Abstract
Background and objectives
Autosomal recessive polycystic kidney disease (ARPKD) is caused by mutations in the PKHD1 gene. The longest open reading frame comprises 66 exons encoding polyductin or fibrocystin, a type I transmembrane protein with 4074 amino acids. Functional investigations are considerably hampered by its large size and lack of expression in tissues that are usually available for analysis such as lymphocytes or fibroblasts.
Design, setting, participants, & measurements
Allegedly strong and clear-cut genotype-phenotype correlations for the type of PKHD1 mutation could be established. Thus far, practically all patients with two truncating mutations showed perinatal or neonatal demise and at least one hypomorphic missense mutation is thought to be indispensable for survival. Mutation analysis of >500 ARPKD families was performed by conventional and next-generation sequencing techniques.
Results
This study presents four unrelated patients with ARPKD with a nonlethal, moderate clinical course despite the burden of two PKHD1 mutations expected to lead to premature termination of translation. In line with parental consanguinity, all mutations occurred in the homozygous state and segregated with the disorder in these families. To try to unravel the mechanisms that underlie this obvious contradiction, these patients were further analyzed in detail by utilizing different methods. In all cases, complex transcriptional alterations were detected. Alternative splicing patterns might disrupt a critical stoichiometric and temporal balance between different protein products and may play a crucial role in defining the phenotype of these patients.
Conclusions
Although these findings represent rare events, they are of importance for genetic counseling and illustrate that some caution is warranted in the interpretation of mutations and their clinical significance. The authors hypothesize that expression of a minimal amount of functional protein is needed for survival of the neonatal period in ARPKD.
Keywords: ADPKD, clinical nephrology, cystic kidney, pediatric nephrology, polycystic, kidney disease
Introduction
Polycystic kidney disease is a clinically and genetically heterogeneous disorder with different modes of inheritance. Autosomal dominant polycystic kidney disease is the most frequent life-threatening genetic disease with a prevalence of one in 500–1000, affecting approximately 12.5 million individuals worldwide. Clinical symptoms typically do not arise until adulthood; however, about 2%–5% of patients with autosomal dominant polycystic kidney disease present with early clinical manifestations that are sometimes indistinguishable from autosomal recessive polycystic kidney disease (ARPKD). ARPKD is much rarer than its dominant counterpart and has a suspected incidence of about one in 20,000 live births (1–3). About one half of patients with ARPKD die shortly after birth from respiratory insufficiency due to pulmonary hypoplasia and thoracic compression by the excessively enlarged kidneys. Although ARPKD clearly represents a pediatric disorder, adult patients have also been described with a relatively mild clinical course (4,5). Patients with ARPKD invariably exhibit histologic liver involvement characterized by defective remodeling of the ductal plate with congenital hepatic fibrosis and biliary duct ectasia (so-called ductal plate malformation). The clinical course of older patients in particular might be dominated by hepatobiliary complications. Progressive hepatic fibrosis and consecutive portal hypertension may cause hypersplenism with pancytopenia and esophageal varices with upper gastrointestinal bleeding. Although most patients show a comparable degree of severity with regard to liver and kidney affection (renal-hepatobiliary morbidity pattern), there is no direct correlation or interdependency between those two organs. Patients with single ARPKD may present with an organ-specific phenotype (i.e., either an [almost] exclusive renal phenotype or a predominant or mere liver phenotype).
The only described gene for ARPKD is PKHD1, a large gene that extends over a genomic segment of almost 500 kb on chromosome 6p12. To date, >700 different PKHD1 mutations are known (5–18). The longest open reading frame of PKHD1 comprises 66 exons and encodes a single-transmembrane protein, called polyductin/fibrocystin, which is mainly expressed in the kidney and liver/cholangiocytes (6,7,19). Polyductin/fibrocystin is localized to primary cilia and basal bodies of the cells and is involved in centrosome duplication and mitotic spindle assembly during cell division (20–25). PKHD1 undergoes a complex pattern of alternative splicing generating several isoproteins with putatively different functions (6,7,19). It might be hypothesized that the balance between full-length protein and different shorter peptides might be crucial for proper polyductin/fibrocystin function.
Genotype-phenotype correlations have been proposed for the type of PKHD1 mutation. These data implicate that no patient who carries truncating mutations on both parental alleles that lead to a premature termination codon is able to survive the perinatal/neonatal period. Thus, at least one hypomorphic (missense) mutation is thought to be indispensable for the survival of newborns (5) (unpublished observations, C.B.). Notably, for some premature termination codons in genes other than PKHD1, activation of a mechanism called nonsense-mediated mRNA decay (NMD) has been described in the literature (26). This control mechanism reduces the amount of truncated, defective protein by the degradation of transcripts harboring a premature termination codon. Another mechanism known for nonsense mutations is nonsense-mediated alternative splicing. In these cases, a shortened, truncated protein with some residual function is produced, which may explain a milder phenotype than generally expected for premature termination of translation.
