Abstract
We characterized a novel group of HCV variants that are genetically related but distinct from each other belonging to genotype 6 (HCV-6). From 26 infected Austronesian-descended aborigines on Hainan Island, China, HCV sequences were determined followed by genetic analyses. Six nearly full-length genomes and 20 E1 sequences of HCV were obtained, which differ from each other and from all known HCV lineages by nucleotides above the intra-subtype level of 13%. Together with subtypes 6g and 6w, they constitute a phylogenetic group sharing a common ancestor dating from the end of the 12th century.
Conclusion
our data indicate the maintenance of an isolated HCV-6 indigenous circulation on Hainan Island at least for six centuries.
Keywords: HCV genotype 6, Austronesian aborigines, evolution, Hainan Island, China
INTRODUCTION
Hepatitis C virus (HCV) belongs to the Hepacivirus genus of the Flaviviridae family [1]. It is one of the major causes of chronic liver disease, leading to cirrhosis, hepatocellular carcinoma, and liver failure [2]. Similar to many RNA viruses, HCV exhibits high genetic heterogeneity, to the extent that seven genotypes and 87 subtypes have been classified. The greatest diversity is observed within HCV-6 [3], of which 24 subtypes (6a–6xa) have been assigned, and for each at least one full length genome has been characterized, in addition to numerous unclassified variants [4].
During our recent investigation of a cohort of HCV-infected individuals in Baisha County on Hainan Island in China, we identified multiple novel variants of HCV-6 that are genetically related but distinct from each other among Austronesian-descended aborigines (unpublished data). Here we report the characterization of nearly full-length HCV genomes from six of these individuals and partial E1 sequences from 20. Our data indicate the maintenance for more than six centuries of a niche HCV-6 circulation in China, which may shed new light on our current understanding of the origin and evolution of HCV.
MATERIALS AND METHODS
Subjects and samples
All participants were members of the Austronesian-descended aboriginal “Li” minority in Baisha County, Hainan Island, China. Serum samples were obtained from 26 individuals, six of whom (designated HK) were selected for ORF (open reading frame) analysis, and 20 (designated HNZL) were selected for E1 analysis. None of these individuals had travelled outside the island, and they had presented at the county hospital with common symptoms of hepatitis. Written informed consent was obtained from all individuals for this study, which was approved by the ethical review committees of the Southern Medical University, the Hainan General Hospital, the Third Affiliated Hospital of Sun Yat-sen University, the Guangzhou Blood Center in China, and the University of Kansas Medical Center in USA.
Sequence amplification and analyses
HCV sequences were characterized using the methods we previously described [5]. Briefly, RNA was extracted from 100 µl of serum using the QiaAmp viral RNA Mini Kit (QIAGEN, Valencia, CA, USA), and cDNA was transcribed using superscript III reverse transcriptase (Invitrogen, Grand Island, NY, USA) and random hexamers (Promega, Madison, WI, USA). Overlapping fragments of HCV genome were amplified using conventional PCR. The expected amplicons were purified using a QIAquick PCR Purification Kit (QIAgen). Sequencing was performed in both directions using the ABI Prism BigDye 3.0 terminators on an ABI Prism 3500 genetic analyzer (PE Applied Biosystems, Foster City, CA, USA). The resulting chromatograms were visually inspected and the sequences were assembled using SeqMan, from which the encoded amino acid sequence was deduced using EditSeq. Sequence alignments were performed using MegAlign. These software programs are contained in the Lasergene 8.1 package (DNASTAR Inc., Madison, WI). Based on the alignments, maximum likelihood (ML) trees were estimated using PHYML under the GTR+I+G4 substitution model [6]. Pairwise p-distances were calculated using MEGA 5.0 [7]. Potential recombination events were excluded using RDP3 [8] with settings modified as previously described [5]. Finally, the possible saturation of nucleotide substitution were assessed using the DAMBE software [9].
