Abstract
Pituitary adenylate cyclase-activating polypeptide (PACAP) promotes neurite outgrowth and inhibits proliferation of rat pheochromocytoma (PC12) cells. Characterizing the PACAP-differentiated PC12 cell transcriptome should provide genetic insight into how these processes occur in these cells, and in neuronal precursors in vivo. For this purpose, RNA samples were collected from PC12 cells before or after a 6-h treatment with PACAP, from which a labeled cDNA was hybridized to a high-density cDNA array containing 15 365 genes. The genomic response to PACAP involves at least 73 genes. Among the genes differentially expressed in the presence of PACAP, 71% were up regulated, and 29% down regulated, 2-fold or more. Sixty-six percent of the messages affected by PACAP code for functionally categorized proteins, most not previously known to be regulated during PC12 cell differentiation. PACAP has been shown to induce PC12 cell neurite outgrowth through the mitogen-activated protein kinase kinase (MEK) pathway independently of protein kinase A (PKA). Therefore treatments were conducted in the absence or presence of the PKA inhibitor H89, or the MEK inhibitor U0126 in order to identify subsets of genes involved in specific aspects of PC12 cell differentiation. Co-treatment of PC12 cells with PACAP plus H89 revealed a cluster of five genes specifically regulated through the PKA pathway and co-treatment of the cells with PACAP and U0126 revealed a cluster of 13 messages specifically activated through the MEK pathway. Many of the known genes regulated by PACAP have been associated with neuritogenesis (i.e. villin 2 or annexin A2) or cell growth (i.e. growth arrest specific 1 or cyclin B2). Thus, some of the expressed sequence tags (ESTs) that exhibit the same regulation pattern (i.e. AU016391 or AW552690) may also be involved in the neuritogenic and anti-mitogenic effects of PACAP in PC12 cells. Among the 73 PACAP regulated genes, 10 are disqualified on pharmacological grounds as actors in PACAP-mediated neurite outgrowth or growth arrest, leaving 63 new PACAP-regulated genes implicated in neuronal differentiation. Thirteen of these are candidates for mediating ERK-dependent neurite outgrowth, and 47 are possibly involved in the ERK-independent growth arrest induced by PACAP.
Keywords: MAP kinase, microarray, neuritogenesis, proliferation
The PC12 cell line was initially developed to study cell differentiation (Greene and Tischler 1976) and has been extensively used ever since (Cowley et al. 1994; Eggert et al. 2000; Rosario et al. 2001; Vaudry et al. 2002c). Neuronal differentiation is a complex process that includes both morphological and biochemical changes. The most obvious are a decrease in cell proliferation, and the emergence of extending processes. During neuronal differentiation, cells also acquire excitability and start to express some chemical coding genes that provide their functional identity (Greene and McGuire 1978).
Nerve growth factor (NGF) is the classical inducer of neuronal differentiation (Greene and Tischler 1976). The adrenomedullary neurotransmitter pituitary adenylate cyclase-activating polypeptide (PACAP), like NGF, promotes growth arrest and causes neuritogenesis in PC12 cells (Deutsch and Sun 1992). PACAP is a 38-amino-acid neuropeptide that belongs to the vasoactive intestinal polypeptide (VIP)/secretin/glucagon/growth hormone-releasing hormone superfamily (Miyata et al. 1989; Vaudry et al. 2000). The sequence of PACAP has been remarkably well conserved during evolution from protochordates to mammals, suggesting that PACAP is involved in the regulation of important biological functions (Vaudry et al. 2000). Indeed, PACAP promotes survival and induces differentiation of cortical and cerebellar neuroblasts both in vitro and in vivo (Gonzalez et al. 1997; Lu and DiCicco-Bloom 1997; Vaudry et al. 1999; Nicot and DiCicco-Bloom 2001). PACAP also functions as an emergency response co-transmitter in the sympathoadrenal axis (Hamelink et al. 2002b). PACAP was initially isolated on the basis of its ability to stimulate cAMP formation in rat anterior pituitary cells (Miyata et al. 1989). Although cAMP analogues are sufficient to induce neuritogenesis in PC12 cells, the neurotrophic effect of PACAP in this cell line is independent of the PKA pathway (Lazarovici et al. 1998). However, one of the key elements that mediates the effect of PACAP on neurite outgrowth in PC12 cells, has been reported to be the ERK/MAP kinase signalling cascade (Barrie et al. 1997; Lazarovici et al. 1998). The MAP kinase pathway has been clearly shown to be important in the regulation of neuronal cell differentiation (Cowley et al. 1994; Kuo et al. 1997; Fukunaga and Miyamoto 1998; York et al. 1998; Vaudry et al. 2002c), but little is known concerning the target genes acting downstream of ERK to promote neuritogenesis. DNA microarray is an efficient technology to comprehensively study the expression patterns of a large number of genes simultaneously within a cell type during cellular changes like neuronal differentiation. In the present report, we have used a microarray DNA high throughput screening method in combination with selective blockade of signalling pathways by the PKA inhibitor H89 and the MEK inhibitor U0126 to identify a cluster of mRNAs potentially involved in PACAP-induced PC12 cell neuritogenesis, and categorize them with respect to the signalling pathways combinatorially activated by PACAP.
Materials and methods
Drugs
The 38-amino acid form of PACAP was purchased from Phoenix Pharmaceuticals (Mountain View, CA, USA). Cycloheximide was obtained from Sigma (St Louis, MO, USA). H89, and U0126 were obtained from Calbiochem (San Diego, CA, USA).
Cell culture and RNA extraction
PC12-G cells (Rausch et al. 1988) were plated at a density of 70 000 cells/mL (140 cells/mm2) on poly-l-lysine coated 500-cm2 plates and cultured at 37°C in 10% CO2 90% air atmosphere. Cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 7% heat-inactivated foetal bovine serum (Sigma), 7% horse serum (Bio Whittaker, Walkersville, MD, USA), 2.5% Hepes (Invitrogen), 1% glutamine (Invitrogen), 100 units/mL penicillin and 100 μg/mL streptomycin antibiotic (Invitrogen). When used, inhibitors were added 30 min before treatment with control medium or PACAP (10−7 M). After 6 h of treatment, RNA was harvested from the cells with Trizol reagent (Invitrogen) and further purified using the RNeasy Maxi kit (Qiagen, Valencia, CA, USA). The integrity of total RNA was confirmed by denaturing gel electrophoresis, and RNA was finally resuspended in RNase-free water at a concentration of 10 μg/μL.
Glass cDNA microarray and probe preparation
The complementary DNA (cDNA) used in this study were obtained from the NIA Mouse 15K cDNA clone set (Tanaka et al. 2000). PCR products generated from these clones were printed onto polylysine coated glass slides. Fluorescently labelled cDNA was synthesized from 100 μg RNA by oligo(dT)-primed reverse transcription in the presence of Cy3- or Cy5-dUTP (Amersham Pharmacia Biotech, Piscataway, NJ, USA), as previously described (Allander et al. 2001; http://www.nhgri.nih.gov/UACORE/index.html). This mouse array was used because it was the most complex available. The high similarity of the mouse and rat genomes provided reliable results as confirmed by Q-RT-PCR control experiments although some targets may have been overlooked due to lower-than-average rat/mouse homology.
Microarray hybridization and data analysis
After denaturation, purified Cy3/Cy5-labelled probes were combined and hybridized in the presence of 2 × Denhart’s solution, 3.2 × saline sodium citrate (SSC) and 0.5% sodium dodecyl sulfate (SDS) in a humidified chamber at 65°C overnight. Prior to scanning (Agilent Technologies, Foster City, CA, USA), slides were successively washed at 22°C in 0.5 × SSC/0.1% SDS for 2 min, 0.5 × SSC/0.01% SDS for 2 min, and 0.06 × SSC for 2 min. Image analyses were performed with the IPLab software (Fairfax, VA, USA). The two fluorescent images (red and green channels) obtained from the scanner constituted the raw data from which differential gene expression ratio and quality control values were calculated. All data were entered into a relational database, using the FileMaker Pro 5 software (Santa Clara, CA, USA).
