Abstract
Global gene expression measured by DNA microarray platforms have been extensively used to classify breast carcinomas correlating with clinical characteristics, including outcome. We generated a breast cancer Serial Analysis of Gene Expression (SAGE) high-resolution database of ~2.7 million tags to perform unsupervised statistical analyses to obtain the molecular classification of breast-invasive ductal carcinomas in correlation with clinicopathologic features. Unsupervised statistical analysis by means of a random forest approach identified two main clusters of breast carcinomas, which differed in their lymph node status (P = 0.01); this suggested that lymph node status leads to globally distinct expression profiles. A total of 245 (55 up-modulated and 190 down-modulated) transcripts were differentially expressed between lymph node (+) and lymph node (−) primary breast tumors (fold change, ≥2; P < 0.05). Various lymph node (+) up-modulated transcripts were validated in independent sets of human breast tumors by means of real-time reverse transcription-PCR (RT-PCR). We validated significant overexpression of transcripts for HOXC10 (P = 0.001), TPD52L1 (P = 0.007), ZFP36L1 (P = 0.011), PLINP1 (P = 0.013), DCTN3 (P = 0.025), DEK (P = 0.031), and CSNK1D (P = 0.04) in lymph node (+) breast carcinomas. Moreover, the DCTN3 (P = 0.022) and RHBDD2 (P = 0.002) transcripts were confirmed to be overexpressed in tumors that recurred within 6 years of follow-up by real-time RT-PCR. In addition, meta-analysis was used to compare SAGE data associated with lymph node (+) status with publicly available breast cancer DNA microarray data sets. We have generated evidence indicating that the pattern of gene expression in primary breast cancers at the time of surgical removal could discriminate those tumors with lymph node metastatic involvement using SAGE to identify specific transcripts that behave as predictors of recurrence as well.
Introduction
Although breast cancer is the most common malignancy in women, the biology of breast cancer remains poorly understood mainly due to the characteristic cellular and molecular heterogeneity of breast tumors. Global gene expression profiling is providing novel information of biological and clinical relevance for the classification of breast cancers.
By means of DNA microarray analyses, various laboratories identified gene expression patterns that correlated with breast cancer patient prognosis (1–9). In spite of the described progress in molecular oncology, invasion into axillary lymph nodes and steroid hormone receptors status still remain as the most reliable prognostic factor for breast cancer patients (10).
Development of metastases (local and distant) requires that a cancer cell must complete a series of steps involving complex interactions with the host microenvironment. This process involves the dysregulation of multiple genes and transcriptional programs. The primary goal of this study was to identify gene expression signatures of relevance for breast cancer subclassification and prognosis. We analyzed a high-resolution Serial Analysis of Gene Expression (SAGE) database obtained from 27 breast-invasive ductal carcinomas (IDCA). A random forest (RF) clustering approach was used for SAGE data analysis (11, 12). This unsupervised analysis of gene expression profiles grouped breast carcinomas predominantly according to their lymph node status. This suggests that lymph node status leads to globally distinct breast cancer gene expression profiles.
The identification of gene expression profiles, individual biomarkers, and biological pathways that contribute to the development of lymph node metastases will be of significant benefit to improve tumor classification and may, in the future, influence clinical decision making and the development of targeted therapies.
Results and Discussion
Generation and Analysis of SAGE Libraries
Breast cancer phenotypic and genetic heterogeneity corresponds to heterogeneity of gene expression profiles. SAGE data were obtained from a set of invasive breast carcinomas at a resolution of 100,000 tags per library. Thus, a breast cancer SAGE high-resolution database of almost 2.7 million tags was generated and analyzed, monitoring the expression behavior of more than 30,000 transcripts.
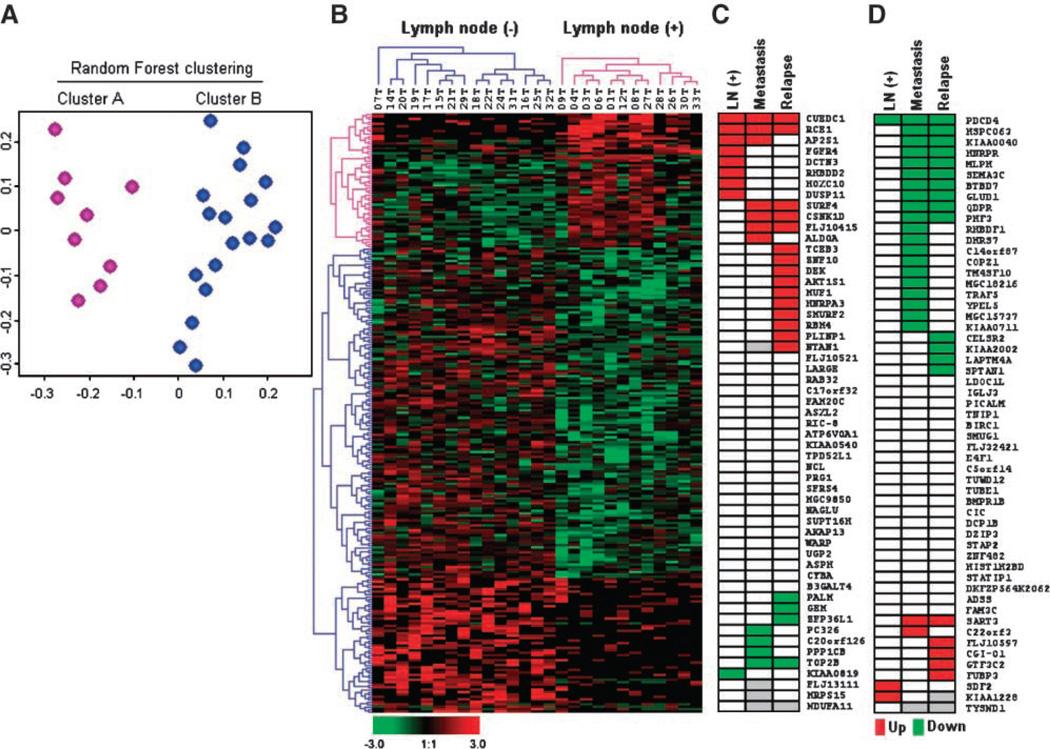
An unsupervised clustering method (RF clustering) allowed us to group the invasive breast carcinomas on the basis of their gene expression pattern. Two dominant clusters were identified (Fig. 1A). To further elucidate the reasons driving the separation of breast carcinomas in two major groups, we analyzed the identified clusters in the light of available histopathologic data (see Table 1). Interestingly, the variable that correlated with the RF clustering results was the lymph node status of tumors (P = 0.01). A total of 7 of 9 breast cancers (78%) in the cluster A are lymph node (+), and 14 of 18 breast tumors (87%) in cluster B are lymph node (−) IDCA (Fig. 1A). Nonstatistically significant differences were detected for ER status, histologic grade, and tumor size (P > 0.05). In contrast with results from previous gene expression studies, in which ER was the major discriminator between breast cancer groups, in our case, we interpret that the lack of spontaneous association between clusters and ER status in this subset of samples is likely due to that ~75% of the SAGE libraries generated derived from ERα(+) stages I and II primary breast carcinomas.
