Abstract
We report on a new concept for profiling genetic mutations of (lung) cancer cells, based on the detection of patterns of volatile organic compounds (VOCs) emitted from cell membranes, using an array of nanomaterial-based sensors. In this in-vitro pilot study we have derived a volatile fingerprint assay for representative genetic mutations in cancer cells that are known to be associated with targeted cancer therapy. Five VOCs were associated with the studied oncogenes, using complementary chemical analysis, and were discussed in terms of possible metabolic pathways. The reported approach could lead to the development of novel methods for guiding treatments, so that patients could benefit from safer, more timely and effective interventions that improve survival and quality of life while avoiding unnecessary invasive procedures. Studying clinical samples (tissue/blood/breath) will be required as next step in order to determine whether this cell-line study can be translated into a clinically useful tool.
Keywords: Lung cancer, Genetic, Mutation, Volatile organic compound, Sensor
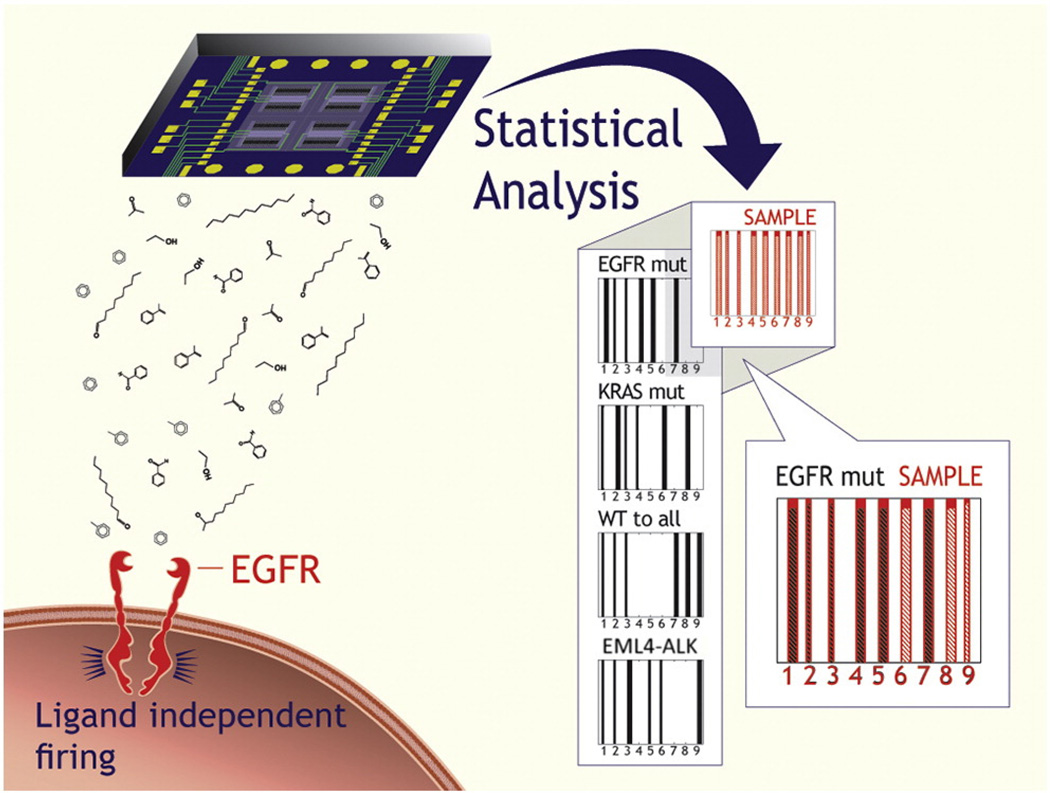
Cancer emergence, aggressiveness and treatment response varies greatly from patient to patient.1 Gene expression profiling2 is currently gaining importance for accurately classifying tumors in individual patients, predicting the response to the available treatments and personalizing cancer therapy.3–6 Results with conventional tests can be obtained in several days. However, invasive tissue sampling from the tumor is required and frequent monitoring is needed to detect changes in the cancer cells over time.1 Here, we report on a new approach for profiling genetic mutations. We focus on representative genetic mutations in cancer cells that are known to be associated with targeted cancer therapy, viz. mutations of the epidermal growth factor receptor (EGFRmut)5, mutations of the v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRASmut), and fusion of the echinoderm microtubule-associated protein-like 4 (EML4) gene to the anaplastic lmphoma kinase (ALK) gene (EML4-ALK). The method is based on detecting and identifying patterns of volatile organic compounds (VOCs; i.e., compounds with a relatively high vapor pressure, typically equal to or greater than 0.1 mmHg)7,8 that are emitted from the cell membranes, using an array of nanomaterial-based sensors (see Figure 1).
Figure 1.
Schematic representation of the oncogene identification in an unknown sample. The nanomaterial-based sensor array is exposed to the cell-line headspace, and the VOC fingerprint is obtained through DFA analysis, using three primary, general DFA models (cf. Figure 2) and, if necessary, six secondary, specific DFA models (cf. Figure 3). The oncogene is identified through comparison with the expected DFA classifications for the EGFRmut, KRASmut and EML4-ALK.
Cancer-specific VOCs can be detected (i) from the headspace of the cancer cells (the gaseous constituents of a closed space above the cell-lines, in-vitro approach); (ii) from exhaled breath: (iii) from blood samples; (iv) from skin excretions; or (v) via tissue sampling. 7–9 For the current study, we have chosen the in-vitro approach as a way to eliminate potential effects of confounding factors that are associated with clinical samples, such as patients’ diet, age, gender, metabolic state etc. Additionally, direct detection of VOCs from cancer cells will provide clear-cut evidence that the findings are associated with the cell per se, rather than with the microenvironment of the tumor or with indirect metabolic pathways in the body.
It was recently shown that arrays of cross-reactive nanomaterial-based sensors combined with statistical pattern recognition methods can identify and separate the VOC patterns of several types of lung cancer (LC) cells with different histology, as well as liver cancer cells with different metastatic potential, based on the analysis of in-vitro headspace samples from cell-lines.10–12 In this pilot study we have adapted the nanomaterial-sensor-technology for in-vitro differentiation between subtle differences in the VOC profiles of genetic LC mutations. LC, which causes most cancer related deaths worldwide and is a major burden to the health care systems,13 was chosen as a representative example of cancerous diseases. However, this approach is expected to be viable for a large variety of cancers. Complementary chemical analysis of the headspace samples identified five headspace VOCs that could distinguish between the studied oncogenes.