In obvious contradiction to published genotype-phenotype data, here we describe four unrelated, only moderately affected patients with ARPKD with homozygous PKHD1 mutations that are each expected to lead to premature termination of translation. To unravel the mechanisms underlying obvious discrepancies with regard to genotype-phenotype correlation, we analyzed these patients in greater detail.
Materials and Methods
Patients
DNA samples and clinical data provided by the caring physician were available for analysis from all affected patients and their parents after informed consent was given. Patients were chosen for analysis if they showed typical clinical and/or histologic features of ARPKD as previously described in detail (2,5,8). We performed PKHD1 mutation analysis in a large cohort of >500 patients with polycystic kidney disease phenotypes using conventional Sanger sequencing or a targeted next-generation sequencing approach. For the latter, all exons and adjacent intronic boundaries of 129 and, more recently, 306 genes (including PKHD1) known or hypothesized to cause ciliopathies were targeted by a custom SeqCap EZ choice sequence capture library (NimbleGen, Madison, WI) and subsequently sequenced on a Roche 454 GS FLX or an Illumina MiSeq or HiSeq platform (2X150 PE) according to the manufacturer’s protocol. Patients were analyzed with an average coverage of 60-fold (GS FLX) or >120-fold (MiSeq and HiSeq), respectively. Bioinformatic analysis was performed using the Roche GS Reference Mapper software (version 2.6), the SeqPilot SeqNext module (version 3.5.2; JSI Medical Systems, Kippenheim, Germany), as well as an in-house bioinformatic pipeline established at Bioscientia. All mutations identified by next-generation sequencing were validated by Sanger sequencing. No further mutation thought to be of pathogenic relevance for the disease phenotype was present among the patients described in this article.
In all four families described in this article, we initially performed linkage analysis for PKHD1 by typing several polymorphic microsatellite markers of which at least one proximally and distally flanking marker was shown to be fully informative. By this, all patients demonstrated homozygous haplotypes for PKHD1. This finding was in line with parental consanguinity present in all pedigrees described and further corroborated the clinical assumption that PKHD1 is the correct and underlying disease locus in these families. After identification of a homozygous PKHD1 mutation in the family’s index patient, we further checked the segregation pattern and analyzed parental DNA samples for the presence of the detected mutation. As expected, these analyses invariably showed heterozygosity for the respective mutation in each parent and thus were in accordance with the expected segregation pattern in all families and the previous results obtained by linkage analysis.
Bioinformatic Analyses
We performed bioinformatic analyses and checked splice databases to predict any effect on the splicing pattern caused by the PKHD1 mutations identified. The following bioinformatic prediction programs were used: Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/splice.html), NetGene2 server (http://www.cbs.dtu.dk/services/NetGene2/), and ESEfinder 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi). The respective exons and about 100 bp of the according intronic sequences were analyzed and compared with the respective wild-type (WT) sequence.
Minigene Constructs
Fragments of PKHD1 comprising exon 29 (patient F719) and its adjacent intronic sequences were amplified from the patient’s genomic DNA. PCR products containing primer-introduced XhoI and BamHI sites at the 5′ and 3′ ends were cloned into an exon trap vector pSPL3 (Invitrogen). A segment harboring exons 43–46 was amplified from patient genomic DNA and cloned into pTargeT Mammalian Expression System (Promega) by TA ligation according to the manufacturer’s protocol (in patients F153 and F259). Plasmids were transformed into Escherichia coli strain JM109 (Promega). Positive clones were confirmed by sequencing according to standard protocols. Purified plasmids were used for transfections of COS7 cells. Cells were grown in 35 mm tissue culture dishes to 95% confluence and were transfected with 800 ng plasmid and 2 μl Lipofectamine 2000 (Invitrogen) per dish. After 48 hours, cells were harvested and prepared for RNA extraction.