Two self-self hybridizations for each treatment group and a dye reversal between control versus treated samples were performed to exclude preferential differences in probe labelling. Genes were identified as differentially regulated only if corrected red/green hybridization signals differed by at least twofold, in five out of seven duplicate experiments. Finally the identity of each gene regulated by PACAP was verified by re-sequencing.
Quantitative RT-PCR
Cells were harvested for total RNA using the RNAeasy Mini Kit (Qiagen). Contaminating DNA was removed by treatment with RNase-free DNase I, and cDNA was synthesized with a SuperScript First Strand Synthesis System for RT-PCR (Invitrogen) by reverse transcription of 0.2–1 μg of total RNA. Real time quantitative- RT-PCR (Q-RT-PCR) was performed on cDNA in the presence of a 1 × Mastermix (Applied Biosystems, Foster City, CA, USA) containing pre-set concentrations of dNTPs, MgCl2, and buffers, along with 90 nm forward and reverse primers and either 100 nm probe or the SYBR green reporter dye, using the Taq-Man 7000 Sequence Detection System (Applied Biosystems). RNA levels were deduced by comparison of cDNA-generated signals in samples with signals generated by a standard curve constructed with known amounts of cDNA, and internally corrected with the GAPDH cDNA signal for variations in amounts of input mRNA. VIP primer set (forward primer, 5′-CACTCATTGGCAAACGAATCA-3′; reverse primer 5′-TTCTTCACAGCCATTTGCTTTC- 3′; probe 6FAM-ATGCAGTCTTCACAGATAACTACACCCGCC- TAMRA); chromogranin A primer set (forward primer, 5′-CGGCAGCATCCAGTTCTCA-3′; reverse primer 5′-AGCCCCTGTCTTTCCATTCA-3′; probe 6FAM-CTGAGACTCCGACTGACCATCATCATCTTTCT- TAMRA); immediate early response 3 primer set (forward primer 5′-GAGGAACCCAACATTGCCAA- 3′; reverse primer 5′-ACCTTCTTCAGCCATCAAAATCTG- 3′); regulator of G-protein signalling 2 primer set (forward primer, 5′-CCGACTTCATCGAGAAGGAA-3′; reverse primer 5′-GCAGCCACTTGTAGCCTCTT-3′); ornithine decarboxylase primer set (forward primer, 5′-CCAGCAGGCTTCTCTGCAA-3′; reverse primer 5′-CACGAAGGTCTCAGGGTCAGTAC-3′; probe 6FAM-ACATGGAAGCTGACACCAATGACATCAATATTTTAMRA); growth arrest specific 1 primer set (forward primer, 5′-AATACAATGTTTAAGGCAGTTTGGAA-3′; reverse primer 5′-AGGTGTGCCCTGTGTAGAAGAAC-3′); protein tyrosine phosphatase 4a1 primer set (forward primer, 5′-TCGTGAA GAACCTGGTTGCTG-3′; reverse primer 5′-TTAATGCTAGGGCAACAAGCAC- 3′); villin 2 primer set (forward primer, 5′-GACGACCGTAACGAGGAGAA- 3′; reverse 5′-CTGGGACAACTCATTGCTCA- 3′); Rodent GAPDH from Applied Biosystems.
Statistical analysis
Data are presented as the mean ± SEM from at least three independent experiments performed in triplicate. Statistical analyses of the data were performed using Student’s t-test.
Results
PACAP causes large-scale changes in PC12 cell gene expression
An incubation of PC12 cells with PACAP for only 1 h is sufficient to elicit full-length neurite outgrowth 48 h later (Vaudry et al. unpublished observations). The anti-mitotic effect of PACAP on cortical precursors has been reported to occur within 4–8 h (Lu and DiCicco-Bloom 1997). Neurite sprouting can be observed in PC12 cells after only 6 h in the presence of PACAP (Figs 1a and b). Taken together, these observations led us to investigate the PC12 cell transcriptome 6 h after treatment with 100 nm PACAP. A typical scatter plot, comparing basal expression levels with those after 6 h of treatment with PACAP is shown in Fig. 2. To assess the reproducibility of probe preparation, replicate experiments were done and used in seven separate hybridizations. Among the 15 365 cDNAs present on the microarray, 73 unique genes were reproducibly altered by twofold or greater (quality control > 0.3) in five experiments out of seven and were unchanged in self-self hybridizations (control vs. control or PACAP vs. PACAP hybridizations). Fifty-two genes (71%) were up-regulated and 21 genes (29%) were down regulated after a 6-h treatment with PACAP. We have identified three genes with an average level of activation by PACAP that exceeded 10-fold and that appeared to be regulated through the ERK pathway (Figs 2 and 4, Table 1). The highest induced message by PACAP (15.9 fold) was immediate early response 3 followed by regulator of G-signalling 2 and ornithine decarboxylase (Table 1).
Fig. 1.
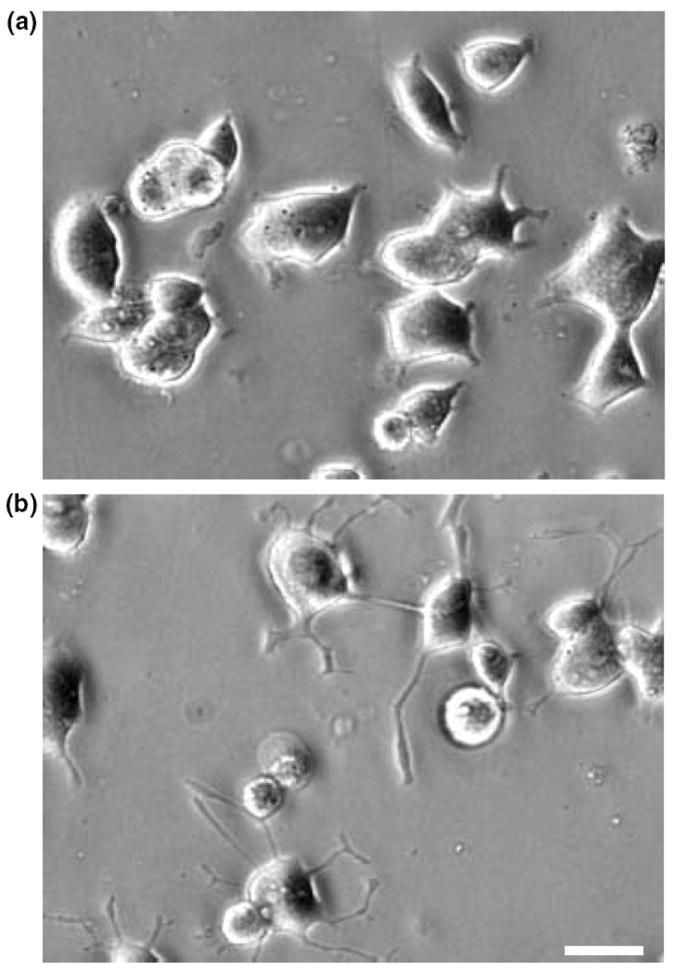
In PC12 cells, PACAP induces rapid morphological changes. Typical photomicrographs illustrating PC12 cells cultured in control conditions (a) or treated with PACAP (10−7 m) for 6 h (b). Scale bar = 15 μm.
Fig. 2.