FIGURE 1.
SAGE profiles of 27 primary invasive breast carcinomas. A. The SAGE profiles of 27 breast carcinomas are visualized in a two-dimensional multidimensional scaling plot where each dot represents one sample and the relative distances between samples are correlated with their RF dissimilarities. Breast carcinomas are colored by their RF clustering memberships: cluster A (fuchsia) composed by 78% of lymph node (+) carcinomas and cluster B (blue) composed of 87% of lymph node (−) breast carcinomas. B. Hierarchical clustering of 245 differentially expressed genes (55 up-modulated transcripts and 190 down-modulated transcripts) according to patient’s lymph node status based on pathologic diagnosis. Color scale at the bottom of the picture is used to represent expression level: low expression is represented by green, and high expression is represented by red. Results of meta-analysis (from publicly available gene expression microarray data sets) of 55 up-modulated (C) and 55 down-modulated transcripts (D) identified by SAGE. Red or green boxes, represent statistically significant agreement between our study and previously published studies not only on lymph node status, but also in association with other progression parameters such as metastasis or relapse. Red, statistically significant P values (P < 0.05) associated with gene overexpression in lymph node (+), metastasis, and relapse (DFS); green, statistically significant down-modulated expression. Gray boxes, Unavailable data.
Table 1.
Histopathologic Characteristics of Primary Breast Carcinomas Analyzed by SAGE
| Number (%) | |
|---|---|
| Histology type | |
| Invasive ductal carcinoma | 27 (100) |
| Tumor size | |
| 1-2 cm | 11 (41) |
| 2-5 cm | 13 (48) |
| >5 cm | 3 (11) |
| Nodal status | |
| N0 | 16 (59) |
| N1 | 11 (41) |
| Nuclear grade | |
| Grade I | 4 (15) |
| Grade II | 14 (52) |
| Grade III | 9 (33) |
| ERα status | |
| Negative | 7 (26) |
| Positive | 20 (74) |
To identify the most representative differentially expressed transcripts between tumor groups, we employed a statistically supervised method previously described by us as a modified t test (13). This analysis revealed 245 genes differentially expressed (P < 0.05) between lymph node (+) and lymph node (−) IDCA (Fig. 1B; Supplementary Data File 1). Among the 245 transcripts, 55 were up-modulated, and 190 were down-modulated transcripts in lymph node (+) tumors.
We used the Expression Analysis Systematic Explorer software (EASE) to annotate the 245 deregulated genes according to the information provided by the GO Consortium (14, 15). We observed that 32% of the transcripts are involved in biological processes related to metabolism, 22% are related to cellular physiologic process, and 14% are related to cell communication. Approximately 25% of these dysregulated genes are related to molecular functions associated with nucleic acid/protein binding, 15% are related to hydrolase/transferase activity, and 4% are related to metal ion-binding functions.
Cross-Platform Gene Expression Profile Comparison
