The human genome consists of more than 2 m of linear DNA, which is packaged into a three-dimensional structure in the nucleus of each cell. To ensure proper cell-type–specific gene regulation, each cell must organize its DNA, RNA, and protein within the nucleus in ways that differ in each cell type. It had long been suspected that RNA itself might be a key organizing factor that shapes this dynamic nuclear floor plan (1), with recent research pointing to a role for nuclear-retained long noncoding RNAs (lncRNAs) in organizing nuclear architecture. Here we provide a perspective on the classical and newly emerging role of RNA in this process.
The nucleus is organized into domains generally associated with shared functional and regulatory roles (2, 3) (see the figure): For example, with ribosomal RNA (rRNA) processing and biogenesis (4), mRNA splicing factors (5), and domains of genes that are regulated by specific transcription factors (6) and chromatin regulators (3).
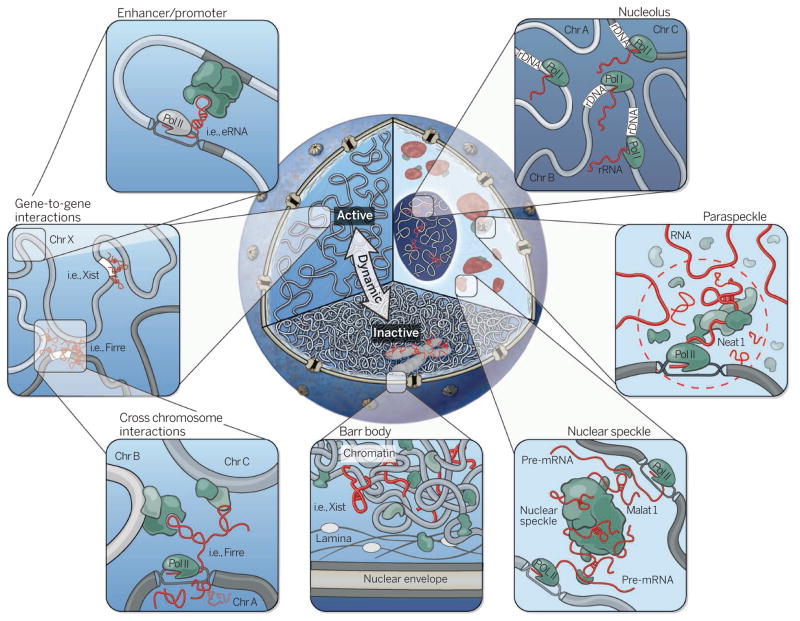
Figure 1. An RNA roadmap to nuclear organization.
Cutaway of the nucleus highlighting three organizational levels: active (left) and inactive (bottom) regions, and nuclear bodies (right). Clockwise from right to left: The nucleolus is formed around actively transcribed rRNA sites; paraspeckles are formed by the Neat1 lncRNA; the Malat1 lncRNA is present within the nuclear speckle, and actively transcribed genes are repositioned close to nuclear speckles; the inactive X chromosome (Barr body) is coated by the Xist lncRNA and dynamically repositioned from the active to inactive compartments where it is localized to the periphery of the nucleus; lncRNAs can mediate gene-gene interactions across chromosomes (bottom panel inset) and within chromosomes (top panel inset).
More than 25 years ago, it was first noted that RNA was associated with the “nuclear matrix” (1). Digesting or stopping the production of RNA, but not protein, resulted in disorganized chromatin regions inside the nucleus (1). Initial insights into the role of this RNA component came from studies of the nucleolus. The nucleolus forms a specific domain where rRNA gene loci, which are spread across multiple chromosomes, coalesce into a spatially organized compartment. Here, the rRNA genes are coordinately transcribed and processed (4). The act of transcribing a rRNA gene is sufficient to reposition the gene locus into the nucleolus (4).
The relationship between transcription and nuclear organization is not restricted to the nucleolus. For example, there are many genomic regions, often on different chromosomes, that when transcribed by a specific transcription factor are brought into spatial proximity within a common “transcription factory” domain (6). In addition, the act of transcription is sufficient to reposition genomic DNA close to the nuclear speckle (5), a nuclear domain enriched with pre-mRNA splicing factors.