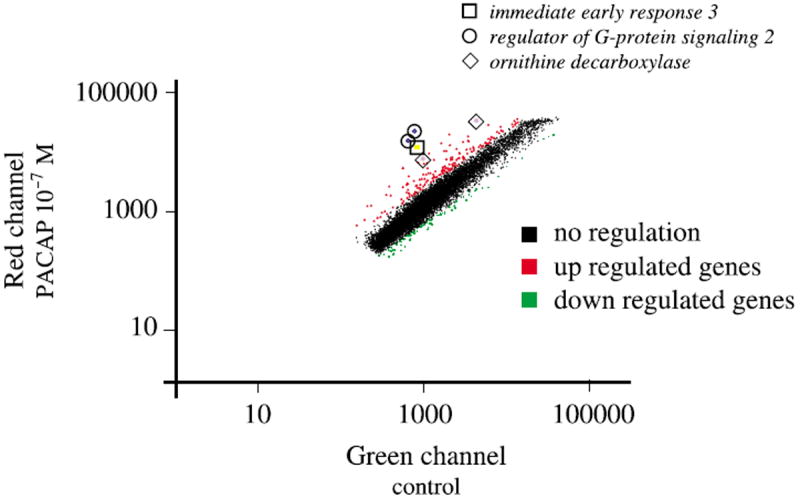
Scatter plots illustrating the effect of PACAP treatment on PC12 cells gene expression profile. The mRNA levels of expression were compared for cells treated in the absence or presence of PACAP (10−7 m) for 6 h (a). Red dots, genes that are up regulated > 2-fold; green dots, genes that are down regulated > 2-fold. Boxes indicate the relative expression of the three top candidates after PACAP treatment; □, immediate early response 3; ○, regulator of G-protein signaling 2; ◇, ornithine decarboxylase. Regulator of G-protein signaling 2 and ornithine decarboxylase are present in duplicate on the array. Only genes with a quality control > 0.3 have been represented on the graph.
Fig. 4.
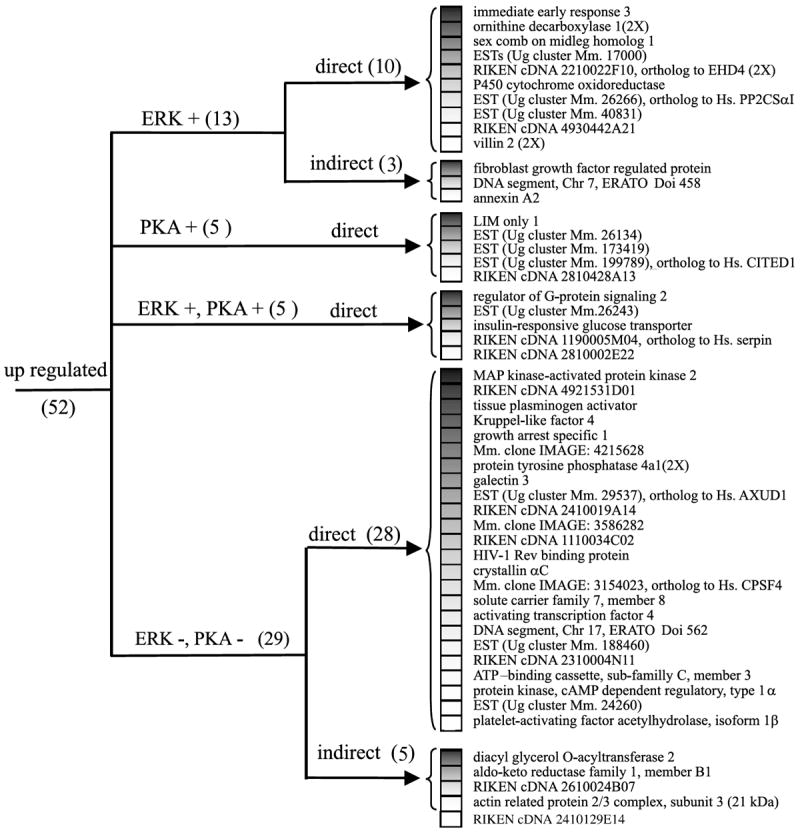
Cluster analysis of the expression pattern of the genes activated after PACAP treatment. Genes were organized according to their regulation expression profile after treatment with H89 (10 μm), U0126 (25 μm) and cycloheximide (5 μm). AXUD1, axin1 up regulated 1; CITED1, CBP/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1; CPSF4, cleavage and polyadenylation specific factor 4, 30 kDa subunit; Hs., homo sapiens; Mm., mouse musculus; PKA, protein kinase A; PP2CSαI, protein phosphatase 2 catalytic subunit α isoform; Ug, unigene.
Table 1.
Functional classification of the genes whose expression is altered after a 6-h exposure to PACAP
| Function | Clone ID | Avg ± SEM | ERK | PKA | Direct | Gene |
|---|---|---|---|---|---|---|
| Neuritogenesis | ||||||
| C86855 | 5.8 ± 0.7 | yes | no | yes | sex comb on midleg homolog 1 | |
| AU020998 | 5.5 ± 1.3 | no | no | yes | tissue plasminogen activator | |
| AU018863 | 5.5 ± 0.9 | no | no | yes | Kruppel-like factor 4 | |
| AU022218 | 4.0 ± 0.4 | no | no | yes | protein tyrosine phosphatase 4a1 (also clone ID AW548730) | |
| AW558398 | 2.9 ± 0.3 | yes | no | yes | villin 2 (also clone ID AW552089) | |
| AW546733 | 2.8 ± 0.3 | no | no | no | actin related protein 2/3 complex, subunit 3 (21 kDa) | |
| AW550463 | 2.8 ± 0.2 | no | no | yes | activating transcription factor 4 | |
| AW551165 | 2.7 ± 0.4 | yes | no | no | annexin A2 | |
| AU020799 | 2.6 ± 0.2 | no | no | – | RIKEN cDNA 2410129E14, ortholog to Hs. tubulin b2 | |
| AU023128 | 2.5 ± 0.1 | no | no | yes | protein kinase, cAMP dependent regulatory, type 1α | |
| AW553335 | 0.5 ± 0.1 | no | no | no | ADP-ribosylation-like 2 | |
| AW559016 | 0.4 ± 0.1 | no | no | no | EST (Ug cluster Mm. 5264), ortholg to Hs. FEZ1 | |
| AW546306 | 0.4 ± 0.1 | no | no | no | high mobility group box 2 | |
| AW551831 | 0.4 ± 0.1 | no | no | no | upregulated by 1,25-dihydroxyvitamin D-3 | |
| Growth arrest | ||||||
| AW549049 | 8.8 ± 1.8 | yes | yes | yes | RIKEN cDNA 1190005M04, ortholog to Hs. Serpin 5 | |
| AW557658 | 6.4 ± 0.6 | no | no | yes | MAP kinase-activated protein kinase 2 | |
| AU015509 | 3.7 ± 0.6 | no | no | yes | EST (Ug cluster Mm. 29537), ortholog to Hs. AXUD1 | |
| Cell growth | ||||||
| C87164 | 15.9 ± 0.9 | yes | no | yes | immediate early response 3 | |
| AU023169 | 14.8 ± 5.1 | yes | yes | yes | regulator of G-protein signaling 2 (also clone ID C85919) | |
| AU020132 | 11.1 ± 1.5 | yes | no | yes | ornithine decarboxylase (also clone ID AW537017) | |
| AU015284 | 4.9 ± 0.9 | no | yes | yes | LIM only 1 | |
| AW554898 | 4.5 ± 0.4 | no | no | yes | growth arrest specific 1 | |
| AW554903 | 3.9 ± 0.5 | yes | no | yes | EST (Ug cluster Mm. 26266), ortholog to Hs. protein phosphatase 2, catalytic subunit, α isoform | |
| AW543680 | 3.8 ± 0.5 | no | no | yes | galectin 3 | |
| AU016354 | 0.5 ± 0.1 | no | no | no | protein tyrosine phosphatase 4a3 | |
| AU045362 | 0.5 ± 0.1 | no | no | no | cyclin B2 | |
| C77364 | 0.5 ± 0.1 | no | no | no | ligase I, DNA, ATP-dependent | |
| AW549381 | 0.4 ± 0.1 | no | no | yes | ral guanine nucleotide dissociation stimulator | |
| AW555913 | 0.4 ± 0.1 | no | no | no | mini chromosome maintenance deficient 6 (S. cerevisiae) | |
| Drug resistance | ||||||
| AU016777 | 3.9 ± 1.2 | yes | no | yes | P450 cytochrome oxidoreductase | |
| C77965 | 3.4 ± 0.3 | yes | no | no | fibroblast growth factor regulated protein | |
| AU018999 | 3.1 ± 0.3 | no | no | yes | crystallin, αC | |
| AU018609 | 2.6 ± 0.2 | no | no | yes | ATP-binding cassette, sub-family C, member 3 | |
| AU022813 | 0.5 ± 0.1 | no | no | no | EST (Ug cluster Mm. 22898), ortholog to Hs. mitogen-activated protein kinase 9 | |
| AU020161 | 0.5 ± 0.1 | no | no | no | X-ray repair complementing defective repair in CHO 1 | |
| Intracellular traffic | ||||||
| AW544709 | 4.4 ± 0.8 | yes | no | yes | RIKEN cDNA 2210022F10 ortholog to Hs. EHD4 (also clone ID AU044505) | |
| AU043959 | 3.1 ± 0.3 | no | no | yes | HIV-1 Rev binding protein | |
| AU020150 | 3.0 ± 0.2 | no | no | yes | Mm. clone IMAGE: 3154023, ortholog to Hs. CPSF4 | |
| AU043420 | 0.5 ± 0.1 | no | no | no | Mm. clone IMAGE: 3491260, ortholog to Hs. hook2 protein | |
