Abstract
Introduction
The goal of personalized medicine is to treat patients with a therapy predicted to be efficacious based on the molecular characteristics of the tumor, thereby sparing the patient futile or toxic therapy. Anaplastic lymphoma kinase (ALK) inhibitors are effective against ALK-positive non–small-cell lung cancer (NSCLC) tumors, but to date the only approved companion diagnostic is a break-apart fluorescence in situ hybridization (FISH) assay. Immunohistochemistry (IHC) is a clinically applicable cost-effective test that is sensitive and specific for ALK protein expression. The purpose of this study was to assemble an international team of expert pathologists to evaluate a new automated standardized ALK IHC assay.
Methods
Archival NSCLC tumor specimens (n =103) previously tested for ALK rearrangement by FISH were provided by the international collaborators. These specimens were stained by IHC with the anti-ALK (D5F3) primary antibody combined with OptiView DAB IHC detection and OptiView amplification (Ventana Medical Systems, Inc., Tucson, AZ). Specimens were scored binarily as positive if strong granular cytoplasmic brown staining was present in tumor cells. IHC results were compared with the FISH results and interevaluator comparisons made.
Results
Overall for the 100 evaluable cases the ALK IHC assay was highly sensitive (90%), specific (95%), and accurate relative (93%) to the ALK FISH results. Similar results were observed using a majority score. IHC negativity was scored by seven of seven and six of seven evaluators on three and two FISH-positive cases, respectively. IHC positivity was scored on two FISH-negative cases by seven of seven readers. There was agreement among seven of seven and six of seven readers on 88% and 96% of the cases before review, respectively, and after review there was agreement among seven of seven and six of seven on 95% and 97% of the cases, respectively.
Conclusions
On the basis of expert evaluation the ALK IHC test is sensitive, specific, and accurate, and a majority score of multiple readers does not improve these results over an individual reader’s score. Excellent inter-reader agreement was observed. These data support the algorithmic use of ALK IHC in the evaluation of NSCLC.
Keywords: Non–small-cell lung cancer, Anaplastic lymphoma kinase, Immunohistochemistry, Fluorescence in situ hybridization, Companion diagnostics, Biomarkers, Crizotinib
Personalized medicine is at the forefront of lung cancer therapy with the goal to reduce the 1.4 million lung cancer deaths per year worldwide.1 The fundamental principal of personalized medicine is treating patients with a therapy predicted to be efficacious based on the molecular characteristics of the tumor, thereby sparing the patient any potential morbidity and mortality associated with ineffective therapy. Non–small-cell lung cancer (NSCLC) accounts for 85% of all lung cancer and the 5-year survival rate for NSCLC is only 16% because of late-stage diagnosis2 and lack of effective systemic therapy. However, treatment for advanced disease has improved recently commensurate with the identification of key oncogenic alternations driving tumorigenesis, such as activating somatic mutations or chromosomal rearrangements targetable with specific therapeutics. 3 Tumors harboring epidermal growth factor receptor mutations are distinctly sensitive to epidermal growth factor receptor tyrosine kinase inhibitors (TKIs) like erlotinib and gefitinib,4–8 whereas tumors containing anaplastic lymphoma kinase (ALK) gene rearrangements are sensitive to the ALK TKI crizotinib.9,10
ALK gene rearrangements were first discovered in NSCLC in 2007 by Soda et al.11 who identified that the 3′ end of ALK was juxtaposed to the 5′ end of echinoderm microtubule- associated protein-like 4 (EML4) gene attributable to an inversion within chromosome 2p. Although EML4 is the main fusion partner in NSCLC, the 2p21 break point (EML4) is variable and other chromosomal fusion partners have been reported, for example, kinesin family member 5B (KIF5B)–ALK, TRK-fused gene (TFG)–ALK, and kinesin light chain (KLC1)–ALK.12–15 The clinical significance of the numerous variant rearrangements has not been defined but is being investigated. The coiled-coil domains in the 5′ fusion partners promote dimerization and oligomerization, which leads to constitutive activation of the ALK kinase domain and downstream signaling pathways culminating in tumor cell proliferation, survival, and oncogenesis.13,16 The incidence of ALK gene rearrangements is approximately 3% to 4% in an unselected NSCLC population, which equates to roughly 40,000 ALK- positive patients/year worldwide.1,17
