Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease that affects a number of different organs and tissues. Interleukin-1 (IL1) and estrogen are considered potential elements in the pathology of SLE. Recently, the variable number of tandem repeats (VNTR) polymorphism in the IL1 receptor antagonist gene (IL1-RN) and PvuII (rs2234693) and XbaI (rs9340799) polymorphisms in the estrogen receptor 1 gene (ESR1) have been associated with a predisposition to SLE. However, the evidence for these associations is inconclusive. We therefore conducted a meta-analysis to validate the roles of these polymorphisms in SLE susceptibility. We searched four databases and identified a total of 17 eligible articles comprising 24 studies. The Newcastle-Ottawa quality assessment scale was used to assess the qualities of the selected studies. We assessed the strengths of the associations using odds ratios (ORs) with 95% confidence intervals (95% CIs). Regarding the IL-1RN VNTR, the 2 allele significantly increased SLE susceptibility (2 vs. L: OR = 1.34, 95% CI = 1.03–1.73, P = 0.03). The ESR1 PvuII CC/CT genotype was also associated with SLE susceptibility (CC/CT vs. TT: OR = 1.25, 95% CI = 1.06–1.47, P = 0.01), and the difference was especially pronounced among Asians (CC/CT vs. TT: OR = 1.33, 95% CI = 1.04–1.69, P = 0.02). No significant association between the ESR1 XbaI polymorphism and SLE susceptibility was observed in the overall analysis. However, a marginally significant association between the GG/GA genotype was found in individuals of Asian descent (GG/GA vs. AA: OR = 1.30, 95% CI = 1.01–1.67, P = 0.04). These results indicate that the IL1-RN VNTR 2 allele, ESR1 PvuII CC/CT genotype and ESR1 XbaI GG/GA genotype may increase SLE susceptibility, especially in Asian individuals.
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease that affects various organs and tissues, involving the production of a range of autoantibodies against serological, intracellular, nucleic acid and cell surface antigens [1]. Although the mechanisms underlying SLE are not fully understood, genetic, environmental and hormonal factors are all thought to impact on the development of the disease [2].
Cytokines are considered to be potential elements in the pathology of SLE. These include interleukin-1 (IL1), which plays a key regulatory role in initiating and modulating immunologic and inflammatory events [3], [4]. Animals with experimental SLE produced increased levels of IL1 throughout the disease course [5]. The IL1 family consists of IL1α, IL1β and IL1 receptor antagonist (IL1-RA) [6]. IL1-RA is an important anti-inflammatory molecule that binds to IL1 receptors in competition with IL1α and IL1β, thus inhibiting their activities and modulating a variety of IL1-related immune and inflammatory activities [7]. The IL1-RA gene (IL1-RN) has a variable number of tandem repeats (VNTR) polymorphism of 86 base pairs (bp) in intron 2. Five alleles correspond to allele 1 (four repeats), allele 2 (two repeats), allele 3 (five repeats), allele 4 (three repeats) and allele 5 (six repeats), which can be further categorized into a long allele (L: 3–6 repeats) and a short allele (2∶2 repeats). The genotypes are therefore classified as LL, 2L and 22 [8]. Blakemore et al. [9] first revealed that the frequency of the IL1-RN VNTR 2 allele was increased in SLE patients, since when mounting studies have explored the relationship between the IL1-RN VNTR polymorphism and SLE susceptibility in different populations; however, the findings have been controversial [10]–[12].
Estrogen is another underlying element in the pathology of SLE. SLE typically presents in women of childbearing age [13] and its morbidity falls remarkably after the menopause, in line with the decline in endogenous estrogen [14]. In an SLE mouse model, female mice had poorer outcomes than male mice, and estrogens exacerbated, while androgens ameliorated, the disease [15]. One possible mechanism is that physiological concentrations of estrogen could affect the secretion of cytokines such as IL1 [16]–[18]. However, the roles of estrogen and IL1 in SLE remain unclear. Estrogen acts on target cells through binding to estrogen receptors (ERs). ERα, encoded by the ER 1 gene (ESR1), is the main form of ER. Two polymorphisms, ESR1 PvuII T/C (rs2234693) and ESR1 XbaI A/G (rs9340799), located in the first intron of the ESR1 gene, have been extensively studied, but the associations between these polymorphisms and SLE susceptibility remain controversial [19], [20].