| Metabolism | ||||||
| AU020528 | 9.3 ± 1.3 | yes | yes | yes | insulin-responsive glucose transporter | |
| AU019144 | 3.8 ± 0.5 | no | yes | yes | EST (Ug cluster Mm. 199789) ortholog to Hs. CITED1 | |
| AU022525 | 2.7 ± 0.3 | no | no | yes | solute carrier family 7, member 8 | |
| AU043052 | 2.7 ± 0.3 | no | no | no | diacylglycerol O-acyltransferase 2 | |
| AW550812 | 2.6 ± 0.1 | no | no | no | aldo-keto reductase family 1, member B1 | |
| AW557657 | 0.5 ± 0.0 | no | no | no | isocitrate dehydrogenase 1 | |
| AU021456 | 0.4 ± 0.2 | no | yes | no | sorbitol dehydrogenase 1 | |
| AW547245 | 0.3 ± 0.1 | no | no | yes | secreted acidic cysteine rich glycoprotein (also clone ID AW536169) | |
| Unknown | ||||||
| AU020865 | 9.4 ± 1.9 | yes | yes | yes | EST (Ug Mm. 26243) | |
| AW553563 | 5.8 ± 0.6 | no | no | yes | RIKEN cDNA 4921531D01 | |
| AU016391 | 4.9 ± 0.8 | yes | no | yes | EST (Ug cluster Mm. 17000) | |
| AU022455 | 4.4 ± 0.6 | no | no | yes | Mm. clone IMAGE: 4215628 | |
| AW550633 | 4.3 ± 0.6 | yes | yes | yes | RIKEN cDNA 2810002E22 | |
| AU018728 | 4.1 ± 0.7 | no | yes | yes | EST (Ug cluster Mm. 26134) | |
| AU019731 | 3.9 ± 0.5 | no | yes | yes | EST (Ug cluster Mm. 173419) | |
| AW554859 | 3.7 ± 0.2 | yes | no | yes | EST (Ug cluster Mm. 40831) | |
| AW555539 | 3.7 ± 0.2 | no | no | yes | RIKEN cDNA 2410019A14 | |
| C86693 | 3.6 ± 0.6 | yes | no | no | DNA segment, Chr 7, ERATO Doi 458 | |
| AU042737 | 3.4 ± 0.4 | no | no | yes | Mm. clone IMAGE: 3586282 | |
| AU019102 | 3.4 ± 0.4 | yes | no | yes | RIKEN cDNA 4930442A21 | |
| AW551294 | 3.3 ± 0.1 | no | yes | yes | RIKEN cDNA 2810428A13 | |
| AU043374 | 3.3 ± 0.4 | no | no | yes | RIKEN cDNA 1110034C02 | |
| AU022753 | 2.8 ± 0.2 | no | no | yes | DNA segment, Chr 17, ERATO Doi 562 | |
| AW543519 | 2.7 ± 0.1 | no | no | yes | EST (Ug cluster Mm. 188460) | |
| AU015210 | 2.6 ± 0.3 | no | no | yes | RIKEN cDNA 2310004N11 | |
| AU024582 | 2.5 ± 0.1 | no | no | yes | EST (Ug cluster Mm. 24260) | |
| AW536239 | 2.4 ± 0.2 | no | no | no | RIKEN cDNA 2610024B07 | |
| AU021752 | 2.3 ± 0.1 | no | no | yes | platelet-activating factor acetylhydrolase, isoform 1β | |
| AA409629 | 0.5 ± 0.1 | no | no | no | EST (Ug cluster Mm. 200884) | |
| AW552690 | 0.5 ± 0.1 | no | no | yes | EST (Ug cluster Mm. 27628) | |
| AW554776 | 0.5 ± 0.0 | no | no | no | RIKEN cDNA 2900070I05 | |
| AU042856 | 0.5 ± 0.1 | no | no | no | RIKEN cDNA 1110068E11 | |
| AW539579 | 0.4 ± 0.1 | no | no | no | RIKEN cDNA 2700059D21 | |
| C86958 | 0.4 ± 0.1 | no | yes | yes | ESTs (Ug cluster Mm. 24807) | |
Direct, a gene is considered to be directly regulated if its regulation by PACAP does not require de novo protein synthesis. AXUD1, axin1 up regulated 1; CITED1, CBP/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1; CPSF4, cleavage and polyadenylation specific factor 4, 30 kDa subunit; EHD4, eps 15 homology domain containing 4; ERK, extracellular regulated kinase; FEZ1, fasciculation and elongation protein zeta 1; Hs., homo sapiens; Mm., mouse musculus; PKA, protein kinase A; PP2CSαI, protein phosphatase 2 catalytic subunit α isoform; Ug, unigene.
Signal transduction pathways involved in gene regulation
After 48 h of treatment, in control conditions, most PC12 cells exhibited a round shape with very few short processes. Exposure of the same cells to PACAP (10−7 m) for 48 h caused a marked increase in the number and the length of the neurites borne by each cell resulting in more than a 10-fold increase of the total length (Fig. 3a). Incubation of cells with the selective PKA inhibitor H89 (10 μm) did not impair the effect of PACAP on neurite outgrowth (Fig. 3a). In contrast, the ERK inhibitor U0126 (25 μm) totally abolished PACAP induced neurite outgrowth.
Fig. 3.
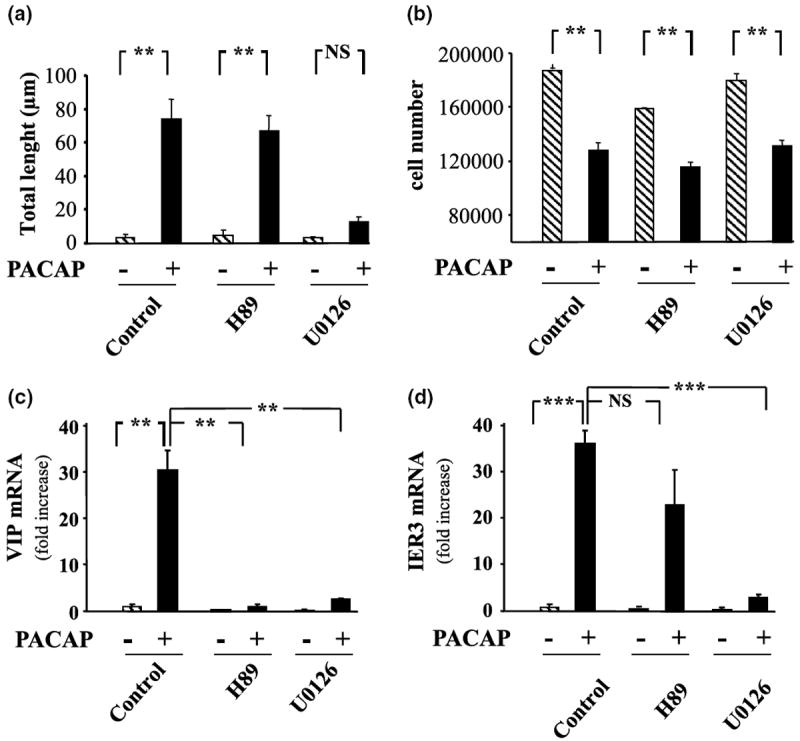
Effect of PACAP on PC12 cell differentiation. (a) Quantification of neurite total length 48 h after treatment with PACAP alone (10−7 m) or in the presence of the selective PKA inhibitor H89 (10 μm) or MEK inhibitor U0126 (25 μm). (b) Quantification of cell number 48 h after treatment with PACAP alone (10−7 m) or in the presence of the selective PKA inhibitor H89 (10 μm) or MEK inhibitor U0126 (25 μm). (c) Quantification of VIP mRNA levels after treatment with PACAP alone (10−7 m) or in the presence of the selective PKA inhibitor H89 (10 μm) or MEK inhibitor U0126 (25 μm) for 6 h. (d) Quantification of immediate early response three mRNA levels after treatment with PACAP alone (10−7 m) or in the presence of the selective PKA inhibitor H89 (10 μm) or MEK inhibitor U0126 (25 μm) for 6 h. Each value represents the mean fold increase (± SEM) compared to the control level of two independent experiments performed in triplicate. Data were corrected using GAPDH signal as internal control. Statistical value significantly different **p < 0.01, ***p < 0.001, from respective control. NS, not significantly different from control.
