Abstract
Nuclear factor E2-related factor-1 (Nrf1) is a basic leucine zipper transcription factor that is known to regulate antioxidant and cytoprotective gene expression. It was recently shown that Nrf1 is regulated by SCF-Fbw7 ubiquitin ligase. However our knowledge of upstream signals that targets Nrf1 for degradation by the UPS is not known. We report here that Nrf1 expression is negatively regulated by glycogen synthase kinase 3 (GSK3) in Fbw7-dependent manner. We show that GSK3 interacts with Nrf1 and phosphorylates the Cdc4 phosphodegron domain (CPD) in Nrf1. Mutation of serine residue in the CPD of Nrf1 to alanine (S350A), blocks Nrf1 from phosphorylation by GSK3, and stabilizes Nrf1. Knockdown of Nrf1 and expression of a constitutively active form of GSK3 results in increased apoptosis in neuronal cells in response to ER stress, while expression of the GSK3 phosphorylation resistant S350A–Nrfl attenuates apoptotic cell death. Together these data suggest that GSK3 regulates Nrf1 expression and cell survival function in response to stress activation.
Keywords: Stress response, Nuclear factor E2-related factor-1, CNC-bZIP, Transcription factor, Fbw7
Introduction
Nuclear factor erythroid derived 2 related factor-1 (Nrf1) is a transcription factor that belongs to the Cap ‘n’ Collar-type basic leucine zipper (CNC-bZIP) protein family. Members of CNC-bZIP family include p45NFE2, Nrf2, Nrf3, Bach1, and Bach2 [1–5]. CNC-bZIP transcription factors form heterodimers with small Maf proteins (Maf G, Maf K, and Maf F) and other bZIP proteins such as activating transcription factor-4 for DNA binding [6,7]. Nrf1 has been implicated in various developmental processes, and it has been found to play an important role in maintaining cellular homeostasis. Germ line loss of Nrf1 function leads to embryonic lethality in mice [8]. Conditional loss of Nrf1 in hepatocytes and neurons leads to apoptosis, and the development of steatohepatitis and neurodegeneration, respectively [9,10]. Moreover, a lack of Nrf1 function in hepatocytes leads to liver tumors in mouse, which suggests that Nrf1 might function as a tumor suppressor [11]. Nrf1 has been shown to control expression of genes involved in cellular stress response through cis acting sequences known as the antioxidant response element (ARE) [12,13]. These elements have been identified in the regulatory regions of genes encoding phase-2 detoxification enzymes and a variety of other cytoprotective genes such as NADPH quinone oxidoreductase, metallothioneins, glutamate–cysteine ligase, as well as genes encoding subunits that make up the proteasome [13–17].
Several Nrf1 isoforms have been reported, and two have been characterized. The 120 kDa isoform (referred to as Nrf1 here) is anchored to the endoplasmic reticulum by its N-terminal domain, and a 65 kDa, N-terminal truncated isoform which is located in the nucleus [18–21]. Available evidence indicates that Nrf1 functions as an activator while the 65 kDa isoform of Nrf1 may function as a transdominant repressor to inhibit ARE-gene expression [19]. Although the precise mechanism that causes translocation of Nrf1 from the ER to the nucleus is unknown, it has been suggested that various stress stimuli result in release of Nrf1 from the ER to enter the nucleus for activation of ARE containing genes.
Glycogen synthase kinase 3 (GSK3) is a multifunctional serine/ threonine kinase found in all eukaryotes. The enzyme is a key regulator of numerous signaling pathways and is involved in a wide range of cellular processes, ranging from glycogen metabolism to cell cycle regulation and proliferation. GSK3 is unusual in that it is normally active in cells and is primarily regulated through inhibition of its activity. It was first isolated and purified as an enzyme capable of phosphorylating and inactivating the enzyme glycogen synthase. We now know that, beyond its role in glycogen metabolism, GSK3 acts as a downstream regulatory switch that determines the output of numerous signaling pathways initiated by diverse stimuli. The pathways in which GSK3 acts as a key regulator, when dysregulated, have been implicated in the development of human diseases such as diabetes, Alzheimer's disease, bipolar disorder and cancer [22,23]. There are two mammalian GSK3 isoforms encoded by distinct genes: GSK3α and GSK3β. GSK3α has a mass of 51 kDa, whereas GSK3β is a protein of 47 kDa. The difference in size is due to a glycine-rich extension at the N-terminus of GSK3α. Although highly homologous within their kinase domains (98% identity), the two gene products share only 36% identity in the last 76 C-terminal residues. Homologs of GSK3 exist in all eukaryotes examined to date and display a high degree of homology. GSK3α and GSK3β, although structurally similar, they are not functionally identical [24]. GSK3β is well studied and has been implicated in regulation of wide variety of cellular processes. Post-translational modification of substrates by GSKβ is frequently associated with ubiquitin mediated substrate turnover as it aids in substrate identification by the ubiquitin proteasome system (UPS) [25].
Recently, we have shown that Nrf1 is an unstable protein is targeted to the UPS by SCF (Skp1–Cul1–F-box)–Fbw7 ring E3 ubiquitin ligase complex [26]. Fbw7 recognizes its substrates by a consensus motif termed the Cdc4 phosphodegron (CPD) that is phosphorylated by serine/threonine kinases. These substrates are phosphorylated by GSK3 for recognition and subsequent degradation by the SCF-Fbw7 ubiquitin ligase complex. In this study, we examined the role of GSK3 in the regulation of Nrf1 degradation in cells. We show that GSK3 negatively regulates Nrf1 in an Fbw7-dependent manner, and negatively impacts Nrf1's anti-apoptotic function in neuronal tissues in response to endoplasmic reticulum (ER) stress induction.
Materials and methods
Reagents
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), streptomycin, penicillin and Lipofectamine were purchased from Invitrogen. F12 and EMEM medium were purchased from Lonza. Cycloheximide and Anti-FLAG (M2) were purchased from Sigma (St. Louis, MO). The Myc tag (9E10) mouse monoclonal antibody was purchased from Genetex. GSK3(5676) antibody, HA tag (6E2) monoclonal antibody, horseradish peroxidase-linked anti-rabbit IgG and anti-mouse IgG antibodies were from Cell Signaling (Beverly, MA). Cdc4/Fbw7/hSel 10 antibody (12292) was purchased from Abeam. Nrf1 antibody has been previously described [18]. APC Annexin V (550474) was purchased from BD Pharmagen. γ32P labeled ATP (10 mCi/ml) was purchased from Perkin Elmer. SB216763 and lithium chloride were purchased from Sigma-Aldrich. Escherichia coli expression vector pGEX-4T-l was a gift from Dr. Phang Lang Chen (University of California, Irvine). GSK3 siRNA was purchased from Life Technologies (Grand Island, NY).
