on our findings, providing a targeted bacterial PCR package for sputum testing and a separate viral package for OPS testing would provide almost the same diagnostic yield as the full spectrum of tests used in the study. This would only be feasible if results of PCR can be available with very short turnaround time. When looking at diagnostic yield, the sputum package could include HRV and RSV testing. While this would lead to a potential diagnosis in a high proportion of CAP patients, a critical appraisal of the added value of the expanding diagnostic packages is needed, given the costs of such procedures. Studies are needed to evaluate the impact of testing algorithms on patient treatment and outcome.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.File TM. Community-acquired pneumonia. Lancet. 2003;362:1991–2001. doi: 10.1016/S0140-6736(03)15021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet. 2011;377:1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson N, Kalin M, Tiveljung-Lindell A, Giske CG, Hedlund J. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50:202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjerre LM, Verheij TJ, Kochen MM. Antibiotics for community acquired pneumonia in adult outpatients. Cochrane Database Syst Rev. 2009;4:CD002109. doi: 10.1002/14651858.CD002109.pub2. [DOI] [PubMed] [Google Scholar]
- 5.Charles PG, Whitby M, Fuller AJ, et al. The etiology of community-acquired pneumonia in Australia: why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin Infect Dis. 2008;46:1513–1521. doi: 10.1086/586749. [DOI] [PubMed] [Google Scholar]
- 6.Cilloniz C, Ewig S, Polverino E, et al. Community-acquired pneumonia in outpatients: etiology and outcomes. Eur Respir J. 2012;40:931–938. doi: 10.1183/09031936.00168811. [DOI] [PubMed] [Google Scholar]
- 7.van de Garde EM, Endeman H, van Hemert RN, et al. Prior outpatient antibiotic use as predictor for microbial aetiology of community-acquired pneumonia: hospital-based study. Eur J Clin Pharmacol. 2008;64:405–410. doi: 10.1007/s00228-007-0407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewig S, Schlochtermeier M, Goke N, Niederman MS. Applying sputum as a diagnostic tool in pneumonia: limited yield, minimal impact on treatment decisions. Chest. 2002;121:1486–1492. doi: 10.1378/chest.121.5.1486. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Vazquez E, Marcos MA, Mensa J, et al. Assessment of the usefulness of sputum culture for diagnosis of community-acquired pneumonia using the PORT predictive scoring system. Arch Intern Med. 2004;164:1807–1811. doi: 10.1001/archinte.164.16.1807. [DOI] [PubMed] [Google Scholar]
- 10.Lidman C, Burman LG, Lagergren A, Ortqvist A. Limited value of routine microbiological diagnostics in patients hospitalized for community-acquired pneumonia. Scand J Infect Dis. 2002;34:873–879. doi: 10.1080/0036554021000026967. [DOI] [PubMed] [Google Scholar]
- 11.Afshar N, Tabas J, Afshar K, Silbergleit R. Blood cultures for community-acquired pneumonia: are they worthy of two quality measures? A systematic review. J Hosp Med. 2009;4:112–123. doi: 10.1002/jhm.382. [DOI] [PubMed] [Google Scholar]
- 12.Angeles Marcos M, Camps M, Pumarola T, et al. The role of viruses in the aetiology of community-acquired pneumonia in adults. Antivir Ther. 2006;11:351–359. [PubMed] [Google Scholar]
- 13.van de Pol AC, van Loon AM, Wolfs TF, et al. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenoviruses with real-time PCR in samples from patients with respiratory symptoms. J Clin Microbiol. 2007;45:2260–2262. doi: 10.1128/JCM.00848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Pol AC, Wolfs TF, Jansen NJ, et al. Human bocavirus and KI/WU polyomaviruses in pediatric intensive care patients. Emerg Infect Dis. 2009;15:454–457. doi: 10.3201/eid1503.081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Pol AC, Wolfs TF, Jansen NJ, van Loon AM, Rossen JW. Diagnostic value of real-time polymerase chain reaction to detect viruses in young children admitted to the paediatric intensive care unit with lower respiratory tract infection. Crit Care. 2006;10:R61. doi: 10.1186/cc4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diederen BM, Kluytmans JA, Vandenbroucke-Grauls CM, Peeters MF. Utility of real-time PCR for diagnosis of Legionnaires' disease in routine clinical practice. J Clin Microbiol. 2008;46:671–677. doi: 10.1128/JCM.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greiner O, Day PJ, Bosshard PP, et al. Quantitative detection of Streptococcus pneumoniae in nasopharyngeal secretions by real-time PCR. J Clin Microbiol. 2001;39:3129–3134. doi: 10.1128/JCM.39.9.3129-3134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heddema ER, Beld MG, de Wever B, et al. Development of an internally controlled real-time PCR assay for detection of Chlamydophila psittaci in the LightCycler 2.0nn system. Clin Microbiol Infect. 2006;12:571–575. doi: 10.1111/j.1469-0691.2006.01417.x. [DOI] [PubMed] [Google Scholar]