Treatment of PC12 cells for 48 h with PACAP (10−7 m) resulted in a robust (~ 40%) inhibition of PC12 cell proliferation (Fig. 3b) through a transduction pathway that is independent of the PKA and the ERK pathways.
Q-RT-PCR experiments indicate that the chemical coding neurotransmitter VIP message is regulated through both PKA and ERK dependent pathways (Fig. 3c) whereas immediate early response 3 induction, like neurite outgrowth is regulated through ERK but not PKA (Fig. 3d). Incubation of PC12 cells with PACAP in the absence or presence of the selective inhibitor H89 (10 μm) combined with the microarray technology revealed that PACAP regulated at least 12 additional genes through the PKA pathway (Figs 4 and 5). Incubation of PC12 cells with PACAP in the absence or presence of the selective inhibitor U0126 (25 μm) identified 18 genes regulated by PACAP through the ERK MAP kinase pathway (Figs 4 and 5). Among all the genes affected by PACAP, five genes were combinatorially regulated by the PKA and the ERK pathways while the others were regulated by either one or the other pathway (Figs 4 and 5).
Fig. 5.
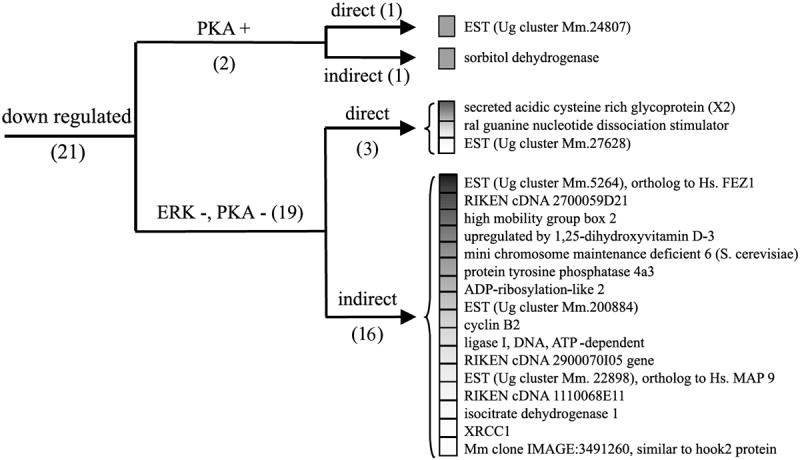
Cluster analysis of the expression pattern of the genes repressed after PACAP treatment. Genes were organized according to their regulation expression profile after treatment with H89 (10 μm), U0126 (25 μm) and cycloheximide (5 μm). ERK, extracellular regulated protein kinase; Hs., homo sapiens; Mm., mouse musculus; PKA, protein kinase A; Ug, unigene; XRCC1, X-ray repair complementing defective repair in CHO 1.
Incubation of PC12 cells with PACAP in the presence of the protein synthesis inhibitor cycloheximide (5 μm) abolished PACAP up-regulation of eight genes out of 52 (Fig. 4, Table 1). In contrast, cycloheximide blocked the ability of PACAP to down-regulate 17 genes out of 21 (Fig. 5, Table 1). A subset of 18 genes including the regulator of G-protein signalling 2 and the villin 2 were superinduced in the presence of cycloheximide while the expression of some other messages such as the immediate early response 3 was not affected.
Functional classification of the genes regulated by PACAP
The functional organization of the messages differentially expressed after treatment with PACAP was carried out using the LocusLink from GenBank and the accession identification numbers provided for each clone. Genes were categorized, based on their reported suggested biological activity, into six broad functional groups, i.e. neuritogenesis, growth arrest, cell growth, proliferation, drug resistance, intracellular traffic and metabolism (Fig. 6, Table 1). Genes like expressed sequence tags (ESTs), clone image and RIKEN cDNA that had no referenced orthologue in other species, or genes with unknown functions, were placed in a seventh group (Fig. 6). Among all the genes identified, only 66% had an inferred biological function. The classification established in Fig. 6 indicates that a majority of the genes regulated by PACAP in PC12 cells with a predicted function code for proteins known or suggested to control cell differentiation (30%) or cell growth (31%).
Fig. 6.
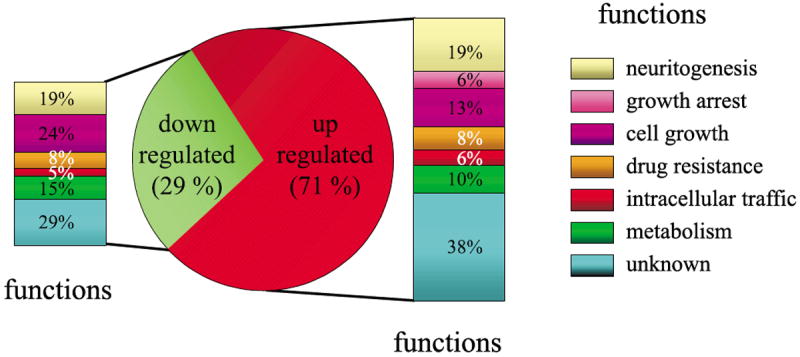
Genes were assigned to a functional class based on their established or suggested function. The pie chart shows the repartition of messengers up or down regulated after 6 h of treatment with PACAP and the histograms indicate the functional repartition these genes.
Target verification by Q-RT-PCR analysis
To verify the alterations in mRNA levels, that were detected by microarray experiments with an independent method, we chose six genes with varying expression profiles for Q-RT-PCR analysis. Figure 7 shows the representative amplicons (Fig. 7a) and quantitative PCR curves (Fig. 7b) for immediate early response 3, regulator of G-protein signalling 2, ornithine decarboxylase, growth arrest specific 1, protein tyrosine phosphatase 4a1 and villin 2. The GAPDH RNA was used as an internal standard. A correlation profile between the microarray data and the Q-RT-PCR results (Fig. 7c) and the summary results (Table 2) indicate that, for the six genes tested, the overall changes in mRNA levels obtained using the Q-RT-PCR technique were consistent with the microarray results. Comparison of the expression level for these six genes estimated by each method suggests that at high levels of expression, microarray experiments may slightly underestimate the fold stimulation induced by PACAP (Fig. 7C and Table 2).
Fig. 7.