Plasmids
PCMVNrf1-myc was generated as previously described. Tag5Amyc-GSK3 WT, Tag5Amyc-GSK3β CA and Tag5Amyc-GSK3β KD was purchased from Addgene. S350A–Nrf1 was generated using Phusion Site Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA) with the following primers CTCTTCGCTCCCGAGGTGGAG and CC TCTTCAGGACTGACGTCGG. pFLAG-Fbw7 was a gift from Guy J. Rosman and A. Dusty Miller (Fred Hutchinson Cancer Center, Seattle, WA). GST-tagged Nrf1-myc, S350A–Nrfl and Nrf1 βCPD was generated by PCR amplification of pCMVNrf1-myc, S350A–Nrf1 and Nrf1β350–354 with CCGGAATTCATGCTTTCTGAAGAAATAT and GAGC GGCCGCTCTTCCTCCGGTCCTTTGGCTT primers in which EcoRI and Not1 restriction enzyme sequence were incorporated in the 5′ and 3′ primers, respectively. The amplified fragment was digested with EcoRI and Not1 and inserted into the EcoRI and Not1 sites of pGEX-4T-1 vector.
Cell culture
HEK293 and SH-SY5Y cells were grown in Dulbecco's modified Eagle's and F12/EMEM (1:1 mixture) medium respectively supplemented with 10% fetal calf serum, 100 µg/ml streptomycin, and 100 units/ml penicillin at 37 °C in a humidified, 5% CO2 atmosphere. The cells were transfected using BioT reagent according to the manufacturer's protocol. Nrf1 knockout MEF cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 µg/ml of each streptomycin, and 100 units/ml penicillin at 37 °C in a humidified, 5% CO2 atmosphere. Cells were transfected using the Neon electroporation system (Life Technologies, Carlsbad, CA, USA), per manufacturer's guidelines.
Immunoblotting
Cells were lysed in cold radioimmune precipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 5 µg/ml aprotinin, 5 µg/ml leupeptin) and cleared by centrifugation for 15 min at 4 °C. Bio-Rad protein assay reagent was used to measure protein concentrations. An equal volume of 2X SDS sample buffer (100 mM Tris, pH 6.8, 25% glycerol, 2% SDS, 0.01% bromphenol blue, 10% 2-mercaptoethanol) was added to cell lysates and boiled for 5 min. The samples were resolved by SDS–PAGE and transferred to nitrocellulose membranes. After blocking with 5% skim milk in TBST (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, and 0.05% Tween 20), the membranes were probed with the indicated antibodies. The antibody-antigen complexes were detected using the ECL system.
Coimmunoprecipitation experiments
Subconfluent HEK293 cells were transfected with expression vectors using BioT, and lysates prepared using radioimmune precipitation assay buffer 48 h after transfection. The lysates were cleared by centrifugation for 15 min at 4 °C followed by overnight incubation with indicated antibodies. The following day, protein-G Sepharose beads were added, and incubated for 1-h in the cold. The beads were collected by centrifugation, and washed extensively with radioimmune precipitation assay buffer. Proteins were eluted in 1 × SDS sample buffer and heating at 95 °C for 5 min. Samples were separated by SDS-PAGE and transferred to nitrocellulose membrane, followed by immunoblotting with indicated primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Detection of peroxidase signal was performed using the enhanced chemiluminescence method.
Protein ubiquitination assays
HEK293 cells were transfected with vectors expressing HA-tagged ubiquitin, pFLAG-Fbw7 and Myc-tagged GSK3. Cell lysates were prepared using RIPA buffer containing 10 mM N-ethylmaleimide and were immunoprecipitated with Nrf1 antibody. Polyubiquitinated species of Nrf1 were visualized by immunoblotting Nrf1 immunoprecipitates with antibody against HA-tag. Immunoprecipitation efficiency was verified by immunoblotting against Nrf1.
Flow Cytometry
Cells were trypsinized and resuspended at 1 × 106 cells/mL in PBS containing 0.5% BSA. To measure apoptosis, 1 × 105 cells in 100 uL suspension were incubated with APC-Annexin V for 15 min at room temperature and washed with PBS. Cells were analyzed on a FACSCalibur (BD Biosciences, San Jose, CA) and data analysis was performed using the FlowJo software (Tree Star Inc. Ashland, OR).
Expression of recombinant GST-tagged Nrf1 and its mutants
The pGEX-4T-1 expression vector was used to express Nrf1 and its mutants as a GST-fusion protein in Escherichia coli cultivated overnight at 37 °C in LB medium. Expression was induced at an optical density A600 = 1 by adding isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 0.5 mM. After 90 min at room temperature, cells were lysed, and GST-fusion proteins were purified using glutathione–Sepharose 4B beads as described by the manufacturer. The protein concentration was measured by Bradford assay.
In-vitro GSK3 kinase assay
In-vitro kinase reactions were performed using 1 µg of Nrf1–GST fusion proteins as substrates in kinase reaction buffer (NEB), 10 µCi of β-p32 labeled ATP and 200 µM ATP and 500 units of GSK3 enzyme (NEB p6040S) at 30 °C for 1 h. Reactions were terminated by addition of SDS sample buffer, and reactions were analyzed by autoradiography following SDS-PAGE.
RT-PCR
MEF cells were harvested 48 h after transfection and total RNA was extracted using UltraSpec RNA (Biotecx). cDNA was synthesized from 10 µg total RNA in 20 µL reactions containing 1 × RT buffer, 1 mM dNTPs, 0.3 µg random hexamer, 40 U of RNase inhibitor, and 250 U of Moloney murine leukemia virus reverse transcriptase. Reverse transcription reactions were incubated at 72 °C for 5 min, 25 °C for 10 min, and followed by 42 °C for 60 min. Quantitative RT-PCR was performed by amplifying cDNA in a Step One Plus PCR machine (Applied Biosystems) using FastStart SyBr Green reagent (Roche) in duplicates in 10 µL reaction volumes. PCR cycling condition consists of 95 °C for 15 min and 45 cycles of 95 °C for 30 s, 60 °C for 30 s, and 68 °C for 45 s. Primer sequences used were described previously [10]. Expression levels were calculated relative to TBP levels as endogenous controls.
Statistical analysis
Data are expressed as means ± SEM, and statistical analysis using Student's t-test was done with Microsoft Excel (Redmond, WA). *indicates P values < 0.05, and considered significant.