Limited sample sizes and inadequate statistical power mean that the results of studies of the relationships between the IL1-RN VNTR, ESR1 PvuII, and ESR1 XbaI polymorphisms and SLE susceptibility remain conflicting, rather than conclusive [9]–[12], [19]–[31]. Given the potentially important roles of these three polymorphisms in the pathological process and the increasing numbers of studies in different populations, we performed a meta-analysis to derive a more precise estimation of the associations between the IL1-RN VNTR, ESR1 PvuII, and ESR1 XbaI polymorphisms and SLE susceptibility.
Materials and Methods
Search strategy
We searched the PubMed, Embase, Wanfang and Chinese National Knowledge Infrastructure databases using the search terms: ‘systemic lupus erythematosus’ or ‘SLE’, ‘polymorphism’ or ‘allele’ or ‘genotype’, ‘interleukin-1 receptor antagonist’ or ‘IL1-RN’ or ‘estrogen receptor’ or ‘ER’. The literature search was updated on December 2013 and there was no date limit. The results were also supplemented with manual searches of references from the final published articles.
Study selection
The inclusion criteria were: (1) case-control design; (2) studies investigating the relationship between the IL1-RN VNTR, ESR1 PvuII or ESR1 XbaI polymorphisms and SLE susceptibility; (3) studies with sufficient data to provide odds ratios (ORs) and 95% confidence intervals (CIs); and (4) diagnosis of SLE patients performed according to the American College of Rheumatology criteria [32], [33]. The exclusion criteria were: (1) studies with overlapping populations; and (2) studies with insufficient data.
Data extraction
The following information was sought from each publication: first author’s surname, year of publication, participants’ country, ethnicity, sex distribution, genotyping methods, the source of control groups (population-based or hospital-based controls) and matching numbers of genotyped cases and controls. The literature search, eligible study selection and data extraction were carefully conducted independently by two reviewers (Cai and Zhang) and consensuses were reached on all items.
Quality appraisal
Two reviewers (Cai and Zhang) independently rated the methodological quality of every included study according to the Newcastle-Ottawa quality assessment scale [34]. This scale contains nine items (1 point for each) in three parts: selection (four items), comparability (two items) and exposure (three items).
Statistical analysis
Statistical manipulations were conducted using Stata 10.0 (Stata Corporation, College Station, TX, USA). A χ2 test for goodness of fit was used to test for Hardy-Weinberg equilibrium (HWE) in the control group, with P<0.05 indicating a deviation from HWE. Crude ORs and 95% CIs were used to assess the strengths of the associations between the IL1-RN and ESR1 polymorphisms and SLE susceptibility. The statistical significance of the pooled ORs was determined by the Z test, with P<0.05 considered significant. Statistical heterogeneity among studies was assessed by the Q-test and the I2 statistic was used to estimate heterogeneity quantitatively [35]. In the absence of heterogeneity (P>0.10 or I2<50%), pooled ORs and 95% CIs were calculated by the fixed-effects model (Mantel-Haenszel method), otherwise the random-effects model (DerSimonian-Laird method) was used. Sensitivity analysis was used to evaluate the stability of the results of the meta-analysis by removing one study at a time to determine the influence of the individual data set on the pooled OR. Potential publication bias was examined by a funnel plot of log OR against its standard error using Begg’s test, and the degree of asymmetry was assessed by Egger’s unweighted regression asymmetry test. Publication bias may be present if P<0.05 [36].