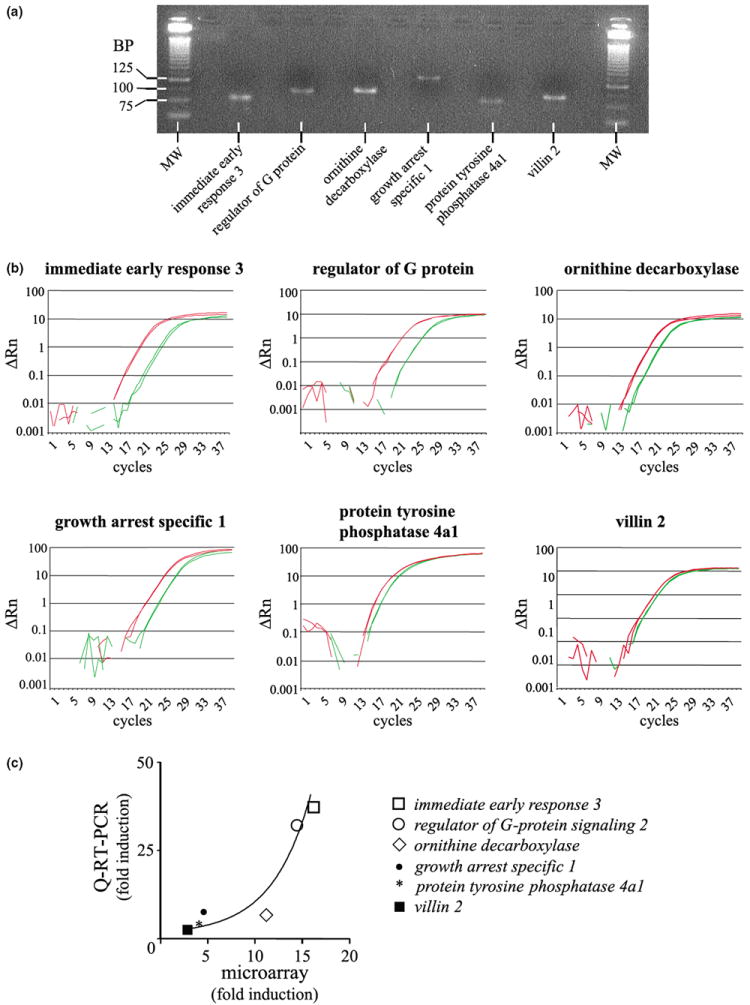
Independent verification of representative genes regulated by PACAP in PC12 cells. (a) Representative amplicons after migration on a 3% agarose gel in TBE. MW, molecular weight; BP, base pair. (b) Quantification of immediate early response 3, regulator of G-protein signaling 2, ornithine decarboxylase, growth arrest specific 1, protein tyrosine phosphatase 4a1 and villin 2 expression level by Q-RT-PCR after 6 h in the absence or presence of PACAP (10−7 m). For each gene of interest, Q-RT-PCR plots of control-treated samples are in green while Q-RT-PCR plots from PACAP-treated samples are in red. (c) Correlation profile of the level of expression observed by microarray and Q-RT-PCR.
Table 2.
Summary results comparing the amplitude of the changes obtained after treatment with PACAP by either Q-RT-PCR or microarray analysis
| Genes | Microarray results Average ± SEM | Q-RT-PCR results Average ± SEM | Microarray versus Q-RT-PCR |
|---|---|---|---|
| Immediate early response 3 | 15.9 ± 0.9 | 38.6 ± 3.4 | *** |
| Regulator of G-protein signaling 2 | 14.8 ± 5.1 | 32.3 ± 1.8 | ** |
| Ornithine decarboxylase | 11.1 ± 1.5 | 7.9 ± 0.4 | NS |
| Growth arrest specific 1 | 4.5 ± 0.4 | 8.7 ± 0.4 | ** |
| Protein tyrosine phosphatase 4a1 | 4.0 ± 0.4 | 4.6 ± 0.1 | NS |
| Villin 2 | 2.9 ± 0.3 | 2.8 ± 0.1 | NS |
p < 0.001;
p < 0.01, significantly higher in Q-RT-PCR determination versus microarray measurements.
NS, not significantly different in Q-RT-PCR determination versus microarray measurements.
Discussion
PACAP has been reported to act on PC12 cells through several pathways singly and in combination. For instance, PACAP induces neuropeptide Y gene expression in a PKA dependent manner (Colbert et al. 1994) and increases the level of PACAP mRNA itself through the MEK pathway (Hashimoto et al. 2000). The neurogenic effect of PACAP involves a MEK dependent/PKA independent pathway (Barrie et al. 1997; Lazarovici et al. 1998). As we show here, regulation of the VIP gene is under control of both the PKA and the ERK pathways, unlike in bovine chromaffin cells where VIP gene regulation by PACAP is PKA independent (Hamelink et al. 2002a). Neither H89 nor U0126 blocked PACAP-induced growth arrest, suggesting that neither the PKA nor the ERK pathway contributes to this process. It should be noted that separate blockade of the PKA and ERK pathways with H89 and U0126 does not exclude their redundant participation in growth arrest initiated by PACAP. However, complete redundancy of the PKA/ERK pathways in PACAP-initiated signalling in PC12 cells seems unlikely, given the presence of multiple target genes for PACAP that are completely independently regulated by each pathway, and indeed multiple target genes that depend on both. Thus, the present observations suggest that genes regulated through PKA are unlikely to be critically involved in the effect of PACAP on neurite outgrowth or proliferation, whereas genes regulated in a MEK-dependent/PKA-independent manner are potential regulators of the morphological changes induced by PACAP in PC12 cells. It should be noted that H89-sensitive differentiation responses to G-protein coupled receptor ligands, including PACAP, have been reported, indicating that signalling initiated by cAMP elevation and proceeding through PKA activation is probably an important alternative pathway for some cell types (Lelièvre et al. 2002).
The results obtained with the different transduction pathway inhibitors led to the regulation profile clustering presented in Figs 4 and 5 which clearly identified a group of 13 genes regulated only through ERK in our experiments. Four of these, based on their predicted protein structure, represent ESTs with no putative function. Assessment of their role in ERK-dependent differentiation events will require functional characterization by antisense mRNA expression in PC12 cells, as has been demonstrated with Annexin A2 involvement in nerve growth factor (NGF)- induced differentiation (Jacovina et al. 2001). It will be of further interest to compare the promoter region of the 10 genes induced exclusively through ERK independently of de novo protein synthesis (Fig. 4) to search for consensus motifs that would help to understand how PACAP regulation of gene transcription through MAP kinase signalling occurs.
Genes up-regulated only transiently, as part of an immediate reponse of PC12 cells to PACAP, may have been overlooked in our screen. For example, fos gene expression was observed in PACAP treated cells at 6 h only after co-treatment with cycloheximide, consistent with negative regulation of fos gene transcription by the Fos protein itself (Rivera et al. 1990). Super-induction of Fos by cycloheximide in the presence of PACAP suggests that the previously reported up-regulation of c-fos by PACAP in PC12 cells (Monnier and Loeffler 1998) is indeed transient, and may precede protein synthesis-dependent transcriptional activation in PC12 cells by PACAP. This is consistent with transient up-regulation of c-fos during PACAP rescue of cerebellar granule cells from apoptosis induced by serum starvation (Vaudry et al. 1998). Some genes, like chromogranin A, previously reported to be regulated by PACAP in PC12 cells were not detected in the present study (Taupenot et al. 1998). Q-RT-PCR verification revealed that chromogranin A up-regulation after 6 h of treatment with PACAP is modest (1.6-fold) which is similar to the twofold increase previously reported (Taupenot et al. 1998). In fact, full induction of chromogranin A mRNA by PACAP is slow and occurs only after 24–48 h of treatment which suggests that late response genes, like chromogranin A, do not represent candidates involved in the neuritogenesis and growth arrest processes.
In addition to their clustering based on regulation by the multiple signalling pathways stimulated by PACAP, the genes specifically induced by PACAP treatment of PC12 cells may be grouped by the potential functions of the proteins they encode. A striking response of PC12 cells to PACAP is growth arrest. Identifying the genes involved in this process is relevant to understanding both regulation of neuroblast proliferation and the neuroblast-neuron transition. Fifteen genes presumed to control cell growth are induced by PACAP in PC12 cells. PACAP induces an 8.8-fold increase of a RIKEN cDNA orthologue to serpin 5, which is found in mammary epithelial cells and codes for a protein with tumour suppressor activity (Zou et al. 1994; Zhang et al. 2000). Cyclin B2 expression is decreased within 6 h after treatment with PACAP. This regulation is indirect, requiring new protein synthesis. Cyclin B2, is a central regulator of progression through the cell division cycle (Maller 1990). Down-regulation of cyclin B2 mRNA could be required for PACAP’s anti-proliferative effects. B-type cyclins appear in S-phase and accumulate in G2 and mitosis before disappearing at transition from metaphase to anaphase (Brandeis and Hunt 1996).