Evolutionary analysis
Based on the determined HCV sequences with addition of the references, multiple sequence alignment was performed, from which three regions (Core, E1, and NS5B) were partitioned and time-scale trees were estimated using the Bayesian Markov Chain Monte Carlo (MCMC) algorithm implemented in the BEAST package (version 1.7.1) [10]. Recent reports on the analysis of HCV sequences in these three regions have indicated that the exponential model is preferable to the lognormal and strict models [11–13]. Therefore, in this study we used the exponential clock model to estimate the trees for sequences in these three regions, all in combination with the GTR+I+⌈ substitution and Bayesian skyline models. However, different molecular rates were used as priors, 1.9 × 10−4 (95% credible interval: 1.0 × 10−4, 2.9 × 10−4), 3.3 × 10−4 (1.6 × 10−4, 5.1 × 10−4), and 7.9 × 10−4 (6.1 × 10−4, 9.9 × 10−4) substitution/site/year for the Core, NS5B, and E1 regions, respectively. These rates were reported by Pybus et al with the former two obtained from the analysis of HCV-6 [11, 14]. Once these parameters were defined, the MCMC procedures were run each for 100 million iterations, during which a tree was logged out every 30,000 iterations. After discarding the first 10% burn-in, the trace was examined for convergence by comparing the statistic of the effective sample size (ESS) using Tracer v1.5 (http://tree.bio.ed.ac.uk). Sufficient sampling was considered to have been achieved when all of the ESS numbers were ≥200. We used TreeAnnotator v1.7.1 (http://tree.bio.ed.ac.uk) to summarize a tree from the resulting set of credible trees, which is called the MCC (Maximum Clade Credibility) tree. Since a molecular clock was incorporated, all the branch lengths and tree node heights were marked in units of years. The tree structure was then deciphered using FigTree v1.4.1 (http://tree.bio.ed.ac.uk).
GenBank accession number
The nucleotide sequences reported in this study were deposited into GenBank with the following accession numbers: KJ470620 – KJ470625.
RESULTS
Characterization of six HK genomes
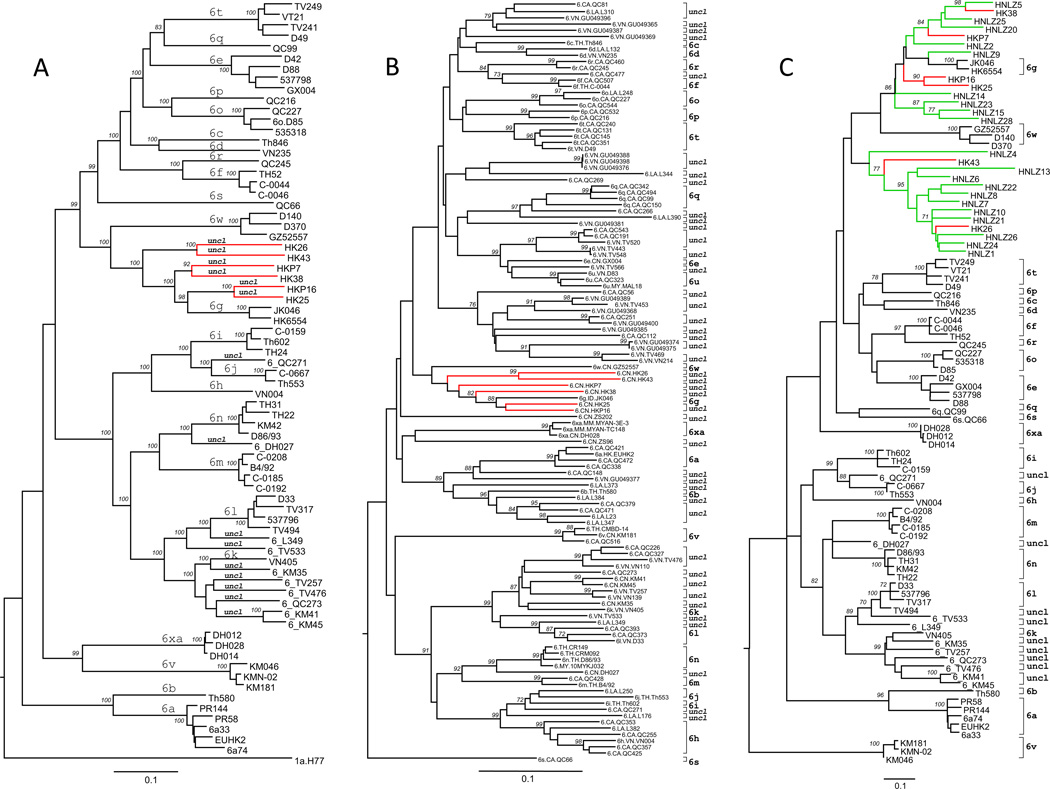
Nearly full-length HCV genomes were characterized from six individuals: HKP7, HKP16, HKP25, HKP26, HKP38, and HKP43. These genomes were between 9430–9475 nucleotides long, starting from the extreme 5'-UTR termini and extending to the stop codon of the ORFs, and corresponding to nucleotides 1–9475 in the reference H77 genome (Table S1). The six genomes were pairwise compared with 66 reference sequences (see Figure 1A) representing all assigned