Comparing data sets generated on different gene expression platforms increases the confidence of specific gene expression classifier data sets (16). By performing a meta-analysis from publicly available breast cancer microarray studies, we provide a robust cross-platform validation of 55 up-regulated and 55 down-regulated (fold change, >3) lymph node (+)-associated transcripts (Fig. 1C and D). Meta-analysis showed that 42% of the transcripts identified by SAGE (46 out of 110) were confirmed as having statistically significant up- or down-modulation in relation to lymph node (+) status (9 genes), distal metastasis (26 genes), and relapse (29 genes; Table 2, Supplementary Data Files 2 and 3). The lack of 100% overlap of findings between the various studies including ours is not surprising when it is considered that these studies have been done with different technologies (cDNA or various oligonucleotide microarrays), different number of genes in the various fixed platforms, different and heterogenous patient populations (with regard to age, tumor staging, hormone receptor status, and treatment). Nevertheless, we show that a significant proportion of lymph node (+)-associated transcripts detected by our SAGE study behave as poor prognostic markers. More importantly, SAGE, an open gene expression platform, also identified novel sets of genes as highly expressed in lymph node (+) primary breast carcinomas not previously reported by others.
Table 2.
Meta-analysis, Cross-Validated SAGE Transcripts as Poor-Prognosis Breast Cancer Biomarkers
| Gene Name | Description | Lymph Node (+) | Metastasis (Yes) | Relapse (Yes) |
|---|---|---|---|---|
| Up-modulated genes positively associated with the variable analyzed | ||||
| CUEDC1 | CUE domain containing 1 | 0.040 | 0.033 | 0.018 |
| RCE1 | RCE1 homologue. prenyl protein protease | 0.038 | 0.0001 | 0.0001 |
| AP2S1 | Adaptor-related protein complex 2 | 0.015 | 0.038 | 0.508 |
| FGFR4 | Fibroblast growth factor receptor 4 | 0.035 | 0.279 | 0.913 |
| DCTN3 | Dynactin 3 (p22) | 0.003 | 0.868 | 0.682 |
| RHBDD2 | Rhomboid domain containing 2 | 0.042 | 0.300 | 0.259 |
| HOXC10 | Homeobox C10 | 0.013 | 0.520 | 0.471 |
| DUSP11 | Dual specificity phosphatase 11 | 0.049 | 0.158 | 0.940 |
| SURF4 | Surfeit 4 | 0.393 | 0.024 | 0.009 |
| CSNK1D | Casein kinase 1 δ | 0.417 | 0.0001 | 0.001 |
| FLJ10415 | Hypothetical protein | 0.029 | 0.0001 | 0.0001 |
| ALDOA | Aldolase A | 0.212 | 0.0001 | 0.109 |
| TCEB3 | Transcription elongation factor B | 0.335 | 0.322 | 0.020 |
| ZNF10 | Zinc finger protein 10 (KOK1) | 0.818 | 0.722 | 0.0001 |
| DEK | DEK oncogene | 0.419 | 0.673 | 0.005 |
| AKT1S1 | AKT1 substrate 1 (proline-rich) | 0.533 | 0.360 | 0.037 |
| MUF1 | MUF1 protein | 0.688 | 0.322 | 0.028 |
| HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 0.977 | 0.140 | 0.004 |
| SMURF2 | SMAD specific E3 ubiquitin protein ligase 2 | 0.704 | 0.058 | 0.0001 |
| RBM4 | RNA binding motif protein 4 | 0.882 | 0.783 | 0.032 |
| PLINP1 | Growth arrest and DNA-damage-inducible | 0.424 | 0.208 | 0.039 |
| NTAN1 | NH2-terminal asparagines amidase | 0.925 | — | 0.005 |
| Down-modulated genes negatively associated with the variables analyzed | ||||