Methods
Cell-cultures and sample preparation
Nineteen (19) human non-small-cell lung carcinoma (NSCLC) cell-lines with long term gene expression analysis were obtained from the Colorado cell bank registry (see Table 1). These included six cell-lines representing the oncogene EGFRmut (H3255, H820, H1650, H1975, HCC4006, HCC2279), four representing KRAS mut (A549, H2009, H460, NE18), one representing EML4-ALK fusion (H2228) and seven representing oncogenes that were wild type (wt) to the three mutations of interest (H322, H1703, H125, H1435, Calu3, HCC15, H520, HCC193). Multiple cell-lines were used per studied oncogene to reflect the natural diversity of LC cells. The samples representing EML4-ALK were obtained from only one cell-line, because of the extreme rarity of this fusion event. The cell-lines were grown in 100 mm cell-culture dishes from seeding (~2 × 106 cells) up to 95% confluence (7 × 106 cells) under standard conditions in a conventional incubator at 37°C in a humidified atmosphere with 95% air/5% CO2, using a two dimensional medium (RPMI 1640 medium + 10% FBS). The same medium was used for all cell-lines. The open 100 mm cell-culture dish was placed in a covered 150 mm dish. Two Ultra II SKC™ badges with Tenax TA as a sorbent (265 mg; SKC Inc.) were placed above the cell-culture, attached to the cover of the 150 mm dish, for absorbing the headspace VOCs during the total growth time (median time 68 hours; range 60–72 h). The cell-lines were grown in several replicas. An empty medium with the same incubation time and conditions, but without cells, served as control. The headspace VOCs were adsorbed to the badges at Sheba Medical Center, Israel or in the University of Colorado Cancer Center, Colorado, US, and tagged with a barcode. All badges were stored under controlled humidity levels and at 4°C at the collection centers until they were send in one shipment (under refrigeration) to the Technion, Israel, where they were analyzed blindly. The results were then conjugated with the relevant clinical data.
Table 1.
Characteristics of the studied samples, taken from different cell lines. Each cell line was grown in several replicas.
| Oncogene | Cell line | Histology | Patient type (source database) | Tissue type | Mutation |
|---|---|---|---|---|---|
| EGFRmut | H3255 | Adeno | Non-smoker; female; Caucasian (Sanger) | Lung | L858R |
| H820 | Adeno | Male; Caucasian (ATCC) | Lymph node | EX19 | |
| H1650 | Adeno | Current-smoker; male; Caucasian (ATCC) | Pleural effusion | EX19 | |
| H1975 | Adeno | Non-smoker; Female (ATCC) | Lung | L858R; T790M | |
| HCC4006 | Adeno | Male; Caucasian (ATCC) | Pleural effusion | EX19 | |
| HCC2279 | Adeno | Unknown (Sanger) | Lung | EX19 | |
| KRASmut | A549 | Adeno | Male; Caucasian (ATCC) | Lung | G12S |
| H2009 | Adeno | Current-smoker; Female; Caucasian (ATCC) | Lymph node | G12A | |
| H460 | Large cell | Male (ATCC) | Pleural effusion | Q61H | |
| NE18 | Squamous | Unknown (Sanger) | Lung | n/aa | |
| EML4-ALK | H2228 | Adeno | Non-smoker; female (ATCC) | Lung | |
| Other oncogenes | H322 | Adeno | Unknown (Sanger) | Lung | |
| (wt to the above) | H1703 | Adeno | Current-smoker; male; Caucasian (ATCC) | Lung | |
| H125 | Adeno | Unknown (Sanger) | Lung | ||
| H1435 | Adeno | Non-smoker; female (ATCC) | Lung | ||
| Calu3 | Adeno | Male; Caucasian (ATCC) | Pleural effusion | ||
| HCC15 | Squamous | Male (Sanger) | Lung | ||
| H520 | Squamous | Male (ATCC) | Lung | ||
| HCC193 | Adeno | Unknown (Sanger) | Lung |
KRAS mutation was identified in NE-18 at the University of Colorado Cancer Center through direct sequencing, but the location of the mutation was not recorded.
Chemical analysis
The headspace samples were analyzed by gas-chromatography combined with mass spectrometry (GC-MS), using a GCMS-QP2010 system (Shimadzu Corporations) with a SLB-5ms capillary column (with 5% phenyl methyl siloxane; 30 m length; 0.25 mm internal diameter; 0.5 µm thickness, column pressure: 23.4 kPa, column flow rate: 0.7 mL/min.); splitless mode. Prior to the GC-MS analysis, the Tenax sorbent material from one Ultra II SKC™ badge was heated in a 350 ml stainless steel thermal desorption device that was pre-heated to 270°C and kept at that temperature for 10 min, in order to release the VOCs into the gas-phase. The VOCs in the 350 ml gaseous samples were then pre-concentrated onto a solid phase microextraction (SPME) fiber assembly of divinylbenzene, carboxen, and polydimethyl-siloxane (DVB/CAR/PDMS; Sigma-Aldrich, Israel). For this purpose a manual SPME holder with the extraction fiber was inserted into the thermal desorption device for 30 min. The fiber was then immediately inserted into the GC injector (direct mode) for thermal desorption (oven temperature profile: 10 min. at 35°C; 4°C/min. until 150°C; 10°C/min. until 300°C; 15 min. at 300°C).
Contaminants of the Tenax sorbent material were identified through analysis of pristine Tenax material from unused Ultra II SKC™ badges.
Compounds were preliminarily identified through spectral library match using the compounds library of the National Institute of Standards and Technology (Gaithersburg, USA). The identity of the compounds was confirmed and quantification was achieved through measurements of external standards: toluene, triethylamine, styrene, benzaldehyde, benzaldehyde-2-hydroxy, 2-ethyl-1-hexanol, phenol (Sigma- Aldrich, Israel); decanal (Holland Moran, Israel), as described in the Supporting Information (SI).
The GC-MS chromatograms were processed using the open source XCMS package version 1.22.1 for R environment (http://metlin.scripps.edu/download/). The VOCs showing significant differences between LC specific mutations were determined from the GC-MS results using the non-parametric Wilcoxon/ Kruskal-Wallis test for populations whose data cannot be assumed to be normally distributed.14,15 Shapiro–Wilk tests confirmed that the null hypothesis for normal distribution of the GC-MS data was not fulfilled.