NSCLC tumors that contain ALK rearrangement are strikingly responsive to the ALK TKI crizotinib (PF-02341066, Xalkori; Pfizer, New York, NY), an adenosine triphosphate (ATP) competitive small- molecule targeting the kinase domain in the ALK protein.18 An international multicenter phase I study initially demonstrated an objective response rate of 60.8% and a disease control rate of 84% among the 143 evaluable patients.9,10 The median duration of response was 49.1 weeks and the median progression-free survival (PFS) was 9.7 months for all, often heavily pretreated patients, but a PFS of 18.3 months was observed for those receiving crizotinib as first-line therapy. The estimated overall survival at 6 and 12 months was 87.9% and 74.8%, but these data are not fully mature. Interim results of an ongoing phase 2 study have shown an overall response rate of 53% and a median PFS of 8.5 months.19 Early results from a phase III study (PROFILE 1007) comparing chemotherapy with crizotinib revealed an increase in PFS from a median of 3.0 months to 7.7 months and an overall response rate of 20% versus 65%.20 Numerous other phase III clinical trials are ongoing. Crizotinib received accelerated approval from the U.S. Food and Drug Administration (FDA) in 2011, but approval was contingent upon documentation of ALK positivity by an FDA-approved diagnostic test, and to date the only FDA-approved companion diagnostic is the fluorescence in situ hybridization (FISH) assay using the Vysis break-apart FISH probe kit (Abbott Molecular, Des Plaines, IL).21
Numerous studies indicate that immunohistochemistry (IHC), under the appropriate conditions, is sensitive and specific for determination of ALK protein expression22–28 and is a practical cost-effective alternative to the ALK FISH assay. In fact, there are even potential clinical benefits to ALK IHC over FISH, as demonstrated by significant clinical improvement from crizotinib in ALK IHC-positive, FISH-negative patients.29,30 However, a standardized ALK IHC diagnostic test is required, as studies comparing IHC with FISH have used different ALK antibody clones, detection systems, antigen retrieval techniques, and scoring methods.
The purpose of this study was to convene an international team of expert pathologists to evaluate the scoring of a new automated standardized ALK IHC assay kit that employs the antibody clone D5F3 (Ventana Medical Systems, Inc., [VMSI], Tucson, AZ), an ultrasensitive detection system (OptiView DAB, VMSI, Tucson, AZ), an interpretation guide and scoring method. The detection system used an extra amplification step that has been shown to improve visualization of low-level ALK protein expression found in NSCLC compared with the same antibody clone and detection system without amplification.31 The ALK IHC positivity or negativity status from each expert reader was compared with the ALK FISH status and inter-reader comparisons were made.
PATIENTS AND METHODS
Patients
Archival NSCLC tumor specimens (n = 103) were provided by the international collaborators. The tumors provided were previously tested for ALK rearrangement by FISH assessment and there were 48 FISH-positive cases, which consisted of five cytology blocks, eight biopsy samples, and 35 surgical resections, whereas there were 55 FISH-negative cases consisting of four cytology blocks, 14 biopsy samples, and 37 resections. The readers of the ALK IHC protein expression assay were blinded to the FISH status until after completion of the IHC scoring.
ALK Protein Expression
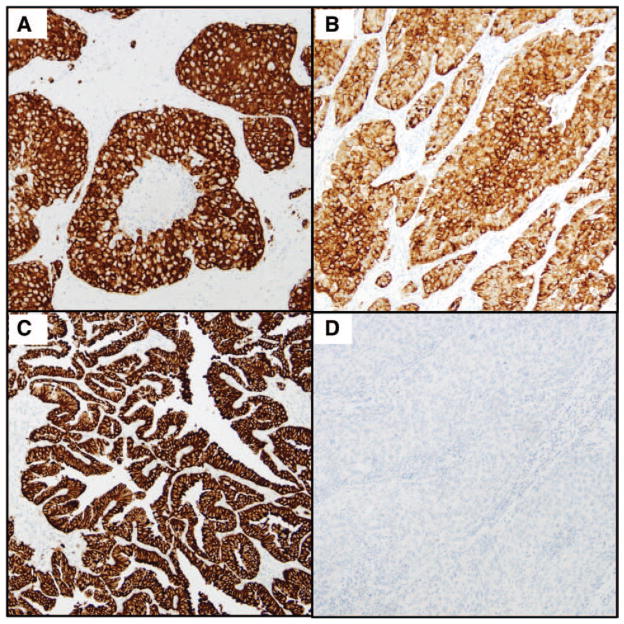
The specimens were sectioned at a thickness of 4 μM and stained by IHC within 3 weeks of the sectioning. ALK IHC staining was performed on a VENTANA BenchMark XT automated slide-processing system at Ventana Medical Systems, Inc.31,32 In brief, slides of NSCLC tumor were subjected to deparaffinization using EZ Prep (VMSI) and extended Cell Conditioning 1. Tissue sections were then incubated with anti-ALK antibody (clone D5F3, VMSI) for 20 minutes. OptiView DAB IHC Detection Kit (VMSI) and OptiView Amplification Kit (VMSI) were used according to the manufacturer’s recommendations for the visualization of the bound primary antibody.31,32 Tissue slides were counterstained with Hematoxylin II (VMSI) and Bluing Reagent (VMSI). Slides were dehydrated and cleared before coverslipping with Tissu-Tek (Sakura Finetek Japan, Tokyo, Japan). Specimens were examined and evaluated by six pathologists and one pathology-trained clinical molecular biologist who assessed only the tumor region of the specimen by using a light microscope. Nontumor (alveolar macrophages, cells of neural origin, glandular epithelial staining, and cells within lymphocytic infiltrate), necrotic, degenerated, crushed, or clearly artifactual (edge, retraction, thermal) tissues were not scored. Specimens were scored positive if strong granular cytoplasmic brown staining in tumor cells (any percentage of positive tumor cells) was present. Homogenous staining of all the tumor cells was not required as long as there were regions with strong cytoplasmic staining. Cases were scored as negative if there was no or only weak cytoplasmic staining. Examples of positive and negative staining are shown in Figure 1. Before the interobserver study commenced the participants were presented with 10 cases, which were scored by the evaluators and then the plenum discussed the positive and negative criteria.