| PDCD4 | Programmed cell death 4 | 0.007 | 0.001 | 0.0001 |
| HSPC063 | HSPC063 protein | 0.185 | 0.0001 | 0.0001 |
| HNRPR | Heterogeneous nuclear ribonucleoprotein R | 0.803 | 0.0001 | 0.0001 |
| KIAA0040 | KIAA0040 protein | 0.624 | 0.003 | 0.005 |
| MLPH | Melanophilin | 0.780 | 0.007 | 0.016 |
| SEMA3C | Sema domain, immunoglobulin domain (Ig) | 0.285 | 0.001 | 0.0001 |
| BTBD7 | BTB (POZ) domain containing 7 | 0.629 | 0.003 | 0.003 |
| GLUD1 | Glutamate dehydrogenase 1 | 0.220 | 0.002 | 0.0001 |
| QDPR | Quinoid dihydropteridine reductase | 0.217 | 0.0001 | 0.0001 |
| PHF3 | PHD finger protein 3 | 0.342 | 0.0001 | 0.001 |
| RHBDF1 | Rhomboid family 1 (Drosophila) | 0.491 | 0.039 | 0.426 |
| DHRS7 | Dehydrogenase/reductase (SDR family) member 7 | 0.584 | 0.047 | 0.167 |
| C14orf87 | Chromosome 14 open reading frame 87 | 0.346 | 0.011 | 0.968 |
| COPZ1 | Coatomer protein complex subunit ζ1 | 0.272 | 0.031 | 0.704 |
| TM4SF10 | Transmembrane 4 superfamily member 10 | 0.570 | 0.026 | 0.293 |
| MGC18216 | Hypothetical protein MGC18216 | 0.731 | 0.002 | 0.508 |
| TRAF5 | Tumor necrosis factor receptor-associated factor 5 | 0.188 | 0.016 | 0.673 |
| YPEL5 | Yippee-like 5 (Drosophila) | 0.188 | 0.001 | 0.212 |
| MGC15737 | Hypothetical protein MGC15737 | 0.841 | 0.019 | 0.365 |
| KIAA0711 | KIAA0711 gene product | 0.936 | 0.003 | 0.841 |
| CELSR2 | Cadherin, EGF LAG seven-pass G-type receptor 2 | 0.494 | 0.384 | 0.009 |
| KIAA2002 | KIAA2002 protein | 0.600 | 0.172 | 0.017 |
| LAPTM4A | Lysosomal-associated protein transmembrane 4 | 0.297 | 0.164 | 0.004 |
| SPTAN1 | Spectrin α, non-erythrocytic 1 | 0.994 | 0.463 | 0.0001 |
Real-time Reverse Transcription-PCR Validation of Lymph Node (+)–Associated Transcripts
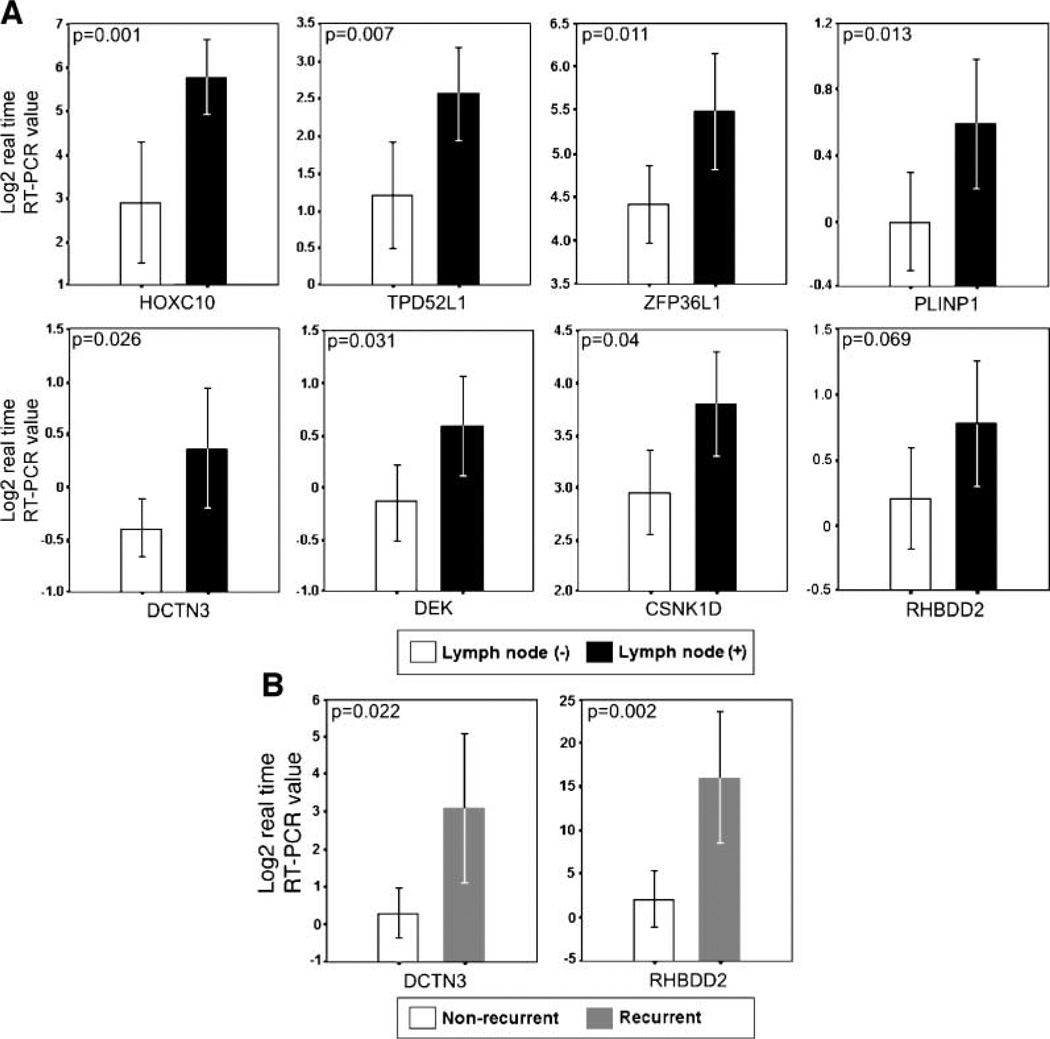
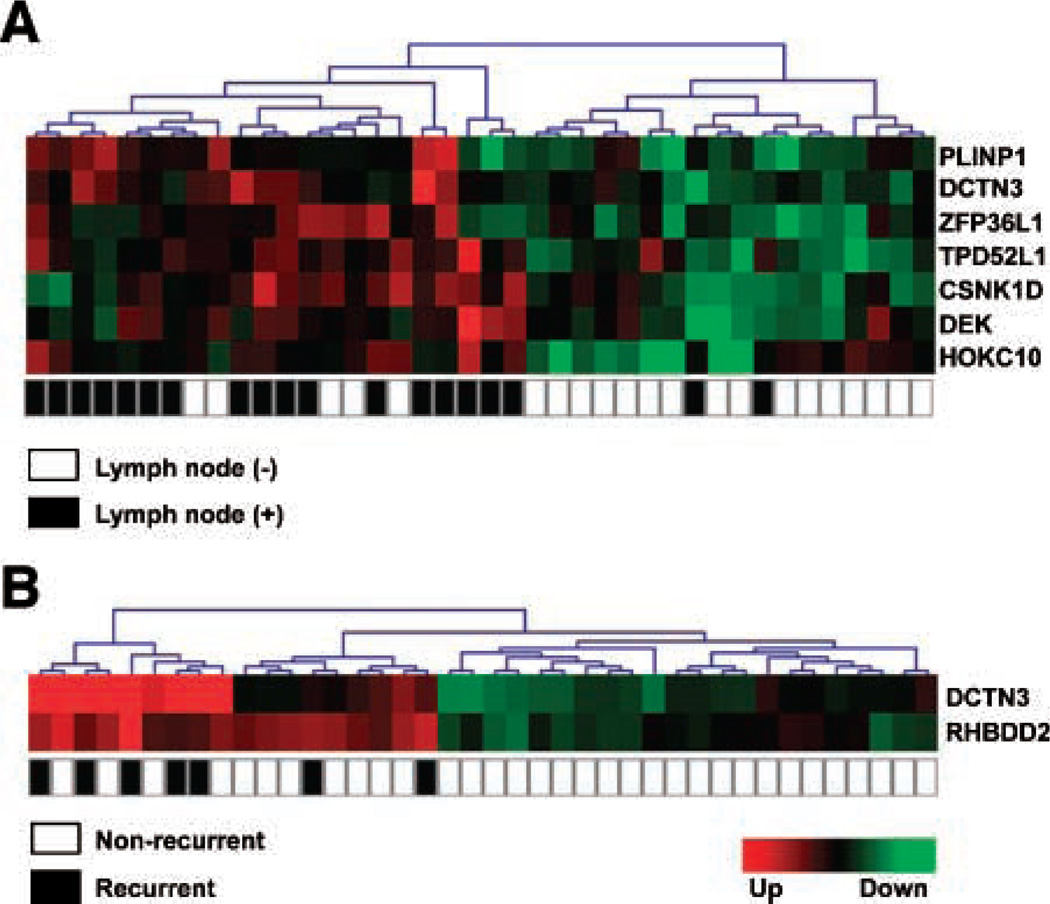
The most commonly dysregulated transcripts between lymph node (+) and lymph node (−) breast IDCA as determined by SAGE are represented in Table 3 (fold change, >3; P < 0.01). To validate our findings, an independent set of 40 breast IDCA was analyzed by means of real-time reverse transcription-PCR (RT-PCR). In agreement with the SAGE data, we detected statistical differences in the overexpression of seven out of eight evaluated transcripts in lymph node (+) breast tumors, including homeobox protein hox-c10 (HOXC10; P = 0.001), tumor protein D52 like-1 (TPD52L1; P = 0.007), zinc finger protein 36 like-1 (ZFP36L1; P = 0.011), p53-responsive gene 6 (PLINP1; P = 0.013), dynactin 3 (DCTN3; P = 0.025), dek oncogene (DEK; P = 0.031), casein kinase 1δ (CSNK1D; P = 0.04; Fig. 2A). A trend of borderline significance was detected for the rhomboid domain containing 2 (RHBDD2; P = 0.069; Fig. 2A). Hierarchical clustering analysis of the validated transcripts successfully classified tumors according to patient’s lymph node status (P < 0.05), distinguishing the lymph node (+) from the lymph node (−) breast carcinomas with an accuracy of 89.5% (2 out of 19 lymph node-positive samples misclassified; Fig. 3A). Nonstatistically significant associations were detected between the expressions of these transcripts and ERα status (P > 0.05).
Table 3.