Characterization with the nanomaterial-based sensors
The Tenax sorbent material from one Ultra II SKC™ badge was heated at 270°C for 10 min. in a pre-heated 750 ml stainless steel TD chamber. Pulses of the gaseous sample from the TD chamber were delivered by a gas sampling system into a stainless steel test chamber containing the sensors. The sensors were based on chemiresistive layers of spherical gold nanoparticles (GNPs; 3–4 nm core diameter) with three different organic ligands (see SI, Table 1S).16 The organic ligands provided broadly cross-reactive adsorption sites for the headspace VOCs.16–20 A schematic representation of the GNP sensors is provided in Figure 1S, SI. The GNPs were synthesized and the sensors were fabricated as described previously. 17–20 The GNP sensors used in this study responded rapidly and reversibly when exposed to simulated, typical headspace VOCs.21,22
The GNP sensors were mounted on a PTFE circuit board inside a stainless steel test chamber with a volume of 100 cm3. The sampling system delivered pulses of the headspace sample from the thermal desorption device to the sensors. The test chamber was evacuated between exposures. An Agilent multifunction switch 34980 was used to measure the resistance of all sensors simultaneously as a function of time.
Typically, the sensors’ baseline responses were recorded for 5 min. in vacuum, followed by 5 min. under headspace sample exposure, followed by another 5 min. baseline response in vacuum. Figure 2S in the SI shows the typical sensing responses of an octadecanethiol-functionalized GNP sensor used in this study to headspace samples from cell-lines with the studied oncogenes. Two sensing features were read for each sensor, as described in the caption of Figure 2S, SI.
Statistical analysis of the sensor array output
The sensing features were analyzed using discriminant factor analysis (DFA).23,24 DFA is a linear, supervised pattern recognition method. The classes to be discriminated are defined before the analysis is performed. DFA determines the linear combinations of the sensing features such that the variance within each class is minimized and the variance between classes is maximized. The DFA output variables (viz. canonical variables) are obtained in mutually orthogonal dimensions, the first canonical variable (CV1) being the most powerful discriminating dimension.
Each sensor responded to all (or to a certain subset) of the VOCs found in the headspace samples and DFA identified the gene-specific VOC patterns that were obtained from the sensing features. Note that DFA was also used as a heuristic to select the most suitable sensing features. The reason for selecting a certain set of sensing features for a particular problem is directly derived from their ability to discriminate between the various classification groups.
The classification success rate of the binary problems was estimated through leave-one-out cross-validation. For this purpose, DFA was computed, using a training data set that excluded one test sample. After the DFA computation, the test sample was projected onto the CV1 axis that was calculated using the training set. Thereby the test sample was “blinded” against the DFA model, so that its class affiliation was unknown. All possibilities of leaving out one sample were tested and the left out sample was classified as true positive (TP), true negative (TN), false positive (FP) or false negative (FN).
Results
Chemical analysis of the headspace samples
The cell-line headspace was analyzed in search of the specific VOCs that make up the volatile fingerprints of the studied oncogenes, as compared to the control headspace obtained from the empty growth medium without any cells. The headspace samples (9 EGFRmut samples; 5 KRASmut; 5 EML4-ALK; 12 that were wt to the oncogenes of interest) were collected from several different cell-lines per oncogene (see Table 1), in order to account for the genetic complexity of LC cells. The control samples were collected from eight separate cell-culture dishes containing only the medium without the cells, which received the same treatment as the cell-cultures (2 headspace samples from each cell-culture dish).
The GC-MS analysis identified over 600 different VOCs per headspace sample. The analysis of pristine Tenax material from unused Ultra II SKC™ badges yielded five VOCs as possible contaminants of the Tenax sorbent material of the collection badges. These VOCs were tentatively identified through spectral library match as naphthalene, L-cysteine sulfonic acid, malonic acid, acetaldehyde and methylene chloride. These VOCs were disregarded in the subsequent comparative analysis (according to main mass and retention time). Two hundred five VOCs that were present in >80% of the samples of the separate study groups were further analyzed. Non-parametric Wilcoxon/Kruskal-Wallis tests identified (after measurement of external standards) a total of five VOCs that were on average significantly elevated or reduced for at least one of the studied oncogenes, as compared to the control medium (see Table 2). The triethylamine (TEA) levels in the EGFRmut and the wt to all samples were completely depleted (i.e. no TEA peak was observed in the GC-MS chromatogram), but showed no significant changes with respect to the empty control medium in the KRASmut and EML4-ALK fusion samples. The aromatic compounds toluene and styrene were found in increased concentration in the headspace of NSCLC cells with certain mutations. An increase in styrene was significant for EGFRmut, whereas toluene levels were significantly increased for EML4-ALK. Both styrene and toluene were increased for the cell-cultures with wt oncogenes. The benzaldehyde concentration in the KRASmut cell-cultures was totally depleted, but remained unchanged for all other oncogenes. Decanal was selectively depleted in EGFRmut cell-cultures and EML4-ALK fusion cell-cultures.
Table 2.
Quantification of the VOC concentrations in the headspace of cell lines having different oncogenes.
| Compound | CAS# | Chemical group |
m/z | Retention time (minimum – maximum) [min.] |
R2 | Concentration [ppbv] |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control: empty medium |
EGFRmut |
KRASmut |
EML4-ALK |
wt to all |
||||||||||||||
| Mean ± SD | Mean ± SD | p-value | Trend | Mean ± SD | p-value | Trend | Mean ± SD | P- value |
Trend | Mean ± SD | p-value | Trend | ||||||
| Triethylamine | 121-44-8 | Amine | 86 | 6.454 – 6.715 | 0.949 | 37.8 ± 15.3 | no peak identified* |
0.0002 | − | 159.9 ± 135.3 | n.s. | 1095.1 ± 1347.5 | n.s. | no peak identified* |
<0.0001 | − | ||
| Toluene | 108-88-3 | Aromatic compounds |
91 | 9.882 – 9.902 | 0.999 | 1322.9 ± 1447.1 | 2881.1 ± 1621.4 |
n.s. | 2121.3 ± 1101.4 | n.s. | 2827.5 ±291.2 | 0.0500 | + | 2944.2 ± 1106.7 |
0.0253 | + | ||
| Styrene | 100-42-5 | 78 | 15.055 – 15.12 | 0.999 | 1545.3 ± 802.9 | 3338.9 ± 2074.5 |
0.0500 | + | 2858.0 ± 1684.3 | n.s. | 5442.2 ± 4952.2 | n.s. | 5394.7 ± 3147.1 |
0.0007 | + | |||
| Benzaldehyde | 100-52-7 | Aldehydes | 106 | 17.831 – 17.926 | 0.989 | 1173.1 ±774.7 | 143.3 ± 205.0 |
n.s. | no peak identified* |
0.0034 | − | 671.3 ± 834.9 | n.s. | 621.3 ± 834.9 | n.s. | |||
| Decanal | 112-31-2 | − | 43 | 25.819 – 25.827 | 0.999 | 50.2 ±31.2 | no peak identified* |
0.0049 | − | 28.1 ± 23.1 | n.s. | no peak identified* |
0.0239 | − | 28.6 ± 24.0 | n.s. | ||
All cell lines were cultures under the same conditions, using the same medium. The trends in VOC concentration are given with respect to the average VOC concentrations in the empty control medium (determined from eight separate dishes containing only the growth medium without any cells that underwent the same processes as the cell growth dishes). An increase in average concentration is marked by (+), a decrease is marked by (−), and changes that were not statistically significant (i.e. p > 0.05) are marked by (n.s.).