FIGURE 1.
Anaplastic lymphoma kinase immunohistochemistry staining. A–C, Examples of positive staining. D, Example of negative staining.
Statistical Analysis
For the calculation of all concordance estimates (IHC versus FISH), only cases recorded as evaluable were used. Individual reader concordance was calculated using FISH as the reference group. The associated 95% confidence intervals were calculated using the score method. The overall concordance for all readers when comparing IHC versus FISH is based on the weighted average of the individual reader concordance estimates; the 95% confidence intervals were calculated using the percentile bootstrap method. The data were recorded in Microsoft Excel spreadsheets, and analyzed using SAS version 9.2.
RESULTS
Agreement between ALK protein expression and ALK break-apart by FISH
A total of 103 NSCLC specimens were blindly evaluated by the panel and it was unanimously determined that three FISH-positive cases no longer contained any tumor tissue and were therefore excluded from any further analysis. Table 1 shows the comparison between the ALK IHC results for each reader and the previously determined FISH results. The FISH status was not re-evaluated, and any differences in the number of cases for each reader reflects that reader’s discretion of evaluability of the IHC. The ALK IHC assay was highly sensitive, specific, and accurate relative to the ALK FISH results. Overall sensitivity, specificity, and accuracy were 90%, 95%, and 93%, respectively, with the ranges being 87% to 91%, 94% to 96%, and 91–94%. After scoring the discrepant cases, both IHC versus FISH scores and inter-reader scores were discussed among the readers, and after this discussion four readers made a total of eight changes. Each of the four readers changed one IHC-positive case to IHC negative and two readers each changed two unevaluable cases to IHC negative. Among the cases that were changed from positive to negative, one was changed because of macrophage staining, two because of weak staining, and one because it was a score-sheet entry input error. Among the unevaluable to negative cases three were related to number of tumor cells in the specimens and one case was a score-sheet entry input error. These changes are detailed in Table 2, which shows that there was no change in overall sensitivity, accuracy changed by less than 1%, and specificity changed from 95% to 96% after the review of the discrepant cases. To determine whether having a majority score improved upon the results of the individual readers, the cases were then classified as either positive or negative based on four of the seven readers agreeing upon the score before consensus review. Table 3 shows that results of this majority score were minimally different than that of any of the individual readers, with the biggest difference being relative to reader 5 who had a sensitivity of 87% (39 IHC+/45 FISH+), whereas the majority score sensitivity was 91% (39 IHC+/43 FISH+). Taken together these data indicate that the ALK IHC test is sensitive, specific, and accurate relative to FISH and that having a majority score of multiple readers does not improve these parameters over an individual reader.
TABLE 1.