Most Highly Deregulated Transcripts in Lymph Node (+) Breast Carcinomas Identified by SAGE (Fold Change, >3; P <0.01)
| Gene Name | Description | Tag | Entrez Gene ID | Expression |
|---|---|---|---|---|
| Regulation of cell proliferation | ||||
| DEK | DEK oncogene | ACAAAAGTGA | 7913 | ▲ |
| TPD52L1 | Tumor protein D52-like 1 | ACTGTCTCCA | 7164 | ▲ |
| GEM | GTP binding protein | GAGCCATCAT | 2669 | ▲ |
| AKAP13 | A kinase (PRKA) anchor protein 13 | GGATGCGCAG | 11214 | ▲ |
| CCRK | Cell cycle related kinase | GGATGATGTC | 23552 | ▼ |
| Regulation of transcription related | ||||
| MGC9850 | Hypothetical protein | TGCTTGACAA | 219404 | ▲ |
| ZFP36L1 | Zinc finger protein 36, C3H type-like 1 | CTTTCTTCCC | 677 | ▲ |
| TP53BP1 | Tumor protein p53 binding protein 1 | ACAGTGCTTG | 7158 | ▼ |
| ATF2 | Activating transcription factor 2 | GTGGATTCAT | 1386 | ▼ |
| CBX4 | Chromobox homologue 4 | AAAGTCTAGA | 8535 | ▼ |
| Signal transduction related | ||||
| CSNK1D | Casein kinase 1, δ | GCTGATCTAC | 1453 | ▲ |
| PPP1CB | Protein phosphatase 1 | AAGATTTTAG | 5500 | ▲ |
| IGFBP4 | Insulin-like growth factor binding prot. 4 | TTTGGAATGT | 3487 | ▼ |
| ARHGAP1 | Rho GTPase activating protein 1 | TGTCTGTGGT | 392 | ▼ |
| FYCO1 | FYVE and coiled-coil domain containing | TTAAATGCAA | 79443 | ▼ |
| P2RY2 | Purinergic receptor P2Y, G-protein | AGTAAACCAT | 5029 | ▼ |
| Cytoskeleton | ||||
| DCTN3 | Dynactin 3 (p22) | CTGCCCGCCT | 11258 | ▲ |
| MYH3 | Myosin, heavy polypeptide 3 | GTCTCATTTC | 4621 | ▼ |
| Protein transport/targeting and biosynthesis | ||||
| AP2S1 | Adaptor-related protein complex 2 | CCGTGGTCAC | 1175 | ▲ |
| SUPT16H | Suppressor of Ty 16 homologue | CCTTGGGCCT | 11198 | ▲ |
| HSPS4 | Hermansky-Pudlak syndrome 4 | TTTGTGACTG | 89781 | ▼ |
| TOMM20 | Translocase of outer mitochondrial memb. | TGTGAGCCCT | 9804 | ▼ |
| Metabolism and miscellaneous | ||||
| ATP6V0A1 | ATPase, H+ transporting | TGGCTGTGAG | 535 | ▲ |
| NTAN1 | NH2-terminal asparagine amidase | AATTACCAAA | 123803 | ▲ |
| NAGLU | N-acetylglucosaminidase, α | GCTGAGCTGG | 4669 | ▲ |
| SMURF2 | SMAD specific E3 ubiquitin protein | ATCTTGAACA | 64750 | ▲ |
| PRG1 | Proteoglycan 1, secretory granule | GCCATAAAAT | 5552 | ▲ |
| CNKSR1 | Connector enhancer of kinase suppres. | TACAGTTCCC | 10256 | ▼ |
| DAP13 | 13 kDa differentiation-associated protein | TGTTATTAAA | 55967 | ▼ |
| MANBAL | Mannosidase, β A, lysosomal-like | CAACTAATTC | 63905 | ▼ |
| ADSS | Adenylosuccinate synthase | GACTACCTTT | 159 | ▼ |
| Function unknown | ||||
| FAM20C | Family with sequence similarity 20, C | CGCCCGTCGT | 56975 | ▲ |
| C20orf126 | Chromosome 20 ORF 1 | GGTGGTTGCT | 81572 | ▲ |
| AKT1S1 | AKT1 substrate 1 (proline-rich) | CGCGCGCTGG | 84335 | ▲ |
| RIC-8 | Likely ortholog of mouse synembryn | ATTTGCCTCT | 60626 | ▲ |
| MUF1 | MUF1 protein | GGCTGCCCAG | 10489 | ▲ |
| WARP | Von Willebrand factor A domain | CCCAGGACAC | 64856 | ▲ |
| PKD1-like | Polycystic kidney disease 1-like | TTGACACTTT | 79932 | ▼ |
| MESDC1 | Mesoderm development candidate 1 | ACAAGAATTG | 59274 | ▼ |
| TTC15 | Tetratricopeptide repeat domain 15 | TTTTACTCAC | 51112 | ▼ |
| BCMP11 | Breast cancer membrane protein 11 | CGGCAGAGCT | 155465 | ▼ |
| MGC10067 | Hypothetical protein | GATGTCTTGT | 134510 | ▼ |
| AMIGO2 | Amphoterin induced gene 2 | CCCCATACTA | 347902 | ▼ |
NOTE: ▲, up-regulated gene in lymph node (+) primary breast carcinomas. ▼, down-regulated gene in lymph node (+) primary breast carcinomas.
FIGURE 2.
Validation assays of SAGE expression profiles in an independent set of primary invasive breast carcinomas (n = 40). A. Real-time RT-PCR of seven up-modulated transcripts (HOXC10, TPD52L1, ZFP36L1, PLINP1, DCTN3, DEK, CSNK1D, RHBDD2) in LN(+) carcinomas. B. Real-time RT-PCR of two up-modulated transcripts (DCTN3, RHBDD2) in recurrent breast carcinomas. Mean ± 2 SE based on log2 transformation of real-time RT-PCR values of the assayed gene relative to 18S rRNA used as normalizing control.
FIGURE 3.
Hierarchical clustering of primary breast carcinomas based on real-time RT-PCR validation data. A. Cluster showing nodes in the basis of lymph node distribution (P = 0.0001). B. Cluster showing nodes in the basis of recurrence status distribution (P = 0.001).
Tumor Protein– and Transcription Factor–Related Genes
The TPD52L1 gene encodes a member of the tumor protein D52 family. This protein contains a coiled-coil motif required for homo- and heteromeric interactions with other D52-like proteins (17). The TPD52 gene was first identified as overexpressed in human breast carcinomas (17, 18). Subsequent studies also indicated that these genes are overexpressed in multiple human cancers such as lung, prostate, ovarian, endometrial, and hepatocellular carcinomas (18). TPD52L1 was reported to be involved in cell proliferation and calcium signaling. It also interacts with the mitogen-activated protein kinase 5 (19).
DEK was originally described as a proto-oncogene and has been implicated in multiple cellular processes, including transcriptional regulation and chromatin remodeling (20). Transcriptional up-regulation of wild-type DEK was discovered in various tumor types, including myeloid leukemia, brain tumors, and hepatocellular carcinoma (21, 22). In addition, DEK overexpression was associated with a number of clinical autoimmune conditions (23, 24). Recently, it has been suggested that DEK up-regulation may be a common event in human carcinogenesis and may reflect its senescence inhibitory function (25). Despite these associations with several human disorders, little is known about how DEK could functionally be involved in these diseases (24).
HOXC10 is one of the highly conserved HOXC family members of transcription factors that play an important role in morphogenesis, cell differentiation, and proliferation (26–28). The HOXC protein levels are controlled during cell differentiation and proliferation. Dysregulation of a variety of HOX genes has been implicated in several human cancers, including leukemias, colorectal, breast, and renal carcinomas, melanomas, and squamous cell carcinomas of the skin (26, 27). Recently, it was shown that the overexpression of HOXC4, HOXC5, HOXC6, and HOXC8 genes in malignant cell lines and prostate carcinomas with lymph node metastases (29). In agreement with these data, we validated the overexpression of HOXC10 gene in primary lymph node (+) breast carcinomas by real-time RT-PCR (P = 0.001; Fig. 2A).
ZFP36L1 also known as C3H type-like 1) is a member of the 12-O-tetradecanoylphorbol-13-acetate (TPA)–inducible sequence 11 (TIS11) family of early-response genes. The encoded protein contains a zinc finger domain with a repeating cys-his motif (30). TIS11 gene expression is induced rapidly and transiently in response to extracellular hormone and growth factor signals. The potential role of this gene in breast carcinogenesis remains unknown.