Results
Study characteristics
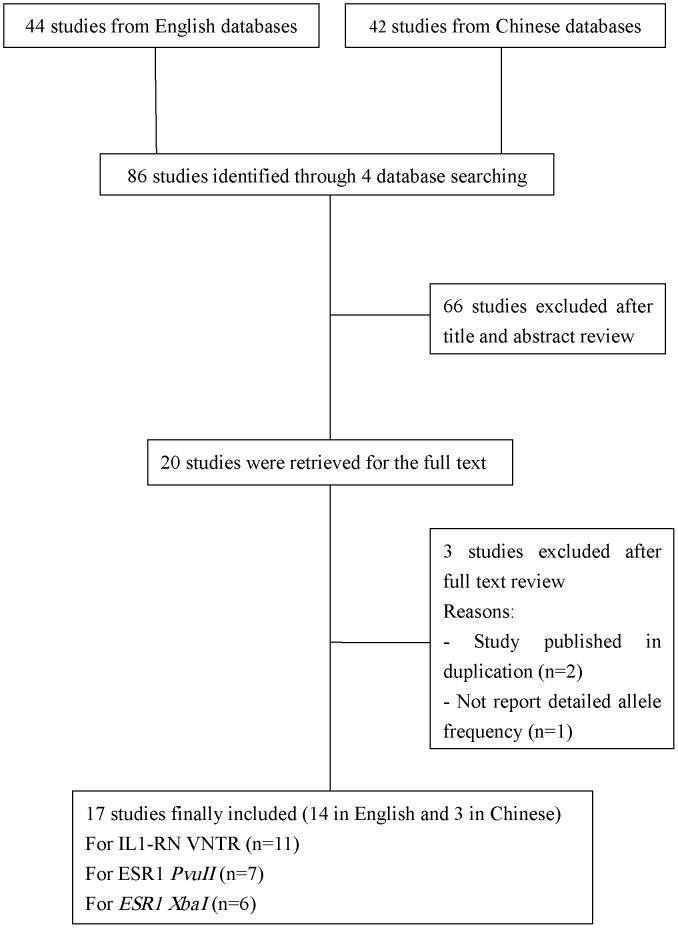
A detailed flow chart of the inclusion and exclusion processes is presented in Figure 1. Overall, 86 studies (44 in English and 42 in Chinese) were retrieved based on the search terms. Of these, 69 studies were excluded according to the inclusion and exclusion criteria and 17 eligible articles representing 24 studies were identified (one article was considered as several separate studies if it involved different populations or different target single nucleotide polymorphisms). Eleven studies including 1171 SLE patients and 1834 controls described IL-1RN VNTR genotypes, seven studies including 1012 SLE patients and 2442 controls described ESR1 PvuII genotypes and six studies including 816 SLE patients and 1478 controls described ESR1 XbaI genotypes. The first author’s surname, publication year, ethnicity, quality score, sex distribution, frequencies of various genotypes in SLE patients and controls and HWE in controls for each study are listed in Tables 1, 2 and 3. The mean score of the quality appraisal was 6.54. In addition, most of the eligible studies were population-based and polymerase chain reaction was performed in all studies. Genotype distributions in the control populations agreed with HWE in all except seven studies [12], [19], [22], [23], [25], [29], [30].
Figure 1. Flow diagram of studies included in the meta-analysis.
Table 1. Characteristics and IL1-RN VNTR polymorphism genotype distributions in studies included in the meta-analysis.