ALK IHC vs. ALK FISH for All Readers and Overall
| Reader (n Evaluable) | FISH+ | FISH− | Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) | |
|---|---|---|---|---|---|---|
| Reader 1 (n = 97) | IHC+ | 39 | 2 | 91 (78–96) | 96 (88–99) | 94 (87–97) |
| IHC− | 4 | 52 | ||||
| Reader 2 (n = 99) | IHC+ | 39 | 2 | 89 (76–95) | 96 (88–99) | 93 (86–97) |
| IHC− | 5 | 53 | ||||
| Reader 3 (n = 99) | IHC+ | 39 | 2 | 89 (76–95) | 96 (88–99) | 93 (86–97) |
| IHC− | 5 | 53 | ||||
| Reader 4 (n = 99) | IHC+ | 42 | 3 | 93 (82–98) | 94 (85–99) | 94 (87–97) |
| IHC− | 3 | 51 | ||||
| Reader 5 (n = 99) | IHC+ | 39 | 3 | 87 (74–94) | 94 (85–98) | 91 (84–95) |
| IHC− | 6 | 51 | ||||
| Reader 6 (n = 97) | IHC+ | 40 | 3 | 91 (79–96) | 94 (85–98) | 93 (86–97) |
| IHC− | 4 | 50 | ||||
| Reader 7 (n = 97) | IHC+ | 40 | 3 | 91 (79–96) | 94 (85–98) | 93 (86–97) |
| IHC− | 4 | 50 | ||||
| Overall (n = 687) | IHC+ | 278 | 18 | 90 (80–98) | 95 (89–99) | 93 (88–97) |
| IHC− | 31 | 360 |
ALK, anaplastic lymphoma kinase; IHC, immunohistochemistry; FISH, fluoroscence in situ hybridization; CI, confidence interval.
TABLE 2.
ALK IHC vs. ALK FISH for All Readers and Overall after Consensus Review
| Reader (n evaluable) | IHC | FISH + | FISH− | Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) |
|---|---|---|---|---|---|---|
| Reader 1 (n = 97) | IHC+ | 39 | 2 | 91 (78–96) | 96 (88–99) | 94 (87–97) |
| IHC− | 4 | 52 | ||||
| Reader 2 (n = 99) | IHC+ | 39 | 2 | 89 (76–95) | 96 (88–99) | 93 (86–97) |
| IHC− | 5 | 53 | ||||
| Reader 3 (n = 99) | IHC+ | 39 | 2 | 89 (76–95) | 96 (88–99) | 93 (86–97) |
| IHC− | 5 | 53 | ||||
| Reader 4 (n = 99) | IHC+ | 42 | 2* | 93 (82–98) | 96* (88–99) | 95* (89–98) |
| IHC− | 3 | 52* | ||||
| Reader 5 (n = 99) | IHC+ | 39 | 2* | 87 (74–94) | 96* (88–99) | 92* (85–96) |
| IHC− | 6 | 52* | ||||
| Reader 6 (n = 99)* | IHC+ | 40 | 2* | 91 (79–96) | 96* (88–99) | 94* (87–97) |
| IHC− | 4 | 53* | ||||
| Reader 7 (n = 99)* | IHC+ | 40 | 2* | 91 (79–96) | 96* (88–99) | 94* (87–97) |
| IHC− | 4 | 53* | ||||
| Overall (n = 691)* | IHC+ | 278 | 14* | 90 (80–98) | 96* (90–100) | 93* (88–98) |
| IHC − | 31 | 368* |
ALK, anaplastic lymphoma kinase; IHC, immunohistochemistry; FISH, fluoroscence in situ hybridization; CI, confidence interval.
indicates changes from Table 1.
TABLE 3.
ALK IHC vs. ALK FISH for Majority Score, Four of Seven Readers in Agreement
| Reader (n evaluable) | IHC | FISH+ | FISH− | Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) |
|---|---|---|---|---|---|---|
| Four-reader agreement | IHC+ | 39 | 2 | 91 (78–96) | 96 (88–99) | 94 (87–97) |
| IHC− | 4 | 53 |
ALK, anaplastic lymphoma kinase; IHC, immunohistochemistry; FISH, fluoroscence in situ hybridization; CI, confidence interval.
Agreement between Readers for ALK Protein Expression
The agreement between the IHC scores for each of the readers was then evaluated for the 100 ALK IHC evaluable cases. Table 4 shows that all seven readers agreed on 48 ALK IHC-negative cases and 40 ALK IHC-positive cases, therefore, there was a striking 100% agreement among all seven readers on 88% of the total cases. Interestingly, three of the 48 cases with 100% agreement on IHC negativity were classified as ALK FISH-positive and two of the 40 IHC-positive cases with 100% reader agreement were deemed FISH negative. Six of seven readers (86%) agreed on seven IHC-negative cases and one IHC positive case. The FISH status of these cases is shown in Table 4 as well as the score from the reader that did not align with the other six readers. For the remaining four cases there was agreement between five of seven and four of seven readers for one IHC-negative cases each and three of seven readers agreement for two IHC-positive cases. The FISH status and other readers’ scores on these cases can be seen in Table 4. Table 5 shows that after the consensus review of the few discrepant cases there was 100% agreement between the seven readers on 95% of the total cases; 55 IHC-negative cases and 30 IHC-positive cases. There were still three cases with seven of seven agreement on IHC negative but FISH positive and two cases with seven of seven agreement on IHC positive but FISH negative. There were two cases with only six of seven agreement, one case with four of seven agreement, and two cases with three of seven agreement; the FISH status of the cases as well as the nonmajority readers’ scores are listed in Table 5. As a whole there was extremely close agreement between all seven expert readers for a majority of the cases.