Recent studies have shown that several lncRNAs actively assemble nuclear domains. For example, the Neat1 lncRNA is necessary for the assembly and maintenance of the paraspeckle (7), a nuclear domain that is thought to be the site of nuclear retention of adenosine-to-inosine edited mRNAs. Moving the Neat1 transcription locus is sufficient to form new paraspeckles at the integration locus (7). Importantly, Neat1 requires active transcription to “tether” the lncRNA to its own transcription locus in order to carry out this role (7).
Another example is the Xist lncRNA, which is essential for silencing, compaction, and repositioning of the X chromosome to the nuclear periphery of the nucleus (8, 9). This large-scale restructuring is dependent on Xist because expression of Xist in male cells or on autosomes, where it is not normally expressed, is sufficient to cause formation of a repressed nuclear compartment. Xist will only form this repressed nuclear compartment in proximity to its integration site (10). The ability of Xist to reposition active genes into this compartment is dependent on the same RNA domain required for silencing transcription (8, 10).
These examples and others (11) suggest that many lncRNAs may function as nuclear organization factors that can establish nuclear domains. lncRNAs may even serve as a layer of specificity—distinguishing the DNA and RNA that will be contained within one nuclear compartment from those that are contained within a different nuclear compartment. Because lncRNAs very often exhibit cell-type and context-specific expression patterns, this may explain how cell-type–specific nuclear domains are established.
In addition, lncRNAs can also use the existing three-dimensional organization of the nucleus to locate specific DNA target sites. For example, Xist spreads across the entire X chromosome by first localizing at genomic sites that are in three-dimensional nuclear proximity to its own transcription locus (10, 12). Simply moving Xist to a different genomic location leads to its relocalization to new genomic target sites that are also defined by their close spatial proximity to the new Xist integration site (10). Other lncRNAs also use spatial proximity to identify target sites (11). This interplay between the spatial position of a lncRNA’s transcription locus and its localization targets is not restricted to interactions on the same chromosome, but can also occur across chromosomes (e.g., the Firre lncRNA) (11, 13, 14).
Many nuclear-retained lncRNAs may work through a proximity-guided search, either within or across chromosomes. This may be a general search strategy for lncRNAs because they can retain “positional identity” by functioning immediately upon transcription in proximity to its encoded genomic locus. Since genomic locations that are on different chromosomes can be in close spatial proximity within the nucleus, this proximity-guided model would explain the observations of both cis- and trans-mediated regulatory effects of lncRNAs. This model would also explain how lncRNAs, which are generally of lower abundance relative to mRNAs, can reliably identify their target genes by searching in spatial proximity—and at a high effective concentration—near their transcription locus.
Collectively, these studies suggest that lncRNAs may shape nuclear organization by using the spatial proximity of their transcription locus as a means to target preexisting local neighborhoods (10, 14). lncRNAs can in turn modify and reshape the organization of these local neighborhoods to establish new nuclear domains by interacting with various protein complexes, including chromatin regulators (7, 10). Once established, a lncRNA can act to maintain these nuclear domains through active transcription and recruitment of interacting proteins to these domains (7, 13). While the mechanism for how lncRNAs establish these domains is not fully understood, it is becoming increasingly clear that lncRNAs are important at all levels of nuclear organization—exploiting, driving, and maintaining nuclear compartmentalization.
Contributor Information
John Rinn, Email: johnrinn@fas.harvard.edu.
Mitchell Guttman, Email: mguttman@caltech.edu.
References
- 1.Nickerson JA, Krochmalnic G, Wan KM, Penman S. Proc Natl Acad Sci USA. 1989;86:177. doi: 10.1073/pnas.86.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misteli T. Cell. 2007;128:787. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Gibcus JH, Dekker J. Mol Cell. 2013;49:773. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mélèse T, Xue Z. Curr Opin Cell Biol. 1995;7:319. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 5.Spector DL, Lamond AI. Cold Spring Harb Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoenfelder S, et al. Nat Genet. 2010;42:53. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao YS, Sunwoo H, Zhang B, Spector DL. Nat Cell Biol. 2011;13:95. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaumeil J, Le Baccon P, Wutz A, Heard E. Genes Dev. 2006;20:2223. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Annu Rev Genet. 2002;36:233. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 10.Engreitz JM, et al. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinodoz S, Guttman M. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon MD, et al. Nature. 2013;504:465. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacisuleyman E, et al. Nat Struct Mol Biol. 2014;21:198. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maass PG, et al. J Clin Invest. 2012;122:3990. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]