subtypes and 10 unclassified variants of HCV-6. When compared with each other, the six genomes displayed nucleotide differences of 13.1%–23.7% over the entire genome and 13.7%–24.6% over the entire ORF. When compared to the 66 references, the nucleotide differences were 18.9%–28.2% over the entire genome and 20.0%–28.8% over the entire ORF. These differences are all above the lower limit of a division gap recently specified for differentiating isolates of the same subtype from those of different subtypes [4]. Based on the six genomes and 66 reference sequences, an ML tree was reconstructed (Figure 1A). The tree showed that the six genomes could be divided into three pairs with each pair displaying a significant bootstrap support of 92%–100%, and that the six genomes diverged between subtypes 6w and 6g. Analysis of the six genomes in the partial NS5B region, which has been used to characterize many novel variants of HCV-6, failed to identify any additional siblings (Figure 1B). On the basis of these data, we concluded that the six genomes represented unique variants. They would represent six new subtypes if each had three closely-related isolates being identified [3]. To exclude potential recent recombination events, pairwise nucleotide similarity curves were plotted to compare the six genomes against each other and against the 66 references, which failed to identify any such evidence (data not shown).
Figure 1.
ML trees estimated based on (A) nearly full-length genome, (B) partial NS5B sequences corresponding to nucleotides 8316–8620 in the reference H77 genome, and (C) the sequences in E1 region corresponding to nucleotides 674–1085. With the exception of the six HK-genomes (red branches) and 20 HNLZ-variants (green branches) determined in this study, references from subtype 6a–6xa and many unclassified variants of HCV-6 (uncl) are also included. In tree (A) the reference H77 genome (GenBank accession number: NC_004102) is used as an outlier group. In both trees (A) and (C), each branch leads to an isolate ID, in which the unclassified variants are each labelled with a prefix 6_before the ID, while in the tree (B), the isolates are shown in the following format: genotype or subtype.country of isolation.isolate ID or GenBank accession number. To allow the isolates to be readily distinguished, assigned subtypes (6a–6xa) and unclassified lineages (uncl) are indicated above the related branches (A) or on the right side with half brackets (B and C). Bootstrap supports are indicated in italics. The scale bars represent 0.10 nucleotide substitutions per site.