TABLE 4.
ALK IHC Reader Agreement before Consensus Review
| Number of Cases by IHC | Readers Agree | FISH Status | Other Readers Score | FISH Status | Other Readers Score |
|---|---|---|---|---|---|
| 48 IHC Neg | 7/7 | 45 FISH Neg | 3 FISH Pos | ||
| 40 IHC Pos | 7/7 | 2 FISH Neg | 38 FISH Pos | ||
| 7 IHC Neg | 6/7 | 6 FISH Neg | 4 IHC Pos, 2 Uneval | 1 FISH Pos | 1 IHC Pos |
| 1 IHC Pos | 6/7 | 1 FISH Pos | 1 IHC Neg | ||
| 1 IHC Neg | 5/7 | 1 FISH Neg | 2 Uneval | ||
| 1 IHC Neg | 4/7 | 1 FISH Neg | 1 IHC Pos, 2 Uneval | ||
| 1 IHC Pos | 3/7 | 1 FISH Pos | 3 IHC Neg, 1 Uneval | ||
| 1 IHC Pos | 3/7 | 1 FISH Pos | 4 Uneval |
ALK, anaplastic lymphoma kinase; IHC, immunohistochemistry; FISH, fluoroscence in situ hybridization, Neg, negative; Pos, positive; Uneval, unevaulable.
TABLE 5.
ALK IHC Reader Agreement after Consensus Review
| Number of Cases by IHC | Readers Agree | FISH Status | Other Readers Score | FISH Status | Other Readers Score |
|---|---|---|---|---|---|
| 55 IHC Neg | 7/7 | 52 FISH Neg | 3 FISH Pos | ||
| 40 IHC Pos | 7/7 | 2 FISH Neg | 38 FISH Pos | ||
| 1 IHC Neg | 6/7 | 1 FISH Pos | 1 IHC Pos | ||
| 1 IHC Pos | 6/7 | 1 FISH Pos | 1 IHC Neg | ||
| 1 IHC Neg | 4/7 | 1 FISH Neg | 1 IHC Pos, 2 Uneval | ||
| 1 IHC Pos | 3/7 | 1 FISH Pos | 3 IHC Neg, 1 Uneval | ||
| 1 IHC Pos | 3/7 | 1 FISH Pos | 4 Uneval |
ALK, anaplastic lymphoma kinase; IHC, immunohistochemistry; FISH, fluoroscence in situ hybridization, Neg, negative; Pos, positive; Uneval, unevaulable.
Assay Background
To evaluate the possibility that the detection system used with this ALK IHC kit, which uses an amplification step, produces some nonspecific background (e.g., nontumor cell or acellular staining) that would hinder the accurate scoring of specimens, six of the seven expert readers assessed staining in the nontumor regions of the specimens. Table 6 shows that high background ranged from 0% to 23% of the cases, with the average being 11% of the cases. Although the readers did report some high background, this did not seem to have a negative impact on the overall ability of the readers to accurately score the specimens, as shown by the overall 90% sensitivity, 95% specificity, and 93% accuracy relative to FISH status (Tables 1 and 2). Likewise, the background did not seem to negatively influence the inter-reader scores, as shown by the high concordance between the readers (Tables 4 and 5) and the 97% agreement (95 of 98 cases) between readers 3 and 4, who reported the widest range of background levels, 0% to 23% of the cases, respectively.
TABLE 6.
ALK IHC Cases with High Background
| Reader | n/N (%) |
|---|---|
| 1 | 4/99 (4.0) |
| 2 | N/A |
| 3 | 0/98 (0) |
| 4 | 23/99 (23.2) |
| 5 | 9/99 (9.1) |
| 6 | 9/97 (9.3) |
| 7 | 19/97 (19.6) |
ALK, anaplastic lymphoma kinase; IHC, immunohistochemistry; N/A, not available.