DCTN3 and RHBDD2 as Predictors or Recurrence
As mentioned, the quantitative RT-PCR analysis validated significant differences between lymph node (+) versus lymph node (−) primary breast carcinoma groups for DCTN3 (P = 0.025), and a trend was detected for RHBDD2 (P = 0.069; Fig. 2A). However, meta-analysis comparisons further confirmed our findings showing statistically significant over-expression of DCTN3 (P = 0.003) and RHBDD2 (P = 0.042) in lymph node (+) compared with lymph node (−) breast IDCA (Fig. 1C, Supplementary Data File 3). More importantly, the DCTN3 (P = 0.022) and RHBDD2 (P = 0.002) transcripts were also observed to be markedly up-modulated in tumors that recurred within 6 years of follow-up (Fig. 2B). Unsupervised hierarchical clustering analysis of these transcripts successfully classified tumors according to recurrence status (P < 0.05; Fig. 3B). These data suggest that overexpression of DCTN3 and RHBDD2 genes could play a role in breast cancer progression.
The DCTN3 gene (also known as DCTN22) encodes the smallest (p22/24) subunit of dynactin, a cytoplasmic motor protein complex involved in organelle trafficking, cytokinesis, spindle formation, chromosome movement, and nuclear positioning (31). Overexpression in mammalian cells of one dynactin subunit (dynamitin) disrupts the complex, resulting in the perturbation of mitosis (32). In addition, DCTN2 over-expression disrupt the dynein-dynactin motor, shifting cellular movement and mitosis with predisposition to mitotic block and polyploidy (33). DCTN3 localizes to the centrosomes during interphase and to kinetochores and spindle poles throughout mitosis. It was also proposed that the dynein-dynactin complex is involved in cytoplasmatic/nuclear transport of p53 (34). The correct balance of dynactin subunits is important for adequate centrosome integrity before centrosome duplication, ultimately governing the G1-S transition.
The RHBDD2 gene (rhomboid domain containing 2) encodes a protein that spans seven-transmembrane domains and is a member of the rhomboid veinlet-like family of genes. Several rhomboid protein members in Drosophila have been implicated in the processing of transforming growth factor-α (TGF-α)–like ligands, and consequent epidermal growth factor (EGF) receptor activation (35). Genetic and molecular studies have revealed that the production of an activated EGF ligand by the signal-sending cell is a key regulatory step in receptor activation (36). Thus, the RHBDD2 protein very likely functions in regulating the response to growth factors. However, the potential role of this protein in breast carcinogenesis remains to be elucidated.
Tissue Microarray Immunohistochemical Analysis of DCTN3 Protein Expression
Because DCTN3 was identified by real-time RT-PCR as distinctively overexpressed in lymph node (+) primary breast carcinomas and in IDCA that recurred within 6 years, we decided to investigate further this gene at protein expression level using a breast cancer progression tissue microarray (Fig. 4).
FIGURE 4.
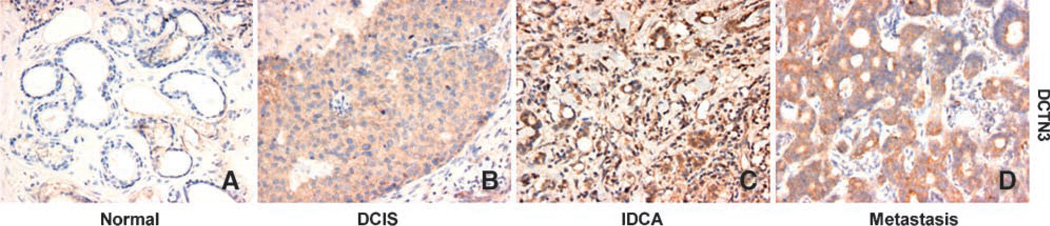
DCTN3 immunohistochemical staining in normal (adjacent tumor), ductal carcinoma in situ (DCIS), invasive ductal carcinoma (IDCA), and metastatic breast samples.
DCTN3 tissue microarray immunohistochemical (TMA-IHC) analysis showed undetectable expression in 72% (13 out of 18) of the normal breast epithelial samples analyzed, whereas strong immunoreactivity for DCTN3 protein was detected in 60% (6 out of 10) of invasive ductal (IDC) and metastatic breast carcinoma tissues analyzed (P trend = 0.001; Table 4). In all positive cases, the DCTN3 inmunostaining had a homogeneous and diffuse pattern that was localized to the cytoplasm. When DCTN3 expression was correlated with lymph node status, we determined that 75% (6 out of 8) of lymph node (+) carcinoma showed strong DCTN3 staining, whereas 67% (12 out of 18) of lymph node (−) breast carcinomas showed negative inmunostaining (P = 0.027; Table 4). These data plus the aforementioned evidence strongly suggest a putative role for DCTN3 mRNA/protein expression and axillary lymph node metastasis and breast cancer progression.
Table 4.
DCTN3 Protein Expression According to Histopathologic Characteristics
| Protein Expression n (%) |
Statistic | |||
|---|---|---|---|---|
| Absent | Moderate | Strong | ||
| Normal epithelium | 13 (72) | 18 (30.5) | 1 (10) | χ2 = 15.3, P = 0.004, P trend = 0.001 |
| DCIS | 1 (5.5) | 20 (34) | 3 (30) | |
| IDCA/Metastasis | 4 (22) | 21 (35.5) | 6 (60) | |
| LN(−) | 12 (67) | 3 (23) | 2 (25) | χ2 = 7.25, P = 0.027, P trend = 0.007 |
| LN(+) | 6 (33) | 10 (77) | 6 (75) | |
Conclusions
The genes that we identified and validated seem to be involved in signaling pathways related to invasion into axillary lymph nodes. Interestingly, deregulated transcripts that correlate with the presence of lymph node metastases at the time of surgery conform a gene expression signature distinguishable to that observed for the lymph node–negative counterparts, suggesting different molecular programs related to the meta-static process.
Gene expression profiling will not necessarily replace classic approaches to predict the outcome; however, it will likely add substantial information that may help in better defining breast cancer outcome classes. The identification of individual proteins is also of high relevance not only for the potential value as prognostic biomarkers but also may provide insight into mechanisms and pathways of relevance in breast cancer progression. Nevertheless, given the molecular heterogeneity of breast cancer, further global and individual gene expression studies are needed to reliably discriminate breast cancer subgroups of value for determining outcome. Results of this study will provide novel insights into the molecular biology underlying breast cancer lymph node metastasis and recurrence.