If no peak corresponding to a specific VOC was identified in the GC-MS chromatograms, the mean concentration was taken to be zero for the statistical analysis.
Identification of VOC-patterns for the LC-specific oncogenes using an array of nanomaterial-based sensors
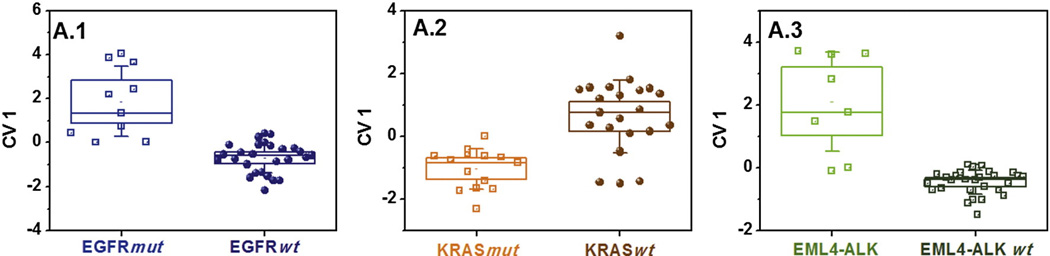
The volatile fingerprints of EGFRmut, KRASmut and EML4-ALK were identified, using an array of three GNP sensors (organic ligands: decanthiol, 2-nitro-4-trifluoro- methylbenzenethiol, octadecanthiol, see Table 1S, SI).16–18,25 Figure 1 shows a schematic representation of the identification of the genetic mutations: The nanomaterial-based sensor-array was exposed to the cell-line headspace, and the VOC fingerprint was obtained through DFA analysis of the sensors’ collective output. Figure 2 represents a set of three primary, general tests comprising three different DFA models for identifying EGFRmut (A.1), KRASmut (A.2) and EML4-ALK (A.3) among a representative, diverse group of mixed LC oncogenes (EGFRmut, KRASmut, EML4-ALK and other mutations that were wt to these three). Panel A.1 of Figure 2 shows that the EGFRmut could be clearly distinguished from the EGFRwt volatile fingerprint (consisting of KRASmut, EML4-ALK genes and wt to all) along the CV1 axis. The CV1 values of the two study groups that were calculated from two sensing features (see Table 1S, SI) formed two well separated clusters (p < 0.0001, see Figure 2). Note that the signatures of the different cell-lines (see Table 1) having the same oncogene overlapped completely and did not form separate sub-clusters within the oncogene-clusters. Leave-one-out cross-validation yielded a classification success of 70% sensitivity, 100% specificity and 92% accuracy (see Table 3). Clear volatile fingerprints were obtained also for KRASmut, using a second DFA model, also based on two sensing features (93% and 78%, respectively, see Table 3 and Figure 2, panel A.2). The method was slightly less sensitive, but highly specific to EML4-ALK (63% and 100%, respectively, see Table 3 and Figure 2, panel A.3). The set of three primary DFA tests (A.1– A.3) shown in Figure 2 should in principle allow classifying any unknown sample. However, due to the realistic limitation in sensitivity and/or specificity, they could sometimes yield ambiguous or wrong classifications.
Figure 2.
Primary test set. DFA plots of CV1 that was calculated from the responses of the GNP sensors to the headspace samples of NSCLC cell-lines having EGFRmut, KRASmut or EML4-ALK oncogenes, or genetic mutations that are wt to the former three. The SD of the CV1 values is represented by the error bars. The boxes represent the 95% CI of the CV1 values, corresponding to 1.96*SE. The subpanels A.1-A.3 represent the three primary, general DFA models that are applied simultaneously to the data and contain all 37 headspace samples. Each point represents one sample. The samples were obtained from multiple cell-lines having the oncogenes of interest (cf. Table 1). Note that the CV1 distribution for the same group of samples may look different for two different DFA models due to different input parameters.
Table 3.
Classification success, estimated through leave-one-out cross validation, of the two tests sets: The primary, general test set comprising three different DFA models, A.1-A.3, to identify EGFRmut, KRASmut and EML4-ALK (cf. Figure 2), and the secondary, specific test set, comprising six further DFA models, B.1-B.6, that distinguish between specific genetic mutations (cf. Figure 3).
| Test | Test group | Control group | TP | TN | FP | FN | Sensitivity [%]a | Specificity [%]b | Accuracy [%]c | |
|---|---|---|---|---|---|---|---|---|---|---|
| Primary (general) tests | A.1 | EGFRmut | KRASmut; EML4-ALK; wt to all | 7 | 27 | 0 | 3 | 70 | 100 | 92 |
| A.2 | KRASmut | EGFRmut; EML4-ALK; wt to all | 13 | 18 | 5 | 1 | 93 | 78 | 84 | |
| A.3 | EML4-ALK | EGFRmut; KRASmut; wt to all | 5 | 29 | 0 | 3 | 63 | 100 | 92 | |
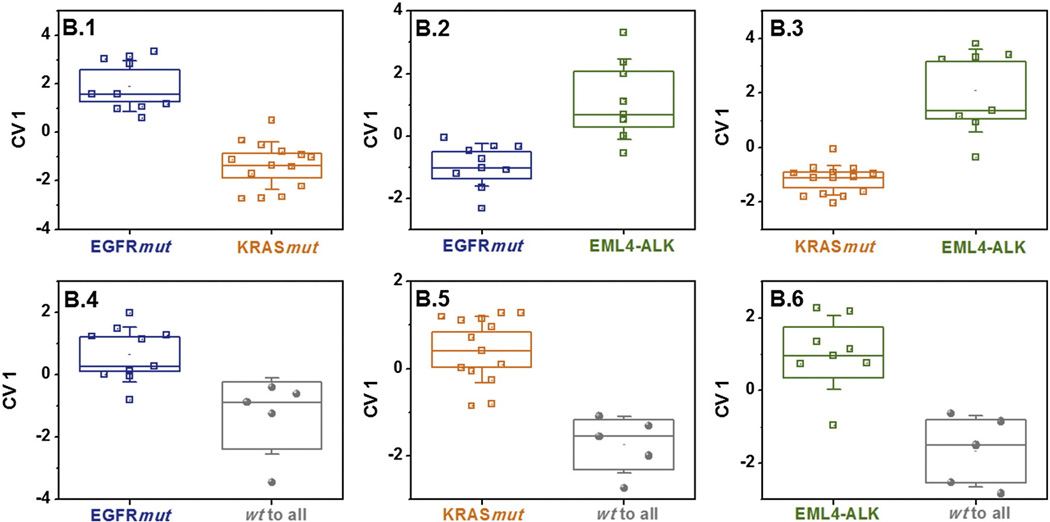
| Secondary (specific) tests | B.1 | EGFRmut | KRASmut | 10 | 13 | 1 | 0 | 100 | 93 | 96 |
| B.2 | EGFRmut | EML4-ALK | 10 | 6 | 2 | 0 | 100 | 75 | 89 | |
| B.3 | KRASmut | EML4-ALK | 14 | 7 | 1 | 0 | 100 | 88 | 96 | |
| B.4 | EGFRmut | wt to all | 9 | 4 | 1 | 1 | 90 | 80 | 87 | |
| B.5 | KRASmut | wt to all | 12 | 4 | 1 | 2 | 86 | 80 | 84 | |
| B.6 | EML4-ALK | wt to all | 7 | 5 | 0 | 1 | 88 | 100 | 92 |
The secondary test set is designed to deliver useful additional information in the case of an ambiguous sample classification through the primary tests.