DISCUSSION
The goal of this study was to bring together for the first time an international team of experts to evaluate a new standardized ALK IHC assay with automated staining. On the basis of the evaluation of 100 cases by ALK IHC and compared with prior ALK FISH results it was observed that the IHC test is sensitive, specific, and accurate compared with FISH and that having a majority score of multiple readers does not improve these results over an individual reader’s score. Even after having a consensus review the sensitivity stayed the same and specificity and accuracy were only marginally improved, indicative of consistency and accuracy with the assay and readers before the consensus review. To achieve a high-quality standardized assay there needs to be high interreader agreement among readers, and this study demonstrated extremely close agreement with 100% agreement among seven readers on 88% of the total cases and only four cases (4%) with a reader agreement of five of seven or lesser. After the consensus review there was 100% agreement among the seven readers on 95% of the cases and there were three cases with a reader agreement of five of seven or lesser. Thus the shift was because of changes from the six of seven agreement to seven of seven agreement. These three cases were all surgical resections where one case had preservation issues and two cases had few tumor cells remaining. There were few cases with heterogeneous staining. There were two cases where the differences in staining we observed seemes to be because of variations in protein expression within the specimens (Fig. 2), similar to what we have observed in the two NSCLC ALK- positive cell lines H2228 and H3122 where some cells within the block have very high expression whereas others have more modest expression (Wynes and Hirsch, unpublished data, 2013). We detected another two cases where there may have been some differences in staining intensity because of fixation issues in the center of the specimen; however, we had no cases that displayed definitive positive staining in one area and negative in other tumor areas. Heterogeneity was not a source of discrepancy among the readers; likewise, there were no discrepancies between these cases and FISH as all were scored positive by IHC as well as FISH. Although some readers did report some background (0%–23% of the cases) depending on reader, this did not affect the sensitivity, specificity, or accuracy relative to FISH or the inter-reader agreement, therefore this seems to be a quality standardized ALK IHC test.
FIGURE 2.
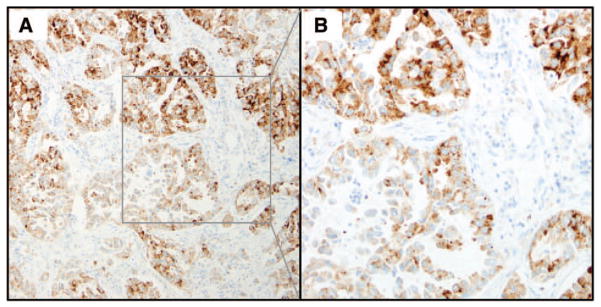
Anaplastic lymphoma kinase heterogeneous immunohistochemistry staining. A, Example of one case that showed heterogeneous staining. B, Higher magnification of the selected region of the specimen shown in (A).
There have been some significant treatment advances for patients with advanced NSCLC in the past few years, which have led to increased PFS and quality of life. These improvements have come through a better understanding that lung cancers are driven by genetic abnormalities and that these genetic abnormalities can be targeted therapeutically.4–10 ALK inhibitors, like crizotinib, but many others in the pipeline, are highly effective in patients that have ALK rearrangement. But to treat these patients there needs to be a validated companion diagnostic that accurately identifies these patients. The only FDA-approved companion diagnostic for ALK inhibitor selection is the Abbott break-apart FISH assay.21 Unfortunately, this assay only indicates whether the ALK gene is broken at the DNA level and does not determine whether there has been a productive rejoining of the DNA creating a continuous open reading frame that results in a complete chimeric mRNA transcript and ultimately a translated functional fusion protein. It may be assumed that an ALK rearrangement detected by FISH always leads to a productive transcript and protein expression. However, it is entirely possible that after a chromosomal rearrangement a stop codon may be generated at the break point or that a 5′ fusion partner without promoter activity or that no fusion partner at all is rejoined to 3′ ALK, all of which would give a FISH break-apart signal but not a productive transcript and functional protein. This may explain the observation that there are some FISH-positive patients who do not receive clinical benefit from crizotinib, for example, 25 of 143 evaluable patients (17.5%), who were all FISH positive, in the international phase I never achieved disease control. 9 Conversely, there are FISH-negative, but IHC-positive, patients who actually do receive significant clinical benefit from crizotinib.29,30 However, these latter examples are limited case reports because all the clinical trials to date required positivity by FISH to be included in a study.
A potentially better assay to select patients to receive an ALK inhibitor is one that detects ALK expression at the protein level. This assay would then allow one to verify that the actual protein target of the inhibitor, that is, the ATP-binding pocket in the kinase domain of ALK, is present, alleviate any concern about unproductive ligation after rearrangement, and detect any expression mediated by any other aberrant nonrearrangement mechanism. ALK is not normally expressed in the lung and any expression would be considered abnormal.