Results
GSK3 modulates Nrf1 expression
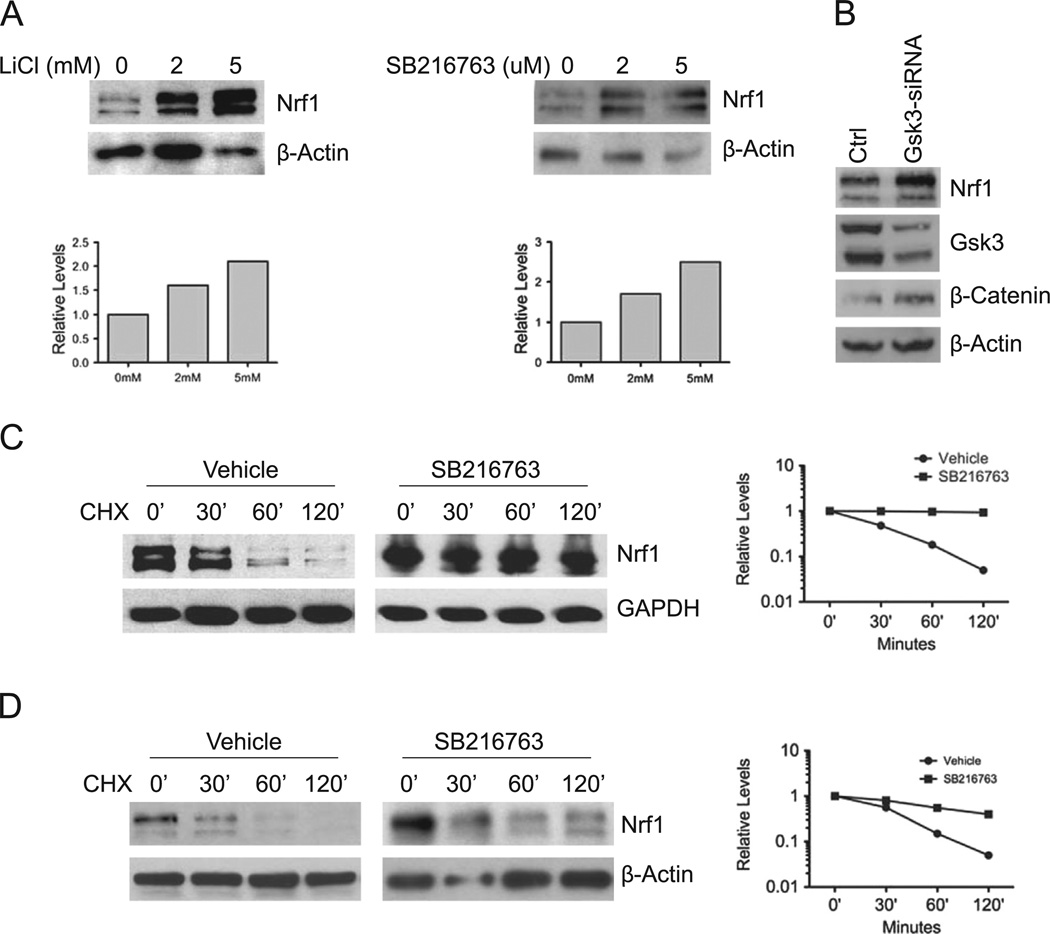
GSK3 has been shown to regulate proteolysis of Fbw7 substrates [25]. Hence, we sought to determine whether GSK3 regulates Nrf1 expression. HEK293 cells were treated with GSK3 inhibitors, SB216763 and lithium chloride, and Nrf1 expression was examined by Western blotting. Both SB216763 and lithium chloride increased Nrf1 levels in a dose dependent manner in HEK293 cells (Fig. 1A). To confirm these results, siRNA mediated knockdown of GSK3 was done. Knockdown of GSK3 resulted in an increase in Nrf1 levels (Fig. 1B). The effectiveness of GSK3 knockdown was confirmed by the increased level of β-catenin (Fig. 1B). Next, we examined the effect of GSK3 inhibition on the stability of Nrf1. HEK293 cells were treated with either vehicle or SB216763, and lysates were collected at the indicated time points after cycloheximide treatment. The half-life of Nrf1 was approximately 30 min in vehicle treated cells, and was increased to greater than 120 min in SB216763 treated cells (Fig. 1C). To determine whether these findings are generalizable to other cells, cycloheximide chase experiment was also carried out in mouse embryonic fibroblast cells (MEF). Similar to results obtained in HEK293 cells, the half-life of Nrf1 was extended in MEF cells treated with SB216763 (Fig. 1D). These results indicate that GSK3 affects Nrf1 stability in cells.
Fig. 1. GSK3 modulates Nrf1 expression.
(A) HEK293 cells were treated with LiC1 or SB216763 at the indicated concentrations for 16 h, and whole cell extracts were immunoblotted with anti-Nrf1 antibody. In accord with previous findings, two major immunoreactive bands representing glycosylated and non-glycosylated forms of Nrf1 were detected [21]. Beta-actin levels were used as loading control. (B) HEK293 cells were transfected with a mixture of the two siRNAs against GSK3. After 48 h, lysates were Western blotted for Nrf1 levels. Western blotting was also done to verify the level of GSK3 knockdown and concomitant increase in β-Catenin protein as control. Beta-actin levels were used as loading control. (C) HEK293 cells were treated with 10 µM SB216763 for 16 h, followed with 50 µg/ml cycloheximide, and cells were harvested at the indicated time points. Whole cell extracts were immunoblotted with anti-Nrf1 antibody. Beta-actin levels were used as loading control. (D) Mouse embryonic fibroblasts were treated with SB216763 for 16 h, followed by 50 µg/ml cycloheximide, and cells were harvested at the indicated time points. Whole cell extracts were prepared and immunoblotted against Nrf1. Beta-actin levels were used as loading control. Graphs show quantitation of protein levels.
GSK3 binds Nrf1
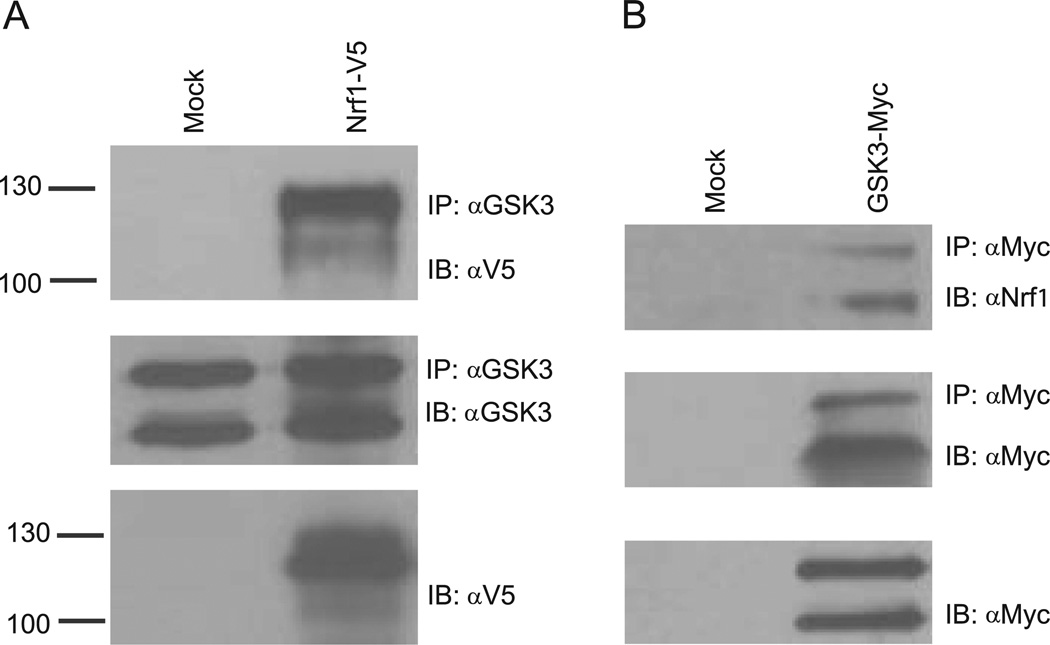
To determine whether Nrf1 interacts with GSK3, co-immunoprecipitation assays were done. HEK293 cells were transiently transfected with V5-tagged Nrf1. Lysates from mock and Nrfl-V5 transfected cells were immunoprecipitated with GSK3 antibody and then probed for Nrfl by V5 antibody. As shown in Fig. 2A, Nrfl-V5 was detected in the anti-GSK3 immune complex. Next, we examined whether endogenous Nrf1 interacts with GSK3. HEK293 cells were transiently transfected with Myc-tagged GSK3β, and lysates were immunoprecipitated with anti-Myc antibody followed by immunoblotting against Nrf1. Western Blotting revealed that endogenous Nrf1 co-immunoprecipitated with Myc-tagged GSK3β (Fig. 2B). Based on these results, we concluded that GSK3 interacts with Nrf1.