- 19.Tilburg JJ, Melchers WJ, Pettersson AM, et al. Interlaboratory evaluation of different extraction and real-time PCR methods for detection of Coxiella burnetii DNA in serum. J Clin Microbiol. 2010;48:3923–3927. doi: 10.1128/JCM.01006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roson B, Carratala J, Verdaguer R, et al. Prospective study of the usefulness of sputum Gram stain in the initial approach to community-acquired pneumonia requiring hospitalization. Clin Infect Dis. 2000;31:869–874. doi: 10.1086/318151. [DOI] [PubMed] [Google Scholar]
- 21.Templeton KE, Scheltinga SA, van den Eeden WC, et al. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41:345–351. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman D, Shimoni A, Shemer-Avni Y, Keren-Naos A, Shtainberg R. Respiratory viruses in adults with community-acquired pneumonia. Chest. 2010;138:811–816. doi: 10.1378/chest.09-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Tabla VO, Masia M, Antequera P, et al. Comparison of combined nose-throat swabs with nasopharyngeal aspirates for detection of pandemic influenza A/H1N1 2009 virus by real-time reverse transcriptase PCR. J Clin Microbiol. 2010;48:3492–3495. doi: 10.1128/JCM.01105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falsey AR, Formica MA, Walsh EE. Yield of sputum for viral detection by reverse transcriptase PCR in adults hospitalized with respiratory illness. J Clin Microbiol. 2012;50:21–24. doi: 10.1128/JCM.05841-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson N, Kalin M, Giske CG, Hedlund J. Quantitative detection of Streptococcus pneumoniae from sputum samples with real-time quantitative polymerase chain reaction for etiologic diagnosis of community-acquired pneumonia. Diagn Microbiol Infect Dis. 2008;60:255–261. doi: 10.1016/j.diagmicrobio.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Abdeldaim G, Herrmann B, Korsgaard J, et al. Is quantitative PCR for the pneumolysin (ply) gene useful for detection of pneumococcal lower respiratory tract infection? Clin Microbiol Infect. 2009;15:565–570. doi: 10.1111/j.1469-0691.2009.02714.x. [DOI] [PubMed] [Google Scholar]
- 27.Kais M, Spindler C, Kalin M, Ortqvist A, Giske CG. Quantitative detection of Streptococcus pneumoniae Haemophilus influenzae, and Moraxella catarrhalis in lower respiratory tract samples by real-time PCR. Diagn Microbiol Infect Dis. 2006;55:169–178. doi: 10.1016/j.diagmicrobio.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Murdoch DR, Anderson TP, Beynon KA, et al. Evaluation of a PCR assay for detection of Streptococcus pneumoniae in respiratory and nonrespiratory samples from adults with community-acquired pneumonia. J Clin Microbiol. 2003;41:63–66. doi: 10.1128/JCM.41.1.63-66.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Werno AM, Anderson TP, Murdoch DR. Association between pneumococcal load and disease severity in adults with pneumonia. J Med Microbiol. 2012;61:1129–1135. doi: 10.1099/jmm.0.044107-0. [DOI] [PubMed] [Google Scholar]
- 30.Albrich WC, Madhi SA, Adrian PV, et al. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin Infect Dis. 2012;54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.File TM., Jr Streptococcus pneumoniae and community-acquired pneumonia: a cause for concern. Am J Med. 2004;117(Suppl 3A):39S–50S. doi: 10.1016/j.amjmed.2004.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 33.Cho MC, Kim H, An D, et al. Comparison of sputum and nasopharyngeal swab specimens for molecular diagnosis of Mycoplasma pneumoniae Chlamydophila pneumoniae, and Legionella pneumophila. Ann Lab Med. 2012;32:133–138. doi: 10.3343/alm.2012.32.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diederen BM, Peeters MF. Are oropharyngeal swabs suitable as samples for Legionella-specific PCR testing? J Clin Microbiol. 2007;45:3482. doi: 10.1128/JCM.01495-07. author reply 3482-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delsing CE, Kullberg BJ, Bleeker-Rovers CP. Q fever in the Netherlands from 2007 to 2010. Neth J Med. 2010;68:382–387. [PubMed] [Google Scholar]
- 36.Jennings LC, Anderson TP, Beynon KA, et al. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63:42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 37.Johnstone J, Majumdar SR, Fox JD, Marrie TJ. Viral infection in adults hospitalized with community-acquired pneumonia: prevalence, pathogens, and presentation. Chest. 2008;134:1141–1148. doi: 10.1378/chest.08-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falsey AR. Respiratory syncytial virus infection in adults. Semin Respir Crit Care Med. 2007;28:171–181. doi: 10.1055/s-2007-976489. [DOI] [PubMed] [Google Scholar]
- 39.van Gageldonk-Lafeber AB, Heijnen ML, Bartelds AI, et al. A case–control study of acute respiratory tract infection in general practice patients in The Netherlands. Clin Infect Dis. 2005;41:490–497. doi: 10.1086/431982. [DOI] [PMC free article] [PubMed] [Google Scholar]