IHC is an assay that would meet all these requirements and it has been in the routine clinical setting for decades, is cost effective, and does not demand unusual expertise to evaluate the specimen. Many studies have shown strong correlations between ALK IHC and FISH, and in an early study comparing ALK IHC with ALK FISH in 153 lung adenocarcinoma (ADC) patients it was shown that there was a difference in sensitivity and specificity relative to FISH, depending on the clone of antibody used with the D5F3 clone generating a sensitivity and specificity of 100% and 99%, respectively, whereas that of the ALK1 antibody was only 67% and 97%, respectively.24 The method of detection here was the Dako EnVision+ system (Dako North America, Inc., Carpinteria, CA). In a study with a large series of 640 patients (450 ADC, 163 squamous cell carcinoma [SCC], 27 other histologies) and using the 5A4 clone with the iView detection system (Ventana) the authors observed a sensitivity of 100% and a specificity of 98% with 10 of 640 patients displaying FISH negativity but IHC positivity.25 A 90% sensitivity and 98% specificity was found when comparing IHC with FISH in a small series of 101 ADC patients using the ALK1 clone and the Dako ADVANCE detection system (Dako North America, Inc.).33 A more recent study by To et al,26 examined 373 ADC patients by ALK IHC and 351 of those by ALK FISH. They found 22 IHC-positive cases of which 20 were FISH positive, giving rise to a sensitivity of 100% and specificity of 99%, however, when the 22 IHC-positive cases were subjected to reverse transcription-polymerase chain reaction (RT-PCR) the two IHC-positive/FISH-negative cases were, in fact, positive by RT-PCR, suggesting FISH false negativity. This study used the clone 5A4 (Abcam, Cambridge, United Kingdom) and the polymer refined detention kit (Leica Microsystems, GmbH, Germany). In a comprehensive study of 377 cases (145 ADCs, 178 SCCs, and 54 other histologies) three antibody clones (5A4, ALK1, and D5F3) were combined with three detection systems, with the two best combinations showing 100% sensitivity, 88% specificity using 5A4 with ADVANCE and 100% sensitivity and 75% specificity using D5F3 with ADVANCE.23 In a report examining only cytology specimens Savic et al.34 reported a 93% sensitivity and 96% sensitivity when comparing immunocytochemistry with FISH and using the 5A4 antibody clone. Interestingly, immunocytochemistry identified two cases as ALK positive, which were originally classified as FISH negative, however, on reanalysis of the FISH these two cases were determined to be in fact FISH positive. Finally, in a study where the D5F3 antibody was used with the OptiView DAB detection and OptiView amplification system, the same as used in our study, they observed 94% sensitivity (31 IHC+/33 FISH+) and 100% specificity (198 IHC−/198 FISH−) when comparing IHC with FISH formalin-fixed, paraffin-embedded tissue. However, the two discrepant cases were subsequently reclassified as FISH negative on ThinPrep specimens from the same patients, thus increasing the sensitivity to 100%.27 Collectively all these studies convey the same message, IHC correlates well with FISH with a very few cases showing discrepancy, but the biggest limitation is that the antibody clones, antigen retrieval, detection systems, and scoring methodology were not standardized.
In the cohort presented in our study there were three FISH-positive cases that were scored IHC negative by seven of seven readers, two FISH-negative cases scored IHC positive by seven of seven readers, and one FISH-positive case scored IHC negative by six of seven readers. The best way to resolve this conflict from an assay detection perspective would be to perform a third assay that would preferably detect more than just a break-apart gene. One such assay would be next-generation sequencing on the genomic DNA, which could detect break-points, an open reading frame, and any potential novel 5′ fusion partner, but would not detect expression. Another approach using RT-PCR combined with sequencing could detect break-apart, if the 5′ partner is known, and verify expression at the mRNA level. Unfortunately there was not enough material to perform any additional assays to further resolve the few discrepancies observed between IHC and FISH in our study.
In the future the only way to fully understand and appreciate how well IHC compares with FISH in predicting response and outcome to ALK inhibitors is to have well-designed prospective clinical trials that evaluate the efficacy of ALK inhibitors in IHC+/FISH− or IHC−/FISH+ patients. The most favorable IHC assay for this screening should be one that has been thoroughly standardized and has exquisitely sensitive detection/amplification systems with limited background to consistently identify all levels of ALK protein expression.
Acknowledgments
This study was funded by Ventana Medical Systems, Inc.