RNA Extraction and Quantitative RT-PCR Experiments
RNA was extracted using the QIAamp RNA Blood Mini Kit (Qiagen) according to the manufacturer’s protocol after frozen tissue was pestled (F304, F730, WT1, and WT2) and transfected cells were harvested (F153, F259, and F719), respectively. Renal tissue (WT1 and WT2) and commercial cDNA (WT) from three different healthy adult individuals and renal tissue from a patient with a severe phenotype and perinatal demise carrying the homozygous truncating PKHD1 mutation c.4256_4257del (p.Arg1419fs) (F304) were used as controls (to note, this patient was specifically chosen as a control because he showed the expected clinical course in view of his genotype with two truncating PKHD1 mutations and because a sufficient amount of renal tissue was available for analysis). RT-PCR was performed using the AMV RT system (Promega) according to the manufacturer’s recommendations. The resulting cDNA was PCR amplified using primers hybridizing to the sequence of the exon trap vector (for detection of the mutation c.3306delT), exons 19 and 25 of the PKHD1 gene (to detect c.2130_2131insTA), and exons 43 and 45 of the PKHD1 gene (to detect c.7215delG), respectively. All products were analyzed by direct sequencing. For quantitative RT-PCRs, the QuantiTect SYBR Green RT-PCR Kit (Qiagen) was used according to the manufacturer’s recommendations. TATA box binding protein TBP (NM_003194) was used as reference gene for PKHD1 gene expression studies. Relative gene expression was measured and analyzed using the ABI Prism sequence detection system (Applied Biosystems). PCR primers and conditions are available on request.
Polyclonal Anti-Polyductin Antibody, Protein Extraction, and Western Blot Analyses
For production of polyclonal polyductin antibodies, a specific peptide comprising the C-terminal part of the protein was synthesized. The peptide was injected into rabbits and purified sera were collected (Pineda Antibody Service, Berlin, Germany). For Western blots, the antibody was used in a 1:500 dilution. Frozen control and patient renal tissue was pestled and lysed in buffer containing 25 mM Tris-HCl, 1% Triton X-100, 150 mM NaCl, and 5 mM EDTA, pH 8.0. Proteins were separated on 3%–8% Tris/acetate gels (Invitrogen) and blotted on nitrocellulose membranes (Pall Life Sciences). After blocking nonspecific binding sites with 5% nonfat dry milk in PBS-T (PBS containing 0.05% Tween 20), blots were incubated with the antibody overnight at 4°C. Because PKHD1 is not expressed in fibroblasts, lysate from human fibroblasts was used as the negative control.
Results
In a large cohort of unrelated ARPKD families, we identified four patients who survived the neonatal period despite each carrying a homozygous PKHD1 mutation expected to lead to premature termination of translation. All of our patients were of Turkish origin and had a consanguineous parental background. All parents displayed normal abdominal ultrasonography in line with autosomal recessive inheritance of polycystic kidney disease. Table 1 summarizes the clinical information on these patients and families. Female patient F719 showed early systemic hypertension and received peritoneal dialysis for about 2.5 years when living kidney transplantation was performed at age 3 years. Molecular genetic testing revealed homozygosity for the nonsense mutation c.3306delT (p.Tyr1102X) in exon 29 of the PKHD1 gene. Both index patients from families F153 and F259 carried the homozygous 1-bp deletion c.7215delG (p.Gln2405fs) in exon 45 of the PKHD1 gene. A common background of these two pedigrees was not reported, although an ancient Turkish founder allele cannot be excluded. The propositus of family F153 showed early arterial hypertension within the first few months of life and displayed enlarged polycystic kidneys by ultrasonography. At the last examination at age 2 years, he was reported stable. The male patient of family F259 presented with abdominal distention in infancy. Kidney transplantation was performed at age 7 years. The female patient of family F730 reached ESRD early after birth and nephrectomy was done after enlarged kidneys with oligohydramnios had already been noted by ultrasonography during pregnancy. Liver transplantation was performed at age 2 years. One year later, peritoneal dialysis failed and renal transplantation was successfully conducted. Sequencing revealed the homozygous nonsense mutation c.2130_2131insTA (p.Asn711X) in exon 21 of the PKHD1 gene.
Table 1.
Synopsis of ARPKD patients/families
| Family/ Patient | Sex | Origin | PKHD1 Mutations | Mutant Exon | Clinical Course |
|---|---|---|---|---|---|
| F719 (C) | Female | Turkey | c.3306delT (p.Tyr1102X) (P) c.3306delT (p.Tyr1102X) (M) | 29 | Early HTN, enlarged polycystic kidneys, PD until RTX at age 3 yr |
| F153 (C) | Male | Turkey | c.7215delG (p.Gln2405fs) (P) c.7215delG (p.Gln2405fs) (M) | 45 | Early HTN, enlarged polycystic kidneys, stable condition at age 2 yr |
| F259 (C) | Male | Turkey | c.7215delG (p.Gln2405fs) (P) c.7215delG (p.Gln2405fs) (M) | 45 | Abdominal distention in infancy, RTX at age 7 yr |
| F730 (C) | Female | Turkey | c.2130_2131insTA (p.Asn711X) (P) c.2130_2131insTA (p.Asn711X) (M) | 21 | OH and enlarged kidneys in late pregnancy, ESRD shortly after birth with nephrectomy, LTX at age 2 yr, RTX at age 3 yr |
C, parental consanguinity; HTN, systemic arterial hypertension; M, maternal; P, paternal; PD, peritoneal dialysis; RTX, renal transplantation; OH, oligohydramnios; LTX, liver transplantation.