Fig. 2. GSK3 binds Nrf1.
(A) HEK293 cells were transfected with vector or Nrfl-V5. After 48 h, whole cell extracts were prepared, and immunoprecipitated with GSK3 antibody followed by immunblotting against V5 and GSK3. (B) HEK293 cells were transfected with vector or GSK3-myc and whole cell extracts were prepared 48 h after and immunoprecipitated with anti-Myc antibody. Immunoprecipitates were then immunoblotted with anti-Myc and anti-Nrf1 antibody.
GSK3 activity is required for Nrf1's turnover by Fbw7
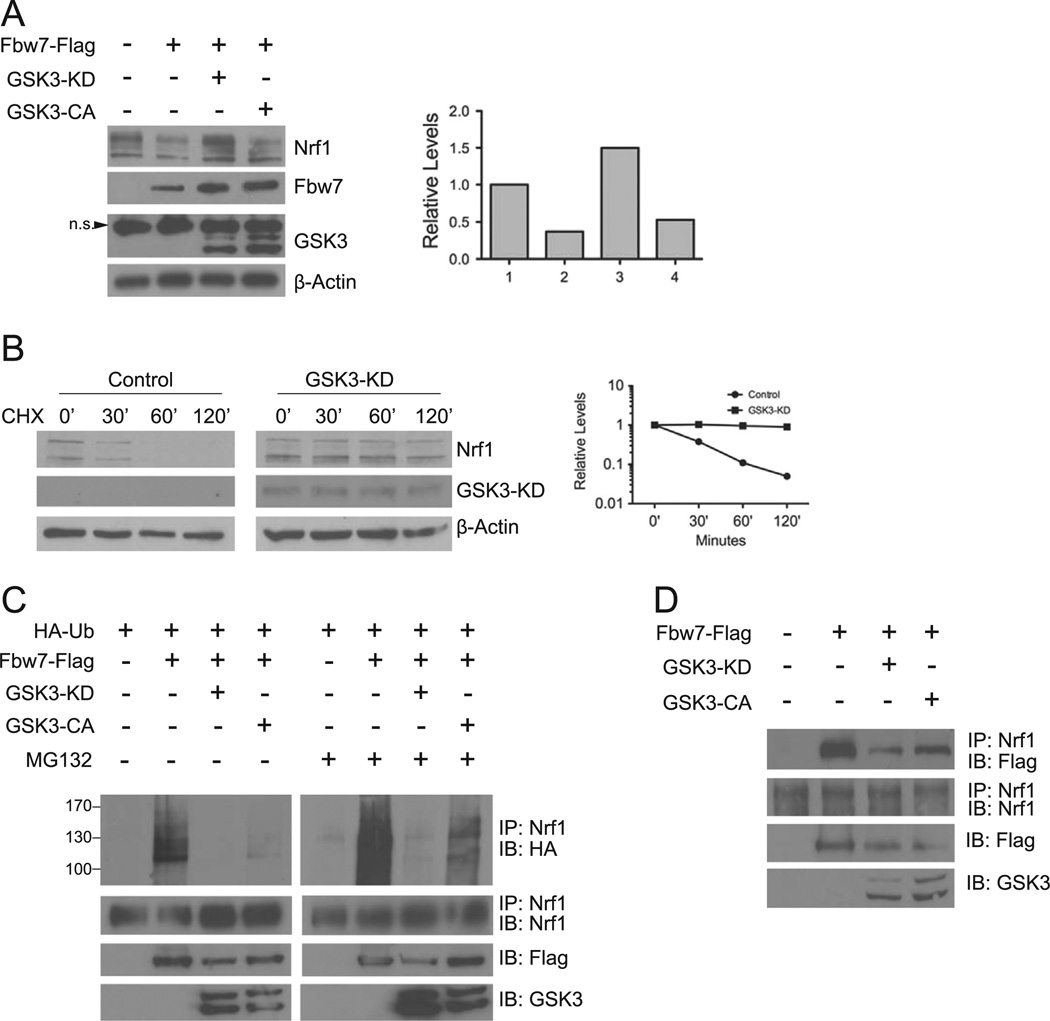
To determine whether Fbw7-mediated degradation of Nrf1 is affected by GSK3 activity, we examined the effects of expression of Fbw7 and either a constitutively active (GSK3β-CA) or inactive (GSK3β-KD) GSK3β on Nrf1. Both Fbw7 and GSK3β-CA reduced Nrf1 in HEK293 cells (Fig. 3A). In contrast, expression of GSK3β- KD attenuated the effects of Fbw7 leading to an increase in Nrf1 levels (Fig. 3A). To further substantiate that GSK3 negatively affects the level of Nrf1, cycloheximide chase experiment was carried out using GSK3β-KD. Fig. 3B shows that the half-life of Nrf1 was extended by expression of GSK3β-KD. These results suggest GSK3 promotes destabilization of Nrf1.
Fig. 3. GSK3 activity is required for Nrf1 turnover by Fbw7.
(A) HEK293 cells were transfected with Fbw7-Flag alone, Fbw7-Flag and GSK3βKD-myc, or Fbw7 and GSK3βCA-myc. Whole cell extracts prepared 48 h after transfection and immunoblotted with Nrf1, Myc, or Flag antibodies. Beta-actin levels were used as loading control. The graph shows quantitation of the Nrf1 expression. Each point represents the mean±SEM of the remaining protein. (B) HEK293 cells were transfected with Nrf1-myc, or Nrf1-myc and GSK3β-KD. After 48 h, cells were treated with 50 µg/ml cycloheximide, and harvested at the indicated time points for Western blot analysis. Beta-actin levels were used to determine loading of each sample. Graph shows quantitation of the protein levels at each time points. Each point represents the mean±SEM of the remaining protein. (C) HEK293 cells were transfected with HA-Ub, HA-Ub and Flag-Fbw7, or HA-Ub, Flag-Fbw7 and GSK3βKD-myc or GSK3βCA-myc. After 48 h, whole cell extracts were immunoprecipitated with anti-Nrf1 antibody followed by immunoblotting with anti-HA antibody. As input control, cell extracts were also immunoblotted with anti-Flag and anti-Myc antibody for expression of Fbw7 and GSK3β, respectively. (D) HEK293 cells were transfected with Flag-Fbw7 alone, Flag-Fbw7 along with GSK3βKD-myc or GSK3βCA-myc. Cell extracts were prepared 48 h after, and immunoprecipitated with Nrf1 antibody. Immunoprecipitates were then Western blotted using anti-Flag and anti-Nrf1 antibodies. As input control, extracts were also probed with anti-Myc antibody for expression of GSK3β.