Characterization of 20 HNLZ sequences
We next determined the E1 sequences of HCV from 20 individuals designated HNLZ and co-analysed these with the sequences referred to above (Fig. 1C). As anticipated, all the 20 HNLZ sequences were more closely related to the six HK genomes, and to subtypes 6g and 6w, than to any other HCV-6 members. All variants displayed the nucleotide p-distances from each other and from the reference sequences that are ≥ 13.4%. However, among themselves they showed no bootstrap supports of 100% and did not cluster at the subtype level. Collectively, these data are strongly suggestive of prolonged and independent evolution of these unique HCV variants in the Austronesian-descended population.
Evolutionary analysis
Before the evolutionary analysis of these sequences, potential substitution saturation was excluded, because we estimated the index of substitution saturation (Iss) significantly smaller than the critical Iss values (the values at which the sequences start to fail to recover the correct tree) that reflect the symmetrical (Iss.c sym) or asymmetrical (Iss.c asym) tree topologies [15]. After that, we estimated three time scaled phylogenetic trees for the Core, E1, and NS5B regions (Figure 2), respectively. The resulting trees showed topologies highly similar to that shown in Figure 1, with the exception that all tips were aligned to the right end, and all nodes and branches were measured by grids corresponding to the timescale displayed in reverse at the base of each tree. As shown in both Core and NS5B trees, the six HK genomes were relative to subtypes 6g and 6w and, as a whole, formed a cluster with a common ancestor dating to approximately 819 (95% highest posterior density credible interval, HPD: 383–1583) and 892 (95% HPD: 426–1773) years ago, respectively. Moreover, in the E1 tree the 20 HNLZ sequences clustered with the six HK genomes, and they collectively formed a group that included subtypes 6g and 6w and had a common ancestor dating to approximately 623 (95% HPD: 393–959) years ago (Figure 2).
Figure 2.
Time-scaled phylogenetic trees estimated based on the sequences in core, E1, and NS5B regions, corresponding to the nucleotides numbered 342–915, 891–1302, and 7602–9308 in the H77 genome, respectively. In each tree, the branch lengths represent the evolutionary time as measured by the grids corresponding to a reverse timescale at the tree base, from the present (right) to the past (left) and a time point indicated by a green circle marks the time (along with its 95% HDP) at which the related lineage had diverged. The six HK genomes are indicated by red branches while the 20 HNLZ sequences are denoted by pink branches. For simplicity, only those posterior scores related to the Hainan variants are shown above the internal nodes.
DISCUSSION
A high degree of genetic diversity was observed among the 26 novel HCV-6 variants sampled on Hainan Island in China. This island is in proximity to Vietnam, a major region of origin for HCV-6 [16–18]. This island is also characterized with a variety of the ethnic aborigines residing in its central southern mountainous area who are Austronesian descendants that resemble Malayans [19–20]. We suggest that this specific combination has given rise to and maintained on this island a unique ecosystem of HCV-6 with its variants strikingly distinct from those found elsewhere.
Analysis of the nearly full-length genomes and partial sequences of these unique variants indicate that they are genetically related but distinct from each other. However, because they showed pairwise nucleotide differences from the closely related subtypes 6g and 6w that are above 15%, these variants should be classified outside the two assigned subtypes based on the criteria of the current HCV classification and nomenclature [3]. Furthermore, because they showed pairwise nucleotide differences from each other that are above the intra-subtype level of 13%, these variants each may represent a novel subtype candidate [4]. The criteria defined that the designation of a new HCV subtype requires the detection of three epidemiologically unrelated isolates, and the isolates should be independently grouped into phylogenetic clusters different from the known subtypes by 13%–15% of nucleotides [3–4]. This is the reason why we characterized these Hainan variants as novel HCV-6 lineages but not assigned to any subtypes. Along with the closely related subtypes 6g and 6w, these variants form a larger group that likely indicates an ancient subtype or a geographic lineage of epidemiological significance. To date, four isolates have been reported for each of the subtypes 6g and 6w, corresponding to three 6g isolates in Indonesia [17] and one 6g isolate in Hong Kong [21], one 6w isolate in Guangzhou, China [22], and three 6w isolates in Taiwan [23]. While no conclusive evidence links these 6g and 6w isolates, their genetic similarity to those Hainan variants is suggestive. They may all have their origins in Hainan Island, because many overseas Chinese in Indonesia came from Hainan Island while the interchanges between Hainan, Hong Kong, and Taiwan have greatly increased during the recent decades. However, given the limited mobility of Chinese populations, such a possibility may be small compared to other scenarios. An alternative situation may be that the observed genetic differences are pre-existed, while the currently used evolutionary analysis only provides an underestimation of the time-span during which these variants have evolved and have been distributed geographically.