Sensitivity = TP/(TP + FN).
Specificity = TN/(TN + FP).
Accuracy = (TP + TN)/(TP + TN + FP + FN).
We therefore developed an additional set of six secondary, specific DFA models (B.1-B.6) that could supplement the primary test A.1-A.3 in cases of ambiguous test results (see Figure 3). Each supplementary test distinguished two specific oncogenes (or one specific oncogene from the mixed group of different oncogenes, as shown in Figure 3, with high accuracy between 84% and 96% (see Table 3). Each studied headspace sample was included in three of the six supplementary tests B1–B6.
Figure 3.
Secondary, specific (auxiliary) test set. DFA plots of CV1 that was calculated from the responses of the GNP sensors to the headspace samples of NSCLC cell-lines having EGFRmut and KRASmut, EML4-ALK genes and genetic mutations that are wt to the former three. The SD of the CV1 values is represented by the error bars. The boxes represent the 95% CI of the CV1 values, corresponding to 1.96*SE. The subpanels B.1-B.6 represent six secondary, auxiliary DFA models that contain specific subsets of the headspace samples, and may be selectively applied if the test results from the primary, general test set (DFA models A. 1-A.3, cf. Figure 2) were inconclusive. Note that the CV1 distribution for the same group of samples may look different for two different DFA models due to different input parameters.
We have examined the classification success of the newly established volatile fingerprints through a global leave-one-out cross-validation procedure. For this purpose, we excluded one sample at a time and calculated the CV1 distribution for the three primary tests A.1–A.3, and for the six supplementary tests B.1– B.6, using the relevant subsets of the remaining, well-defined samples as training sets. The left-out, blinded sample was then projected onto all nine DFA maps A.1–A.3 and B.1–B.6. The set of three primary tests A.1–A.3 that would in principle allow classifying all the samples, classified 27 of the 37 samples correctly, due to the realistic limitation in sensitivity and/or specificity. Seven sample classifications were ambiguous (more than one positive classification), and three samples were unambiguously misclassified, due to two or more misclassifications in the primary tests. The seven samples with ambiguous primary classification could all be correctly classified through the relevant supplementary tests. For example, a sample that was positive both for EGFRmut and KRASmut could be correctly classified through the secondary test B.1 that separated the EGFRmut from the KRASmut. Furthermore, the supplementary tests did not yield consistent results for the three samples that were falsely identified in the primary test set.
Discussion
In-vitro cancer cell-line studies versus clinical studies
This in-vitro pilot study has provided first evidence for the existence of measurable VOC profiles of the most important genetic mutations associated with targeted LC therapy (EGFR mut/KRASmut/EML4-ALK). The three studied oncogenes are usually mutually exclusive, and, hence, are considered distinct genetic subtypes of LC.26 However, owing to the extreme complexity of LC, the studied cell-lines could have additional genetic mutations that could overlap between the categories and could, in principle, affect the VOC output. In order to account for the genetic complexity of LC cells, we have collected headspace samples from 6 different EGFRmut and 4 KRASmut cell-lines (cf. Table 1). EML4-ALK was represented only by a single cell-line due to the extreme rarity of this fusion event. A group of eight different LC cell-lines with genetic mutations that were wt to the three mutations of interest was included as an additional control group. The cell-lines in this study originated from different types of tissue (lung, lymph node, pleural effusion). The differences in origin could, in principle affect the VOC signatures of the cell-lines. However, the goal in personalized medicine is to treat patients by the tumor’s genetic profile, rather than the origin of the tissue/tumor. The observed common signals for the same molecular profile for cell-lines that stem from different tissue types therefore supports our approach.
Studying the metabolic activity of isolated LC cells by analyzing their headspace VOCs avoids the human body’s confounding factors (e.g. variations in the patients' age, gender, lifestyle, medication and other chronic diseases). Cell-lines provide an abundant number of cells with similar characteristics, while avoiding variation between individuals and bypassing ethical issues associated with human experiments. On the other hand, in-vitro studies may fail to replicate the precise cellular conditions in the lung tissue, because they disregard the synergetic effect of the tumor on the whole organism. Hence, the results of this cell-line study can only be considered as indicative; their in-vivo translation will be far from trivial. Consequently, the translation to human samples that would provide useful data for clinical application might seriously differ from the data obtained from this cell-line study. Cancer specific VOCs can be excreted, for instance, through the exhaled breath via the respiratory system that controls gas exchange in the human body.16–25,27–29 These easily (in-vivo) accessible breath VOCs could be products of the metabolic activity of the tumor itself, or by-products of bacteria and necrotic reactions caused by local inflammation in the microenvironment of the tumor, or else they could be partially re-emitted environmental toxins that were previously adsorbed to the body.28 In addition, systemic breath VOCs could be produced or consumed because of cancer-related changes elsewhere in the body, affecting the blood chemistry, and eventually being expired via the respiratory system.28
In particular, the genetic mutations of cancer cells are strongly related to the tumor environment and may affect the interaction between the tumor and the host. Hence, the detected VOC signatures of the genetic mutations of LC cells must ultimately be verified through in-vivo clinical studies in humans, for example through exhaled breath analysis, blood or tissue sampling.