To determine whether GSIGβ modulates Nrf1 ubiquitination, we carried out in-vivo ubiquitination assay. HA-tagged ubiquitin expression vector was transfected into HEK293 cells along with expression constructs encoding Flag-tagged Fbw7, and GSK3β-KD or GSK3β-CA Lysates were immunoprecipitated with Nrf1 antibody followed by immunoblotting using anti-HA antibody. Transfection of Fbw7 and GSK3β-CA resulted in an increase in ubiquitinated Nrf1, whereas transfection of GSK3β-KD reduced the levels of ubiquitinated Nrf1 (Fig. 3C). Ubiquitin-conjugated forms of Nrf1 were stabilized by proteasome inhibition, as shown by accumulation of ubiquitinated Nrfl in cells treated with MG132 (Fig. 3C). These results demonstrate that GSK3 promotes ubiquitination of Nrf1.
Next, we examined whether GSK3β expression affects the interaction between Nrf1 and Fbw7. HEK293 cells were transfected with Flag-tagged Fbw7 with or without expression vector encoding GSK3β-KD, and lysates were prepared for immunopre-cipitation by anti-Nrf1 and blotted against for Flag-tagged Fbw7. Expression of GSK3β-KD reduced the amount of Fbw7 recovered in the anti-Nrf1 immune complex (Fig. 3D) suggesting that the interaction was weakened by inhibiting GSK3β activity. These results suggest that the GSK3-mediated phosphorylation of Nrf1 promotes its interaction with Fbw7.
GSK3 phosphorylates serine residues within the CPD of Nrf1
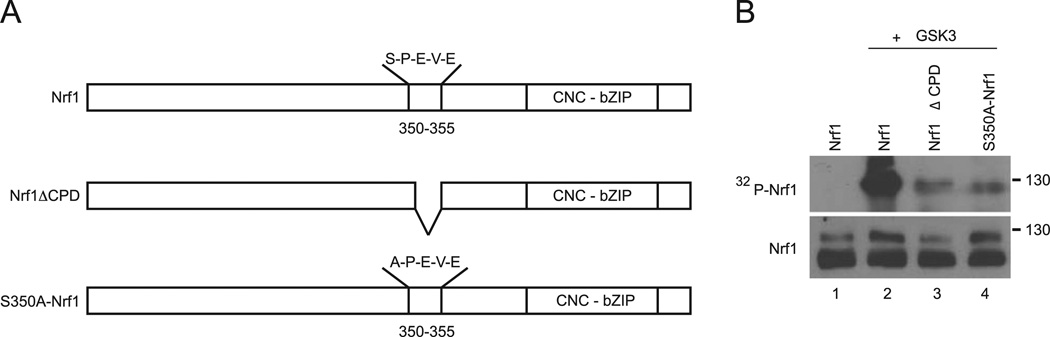
Nrf1 contains a putative CPD motif (S–P–E–V–E) located at protein sequences 350–354 that is highly conserved across species. Previously, we have shown that deletion of the CPD motif in Nrf1 abolished interaction with Fbw7. To demonstrate that GSK3 directly phosphorylates the CPD domain of Nrf1, in-vitro kinase assays were performed on GST fusion proteins. GST tagged wild type Nrf1 (Nrf1), and mutant Nrf1 constructs where the serine residue of the CPD is mutated to alanine (S350A–Nrf1) or the CPD is completely deleted (Nrf1ΔCPD) were generated (Fig. 4A). Fusion proteins were purified from bacteria and they were incubated with recombinant GSK3 in the presence γ32P-ATP. Wild type Nrf1 was efficiently phosphorylated by recombinant GSK3 (Fig. 4B, lane 2). In contrast, phosphorylation of S350A–Nrf1 and Nrf1ACPD was markedly attenuated (Fig. 4B, lanes 3 and 4). These results indicate that the CPD domain of Nrf1 is a GSK3 phosphorylation site in-vitro.
Fig. 4. GSK3 Phosphorylates Nrf1 in-vitro.
(A) Purified GST-Nrf1, GST-S350A–Nrf1, GST-Nrf1CPD were subjected to in-vitro ldnase assay with recombinant GSK3β in the presence of γ-32P ATP. Reaction products were separated on SDS-PAGE and subjected to autoradiography (upper panel). As input control, equal amounts of purified fusion products were immunoblotted against Nrf1 (lower panel).
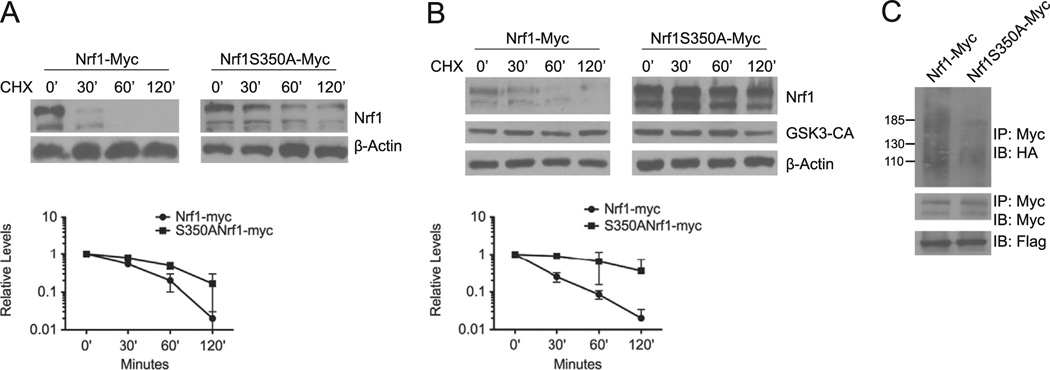
GSK3 mediated degradation of Nrf1 is attenuated by serine 350 to alanine mutation in Nrf1
To determine whether the CPD motif of Nrf1 mediates degradation via GSK3, we investigated the effect of serine 350 to alanine mutation in Nrf1 on its stability. HEK293 cells were transfected with Myc-tagged wild type Nrf1 (Nrf1-myc) or Nrf1 containing the S350A mutation (S350A–Nrf1-myc). Protein lysates were prepared at the indicated time points after cycloheximide treatment for Western blot analysis. Fig. 5A shows the half-life of Nrf1 was increased by the S350A mutation. Consistent with our observations shown in Fig. 3A, wild type Nrf1 was destabilized by GSK3β-CA (Fig. 5B, left panel). However, S350A–Nrf1-myc levels were not significantly altered by co-expression of GSK3β-CA in HEK293 cells (Fig. 5B, right panel). To determine the effect of S350A mutation on Nrf1 ubiquitination, in-vivo ubiquitination assay was done. HEK293 cells were transfected with Nrf1-myc or S350A–Nrf1-myc, along with HA-tagged ubiquitin expression vector. Lysates were immunoprecipitated with anti-Myc antibody, followed by Western blotting against the HA-tagged ubiquitin. As shown in Fig. 5C, the S350A mutation markedly reduced ubiquitination of Nrf1. We conclude from these results that S350 site is important for GSK3-mediated regulation of Nrf1.