We have estimated that the common ancestor of these Hainan variants and the 6g and 6w subtypes dates to approximately 623–892 years ago. Although their earliest origin remains unclear, we speculate that it may be related to a certain group of the Austronesian people. The ancestors of these people may have carried the earliest HCV-6 strains when they sailed across the Indian Ocean to the Pacific Ocean and settled in the Indochina peninsula and on the myriad islands of Southeast Asia, where HCV-6 is indigenous [3]. Hainan Island is located in the Tonkin Bay between East Asia and Southeast Asia. During the last Ice Age, when the sea level was much lower than it is today, the island was connected to the continent and lay on a human migratory path between Southeast Asia and East Asia. A variety of aboriginal tribes descended from these migrants now remain isolated in the central southern mountainous area of this island [19, 20, 24]. This isolation has likely facilitated the transmission from a common ancestor of this novel group of genetically related but distinct variants of HCV-6 that have undergone substantial selection through niche circulation.
Given the millennia that these aborigines have resided on this island, it was anticipated that the ancestor of the related HCV-6 variants would be correspondingly aged. However, our analysis provided the lower-than-expected estimates but largely consistent with those reported by Pybus et al. that were mainly based on the HCV-6 variants sampled in Laos [11]. Because no saturation of nucleotide substitution was detected among the sequences analysed in the present study, the estimates may alternatively reflect the GORS (genome-scale ordered RNA structure) constriction on the HCV genomic variations [18] or a situation that more ancestral and diverse variants have not been identified. With the GORS finding, it has been argued that an estimated HCV origin of no earlier than 1,000 years ago is still too recent [18]. Namely, an estimated origin in approximately 2,000–3,000 years ago would be quite compatible with the uncertainty in dates that emerged from the analysis in the present study.
Previous studies have revealed that multiple lineages of HCV-6 are exclusively detected in Southeast Asia or among the expatriates from this region [25]. In contrast, the endemic subtypes of HCV genotypes 1, 2, and 4 have been found mainly in populations in geographically restricted areas in the sub-Saharan Africa or among African descendants in other geographic regions, but showing persistence for fewer centuries than that we here estimated for those novel Hainan variants [26–29]. The best example of such can be seen in a recent report that has shown the existence of multiple HCV-2 “migrant clusters” moved at some point in the past from West Africa to the other parts of the world. These included HCV-2e and 2f to Indonesia; HCV-2i to Morocco, France, Vietnam, and Quebec; HCV-2j to Venezuela; HCV-2k to Martinique and France; HCV-2m to Vietnam; HCV-2r to Haiti and the Dominican Republic; and numerous unclassified diverse HCV-2 lineages to Surinam [30]. Analogous migration patterns have been also indicated in our recent studies based on the isolates of HCV genotypes 1, 3 and 4 [31–32]. Together with the data from other reports, it is suggestive that the colonial activities of European countries played a major role in the dissemination of these HCV genotypes, predominantly to the Americas and to former colonial territories in Asia as well. The introduction was made mainly by the trans-Atlantic slave trading and also by travels between colonies with an estimated median time window dated to approximately 150 years ago [30]. Correspondingly, younger common ancestors were estimated for these individual genotypes of HCV that have occurred in approximately 400–500 years ago [28, 30, 31–32] than that have occurred in 600–800 years ago we here estimated for those Hainan HCV-6 strains. Different historical events should have determined the differences in their divergence time and hence their distribution patterns and the epidemic behaviours.