Identification of oncogene-specific VOCs
Five distinguishing VOCs were identified during the comparative studies (see Table 2). The VOC profiles were distinct for the four studied groups. Some of the distinguishing VOCs may have logical explanations, while the origin of others is not yet well understood.9
Triethylamine, benzaldehyde and decanal were selectively reduced or completely depleted in the headspace of the NSCLC cells with specific oncogenes, as compared with the empty control medium. This could indicate the selective consumption of these VOCs through the mutated NSCLC cells. For example, triethylamine was depleted for EGFRmut and wt to all, but showed no significant change for KRASmut and EML4-ALK fusion. Oxidative stress in cancer cells may lead to protein peroxidation, which could consume amines such as triethylamine, leading to the observed reduced concentration levels in the headspace.30,31 On the other hand, it is reasonable to assume that the amino acids from the medium would be consumed through the metabolic activity of the cancer cells during cell growth, so that triethylamine and other amines could indeed be released as by- products into the headspace. Possibly, the different observed trends could indicate that the interplay of metabolic activity and/ or oxidative stress differs for different oncogenes. The benzaldehyde concentration in the KRASmut cell-cultures was totally depleted, but remained unchanged for all other oncogenes, while decanal was selectively depleted in the EGFRmut and the EML4-ALK cell-cultures. The observed decrease in the concentration of the aldehydes benzaldehyde and decanal in the headspace of NSCLC cell-lines with specific mutations, as compared to the control medium may be due to enhanced activity of aldehyde dehydrogenase (ALDH) in NSCLC cell-lines,9,32 especially of the enzymes ALDH1A1 and ALDH3A1.33 Again, the different trends for the studied oncogenes could possibly reflect their different metabolic activity. In this study, we observed that the selective reduction/depletion of triethylamine, benzaldehyde and decanal was unique for the three investigated oncogenes: EGFRmut cell-lines showed depleted levels of triethylamine and decanal, KRASmut cells showed significantly reduced levels of triethylamine and depletion of benzaldehyde, and EML4-ALK cells showed depletion of decanal alone. The headspace of the cells that were wt to these three oncogenes was only depleted of triethylamine, but showed no significant changes of benzaldehyde and decanal. Triethylamine, benzaldehyde and decanal together could therefore hold potential as specific biomarker-set for identifying and distinguishing EGFRmut, KRASmut, and EML4-ALK.
The aromatic compounds toluene and styrene were found in increased concentration in the headspace of NSCLC cells with certain oncogenes. An increase in styrene was significant for EGFRmut, whereas toluene levels were significantly increased in the headspace samples of the EML4-ALK fusion cell-line. Both styrene and toluene were on average significantly increased in the headspace of the cell-cultures with the wt oncogenes. Aromatic compounds such as styrene and toluene are considered to be of exogenous origin, e.g. stemming from tobacco smoke9,34–37 and could therefore be increased in LC cell-lines. Possibly, styrene and toluene had been absorbed to the lung tissue of the donors from exogenous sources, before the tissue was removed, from which the studied cell-lines were derived. LC specific genetic mutations are expected to alter the cell metabolism and, hence, they may affect the excretion of VOCs of exogenous origin that were previously absorbed from the environment.7
Note, however, that the sample size in this pilot study is limited and general conclusions about VOC oncogene-markers cannot be drawn from these results. Wider-scale studies on more cell-line samples, including genetically defined, isogenic human cell-line pairs with highly specific targeted knockouts of EGFR and KRAS genes, are underway to extend and further validate these results.
Patterns of oncogenes from nanomaterial-based sensors
The presented results could lead to a nanomaterial-based in-vitro test for the three most important genetic mutations that are associated with targeted therapy in LC. A schematic representation of a possible future test is shown in Figure 1: A nanomaterial-based sensor array would be exposed to the cell-line headspace, and the VOC fingerprint would be obtained through DFA analysis, using three primary, general DFA models (cf. Figure 2) and, if necessary, six secondary, specific DFA models (cf. Figure 3). The genetic mutation would be identified through comparison with the expected DFA classifications for the EGFRmut and KRASmut, EML4-ALK fusion and wt to all.
We have demonstrated that the diagnostic yield could be improved by routinely conducting the six supplementary tests, in addition to the three primary tests for identifying the oncogenes of interest. The three primary tests A.1-A.3 classified only 27 of the 37 samples correctly. The entire seven ambiguous primary classifications could be correctly classified through the relevant supplementary tests. Furthermore, the supplementary test set could falsify the three unambiguous misclassifications of the primary test set, by yielding inconsistent results.
The simultaneous identification or exclusion of EGFRmut, KRASmut and EML4-ALK genes in the same diagnostic test could be highly relevant for selecting the most effective type of therapy. For example, advanced stage NSCLC patients with EGFR mutations could benefit from first-line EGFR TKI treatment, which compares favorably to chemotherapy in terms of efficacy, toxicity and quality of life.38,39 On the other hand, patients with EML4-ALK rearrangement might benefit from crizotinib,40 but not from EGFR TKI. The volatile fingerprint approach holds particularly high potential for monitoring mutation shifts, as many tumors may develop a resistance to the applied therapy, requiring careful monitoring and swift changes in the therapeutic approach, if indicated.41 The EML4-ALK rearrangement, which has been observed in 5–13% of all LC patients,26,40,42 shows potent oncogenic activity both in-vitro and in-vivo.43 This activity can be effectively blocked by small molecule inhibitors that target the ALK.43 Patients with EML4-ALK fusion do not respond to EGFR TKIs, but do respond to crizotinib.44
Wider-scale in-vitro studies, including an investigation for predicting therapy response of EGFR TKI sensitive cell-lines to Gefitinib via volatile fingerprints, as well as studies on genetically defined, isogenic human cell-line pairs with highly specific targeted knockouts of EGFR and KRAS genes are underway to extend and further validate these results.
Conclusion and future prospect
In this pilot in-vitro study we have identified the volatile fingerprints that are associated with EGFRmut, KRASmut and EML4-ALK rearrangement genes, using LC cell-lines. We have presented a comprehensive and highly accurate volatile fingerprint assay based on a set of three primary tests and an additional set of six supplementary tests that could correctly identify the oncogenes of the studied cell-lines in a blind test. Five VOCs could be associated with each of the genetic mutations of interest through complementary chemical analysis. Triethylamine, benzaldehyde and decanal were selectively reduced or completely depleted in the headspace of LC cells with specific oncogenes, and could therefore hold potential as specific biomarker-set for identifying and distinguishing EGFRmut, KRASmut and EML4-ALK. The aromatic compounds toluene and styrene were selectively increased for the studied oncogenes, and were most probably of exogenous origin.