Materials and Methods
SAGE Libraries
We did a comparative analysis of the gene expression profiles of 27 IDCA using SAGE. Libraries were generated at our laboratory (~100,000 SAGE tags per library). Table 1 shows histopathologic characteristics of the specimens analyzed. For the generation of SAGE libraries, snap-frozen samples were obtained from the M.D. Anderson Breast Cancer Tumor Bank, and SAGE analysis was done as previously described (37, 38).
Data Processing and Statistical Analysis of SAGE Libraries
SAGE tag extraction from sequencing files was done using the SAGE2000 software version 4.0 (kindly provided by Dr. K. Kinzler, John Hopkins University). SAGE data management, tag to gene matching, as well as additional gene annotations and links to publicly available resources such as GO, UniGene, RefSeq, were done using a suite of Web-based SAGE library tools developed by us.5
Our analysis of data involved the following steps: (a) use of unsupervised RF clustering to group the patients based on their SAGE expression profiles; (b) investigate potential associations with multiple histopathologic variables; (c) identification of differentially expressed transcripts between clusters; (d) gene ontology analysis of the resulting transcripts.
We propose to use the RF clustering for SAGE data analysis because it has several relevant theoretical advantages. First, the RF dissimilarity approach handles mixed covariate types well, i.e., it can handle ordinal and continuous covariates in an unbiased way: the more related the covariate is to other covariates, the more it will affect the definition of the RF dissimilarity. Second, the clustering results do not change when one or more covariates are monotonically transformed because the dissimilarity only depends on the feature ranks. Third, the RF dissimilarity does not require the user to specify threshold values for dichotomizing tumor expressions. For the detailed description of RF clustering algorithm, consult Breiman (11) and Shi and Horvath (12). Briefly, the RF clustering procedure is carried out as follows. The RF dissimilarity is used to represent each patient as a point in a two-dimensional space with the aid of multidimensional scaling (39, 40). The distances between the points are used in partitioning around medoids clustering. The number of clusters is chosen by visually inspecting multidimensional scaling plots.
We tested whether variables differed across groups using the Fisher’s exact test. All P values were two sided, and P < 0.05 was considered significant. RF clustering and the analyses described above were carried out with the freely available software R (41).
To identify differentially expressed transcripts between clusters, we used a modified t test. This test is based on a beta binomial sampling model that takes into account both the intra-library and the inter-library variability, thus identifying common patterns of SAGE transcript tag changes systematically occurring across samples (13).6
For automated functional annotation and classification of genes of interest based on GO terms, we used the EASE Web-based software resource (14).7
Meta-analysis of Breast Cancer Microarray Data Sets
To identify and validate the most reliable set of genes able to discriminate primary breast carcinomas based on their lymph node status, we did a cross-platform comparison between the described SAGE data set with previously reported breast cancer studies based on DNA microarray methods (1–3, 5–8, 42–45). The Oncomine cancer microarray database was employed for data collection and to investigate histopathologic associations (46). The Oncomine database is an integrated bioinformatic resource providing data collection, processing, and storage of all publicly available cancer microarray studies. All data are log transformed, median centered per array, and SD normalized to one per array. Gene module application lists all differential expression analyses in which the target genes were included and allows the user to select studies of interest, providing comparative statistical analyses. Selected comparisons of interest for meta-analysis included lymph node (−) versus lymph node (+) status, non-metastasis versus metastasis (5 years of follow-up), non-disease versus relapse (5 years of follow-up). The 55 up-modulated and 55 most down-regulated genes in lymph node (+) primary breast carcinomas were included for meta-analysis comparison. Data processing was carried out using comprehensive meta-analysis software v2 (Biostat, 2006). Standardized mean difference measures as scale-free indices and fixed effects analyses were employed for statistical integration. To enable visualization of meta-analysis results, we used The Institute for Genomic Research MultiExperiment Viewer (MeV 3.0) software. This tool was employed for average clustering of the P values obtained from each gene analyzed. When statistically significant coincidence among studies (i.e., SAGE and microarray studies) was observed on the behavior of specific transcripts, this was represented by colored boxes (red or green). Other progression parameters such as metastasis and disease-free survival (DFS) were also compared with the SAGE lymph node status findings. Statistically significant P values (P < 0.05) associated with gene overexpression in lymph node (+), metastasis, and relapse (DFS) are represented in red; statistically significant down-modulated expression is represented in green color.
Real-time RT-PCR Analysis
Template cDNAs were synthesized on mRNAs isolated from snap-frozen samples from an independent set of 40 stages I to II human breast carcinomas [21 lymph node (−) and 19 lymph node (+) IDCA samples]. Primers and probes were obtained from TaqMan Assays-on-Demand Gene Expression Products (Applied Biosystems). All the PCR reactions were done using the TaqMan PCR Core Reagents kit and the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Experiments were done in triplicate, and each data point and 18S rRNA were used as control. Results were expressed as mean ± 2 SE based on log2 transformation of normalized real-time RT-PCR values of the assayed genes. We used the t test to compare the gene expression levels of validated genes between lymph node (+) and lymph node (−) breast tumors (P < 0.05).
DCTN3 Antibodies Production
Polyclonal antibody against DCTN3 (a kind gift of Dr. Kevin Pfister, Department of Cell Biology, University of Virginia, Charlotesville, VA) was generated according to standard procedures. Briefly, we obtained rabbit serum from animals previously immunized with DCTN3 peptides as antigen. After generation of GST-DCTN3 fusion protein, we did an antibody affinity purification of such serum. The antibodies obtained, which were known to work in Western blots, were optimized for immunohistochemical analysis on paraffin sections (47).
Tissue Microarray and Immunohistochemical Analyses
A breast cancer progression TMA was obtained from the M. D. Anderson Cancer Center (Houston, TX), and we were able to analyze a total of 87 cases representative of normal breast epithelium, ductal carcinoma in situ, invasive breast carcinoma, and metastatic tissues. Before immunostaining, endogenous peroxidase activity was blocked with 3% H2O2 in water for 10 min. Heat-induced epitope retrieval was done with 1.0 mmol/L EDTA buffer (pH 8.0) for 10 min in a microwave oven followed by a 20-min cool down. To block nonspecific antibody binding, the slides were incubated with 10% goat serum in PBS for 30 min. DCTN3 protein was detected using primary anti-DCTN3 polyclonal antibody (1:100 dilution), and horseradish peroxidase–conjugated anti-rabbit secondary antibody. Staining development was done with 3,3′-diaminobenzidine (DAB), and the slides were then counterstained with hematoxylin. DCTN3 protein expression were measured using a Chromavision Automated Cellular Imaging System (ACIS) by means of the generic DAB software application. The software determines brown intensity regardless of the area covered by the positive cells.