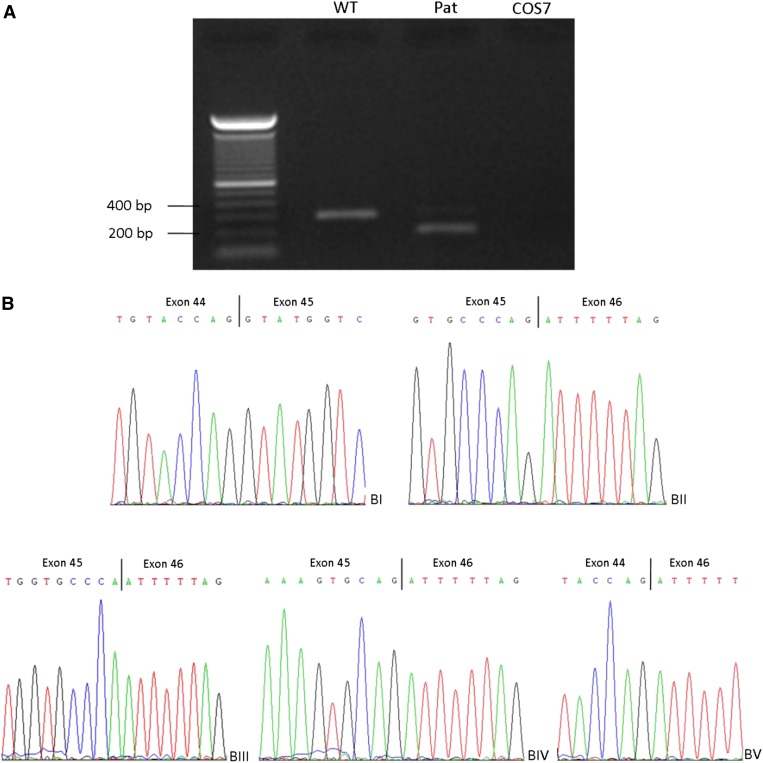
To disclose the mechanisms that underlie those supposed contradictions in genotype-phenotype correlation, we performed bioinformatic analyses and checked splice databases for the respective mutations (Table 2). In silico data demonstrated a strong effect on splicing for the identified 1-bp deletion c.7215delG due to (considerable) weakening of the 5′ donor splice site (Table 2). The mutation c.7215delG affects the last nucleotide of exon 45 and the results of the minigene experiments are shown in Figure 1, A and B. Sequencing of the patient’s WT-sized band revealed a normally spliced transcript missing the last nucleotide of exon 45 reflecting the mutation found on the DNA level. However, an additional smaller band was also detected in the patient. Subcloning revealed either a deletion of only the last residue, deletion of the last 11 nucleotides of exon 45, or skipping of the whole exon 45.
Table 2.
In silico analysis for PKHD1 mutations
| Mutation | NetGene 2 Server | Berkeley Drosophila Genome Project | ESEfinder 3.0 |
|---|---|---|---|
| c.2130_2131insTA | Only mild strengthening of 5′SS (0.93–0.94) | Only mild strengthening of 5′SS (0.94–0.95) | Splice sites: mild weakening of 5′SS (9.12–8.35) |
| SR proteins: loss of SRp55 binding site | |||
| Generation of SF2/ASF binding site: weakening of SRp40 binding site | |||
| c.3306delT | Only mild weakening of 5′SS (0.46–0.41) and 3′SS (0.26–0.25) | Mild weakening of 5′SS (0.46–0.41) | Splice sites: generation of 3′SS in exon 29 |
| SR proteins: loss of SP55 binding site | |||
| c.7215delG | Weakening of the 5′SS (1.00–0.77) | Considerable weakening of 5′SS (0.87–0.32) | Splice sites: loss of 5′SS SR proteins |
| Weakening of SF2/ASF and SF2/ASF (IgM-BRCA1) binding site | |||
| Generation of SC35 binding site |
Data were obtained from the NetGene 2 Server (http://genome.cbs.dtu.dk/services/NetGene2/), Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/splice.html), and ESEfinder 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi). 5′SS, 5′-donor splice site; 3′SS, 3′-acceptor splice site.
Figure 1.