Fig. 5. GSK3 mediated degradation is attenuated by S350A substitution in Nrf1.
(A) HEK293 cells were transfected with Nrf1-myc or S350A–Nrf1-myc. Cells were then treated with 50 µg/ml cycloheximide, and harvested at the indicated time points for Western blotting using anti-Myc antibody. Beta-actin levels were used to determine loading of each sample. The graph shows quantitation of protein levels. Each point represents the mean±SEM of remaining protein. (B) HEK293 cells were co-transfected with Nrf1-myc, or S350A–Nrf1-myc and GSK3β-CA After 48 h, cells were treated with 50 µg/ml cycloheximide, and harvested at the indicated time points for Western blot analysis. Beta-actin levels were used to determine loading of each sample. Graph shows quantitation of the protein levels at each time points. Each point represents the mean±SEM of the remaining protein. (C) HEK293 cells were transfected with Nrf1-Myc or S350A–Nrfl-myc along with HA-ubiquitin and Flag-Fbw7. 48 h after transfections, cell extracts were prepared and immunoprecipitated with anti-Myc antibody, followed by immunoblotting with anti-HA antibody. For input control, the filter was stripped and probed with anti-Myc antibody. Cell extracts were also immunoblotted with anti-Flag antibody to determine Fbw7 expression.
GSK3 modulates Nrf1-mediated stress response in neuronal cells
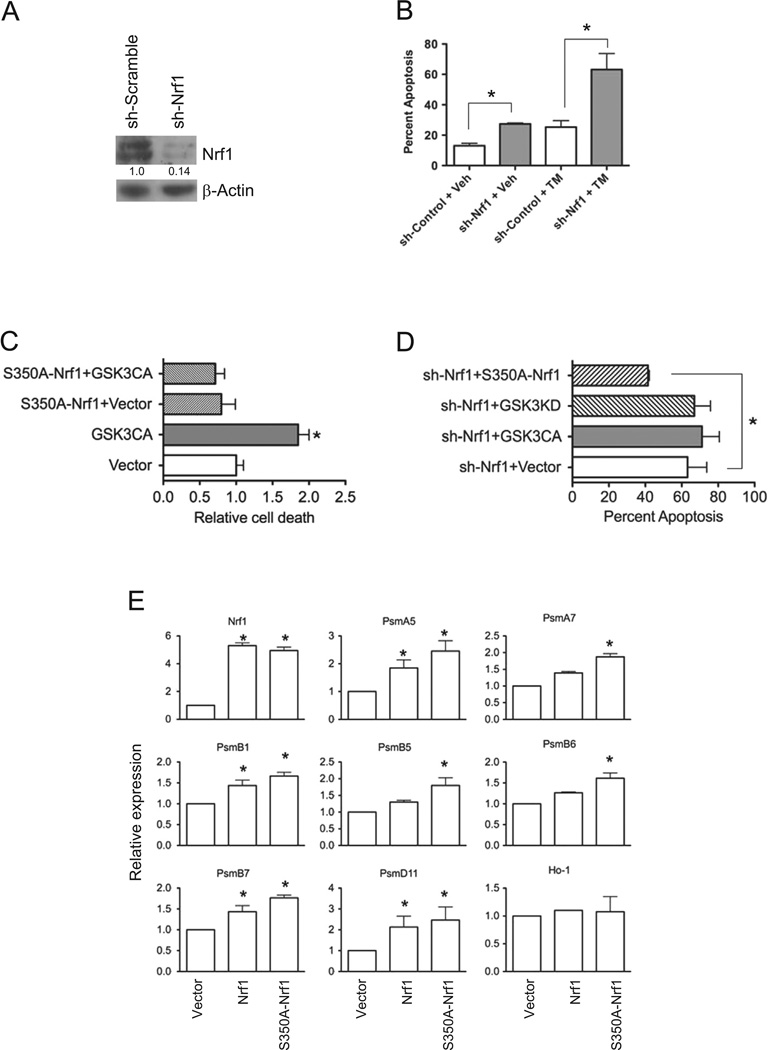
It was recently shown that brain-specific knockout of Nrf1 mice leads to neuronal apoptosis [10]. Hence, we sought to determine whether GSK3 could modulate the neuroprotective effects of Nrf1 by evaluating the effects of ER stress-induced apoptosis in Nrf1 knockdown and scrambled control SH-SY5Y neuronal cells. Western blotting showed that Nrf1 expression was markedly reduced in knockdown cells compared to scramble control cells (Fig. 6A). The knockdown of Nrf1 resulted in a 2-fold increase in apoptosis compared with scrambled control cells (Fig. 6B). To examine the effects of ER stress, cells were treated with tunicamycin. Nrf1 knockdown cells showed a 3-fold increase in apoptosis compared to control cells after treatment (Fig. 6B). These results indicate that a deficiency in Nrf1 sensitizes SH-SY5Y neuronal cells to ER-stress induced cell death.
Fig. 6. GSK3 modulates Nrf1 mediated stress response in neuronal cells.
(A) Western blot analysis of Nrf1 in SH-SY5Y cells transduced with shScramble and shNrf1 virus. Densitometric quantitations of Nrf1 normalized against beta-Actin are shown. (B) shScramble- and shNrf1-transduced SH-SY5Y cells were treated with DMSO, or tunicamycin, and apoptosis was determined by Annexin V staining and flow cytometric analysis. Data represents means±SEM, n=3, *, P<0.05. (C) shScramble control cells co-transfected with EGFP and GSK3β-CA or EGFP, S350A–Nrf1 and GSK3β-CA were treated with tunicamycin, and cell death assessed by flow cytometric analysis of Annexin V staining in GFP-positive gated cells. Data represents means ± SEM, n=3, *, P<0.05. (D) shNrf1-transduced SH-SY5Y cells transfected with E-GFP and GSK3β-CA EGFP and GSK3β-KD, or EGFP and S350A–Nrfl were treated with tunicamycin. After 16 h, cell death assessed by flow cytometric analysis of Annexin V staining in GFP-positive gated cells. Data represents means±SEM, n=3, *, P<0.05. (E) Real time RTPCR analysis of gene expression in Nrf1 knockout MEF cells transfected with vector, Nrf1 or S350A–Nrf1. Expression levels of genes were quantitated relative to endogenous TBP levels as an internal reference, and calculated as 2(ct test gene-Ct TBP). Bars represent mean values±SEM of 3 experiments, and significance was assessed by Student’s t-test (*P<0.05).