While the reported concept was obtained through in-vitro studies, as a way to eliminate confounding factors that are associated with clinical samples, it is reasonable to assume that similar VOC patterns could be detected directly from blood or exhaled breath samples.7,8 This is because the volatile fingerprints have a propensity to escape to the blood and from there to the exhaled breath through exchange via the lungs.7,8,16–18 Sampling blood and breath would therefore allow immediate testing to predict a clinical benefit from targeted therapy. These clinical samples are minimally invasive or non-invasive, always available, do not require any preparation and could provide immediate important genetic information before and during the treatment plan, since they could also detect any genetic alteration in the course of the treatment. Detecting and monitoring the metabolic signature associated with cancer specific genetic mutations could be faster and easier than conventional gene-profiling methods. This feature would help to improve drug selection and detect resistance, thereby increasing the clinical benefit for the patients through safer, more timely and effective interventions that improve survival and quality of life. At the same time, hospitalizations caused by unnecessary invasive procedures could be reduced.8 However, cell-line studies cannot be translated directly into a clinically useful tool. Direct studies of the clinical samples of interest (fresh tissue/ blood/ breath) are required in order to confirm that the reported approach would indeed be useful for clinical practice.
Acknowledgments
Sources of Support: The research leading to these results has received funding from the FP7-Health Program under the LCAOS (grant agreement no. 258868; H.H. and N.P.), FP7’s ERC grant under DIAG-CANCER (grant agreement no. 256639; H.H.), the IASLC (NP) and the Fulbright Israel-US foundation (NP), and the University of Colorado SPORE Grant NCI P50 CA 058187.
Abbreviations
- ALK
Anaplastic lmphoma kinase
- CV1
First canonical variable
- DFA
Discriminant factor analysis
- EGFR
Epidermal growth factor receptor
- EGFRmut
mutated EGFR gene
- EML4
Echinoderm microtubule-associated protein-like 4
- EML4-ALK
fusion of the EML4 gene to the ALK gene
- GC-MS
Gas-chromatography/mass-spectrometry
- GNP
Gold nanoparticle
- KRAS
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
- KRASmut
mutated KRAS gene
- LC
Lung cancer
- NSCLC
Non-small cell lung carcinoma
- TKI
Tyrosine kinase inhibitor
- VOC
Volatile organic compound
- wt
Wild type
Footnotes
From the Clinical Editor: In this novel study, a new concept for profiling genetic mutations of (lung) cancer cells is described, based on the detection of patterns of volatile organic compounds emitted from cell membranes, using an array of nano-gold based sensors.
Author Contributions: H.H., F.R.H., P.A.B. Jr.R and N.P. conceived and designed the experiments: N.P., M.I., and J.M. grew the cell-lines and collected the headspace VOCs: O.B. conducted and in part analyzed the sensing and GC-MS experiments: R.I. analyzed the signals from the nanomaterial-based sensor array: Y.Y.B. analyzed part of the GC-MS data: U.T. and H.H. oversaw the data analysis and designed and wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict of Interest Statement: N.P, O.B., U.T., R. I, Y.Y.B., N.I., J. M., P.A.B. Jr.R and H.H. have no conflict to declare related to the study. F.H: Consultant: Roche/Genentech, Boehringer-Ingelheim, Lilly/Imclone, Pfizer, Celgene, BMS; Research Funding: Genentech, morphotek, Celgene, Imclone/Lilly.
Appendix A. Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nano.2013.01.008.
References
- 1.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Callaghan DS, Savale L, Montani D, Jaïs X, Sitbon O, Simonneau G, et al. Treatment of pulmonary arterial hypertension with targeted therapies. Nat Rev Cardiol. 2011;8:526–538. doi: 10.1038/nrcardio.2011.104. [DOI] [PubMed] [Google Scholar]
- 3.Shedden K, Taylor JMG, Enkemann SA, Tsao M-S, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Einhorn LH. Will we ever have personalized medicine for non-small cell lung cancer? J Thorac Oncol. 2006;1:737–739. [PubMed] [Google Scholar]
- 5.Merlo V, Longo M, Novello S, Scagliotti GV. EGFR pathway in advanced non-small cell lung cancer. Front Biosci. 2011;3:501–517. doi: 10.2741/s168. [DOI] [PubMed] [Google Scholar]
- 6.Kato Y, Peled N, Wynes MW, Yoshida K, Pardo M, Mascaux C, et al. Novel epidermal growth factor receptor mutation-specific antibodies for non-small cell lung cancer: Immunohistochemistry as a possible screening method for epidermal growth factor receptor mutations. J Thorac Oncol. 2010;5:1551–1558. doi: 10.1097/JTO.0b013e3181e9da60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amann A, Corradi M, Mazzone P, Mutti A. Lung cancer biomarkers in exhaled breath. Expert Rev Mol Diagn. 2011;11:207–217. doi: 10.1586/erm.10.112. [DOI] [PubMed] [Google Scholar]
- 8.Modak AS. Breath biomarkers for personalized medicine. Person Med. 2010;7:643–653. doi: 10.2217/pme.10.61. [DOI] [PubMed] [Google Scholar]
- 9.Hakim M, Broza YY, Barash O, Peled N, Phillips M, Amann A, et al. Volatile organic compounds of lung cancer and possible biochemical pathways. Chem Rev. 2012;112:5949–5966. doi: 10.1021/cr300174a. [DOI] [PubMed] [Google Scholar]
- 10.Barash O, Peled N, Hirsch FR, Haick H. Sniffing the unique “odor print” of non-small-cell lung cancer with gold nanoparticles. Small (Weinheim an der Bergstrasse, Germany) 2009;5:2618–2624. doi: 10.1002/smll.200900937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barash O, Peled N, Tisch U, Bunn PAJ, Hirsch FR, Haick H. Classification of lung cancer histology by gold nanoparticle sensors. Nanomedicine. 2012;8:580–589. doi: 10.1016/j.nano.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amal H, Ding L, Liu BB, Tisch U, Xu ZQ, Shi DY, et al. The scent fingerprint of hepatocarcinoma: in-vitro metastasis prediction with volatile organic compounds (VOCs) Int J Nanomedicine. 2012;2012:4135–4146. doi: 10.2147/IJN.S32680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unger M. A pause, progress, and reassessment in lung cancer screening. N Engl J Med. 2006;355:1822–1824. doi: 10.1056/NEJMe068207. [DOI] [PubMed] [Google Scholar]