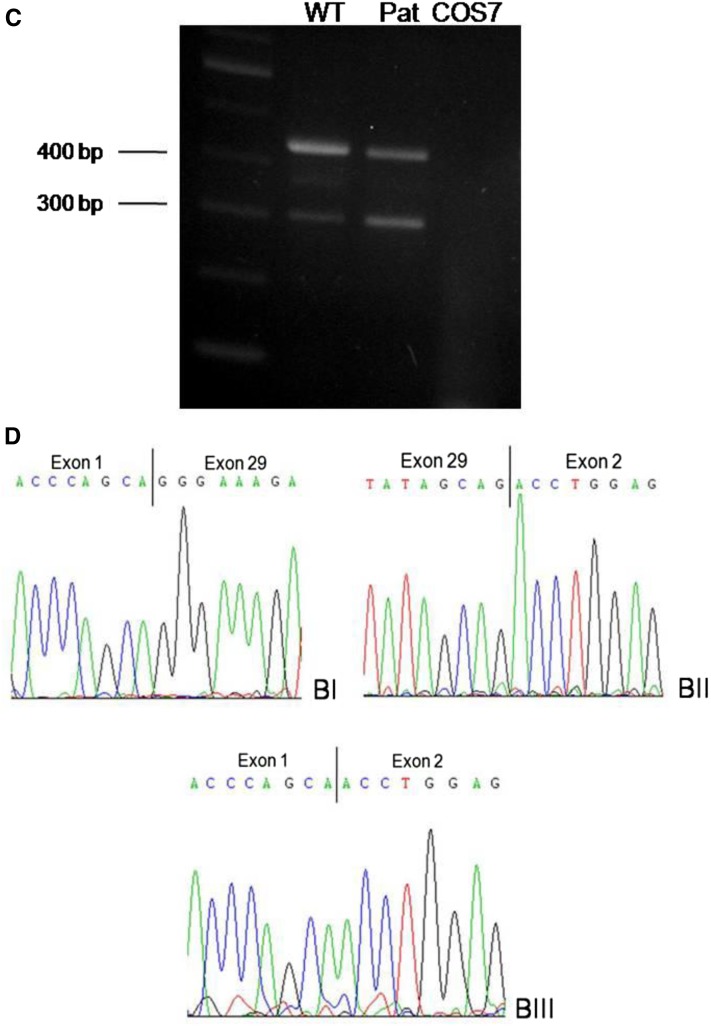
Results of minigene experiments for PKHD1 mutations c.7215delG and c.3306delT. (A and B) After transfection of wild-type (WT) and mutant constructs into COS7 cells, transcripts were analyzed by RT-PCR and cDNA sequencing using primers in exons 44 and 46 of the PKHD1 gene. One band with a length of approximately 320 bp was present in the WT construct as well as in the patient (A) that corresponds to the WT full-length cDNA (BI and BII). Sequencing of the patient’s approximately 320-bp band revealed a normally spliced transcript missing the last nucleotide of exon 45 (BIII), reflecting the mutation found on the DNA level. In the patient, an additional smaller band of approximately 200 bp was also detected (A). After we could not obtain a clear result by sequencing of this band, we subcloned it after gel extraction and sequenced 23 clones. Eleven of those clones showed the WT transcript missing the last nucleotide (BIII) and 10 clones showed skipping of exon 45 (BV). In two cases, the last 11 nucleotides of exon 45 were missing (BIV). (C and D) Results of minigene experiments for mutation c.3306delT using the TRAP vector pSPL3. After transient transfection of WT and mutant constructs into COS7 cells, transcripts were analyzed by RT-PCR and cDNA sequencing using primers in exons 1 and 2 of the pSPL3 vector. Two bands were detected in the WT as well as in the patient’s sample (A). The larger one (approximately 430 bp) corresponds to the expected fragment containing exon 1 and 2 of the pSPL3 vector and in between the PKHD1 exon 29 (BI and II).The smaller band of approximately 300 bp represents a transcript resulting from skipping of PKHD1 exon 29 (BIII).
Two of three in silico analyses for c.3306delT revealed a mild weakening of the 3′ acceptor splice site, whereas the third program did not recognize this splice site, neither for the mutant nor for the WT sequence. Minigene experiments done for this mutation revealed quantitative splicing abnormalities (Figure 1, C and D). Two bands were detected in the WT as well as in the patient’s sample, respectively. Whereas the larger one corresponds to the expected fragment containing exon 1 and 2 of the pSPL3 vector and in between exon 29 of the PKHD1 gene, the smaller band represents a transcript resulting from skipping of exon 29. Our minigene experiments support the predicted in silico findings with a weak character of the acceptor splice site of intron 28 in which the WT construct produces a transcript lacking exon 29.