To determine the effect of GSK3 on ER stress-induced apoptosis, SH-SY5Y cells were transfected with GSK3β-CA. Expression of the constitutively active GSK3β resulted in a 2-fold increase in apoptosis (Fig. 6C). To examine whether the proapoptotic effects of GSK3β-CA is mediated via down regulation of Nrf1, SH-SY5Y cells were co-transfected with S350A–Nrf1. Expression of S350A–Nrf1 rescues SH-SY5Y cells from the effects of GSK3β-CA on ER-stress induced apoptosis (Fig. 6C). To further demonstrate the apoptotic effect of GSK3 can be modulated through Nrf1, we evaluated ER stress-induced apoptosis in Nrf1 knockdown cells expressing a constitutively active or inactive GSK3. No difference was observed in the extent of apoptosis in Nrf1 knockdown cells expressing either GSK3β-CA, GSK3β-KD. However, expression of S350A–Nrf1 resulted in a reduction in apoptotic cell death by 1.5 fold in Nrf1 knockdown cells (Fig. 6D). To investigate whether the effect of S350A–Nrf1 on apoptosis is associated with increased expression of Nrf1 target genes, Nrf1 knockout MEF cells were transfected with Nrf1, S350A–Nrf1 or empty expression vector, and mRNA extracted for qRT-PCR analysis of endogenous proteasome genes. As shown in Fig. 6E, enforced expression of Nrf1 resulted in up-regulation of various proteasome genes. In comparison to Nrf1 transfected cells, expression of proteasome genes were higher in MEF cells transfected with S350A–Nrf1 (Fig. 6E). However, expression of Ho-1, and Nrf2 target gene, was not altered. Together, these results suggest GSK3 can modulate stress response mediated by Nrf1 in neuronal cells.
Discussion
Previously, we have shown that Fbw7 targets Nrf1 for UPS mediated degradation, but the upstream signal that regulates Nrf1 turnover through the UPS is not yet established. In this study we have elucidated that GSK3 kinase regulates Nrf1 stability. Our results show that (i) GSK3 inhibition stabilizes Nrf1, (ii) Nrf1 binds to GSK3, (iii) GSK3 phosphorylates specific residues in Nrf1 CPD and regulates Nrf1 turnover in Fbw7-dependent manner, (iv) Nrf1 with mutated CPD is resistant to GSK3 mediated degradation, (v) protection from ER stress-induced apoptosis conferred by Nrf1 is inhibited by GSK3.
Unlike other protein kinases, GSK3 is constitutively active and its function is inhibited by phosphorylation of specific serine residues on the enzyme by various signaling pathways [22,23]. GSK3 is a downstream target of several cellular signaling pathways, including ones that regulate cell proliferation and survival such as PI3K/AKT and MAPK [27,28]. GSK3 also sits at the convergence of pathways critical for neuronal survival and function, and its activity correlates inversely with neuronal viability [29]. GSK3 is activated by apoptotic stimuli such as ischemia, stress, and amyloid beta peptide accumulation in the brain, while neuroprotective signaling by PI3K/AKT lead to its inactivation. Recent reports have also demonstrated that proapoptotic stimuli increases the levels of GSK3 protein in the nucleus of SH-SY5Y neuroblastoma cells prior to activation of caspase-9 and caspase-3, suggesting that nuclear accumulation of GSK3 triggers cellular events that leads to apoptosis [30]. Our preceding data show Nrf1 regulating proteasome gene expression in neuronal cells [10]. Brain-specific Nrf1 knockout mice show evidence of age related brain atrophy due to increased neuronal apoptosis. Contrary to our expectations, Nrf1 deficiency in neural cells did not induce oxidative stress in brain tissues of Nrf1BKO mice. Instead, neuronal apoptosis in Nrf1BKO brains has been attributed ER stress stemming from proteasomal dysfunction [10,31,32]. In our study here, we have shown that Nrf1 provides protection from ER stress mediated apoptosis in neuronal cells. This protective function of Nrf1 is lost in the presence of GSK3 overexpression due to negative regulation of Nrf1 by GSK3 in SH-SY5Y neuronal cells. Hence GSK3 mediated Nrf1 degradation can serve as an important mechanistic model to understand GSk3's role in neurodegeneration.
In addition to Fbw7, Nrf1 has also been shown to be regulated by β-TRCP ubiquitin ligase [33], and Nrf1 located in the endoplasmic reticulum is targeted to the ER associated degradation (ERAD) pathway through Hrd1 ubiquitin ligase, [26,33,34]. It is interesting to note that Nrf2 stability is also regulated by β-TRCP in a GSK3 dependent manner [35]. Thus, the possibility that β-TRCP-mediated turnover of Nrf1 also involves GSK3 warrants further experimentation. In this regard, it is also important to note that deletion of the CPD and mutation of S350 to alanine in Nrf1 did not completely abolish phosphorylation by GSK3 indicating that other sites outside of the CPD can be phosphorylated by GSK3. These findings suggest that Nrf1 may be subject to multiple pathways of regulation that is in keeping with the diverse cellular processes that has been implicated for Nrf1. It is possible that Nrf1, through regulation via β–RCP, Fbw7 and Hrd1, directs expression of distinct subsets of target genes. Alternately, the involvement of multiple ligases ensures that Nrf1 levels are tightly controlled to prevent deregulated expression of Nrf1 target genes encoding antioxidant proteins and proteasome subunits. While transcriptional activation of cytoprotective genes is required for maintaining cellular homeostasis, if unchecked, persistent activation can promote tumorigenesis and other pathological conditions by enhancing survival of cancerous cells through increased drug resistance, stress tolerance, and other mechanisms [36,37]. For example, it has been demonstrated that inappropriate activation of the oxidative stress response by somatic mutations in the Nrf2–KEAP1 pathway provides growth advantage and anticancer drug resistance in human lung cancers [37–39]. Thus, regulated Nrf1 expression is likely to be vital for proper cell function, and raises the possibility that aberrant Nrf1 expression is involved in cancer development. Consistent with this idea, gene expression profiling studies showed that Nrf1 expression is increased in a variety of cancers including pituitary adenomas and breast carcinomas [40,41]. Given that Fbw7 is a well-known tumor suppressor with multiple oncogene targets that are frequently inactivated in various cancers [25,26], it would be of interest to determine whether elevated Nrf1 expression in these examples involves mechanisms that allow escape from degradation mediated by the GSK3–Fbw7 pathway.
In summary, we have shown that GSK3 regulates Nrf1 turnover, and that GSK3 mediated phosphorylation of Nrf1 is essential for its recognition and subsequent degradation by the SCF–Fbw7 E3 ubiquitin ligase complex. Our data also reveals that GSK3 inhibits the anti-apoptotic function of Nrf1 following stress induction. Together these results suggest that Nrf1 is an in-vivo substrate of GSK3, and Nrf1 may play a role in other GSK3 regulated cellular processes.
Acknowledgments
Funding
This research was supported by the NIH Grants CA091907 and NS065223.