Supplementary Material
Acknowledgments
Grant support: NIH-National Cancer Institute grant 1U19 CA84978-1A1 (C.M. Aldaz), center grant ES-07784, and by the University of California at Los Angeles Integrative Graduate Education and Research Training Bioinformatics Program funded by NSF DGE 9987641 (T. Shi).
Footnotes
All raw SAGE data reported as Supplementary Tables in this manuscript is publicly available at http://sciencepark.mdanderson.org/labs/ggeg/SAGE_Proj_11.htm.
Available at the Database for Annotation, Visualization and Integrated Discovery (DAVID) at http://david.niaid.nih.gov/david (15).
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van’t Veer LJ, Dai Hongyue, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 4.Ahr A, Karn T, Solbach C, et al. Identification of high risk breast-cancer patients by gene expression profiling. Lancet. 2002;359:131–132. doi: 10.1016/S0140-6736(02)07337-3. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sotiriou C, Neo S, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;18:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang E, Cheng SH, Dressman H, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Klijn JGM, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 9.Chang HY, Nuyten DSA, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krag D, Weaver D, Ashikaga T, et al. The sentinel node in breast cancer—a multicenter validation study. N Engl J Med. 1998;339:941–946. doi: 10.1056/NEJM199810013391401. [DOI] [PubMed] [Google Scholar]
- 11.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 12.Shi T, Horvath S. Unsupervised learning with random forest predictors. J Comput Graph Stat. 2006;15:118–138. [Google Scholar]
- 13.Baggerly KA, Deng L, Morris JS, Aldaz CM. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics. 2003;19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- 14.Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis G, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:r60. [PubMed] [Google Scholar]
- 16.Detours V, Dumont JE, Bersini H, Maenhaut C. Integration and cross-validation of high-throughput gene expression data: comparing heterogeneous data sets. FEBS Lett. 2003;546:98–102. doi: 10.1016/s0014-5793(03)00522-2. [DOI] [PubMed] [Google Scholar]
- 17.Byrne JA, Mattei MG, Basset P. Definition of the tumor protein D52 (TPD52) gene family through cloning of D52 homologues in human (hD53) and mouse (mD52) Genomics. 1996;35:523–532. doi: 10.1006/geno.1996.0393. [DOI] [PubMed] [Google Scholar]
- 18.Boutros R, Fanayan S, Shehata M, Byrne JA. The tumor protein D52 family: many pieces, many puzzles. Biochem Biophys Res Commun. 2004;325:1115–1121. doi: 10.1016/j.bbrc.2004.10.112. [DOI] [PubMed] [Google Scholar]
- 19.Boutros R, Byrne JA. D53 (TPD52L1) is a cell cycle-regulated protein maximally expressed at the G2-M transition in breast cancer cells. Exp Cell Res. 2005;310:152–165. doi: 10.1016/j.yexcr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein: an abundant and ubiquitous constitutent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 21.Kondoh N, Wakatsuki T, Ryo A, et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–4996. [PubMed] [Google Scholar]
- 22.Kroes RA, Jastrow A, Mclone MG, et al. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156:191–198. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- 23.Dong X, Wang J, Kabir FN, et al. Autoantibodies to DEK oncoprotein in human inflammatory disease. Arthritis Rheum. 2000;43:85–93. doi: 10.1002/1529-0131(200001)43:1<85::AID-ANR11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 24.Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wise-Draper TM, Allen HV, Thobe MN, et al. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- 27.Abate-Shen C. Deregulated Homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 28.Gabellini D, Colaluca IN, Vodermaier HC, et al. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 2003;22:3715–3724. doi: 10.1093/emboj/cdg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller GJ, Miller HL, van Bokhoven A, et al. Aberrant hoxc expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 30.Varnum BC, Ma QF, Chi TH, Fletcher B, Herschman HR. The TIS11 primary response gene is a member of gene family that encodes proteins with a highly conserved sequence containing an unusual cys-his repeat. Mol Cell Biol. 1991;11:1754–1758. doi: 10.1128/mcb.11.3.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karki S, LaMOnte B, Holzbaur ELF. Characterization of p22 subunit of dynactin reveals the localization of cytoplasmic dynein and dynactin to the midbody of dividing cells. Cell Biol. 1998;142:1023–1034. doi: 10.1083/jcb.142.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burkhardt JK, Echeverri CJ, Nisson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle disruption. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bransfield KL, Askham JM, Leek JP, Robinson PA, Miqhell AJ. Phenotypic changes associated with dynactin-2 (DCTN2) over expression characterize SJSA-1 osteosarcoma cells. Mol Carcinog. 2006;45:157–163. doi: 10.1002/mc.20151. [DOI] [PubMed] [Google Scholar]
- 34.Galigniana MD, Harrell JM, O’Hagen HM, Ljungman M, Pratt WB. HSP90- binding immunophilins link p53 to Dynein during p53 transport to the nucleus. J Biol Chem. 2004;279:22483–22489. doi: 10.1074/jbc.M402223200. [DOI] [PubMed] [Google Scholar]
- 35.Pascall JC, Luck JE, Brown KD. Expression in mammalian cell cultures reveals interdependent, but distinct, functions for star and rhomboid proteins in the processing of the Drosophila transforming-growth-factor-a homologue Spitz. Biochem J. 2002;363:347–352. doi: 10.1042/0264-6021:3630347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urban S, Lee JR, Freeman M. A family of rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 2002;21:4277–4286. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charpentier AH, Bednarek AK, Daniel RL, et al. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 2000;60:5977–5983. [PubMed] [Google Scholar]
- 38.Hu Y, Sun H, Drake J, et al. From mice to human: identification of commonly deregulated genes in mammary cancer via comparative SAGE studies. Cancer Res. 2004;64:7748–7755. doi: 10.1158/0008-5472.CAN-04-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venables WN, Ripley BD. Modern applied statistic with S-Plus. New York: Springer; 1999. [Google Scholar]
- 40.Cox TF, Cox MAA. Multidimensional scaling. United Kingdom: CRC Press; 2001. [Google Scholar]
- 41.R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing; ISBN 3 – 900051 – 07 – 0. Available from: http://www.r-project.org/. [Google Scholar]
- 42.Zhao H, Langerod A, Ji Y, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:742–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 44.West M, Blanchette C, Dressman H, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma X, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfister KK, Benashski SE, Dillman JF, Patel-King RS, King SM. Identification and molecular characterization of the p24 dynactin ligh chain. Cell Motil Cytoskeleton. 1998;41:154–167. doi: 10.1002/(SICI)1097-0169(1998)41:2<154::AID-CM6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.