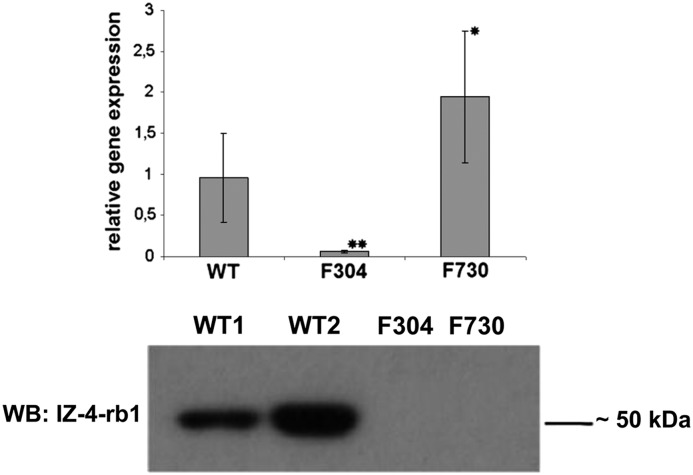
We had kidney tissue from the patient bearing the homozygous PKHD1 mutation c.2130_2131insTA (F730). Thus, we were able to perform real-time PCR experiments and Western blot experiments for this patient. Relative gene expression of PKHD1 in the patient was significantly higher than the expression of WT tissue, which can most likely be explained by the different developmental ages of the samples. Age-matched kidney tissue was not available for analysis. As expected, renal tissue from a severely affected, perinatally demised patient carrying a homozygous PKHD1 frameshift mutation demonstrated a highly significant reduction in PKHD1 expression (Figure 2, top, Supplemental Table 1). Experiments on the protein level using an antibody against the C-terminal end of polyductin/fibrocystin revealed a strong band at about 50 kDa in WT tissue that was lacking in the patients’ samples (Figure 2, bottom).
Figure 2.
PKHD1 expression in kidney tissue. The top panel shows relative PKHD1 expression in moderately affected patient F730 compared with the expression pattern in WT and kidney tissue from a patient with a truncating PKHD1 mutation and a severe form of autosomal recessive polycystic kidney disease (F304). *P<0.05; **P<0.001 compared with WT. The bottom panel shows the results of Western blot (WB) analysis using an antibody against the C-terminal end of polyductin (anti-polyductin, IZ-4-rb1) in renal tissue in the moderately affected patient carrying the homozygous mutation c.2130_2131insTA (F730) compared with the expression of two different wild-type tissues (WT1 and WT2, each different from the one used for RT-PCR experiments) and kidney tissue from a perinatally demised patient carrying the homozygous PKHD1 deletion c.4256_4257del (p.Arg1419fs) (F304).
Discussion
By our long-standing efforts and work on a large cohort of ARPKD families, we were able to identify four unrelated surviving patients with ARPKD with three homozygous PKHD1 mutations that were each expected to lead to premature termination of translation. These results stand in obvious contrast with published data on genotype-phenotype correlations in ARPKD. Overall, these patients represent exceptions to the rule and are the result of continuous work in our laboratories on >500 ARPKD families, which represents one of the largest patient cohorts for ARPKD. Although these are most likely very rare events, these patients tell us a lot about the disease and its pathology.
To disclose the mechanisms that underlie the supposed contradictions in genotype-phenotype correlation, we first performed bioinformatic analyses and checked splice databases for the respective mutations (Table 2). The bioinformatic in silico tools we used are generally thought to predict any effect on the splicing pattern that is caused by the respective nucleotide exchange. Although one definitely should not overvalue these programs and should be aware of their limitations, these software tools are usually regarded to be quite helpful for the decision process and further interpretation. Functional investigations in PKHD1 are considerably hampered by the large size and restricted expression pattern of PKHD1/polyductin. Although kidney tissue was available for analysis in patient F730, no tissue with sufficient protein expression was available for analysis in the other patients. Therefore, we constructed minigenes for the mutations c.3306delT and c.7215delG to perform RNA analysis on a cDNA level after transient transfection of those minigenes into COS7 cells, respectively. Considerable splicing abnormalities were detected for both mutations (Figure 1). Overall, in silico predictions were largely in accordance with our minigene experiments. For c.7215delG, subcloning revealed a much more complex transcriptional pattern than previously expected. For the mutation c.3306delT, splicing primarily resulted in a transcript with exon 29 skipped. The findings obtained by real-time PCR experiments in our study may suggest circumvention of NMD as a plausible explanation for the observed clinical courses. Further experiments for NMD would have been clearly desirable, but could not be performed due to lack of patient material such as viable cells. Thus, we are aware of the limited character of these data and that any conclusions should be drawn with some circumspection. The same applies for the Western blot data. While still speculative, it is conceivable that in our experiment on protein level the detected band in the WT lane represents the part of the protein that remains in the membrane after “ectodomain shedding” has taken place as described by Kaimori et al. (24). The full-length transcript may not have been detected due to the low t1/2 of this isoform or technical problems concerning the transfer of this huge protein. Overall, the mutation’s truncating character was verified and reinitiation of translation of a protein that comprises the C-terminal part of polyductin/fibrocystin could be excluded.