Supplementary Material
We characterized 26 novel variants of HCV genotype 6 on Hainan Island in China.
They were from Austronesian aborigines living in the central southern mountains.
They differ from each other and from all known HCV members by nucleotides over 13%.
Grouped with subtypes 6g and 6w, they may represent an ancient geographic lineage.
They indicate an isolated HCV ecosystem maintained on the island for many centuries.
Acknowledgments
Financial support. The study was supported by a grant from NIAID/NIH (5 R01 AI080734-03A).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contribution. L. L., G. C., Y. Z., and Y. F. conceived the question and designed the study. L. L. obtained funding for the study. T. W. and M. W. collected the serum samples from the patients. Y. A. completed the experiments. L. L., Y. Z., Y. A., and T. W. participated in collection and interpretation of the data. C. L. conducted the data analyses. All authors participated in preparation of the article and approved the final draft for submission.
Potential conflicts of interest. All authors: No reported conflicts.
REFERENCES
- 1.Thiel HJ, Collett MS, Gould EA, et al. Virus Taxonomy, VIIIth Report of the ICTV. 2005:993–998. [Google Scholar]
- 2.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 3.Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 4.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C Virus into 7 genotypes and 67 Subtypes : updated criteria and assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu L, Li C, Fu Y, et al. Complete genomes for hepatitis C virus subtypes 6f, 6i, 6j and 6m: viral genetic diversity among Thai blood donors and infected spouses. J Gen Virol. 2007;88:1505–1518. doi: 10.1099/vir.0.82604-0. [DOI] [PubMed] [Google Scholar]
- 6.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 7.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia X. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 2013;30:1720–1728. doi: 10.1093/molbev/mst064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, et al. Genetic history of hepatitis C virus in East Asia. J Virol. 2009;83:1071–1082. doi: 10.1128/JVI.01501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markov PV, van de Laar TJ, Thomas XV, Aronson SJ, Weegink CJ, van den Berk GE, Prins M, Pybus OG, Schinkel J. Colonial history and contemporary transmission shape the genetic diversity of hepatitis C virus genotype 2 in Amsterdam. J Virol. 2012;86:7677–7687. doi: 10.1128/JVI.06910-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan M, Lu T, Li C, Lu L. The evolutionary rates of HCV estimated with subtype 1a and 1b sequences over the ORF length and in different genomic regions. PloS one. 2013;8:e64698. doi: 10.1371/journal.pone.0064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pybus OG, Charleston MA, Gupta S, Rambaut A, Holmes EC, Harvey PH. The epidemic behavior of the hepatitis C virus. Science. 2001;292:2323–2325. doi: 10.1126/science.1058321. [DOI] [PubMed] [Google Scholar]
- 15.Xia X, Lemey P. Assessing substitution saturation with DAMBE. In: Lemey P, editor. The Phylogenetic Handbook. Cambridge: Cambridge University Press; 2009. pp. 611–626. [Google Scholar]
- 16.Dunford LCM, Dean J, Waters A, et al. Hepatitis C Virus (HCV) in Vietnam: High Prevalence of Infection in Dialysis and Multi transfused Patients Involving Diverse and Novel Virus Variants. PloS one. 2012;7:e41266. doi: 10.1371/journal.pone.0041266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokita H, Okamoto H, Iizuka H, Kishimoto J, Tsuda F, Miyakawa Y, Mayumi M. The entire nucleotide sequences of three hepatitis C virus isolates in genetic groups 7–9 and comparison with those in the other eight genetic groups. J Gen Virol. 1998;79:1847–1857. doi: 10.1099/0022-1317-79-8-1847. [DOI] [PubMed] [Google Scholar]