In PC12 cells, PACAP may also decrease the fraction of cells crossing the G1/S boundary as recently reported in cortical precursors (Carey et al. 2002) because it causes a twofold decrease of the ral guanine nucleotide dissociation stimulator: This factor catalyzes the exchange of bound GDP for GTP to activate Ral proteins. Ral proteins are small GTPases implicated in ras-mediated oncogenic transformation (Feig et al. 1996) which have now been established to activate the expression of cyclin D1 (Henry et al. 2000), which is involved in G1- to S-phase progression (Kato 1999).
It is noteworthy that PACAP inhibits cyclin B2 expression in a PKA and MEK independent manner, reinforcing the idea that a pathway involving neither PKA nor MAPK controls at least in part the anti-proliferative effect of PACAP in PC12 cells. Several ESTs with unknown functions are regulated by PACAP in a PKA and MEK independent manner (AW543519, AW536239 or AW553563 for example) and could thus represent other key controllers of PC12 cell proliferation. The involvement of a PKA- and MEK-independent pathway in PC12 cell growth is also supported by the regulation pattern of growth arrest specific-1 which has been specifically isolated based on its ability to inhibit NIH3T3 cell proliferation (Schneider et al. 1988).
Among all the genes with inferred function, the most abundant category represents those controlling cell differentiation (30%). None of these are regulated through the PKA pathway, which is in agreement with the fact that PKA is not involved in PC12 cell neuritogenesis and growth arrest induced by PACAP. Although the MEK pathway is required for neurite outgrowth in PC12 cells, only 28% of the genes categorized as involved in cell differentiation are regulated through the ERK/MAP kinase pathway. This suggests that although the ERK/MAP kinase pathway is involved in neurite outgrowth, it does not control all the aspects of PC12 cell differentiation.
The annexin A2 and the tissue plasminogen activator, genes induced by PACAP, are also required for NGF-induced differentiation of PC12 cells (Jacovina et al. 2001). The specific role of additional genes associated with cell differentiation and growth arrest must now be established in PC12 cells by functional analysis. This is especially true as cell proliferation is often intimately coupled to cell differentiation and the function of certain genes can vary from enhancement of proliferation to differentiation depending on the cell type. For instance, recent experiments indicate that growth arrest specfic 1 may act as a growth-inducing gene in the brain (Liu et al. 2001), in contrast to its function as a gene specifically involved in growth arrest (Schneider et al. 1988; Evdokiou and Cowled 1998).
Four genes, including cytochrome P450 and fibroblast growth factor regulated protein known to be associated with resistance to neurotoxic agents are induced by PACAP. Theses genes may mediate the anti-apoptotic effect of PACAP demonstrated after treatment of PC12 cells with ceramide (Hartfield et al. 1998) or exposure of cerebellar granule cells to 4-hydroxynonenal (Ito et al. 1999), ethanol (Vaudry et al. 2002b) and hydrogen peroxide (Vaudry et al. 2002a). It is worthwhile to note that PACAP concomitantly inhibits the expression of an EST orthologue to the human mitogen-activated protein kinase 9 given the fact that some MEK can cause sustained activation of jun kinases which in turn induce apoptosis of PC12 cells (Xia et al. 1995).
In the present study, we have identified 73 genes affected by a 6-h treatment with PACAP. Blockade of the PKA and ERK pathways during PACAP treatment led to the identification of a subset of 13 genes potentially implicated in PACAP-mediated neurite outgrowth and another population of 48 genes more likely to play a role in the control of growth arrest. The functional classification that has been conducted indicates that some of the genes affected by PACAP like annexin A2, cyclin B2, growth arrest specific 1, immediate early response 3 and tissue plasminogen activator, are very likely to be involved in neuritogenesis and/or growth arrest initiated by PACAP. Epistatic analysis of the transduction cascade(s) regulating these genes, and demonstration of their functional roles in PC12 cell differentiation, will ultimately provide a comprehensive view of the mechanisms involved in the differentiation of PC12 cells by PACAP.
Abbreviations used
- cAMP
cyclic AMP
- cDNA
complementary DNA
- Cy
cyanine
- dUTP
2′-deoxyuridine 5′-triphosphate
- EGF
epidermal growth factor
- ERK
extracellular regulated kinase
- EST
expressed sequence tag
- MAP
mitogen-activated protein kinase
- MEK
mitogen-activated protein kinase kinase
- PACAP
pituitary adenylate cyclase-activating polypeptide
- NGF
nerve growth factor
- PC12
pheochromocytoma cells 12
- PKA
protein kinase A
- Q-RT-PCR
quantitative-reverse transcription-polymerase chain reaction
- SDS
sodium dodecyl sulfate
- SSC
saline sodium citrate
- VIP
vasoactive intestinal polypeptide
References
- Allander SV, Nupponen NN, Ringner M, Hostetter G, Maher GW, Goldberger N, Chen Y, Carpten J, Elkahloun AG, Meltzer PS. Gastrointestinal stromal tumors with KIT mutations exhibit a remarkably homogeneous gene expression profile. Cancer Res. 2001;61:8624–8628. [PubMed] [Google Scholar]
- Barrie AP, Clohessy AM, Buensuceso CS, Rogers MV, Allen JM. Pituitary adenylyl cyclase-activating peptide stimulates extracellular signal-regulated kinase 1 or 2 (ERK1/2) activity in a Ras-independent, mitogen-activated protein Kinase/ERK kinase 1 or 2-dependent manner in PC12 cells. J Biol Chem. 1997;272:19666–19671. doi: 10.1074/jbc.272.32.19666. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- Carey RG, Li B, DiCicco-Bloom E. Pituitary adenylate cyclase activating polypeptide anti-mitogenic signaling in cerebral cortical progenitors is regulated by p57Kip2-dependent CDK2 activity. J Neurosci. 2002;22:1583–1591. doi: 10.1523/JNEUROSCI.22-05-01583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert RA, Balbi D, Johnson A, Bailey JA, Allen JM. Vasoactive intestinal peptide stimulates neuropeptide Y gene expression and causes neurite extension in PC12 cells through independent mechanisms. J Neurosci. 1994;14:7141–7147. doi: 10.1523/JNEUROSCI.14-11-07141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley S, Paterson H, Kemp P, Marshall CJ. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Deutsch PJ, Sun Y. The 38-amino acid form of pituitary adenylate cyclase-activating polypeptide stimulates dual signaling cascades in PC12 cells and promotes neurite outgrowth. J Biol Chem. 1992;267:5108–5113. [PubMed] [Google Scholar]
- Eggert A, Ikegaki N, Liu X, Chou TT, Lee VM, Trojanowski JQ, Brodeur GM. Molecular dissection of TrkA signal transduction pathways mediating differentiation in human neuroblastoma cells. Oncogene. 2000;19:2043–2051. doi: 10.1038/sj.onc.1203518. [DOI] [PubMed] [Google Scholar]
- Evdokiou A, Cowled PA. Growth-regulatory activity of the growth arrest-specific gene, GAS1, in NIH3T3 fibroblasts. Exp Cell Res. 1998;240:359–367. doi: 10.1006/excr.1998.4011. [DOI] [PubMed] [Google Scholar]
- Feig LA, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- Fukunaga K, Miyamoto E. Role of MAP kinase in neurons. Mol Neurobiol. 1998;16:79–95. doi: 10.1007/BF02740604. [DOI] [PubMed] [Google Scholar]
- Gonzalez BJ, Basille M, Vaudry D, Fournier A, Vaudry H. Pituitary adenylate cyclase-activating polypeptide promotes cell survival and neurite outgrowth in rat cerebellar neuroblasts. Neuroscience. 1997;78:419–430. doi: 10.1016/s0306-4522(96)00617-3. [DOI] [PubMed] [Google Scholar]