In conclusion, all analyzed mutations were demonstrated to lead to an alteration on the transcript level. Our data indicate that some mutations might disrupt the assumed critical functional stoichiometric or temporal balance between the different PKHD1 protein products due to an alternative splicing pattern. In addition, results obtained by RT-PCR may provide a first tentative hint that NMD may play a role in defining the phenotype of patients with ARPKD. Although we are aware of the limitations of some of our findings (which is due to the described challenges of research in ARPKD), it can be hypothesized that the majority of nonsense mutations in PKHD1 will lead to NMD and subsequent loss of protein function with the consequence of a severe phenotype with perinatal or neonatal death. In a very few cases, it can be speculated that a shortened polyductin/fibrocystin isoform with residual protein function might be translated due to alternative splicing and possibly circumvention of NMD that may give rise to a surprisingly mild/moderate phenotype of those patients.
Disclosures
V.F. and C.B. are employees of Bioscientia/Sonic Healthcare. In addition, C.B. holds a part-time faculty appointment at the University of Freiburg.
Supplementary Material
Acknowledgments
This work was funded by grants from the German Research Foundation (to K.Z. and C.B.), the Germany Kidney Foundation (C.B.), and the PKD Foundation (C.B.).
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.00920114/-/DCSupplemental.
References
- 1.Guay-Woodford LM: Autosomal recessive polycystic kidney disease: Clinical and genetic profiles. In: Polycystic Kidney Disease, edited by Watson ML, Torres VE, Oxford, Oxford University Press, 1996, pp 237–266 [Google Scholar]
- 2.Bergmann C: Autosomal recessive polycystic kidney disease. In: Ciliopathies: A Reference for Clinicians, edited by Kenny T, Beales P, Oxford, Oxford University Press, 2014, pp 194–217 [Google Scholar]
- 3.Zerres K, Rudnik-Schöneborn S, Senderek J, Eggermann T, Bergmann C: Autosomal recessive polycystic kidney disease (ARPKD). J Nephrol 16: 453–458, 2003 [PubMed] [Google Scholar]
- 4.Guay-Woodford LM, Desmond RA: Autosomal recessive polycystic kidney disease: The clinical experience in North America. Pediatrics 111: 1072–1080, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Bergmann C, Senderek J, Windelen E, Küpper F, Middeldorf I, Schneider F, Dornia C, Rudnik-Schöneborn S, Konrad M, Schmitt CP, Seeman T, Neuhaus TJ, Vester U, Kirfel J, Büttner R, Zerres K, APN (Arbeitsgemeinschaft für Pädiatrische Nephrologie) : Clinical consequences of PKHD1 mutations in 164 patients with autosomal-recessive polycystic kidney disease (ARPKD). Kidney Int 67: 829–848, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, Milliner DS, Torres VE, Harris PC: The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 30: 259–269, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, Bergmann C, Senderek J, Esquivel E, Zeltner R, Rudnik-Schöneborn S, Mrug M, Sweeney W, Avner ED, Zerres K, Guay-Woodford LM, Somlo S, Germino GG: PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am J Hum Genet 70: 1305–1317, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergmann C, Senderek J, Sedlacek B, Pegiazoglou I, Puglia P, Eggermann T, Rudnik-Schöneborn S, Furu L, Onuchic LF, De Baca M, Germino GG, Guay-Woodford L, Somlo S, Moser M, Büttner R, Zerres K: Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1). J Am Soc Nephrol 14: 76–89, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Furu L, Onuchic LF, Gharavi A, Hou X, Esquivel EL, Nagasawa Y, Bergmann C, Senderek J, Avner E, Zerres K, Germino GG, Guay-Woodford LM, Somlo S: Milder presentation of recessive polycystic kidney disease requires presence of amino acid substitution mutations. J Am Soc Nephrol 14: 2004–2014, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Rossetti S, Torra R, Coto E, Consugar M, Kubly V, Málaga S, Navarro M, El-Youssef M, Torres VE, Harris PC: A complete mutation screen of PKHD1 in autosomal-recessive polycystic kidney disease (ARPKD) pedigrees. Kidney Int 64: 391–403, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Bergmann C, Senderek J, Küpper F, Schneider F, Dornia C, Windelen E, Eggermann T, Rudnik-Schöneborn S, Kirfel J, Furu L, Onuchic LF, Rossetti S, Harris PC, Somlo S, Guay-Woodford L, Germino GG, Moser M, Büttner R, Zerres K: PKHD1 mutations in autosomal recessive polycystic kidney disease (ARPKD). Hum Mutat 23: 453–463, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Bergmann C, Senderek J, Schneider F, Dornia C, Küpper F, Eggermann