- 18.Simmonds P. Genetic diversity and evolution of hepatitis C virus - 15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Li H, Ou C, et al. Paternal genetic structure of Hainan aborigines isolated at the entrance to East Asia. PLoS One. 2008;3:e2168. doi: 10.1371/journal.pone.0002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng MS, He JD, Liu HX, Zhang YP. Tracing the legacy of the early Hainan Islanders--a perspective from mitochondrial DNA. BMC Evol Biol. 2011;11:46. doi: 10.1186/1471-2148-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Fu Y, Lu L, Hagedorn CH. Complete genome sequences for hepatitis C virus subtypes 6e and 6g isolated from Chinese patients with injection drug use and HIV-1 co-infection. J Med Virol. 2006;78:1061–1069. doi: 10.1002/jmv.20663. [DOI] [PubMed] [Google Scholar]
- 22.Lu L, Nakano T, Li C, et al. Hepatitis C virus complete genome sequences identified from China representing subtypes 6k and 6n and a novel yet unassigned subtype within genotype 6. J Gen Virol. 2006;87:629–634. doi: 10.1099/vir.0.81400-0. [DOI] [PubMed] [Google Scholar]
- 23.Lee YM, Lin HJ, Chen YJ, et al. Molecular epidemiology of HCV genotypes among injection drug users in Taiwan: Full-length sequences of two new subtype 6w strains and a recombinant form_2b6w. J Med Virol. 2010;82:57–68. doi: 10.1002/jmv.21658. [DOI] [PubMed] [Google Scholar]
- 24.Gray RD, Jordan FM. Language trees support the express-train sequence of Austronesian expansion. Nature. 2000;405:1052–1055. doi: 10.1038/35016575. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Barnes E, Yuan M, et al. Eight novel hepatitis C virus genomes reveal the changing taxonomic structure of genotype 6. J Gen Virol. 2013;94:76–80. doi: 10.1099/vir.0.047506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasquier C, Njouom R, Ayouba A, et al. Distribution and heterogeneity of hepatitis C genotypes in hepatitis patients in Cameroon. J Med Virol. 2005;77:390–398. doi: 10.1002/jmv.20468. [DOI] [PubMed] [Google Scholar]
- 27.Ndjomou J, Pybus OG, Matz B. Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J Gen Virol. 2003;84:2333–2341. doi: 10.1099/vir.0.19240-0. [DOI] [PubMed] [Google Scholar]
- 28.Njouom R, Frost E, Deslandes S, et al. Predominance of hepatitis C virus genotype 4 infection and rapid transmission between 1935 and 1965 in the Central African Republic. J Gen Virol. 2009;90:2452–2456. doi: 10.1099/vir.0.011981-0. [DOI] [PubMed] [Google Scholar]
- 29.Njouom R, Caron M, Besson G, et al. Phylogeography, risk factors and genetic history of hepatitis C virus in Gabon, central Africa. PLoS One. 2012;7:e42002. doi: 10.1371/journal.pone.0042002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markov PV, van de Laar TJ, Thomas XV, et al. Colonial history and contemporary transmission shape the genetic diversity of hepatitis C virus genotype 2 in Amsterdam. J Virol. 2012;86:7677–7687. doi: 10.1128/JVI.06910-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu L, Li C, Xu Y, Murphy DG. Full-length genomes of 16 HCV genotype 1 isolates representing subtypes 1c, 1d, 1e, 1g, 1h, 1i, 1j, 1k, and two new subtypes 1m and 1n, and four unclassified variants reveals ancestral relationships among subtypes. J Gen Virol. 2014 doi: 10.1099/vir.0.064980-0. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Lu L, Murphy DG, Negro F, Okamoto H. The origin of HCV genotype 3 in Africa as estimated through an evolutionary analysis of the full-length genomes of nine subtypes, including the newly sequenced 3d and 3e. J Gen Virol. 2014 doi: 10.1099/vir.0.065128-0. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.