- 14.Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bull. 1945;1:80–83. [Google Scholar]
- 15.Siegel S. Nonparametric statistics. Am Statistician. 1957;11:13–19. [Google Scholar]
- 16.Hakim M, Billan S, Tisch U, Peng G, Dvrokind I, Marom O, et al. Diagnosis of head-and-neck cancer from exhaled breath. Br J Cancer. 2011;104:1649–1655. doi: 10.1038/bjc.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng G, Hakim M, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, et al. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br J Cancer. 2010;103:542–551. doi: 10.1038/sj.bjc.6605810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng G, Tisch U, Adams O, Hakim M, Shehada N, Broza YY, et al. Diagnosing lung cancer in exhaled breath using gold nanoparticles. Nat Nanotechnol. 2009;4:669–673. doi: 10.1038/nnano.2009.235. [DOI] [PubMed] [Google Scholar]
- 19.Dovgolevsky E, Konvalina G, Tisch U, Haick H. Monolayer-capped cubic platinum nanoparticles for sensing nonpolar analytes in highly humid atmospheres. J Phys Chem C. 2010;114:14042–14049. [Google Scholar]
- 20.Dovgolevsky E, Tisch U, Haick H. Chemically sensitive resistors based on monolayer-capped cubic nanoparticles: Towards configurable nanoporous sensors. Small (Weinheim an der Bergstrasse, Germany) 2009;5:1158–1161. doi: 10.1002/smll.200801831. [DOI] [PubMed] [Google Scholar]
- 21.Peng G, Trock E, Haick H. Detecting simulated patterns of lung cancer biomarkers by random network of single-walled carbon nanotubes coated with nonpolymeric organic materials. Nano Lett. 2008;8:3631–3635. doi: 10.1021/nl801577u. [DOI] [PubMed] [Google Scholar]
- 22.Konvalina G, Haick H. Effect of humidity on nanoparticle-based chemiresistors: a comparison between synthetic and real-world samples. ACS Appl Mater Interfaces. 2012;4:317–325. doi: 10.1021/am2013695. [DOI] [PubMed] [Google Scholar]
- 23.Ionescu R, Llobet E, Vilanova X, Brezmes J, Sueiras JE, Caldererc J, et al. Quantitative analysis of NO2 in the presence of CO using a single tungsten oxide semiconductor sensor and dynamic signal processing. Analyst. 2002;127:1237–1246. doi: 10.1039/b205009a. [DOI] [PubMed] [Google Scholar]
- 24.Brereton RG. Chemometrics, Application of Mathematics Statistics to Laboratory Systems. Chichester: Ellis Horwood; 1990. [Google Scholar]
- 25.Tisch U, Haick H. Arrays of chemisensitive monolayer-capped metallic nanoparticles for diagnostic breath testing. Rev Chem Eng. 2011;26:171–179. [Google Scholar]
- 26.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 27.Horváth I, Lázár Z, Gyulai N, Kollai M, Losonczy G. Exhaled biomarkers in lung cancer. Eur Respir J. 2009;34:261–275. doi: 10.1183/09031936.00142508. [DOI] [PubMed] [Google Scholar]
- 28.Tisch U, Billan S, Ilouze M, Phillips M, Peled N, Haick H. Volatile organic compounds in exhaled breath as biomarkers for the early detection and screening of lung cancer. CML Lung Cancer. 2012;5:107–117. [Google Scholar]
- 29.Amann A, Spaně P, Smith D. Breath analysis: the approach towards clinical applications. Mini Rev Med Chem. 2007;7:115–129. doi: 10.2174/138955707779802606. [DOI] [PubMed] [Google Scholar]
- 30.Kneepkens CMF, Lepage G, Roy CC. The potential of the hydrocarbon breath test as a measure of lipid peroxidation. Free Radical Biol Med. 1994;17:127–160. doi: 10.1016/0891-5849(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 31.Belonogov RN, Titova NM, Lapeshin PV, Ivanova YR, Shevtsova AO, Pokrovskii AA. Changes in the content of protein and lipid oxidative modification products in tumor tissue at different stages of lung cancer. Bull Exp Biol Med. 2009;147:630–631. doi: 10.1007/s10517-009-0565-4. [DOI] [PubMed] [Google Scholar]
- 32.Sponring A, Filipiak W, Ager C, Schubert JK, Miekisch W, Amann A, et al. Analysis of volatile organic compounds (VOCs) in the headspace of NCI-H1666 lung cancer cells. Cancer Biomark. 2010;7:153–161. doi: 10.3233/CBM-2010-0182. [DOI] [PubMed] [Google Scholar]
- 33.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 34.Poli D, Goldoni M, Corradi M, Acampa O, Carbognani P, Internullo E, et al. Determination of aldehydes in exhaled breath of patients with lung cancer by means of on-fiber-derivatisation SPME-GC/MS. J Chromatogr B. 2010;878:2643–2651. doi: 10.1016/j.jchromb.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 35.Bajtarevic A, Ager C, Pienz M, Klieber M, Schwarz K, Ligor M, et al. Noninvasive detection of lung cancer by analysis of exhaled breath. BMC Cancer. 2009;9:348. doi: 10.1186/1471-2407-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kischkel S, Miekisch W, Sawacki A, Straker EM, Trefz P, Amann A, et al. Breath biomarkers for lung cancer detection and assessment of smoking related effects-confounding variables, influence of normalization and statistical algorithms. Clin Chim Acta. 2010;411:1637–1644. doi: 10.1016/j.cca.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Poli D, Carbognani P, Corradi M, Goldoni M, Acampa O, Balbi B, et al. Exhaled volatile organic compounds in patients with non-small cell lung cancer: cross sectional and nested short-term follow-up study. Respir Res. 2005;6:71. doi: 10.1186/1465-9921-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 39.Cappuzzo F, Ligorio C, Toschi L, Rossi E, Trisolini R, Paioli D, et al. EGFR and HER2 gene copy number and response to first-line chemotherapy in patients with advanced non-small cell lung cancer (NSCLC) J Thorac Oncol. 2007;2:423–429. doi: 10.1097/01.JTO.0000268676.79872.9b. [DOI] [PubMed] [Google Scholar]
- 40.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doebele RC, Oton AB, Peled N, Camidge DR, Bunn PAJ. New strategies to overcome limitations of reversible EGFR tyrosine kinase inhibitor therapy in non-small cell lung cancer. Lung Cancer. 2010;69:1–12. doi: 10.1016/j.lungcan.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Mano H. Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci. 2008;99:2349–2355. doi: 10.1111/j.1349-7006.2008.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, Ueno T, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res. 2011;17:2081–2086. doi: 10.1158/1078-0432.CCR-10-1591. [DOI] [PubMed] [Google Scholar]