T, Rudnik-Schöneborn S, Kirfel J, Moser M, Büttner R, Zerres K: PKHD1 mutations in families requesting prenatal diagnosis for autosomal recessive polycystic kidney disease (ARPKD). Hum Mutat 23: 487–495, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Sharp AM, Messiaen LM, Page G, Antignac C, Gubler MC, Onuchic LF, Somlo S, Germino GG, Guay-Woodford LM: Comprehensive genomic analysis of PKHD1 mutations in ARPKD cohorts. J Med Genet 42: 336–349, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Losekoot M, Haarloo C, Ruivenkamp C, White SJ, Breuning MH, Peters DJ: Analysis of missense variants in the PKHD1-gene in patients with autosomal recessive polycystic kidney disease (ARPKD). Hum Genet 118: 185–206, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Adeva M, El-Youssef M, Rossetti S, Kamath PS, Kubly V, Consugar MB, Milliner DM, King BF, Torres VE, Harris PC: Clinical and molecular characterization defines a broadened spectrum of autosomal recessive polycystic kidney disease (ARPKD). Medicine (Baltimore) 85: 1–21, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Gunay-Aygun M, Tuchman M, Font-Montgomery E, Lukose L, Edwards H, Garcia A, Ausavarat S, Ziegler SG, Piwnica-Worms K, Bryant J, Bernardini I, Fischer R, Huizing M, Guay-Woodford L, Gahl WA: PKHD1 sequence variations in 78 children and adults with autosomal recessive polycystic kidney disease and congenital hepatic fibrosis. Mol Genet Metab 99: 160–173, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denamur E, Delezoide AL, Alberti C, Bourillon A, Gubler MC, Bouvier R, Pascaud O, Elion J, Grandchamp B, Michel-Calemard L, Missy P, Zaccaria I, Le Nagard H, Gerard B, Loirat C, Barbet J, Beaufrère AM, Berchel C, Bessières B, Boudjemaa S, Buenerd A, Carles D, Clemenson A, Dechelotte P, Devisme L, Dijoud F, Espérandieu O, Fallet C, Gonzalès M, Hillion Y, Jacob B, Joubert M, Kermanach P, Lallemand A, Laquerrière A, Laurent N, Liprandi A, Loeuillet L, Loget P, Martinovic J, Ménez F, Narcy F, Roux JJ, Rouleau-Dubois C, Sinico M, Tantau J, Wann AR, Société Française de Foetopathologie : Genotype-phenotype correlations in fetuses and neonates with autosomal recessive polycystic kidney disease. Kidney Int 77: 350–358, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Zvereff V, Yao S, Ramsey J, Mikhail FM, Vijzelaar R, Messiaen L: Identification of PKHD1 multiexon deletions using multiplex ligation-dependent probe amplification and quantitative polymerase chain reaction. Genet Test Mol Biomarkers 14: 505–510, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Nagasawa Y, Matthiesen S, Onuchic LF, Hou X, Bergmann C, Esquivel E, Senderek J, Ren Z, Zeltner R, Furu L, Avner E, Moser M, Somlo S, Guay-Woodford L, Büttner R, Zerres K, Germino GG: Identification and characterization of Pkhd1, the mouse orthologue of the human ARPKD gene. J Am Soc Nephrol 13: 2246–2258, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Ward CJ, Yuan D, Masyuk TV, Wang X, Punyashthiti R, Whelan S, Bacallao R, Torra R, LaRusso NF, Torres VE, Harris PC: Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum Mol Genet 12: 2703–2710, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Zhang MZ, Mai W, Li C, Cho SY, Hao C, Moeckel G, Zhao R, Kim I, Wang J, Xiong H, Wang H, Sato Y, Wu Y, Nakanuma Y, Lilova M, Pei Y, Harris RC, Li S, Coffey RJ, Sun L, Wu D, Chen XZ, Breyer MD, Zhao ZJ, McKanna JA, Wu G: PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc Natl Acad Sci U S A 101: 2311–2316, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, Luo Y, Wilson PD, Witman GB, Zhou J: The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J Am Soc Nephrol 15: 592–602, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Menezes LF, Cai Y, Nagasawa Y, Silva AM, Watkins ML, Da Silva AM, Somlo S, Guay-Woodford LM, Germino GG, Onuchic LF: Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int 66: 1345–1355, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Kaimori JY, Nagasawa Y, Menezes LF, Garcia-Gonzalez MA, Deng J, Imai E, Onuchic LF, Guay-Woodford LM, Germino GG: Polyductin undergoes notch-like processing and regulated release from primary cilia. Hum Mol Genet 16: 942–956, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Wu M, Wang S, Shah JV, Wilson PD, Zhou J: Polycystic kidney disease protein fibrocystin localizes to the mitotic spindle and regulates spindle bipolarity. Hum Mol Genet 19: 3306–3319, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cartegni L, Chew SL, Krainer AR: Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298, 2002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.