Footnotes
Disclosure: The authors declare no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2010. National Cancer Institute; Apr, 2013. [Accessed May 15, 2013]. November 2012 SEER data submission, posted to the SEER web site. Available at: http://seer.cancer.gov/csr/1975_2010/ [Google Scholar]
- 3.Kris MG, Johnson BE, Kwiatkowski DJ, et al. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: the NCI’s Lung Cancer Mutation Consortium (LCMC) ASCO Meeting Abstracts. 2011;29:CRA7506. [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, et al. North-East Japan Study Group. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T, Morita S, Yatabe Y, et al. West Japan Oncology Group. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, Carcereny E, Gervais R, et al. Spanish Lung Cancer Group in collaboration with Groupe Français de Pneumo-Cancérologie and Associazione Italiana Oncologia Toracica. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 8.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non- small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 9.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13:1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 12.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki T, Rodig SJ, Chirieac LR, Jänne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010;46:1773–1780. doi: 10.1016/j.ejca.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 15.Togashi Y, Soda M, Sakata S, et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One. 2012;7:e31323. doi: 10.1371/journal.pone.0031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009;420:345–361. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol. 2009;4:1450–1454. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- 18.Cui JJ, Tran-Dubé M, Shen H, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal- epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J Med Chem. 2011;54:6342–6363. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 19.Kim D-W, Ahn M-J, Shi Y, et al. Results of a global phase II study with crizotinib in advanced ALK-positive non-small cell lung cancer (NSCLC) ASCO Meeting Abstracts. 2012;30:7533. [Google Scholar]
- 20.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 21.FDA Approves Xalkori with Companion Diagnostic for a Type of Late- Stage Lung Cancer. Silver Spring, MD: United States Food and Drug Administration; 2011. [Accessed February 2014]. Available at: http://www.fda.gov/aboutfda/centersoffices/officeofmedicalproductsandtobacco/cder/ucm270058.htm. [Google Scholar]
- 22.Boland JM, Erdogan S, Vasmatzis G, et al. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol. 2009;40:1152–1158. doi: 10.1016/j.humpath.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Conklin CM, Craddock KJ, Have C, Laskin J, Couture C, Ionescu DN. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol. 2013;8:45–51. doi: 10.1097/JTO.0b013e318274a83e. [DOI] [PubMed] [Google Scholar]
- 24.Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011;6:466–472. doi: 10.1097/JTO.0b013e31820b82e8. [DOI] [PubMed] [Google Scholar]
- 26.To KF, Tong JH, Yeung KS, et al. Detection of ALK rearrangement by immunohistochemistry in lung adenocarcinoma and the identification of a novel EML4-ALK variant. J Thorac Oncol. 2013;8:883–891. doi: 10.1097/JTO.0b013e3182904e22. [DOI] [PubMed] [Google Scholar]
- 27.Minca EC, Portier BP, Wang Z, et al. ALK status testing in non-small cell lung carcinoma: correlation between ultrasensitive IHC and FISH. J Mol Diagn. 2013;15:341–346. doi: 10.1016/j.jmoldx.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 28.McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thorac Oncol. 2012;7:348–354. doi: 10.1097/JTO.0b013e3182381535. [DOI] [PubMed] [Google Scholar]
- 29.Peled N, Palmer G, Hirsch FR, et al. Next-generation sequencing identifies and immunohistochemistry confirms a novel crizotinib-sensitive ALK rearrangement in a patient with metastatic non-small-cell lung cancer. J Thorac Oncol. 2012;7:e14–e16. doi: 10.1097/JTO.0b013e3182614ab5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun JM, Choi YL, Won JK, et al. A dramatic response to crizotinib in a non-small-cell lung cancer patient with IHC-positive and FISH-negative ALK. J Thorac Oncol. 2012;7:e36–e38. doi: 10.1097/JTO.0b013e318274694e. [DOI] [PubMed] [Google Scholar]
- 31.Nitta H, Tsuta K, Yoshida A, et al. New methods for ALK status diagnosis in non-small-cell lung cancer: an improved ALK immunohistochemical assay and a new, Brightfield, dual ALK IHC-in situ hybridization assay. J Thorac Oncol. 2013;8:1019–1031. doi: 10.1097/JTO.0b013e31829ebb4d. [DOI] [PubMed] [Google Scholar]
- 32.Ying J, Guo L, Qiu T, et al. Diagnostic value of a novel fully automated immunochemistry assay for detection of ALK rearrangement in primary lung adenocarcinoma. Ann Oncol. 2013;24:2589–2593. doi: 10.1093/annonc/mdt295. [DOI] [PubMed] [Google Scholar]
- 33.Yi ES, Boland JM, Maleszewski JJ, et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol. 2011;6:459–465. doi: 10.1097/JTO.0b013e318209edb9. [DOI] [PubMed] [Google Scholar]
- 34.Savic S, Bode B, Diebold J, et al. Detection of ALK-positive non- small-cell lung cancers on cytological specimens: high accuracy of immunocytochemistry with the 5A4 clone. J Thorac Oncol. 2013;8:1004–1011. doi: 10.1097/JTO.0b013e3182936ca9. [DOI] [PubMed] [Google Scholar]