- Greene LA, McGuire JC. Induction of ornithine decarboxylase by nerve growth factor dissociated from effects on survival and neurite outgrowth. Nature. 1978;276:191–194. doi: 10.1038/276191a0. [DOI] [PubMed] [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelink C, Lee HW, Chen Y, Grimaldi M, Eiden LE. Coincident elevation of cAMP and calcium influx by PACAP-27 synergistically regulates vasoactive intestinal polypeptide gene transcription through a novel PKA-independent signaling pathway. J Neurosci. 2002a;22:5310–5320. doi: 10.1523/JNEUROSCI.22-13-05310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelink C, Tjurmina O, Damadzic R, Young WS, Weihe E, Lee HW, Eiden LE. Pituitary adenylate cyclase-activating polypeptide is a sympathoadrenal neurotransmitter involved in catecholamine regulation and glucohomeostasis. Proc Natl Acad Sci USA. 2002b;99:461–466. doi: 10.1073/pnas.012608999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfield PJ, Bilney AJ, Murray AW. Neurotrophic factors prevent ceramide-induced apoptosis downstream of c-Jun N-terminal kinase activation in PC12 cells. J Neurochem. 1998;71:161–169. doi: 10.1046/j.1471-4159.1998.71010161.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Hagihara N, Koga K, Yamamoto K, Shintani N, Tomimoto S, Mori W, Koyama Y, Matsuda T, Baba A. Synergistic induction of pituitary adenylate cyclase-activating polypeptide (PACAP) gene expression by nerve growth factor and PACAP in PC12 cells. J Neurochem. 2000;74:501–507. doi: 10.1046/j.1471-4159.2000.740501.x. [DOI] [PubMed] [Google Scholar]
- Henry DO, Moskalenko SA, Kaur KJ, Fu M, Pestell RG, Camonis JH, White MA. Ral GTPases contribute to regulation of cyclin D1 through activation of NF-kappaB. Mol Cell Biol. 2000;20:8084–8092. doi: 10.1128/mcb.20.21.8084-8092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Arakawa M, Ishige K, Fukuda H. Comparative study of survival signal withdrawal- and 4-hydroxynonenal-induced cell death in cerebellar granule cells. Neurosci Res. 1999;35:321–327. doi: 10.1016/s0168-0102(99)00097-8. [DOI] [PubMed] [Google Scholar]
- Jacovina AT, Zhong F, Khazanova E, Lev E, Deora AB, Hajjar KA. Neuritogenesis and the nerve growth factor-induced differentiation of PC-12 cells requires annexin II-mediated plasmin generation. J Biol Chem. 2001;276:49350–49358. doi: 10.1074/jbc.M106289200. [DOI] [PubMed] [Google Scholar]
- Kato J. Induction of S phase by G1 regulatory factors. Front Biosci. 1999;4:D787–D792. doi: 10.2741/kato. [DOI] [PubMed] [Google Scholar]
- Kuo WL, Chung KC, Rosner MR. Differentiation of central nervous system neuronal cells by fibroblast-derived growth factor requires at least two signaling pathways: roles for Ras and Src. Mol Cell Biol. 1997;17:4633–4643. doi: 10.1128/mcb.17.8.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarovici P, Jiang H, Fink D. The 38-amino-acid form of pituitary adenylate cyclase-activating polypeptide induces neurite outgrowth in PC12 cells that is dependent on protein kinase C and extracellular signal-regulated kinase but not on protein kinase A, nerve growth factor receptor tyrosine kinase, p21 (ras) G protein, and pp60 (c-src) cytoplasmic tyrosine kinase. Mol Pharmacol. 1998;54:547–558. doi: 10.1124/mol.54.3.547. [DOI] [PubMed] [Google Scholar]
- Lelièvre V, Hu Z, Byun JY, Ioffe Y, Waschek JA. Fibroblast growth factor-2 converts PACAP growth action on embryonic hindbrain precursors from stimulation to inhibition. J Neurosci Res. 2002;67:566–573. doi: 10.1002/jnr.10153. [DOI] [PubMed] [Google Scholar]
- Liu Y, May NR, Fan CM. Growth arrest specific gene 1 is a positive growth regulator for the cerebellum. Dev Biol. 2001;236:30–45. doi: 10.1006/dbio.2000.0146. [DOI] [PubMed] [Google Scholar]
- Lu N, DiCicco-Bloom E. Pituitary adenylate cyclase-activating polypeptide is an autocrine inhibitor of mitosis in cultured cortical precursor cells. Proc Natl Acad Sci USA. 1997;94:3357–3362. doi: 10.1073/pnas.94.7.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller JL. Xenopus oocytes and the biochemistry of cell division. Biochemistry. 1990;29:3157–3166. doi: 10.1021/bi00465a001. [DOI] [PubMed] [Google Scholar]
- Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, Jiang L, Culler MD, Coy DH. Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun. 1989;164:567–574. doi: 10.1016/0006-291x(89)91757-9. [DOI] [PubMed] [Google Scholar]
- Monnier D, Loeffler JP. Pituitary adenylate cyclase-activating polypeptide stimulates proenkephalin gene transcription through AP1- and CREB-dependent mechanisms. DNA Cell Biol. 1998;17:151–159. doi: 10.1089/dna.1998.17.151. [DOI] [PubMed] [Google Scholar]
- Nicot A, DiCicco-Bloom E. Regulation of neuroblast mitosis is determined by PACAP receptor isoform expression. Proc Natl Acad Sci USA. 2001;98:4758–4763. doi: 10.1073/pnas.071465398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch DM, Iacangelo AL, Eiden LE. Glucocorticoid- and nerve growth factor-induced changes in chromogranin A expression define two different neuronal phenotypes in PC12 cells. Mol Endocrinol. 1988;2:921–927. doi: 10.1210/mend-2-10-921. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Sheng M, Greenberg ME. The inner core of the serum response element mediates both the rapid induction and subsequent repression of c-fos transcription following serum stimulation. Genes Dev. 1990;4:255–268. doi: 10.1101/gad.4.2.255. [DOI] [PubMed] [Google Scholar]
- Rosario M, Paterson HF, Marshall CJ. Activation of the Ral and phosphatidylinositol 3′ kinase signaling pathways by the ras-related protein TC21. Mol Cell Biol. 2001;21:3750–3762. doi: 10.1128/MCB.21.11.3750-3762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, 3rd, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taupenot L, Mahata SK, Wu H, O’Connor D. Peptidergic activation of transcription and secretion in chromaffin cells: cis and trans signaling determinants of pituitary adenylyl cyclase-activating polypeptide (PACAP) J Clin Inv. 1998;101:863–876. doi: 10.1172/JCI1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry D, Gonzalez BJ, Basille M, Anouar Y, Fournier A, Vaudry H. Pituitary adenylate cyclase-activating polypeptide stimulates both c-fos gene expression and cell survival in rat cerebellar granule neurons through activation of the protein kinase A pathway. Neuroscience. 1998;84:801–812. doi: 10.1016/s0306-4522(97)00545-9. [DOI] [PubMed] [Google Scholar]
- Vaudry D, Gonzalez BJ, Basille M, Fournier A, Vaudry H. Neurotrophic activity of pituitary adenylate cyclase-activating polypeptide on rat cerebellar cortex during development. Proc Natl Acad Sci USA. 1999;96:9415–9420. doi: 10.1073/pnas.96.16.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry D, Gonzalez BJ, Basille M, Yon L, Fournier A, Vaudry H. Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol Rev. 2000;52:269–324. [PubMed] [Google Scholar]
- Vaudry D, Pamantung TF, Basille M, Rousselle C, Fournier A, Vaudry H, Beauvillain JC, Gonzalez BJ. PACAP protects cerebellar granule neurons against oxidative stress-induced apoptosis. Eur J Neurosci. 2002a;15:1451–1460. doi: 10.1046/j.1460-9568.2002.01981.x. [DOI] [PubMed] [Google Scholar]
- Vaudry D, Rousselle C, Basille M, Falluel-Morel A, Pamantung TF, Fontaine M, Fournier A, Vaudry H, Gonzalez BJ. Pituitary adenylate cyclase-activating polypeptide protects rat cerebellar granule neurons against ethanol-induced apoptotic cell death. Proc Natl Acad Sci USA. 2002b;99:6398–6403. doi: 10.1073/pnas.082112699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry D, Stork PJ, Lazarovici P, Eiden LE. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 2002c;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- York RD, Yao H, Dillon T, Ellig CL, Eckert SP, McCleskey EW, Stork PJ. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]