Abbreviations
- ARE
antioxidant response element
- CNC-BZIP
Cap ‘n’ Collar-type basic leucine zipper
- CPD
Cdc4 phosphodegron domain
- ER
endoplasmic reticulum
- GSK3
glycogen synthase kinase 3
- Nrf1
nuclear factor E2-related factor-1
- SCF
Skp1-Cu1l–F-box
- UPS
ubiquitin proteasome system
Footnotes
Conflict of interest
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH, et al. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 2.Chan JY, Han XX, Kan YW, et al. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi A, Ito E, Told T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M, et al. Molecular cloning and functional characterization of a new Cap ‘n’ collar family transcription factor Nrf3. J. Biol. Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 4.Moi P, Chan K, Asunis I, Cao A, Kan YW, et al. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JY, Cheung MC, Moi P, Chan K, Kan YW, et al. Chromosomal localization of the human NF-E2 family of bZIP transcription factors by fluorescence in situ hybridization. Hum. Genet. 1995;95:265–269. doi: 10.1007/BF00225191. [DOI] [PubMed] [Google Scholar]
- 7.Motohashi H, O'Connor T, Katsuoka F, Engel JD, Yamamoto M, et al. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 8.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW, et al. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Kwong M, Lu R, Ginzinger D, Lee C, Leung L, Chan JY, et al. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell Biol. 2003;23:4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CS, Lee C, Hu T, Nguyen JM, Zhang J, Martin MV, Vawter MP, Huang EJ, Chan JY, et al. Loss of nuclear factor E2-related factor-1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc. Natl. Acad. Sci. USA. 2011;108:8408–8413. doi: 10.1073/pnas.1019209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY, et al. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc. Natl. Acad. Sci. USA. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnsen O, Murphy P, Prydz H, Kolsto AB, et al. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong M, Kan YW, Chan JY, et al. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in gamma-gcs(l) and gss expression in mouse fibroblasts. J. Biol. Chem. 1999;274:37491–37498. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- 14.Myhrstad MC, Husberg C, Murphy P, Nordstrom O, Blomhoff R, Moskaug JO, Kolsto AB, et al. TCF11/Nrf1 overexpression increases the intracellular glutathione level and can transactivate the gamma-glutamylcysteine synthetase (GCS) heavy subunit promoter. Biochim. Biophys. Acta. 2001;1517:212–219. doi: 10.1016/s0167-4781(00)00276-1. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M, et al. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ, et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venugopal R, Jaiswal AK, et al. Nrf1 and Nrf2 positively and c-Fos and Fral negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductasel gene. Proc. Natl. Acad. Sci. USA. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Chan JY, et al. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain Inhibition of nuclear translocation and transacting function. J. Biol. Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Kwok AM, Chan JY, et al. The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription. J. Biol. Chem. 2007;282:24670–24678. doi: 10.1074/jbc.M700159200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Crouch DH, Yamamoto M, Hayes JD, et al. Negative regulation of the Nrfl transcription factor by its N-terminal domain is independent of Keapl: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J. 2006;399:373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Lucocq JM, Yamamoto M, Hayes JD, et al. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrfl is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. Biochem. J. 2007;408:161–172. doi: 10.1042/BJ20070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen P, Frame S, et al. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 23.Frame S, Cohen P, et al. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rayasam GV, Tulasi VK, Sodhi R, Davis JA, Ray A, et al. Glycogen synthase kinase 3: more than a namesake. Br. J. Pharmacol. 2009;156:885–898. doi: 10.1111/j.1476-5381.2008.00085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welcker M, Clurman BE, et al. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 26.Biswas M, Phan D, Watanabe M, Chan JY, et al. The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. J. Biol. Chem. 2011;286:39282–39289. doi: 10.1074/jbc.M111.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Dan HC, Park S, Yang L, Liu Q, Kaneko S, Ning J, He L, Yang H, Sun M, Nicosia SV, Cheng JQ, et al. AKT/PKB signaling mechanisms in cancer and chemoresistance. Front. Biosci. 2005;10:975–987. doi: 10.2741/1592. [DOI] [PubMed] [Google Scholar]
- 28.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin RA, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta. 1773;2007:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaytor MD, Orr HT, et al. The GSK3 beta signaling cascade and neurodegenerative disease. Curr. Opinions Neurobiol. 2002;12:275–278. doi: 10.1016/s0959-4388(02)00320-3. [DOI] [PubMed] [Google Scholar]
- 30.Bijur GN, Jope RS, et al. Proapoptotic stimuli induce nuclear accumulation of glycogen synthase kinase-3 beta. J. Biol. Chem. 2001;276:37436–37442. doi: 10.1074/jbc.M105725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Boise LH, et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szokalska A, Makowski M, Nowis D, Wilczynski GM, Kujawa M, Wojcik C, Mlynarczuk-Bialy I, Salwa P, Bil J, Janowska S, Agostinis P, Verfaillie T, Bugajski M, Gietka J, Issat T, Glodkowska E, Mrowka P, Stoklosa T, Hamblin MR, Mroz P, Jakobisiak M, Golab J, et al. Proteasome inhibition potentiates antitumor effects of photodynamic therapy in mice through induction of endoplasmic reticulum stress and unfolded protein response. Cancer Res. 2009;69:4235–4243. doi: 10.1158/0008-5472.CAN-08-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchiya Y, Morita T, Kim M, Iemura S, Natsume T, Yamamoto M, Kobayashi A, et al. Dual regulation of the transcriptional activity of Nrfl by beta-TrCP- and Hrdl-dependent degradation mechanisms. Mol. Cell Biol. 2011;31:4500–4512. doi: 10.1128/MCB.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steffen J, Seeger M, Koch A, Kruger E, et al. Proteasomal degradation is transcriptionally controlled byTCFll via an ERAD-dependent feedback loop. Mol. Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Rada P, Rojo Al, Chowdhry S, McMahon M, Hayes JD, Cuadrado A, et al. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keapl-independent manner. Mol. Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, Zhang DD, et al. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010;70:5486–5496. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S, et al. Cancer related mutations in NRF2 impair its recognition by Keapl-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S, et al. Loss of Keapl function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 39.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galland F, Lacroix L, Saulnier P, Dessen P, Meduri G, Bernier M, Gaillard S, Guibourdenche J, Fournier T, Evain-Brion D, Bidart JM, Chanson P, et al. Differential gene expression profiles of invasive and non-invasive non-functioning pituitary adenomas based on microarray analysis. Endocr. Relat. Cancer. 2010;17:361–371. doi: 10.1677/ERC-10-0018. [DOI] [PubMed] [Google Scholar]
- 41.Vegran F, Boidot R, Coudert B, Fumoleau P, Arnould L, Garnier J, Causeret S, Fraise J, Dembele D, Lizard S. Gene expression profile and response to trastuzumab-docetaxel-based treatment in breast carcinoma. Br. J. Cancer. 2009;101:1357–1364. doi: 10.1038/sj.bjc.6605310. [DOI] [PMC free article] [PubMed] [Google Scholar]