| Author, year | Ethnicity | Quality scorea | Control | Case | P HWE | ||||||||
| LL | 2L | 22 | L | 2 | LL | 2L | 22 | L | 2 | ||||
| Tsai 2006 [10] | Taiwan(Asian) | 6 | – | – | – | 142 | 6 | – | – | – | 198 | 10 | – |
| Lee 2004 [11] | Korean(Asian) | 7 | 109 | 18 | 0 | 236 | 18 | 83 | 10 | 0 | 176 | 10 | 0.39 |
| Parks 2004 [12] | United States(Caucasian) | 7 | 169 | 18 | 15 | 356 | 48 | 66 | 12 | 8 | 144 | 28 | <0.01 |
| Parks 2004 [12] | United States(African-American) | 7 | 69 | 3 | 0 | 141 | 3 | 137 | 6 | 1 | 280 | 8 | 0. 86 |
| Jonsen2004 [21] | Sweden(Caucasian) | 6 | 111 | 75 | 14 | 297 | 103 | 86 | 38 | 14 | 210 | 66 | 0.78 |
| Huang 2002 [22] | Taiwan(Asian) | 7 | 96 | 6 | 1 | 198 | 8 | 43 | 8 | 1 | 94 | 10 | 0.03 |
| Zhu 2000 [23] | China(Asian) | 5 | 15 | 31 | 4 | 61 | 39 | 26 | 52 | 2 | 104 | 56 | 0.03 |
| Tjernstrom 1999 [24] | Sweden(Caucasian) | 7 | – | – | – | 339 | 39 | – | – | – | 130 | 32 | – |
| Heward 1999 [25] | Caucasian(Caucasian) | 4 | 312 | 7 | 19 | 631 | 45 | 106 | 4 | 6 | 216 | 16 | <0.01 |
| Suzuki 1997 [26] | Japan(Asian) | 4 | – | – | – | 418 | 18 | – | – | – | 354 | 38 | – |
| Blakemore 1994 [9] | England(Caucasian) | 7 | 152 | 92 | 17 | 396 | 126 | 39 | 31 | 11 | 109 | 53 | 0.54 |
The quality score was determinded by using the Newcastle-Ottawa quality assessment scale.
IL1-RN: Interleukin-1 receptor antagonist gene; VNTR: variable number of tandem repeats; HWE: Hardy-Weinberg equilibrium.
Table 2. Characteristics and ESR1 PvuII polymorphism genotype distributions in studies included in the meta-analysis.
| Author, year | Ethnicity | Quality scorea | Gender (M/F) | Control | Case | P HWE | |||||||||
| Control | Case | TT | TC | CC | T | C | TT | TC | CC | T | C | ||||
| Kisiel 2011 [20] | Poland (Caucasian) | 6 | 482/482 | 14/182 | 270 | 467 | 227 | 1007 | 921 | 44 | 101 | 51 | 189 | 203 | 0.36 |
| Wang 2010 [23] | United States(Mixed) | 8 | 0/102 | 0/46 | 38 | 48 | 15 | 124 | 78 | 9 | 26 | 11 | 44 | 48 | 0.98 |
| Lu 2009 [28] | China(Asian) | 7 | 0/157 | 0/221 | 83 | 56 | 18 | 222 | 92 | 95 | 92 | 34 | 282 | 160 | 0.08 |
| Li 2008 [29] | China(Asian) | 6 | 0/200 | 0/70 | 86 | 82 | 32 | 254 | 146 | 23 | 39 | 8 | 85 | 55 | 0.10 |
| Chen 2008 [30] | China(Asian) | 5 | 36/46 | 6/76 | 30 | 31 | 21 | 91 | 73 | 37 | 30 | 15 | 104 | 60 | 0.03 |
| Johansson 2005 [31] | Sweden(Caucasian) | 9 | 180/490 | 40/220 | 208 | 332 | 130 | 748 | 592 | 83 | 132 | 45 | 298 | 222 | 0.90 |
| Lee 2004 [19] | Korean(Asian) | 7 | 0/268 | 0/137 | 114 | 110 | 44 | 338 | 198 | 46 | 76 | 15 | 106 | 168 | 0.05 |
The quality score was determinded by using the Newcastle-Ottawa quality assessment scale.
ESR1: estrogen receptor 1 gene; M: Male; F: Female; HWE: Hardy-Weinberg equilibrium.
Table 3. Characteristics and ESR1 XbaI polymorphism genotype distributions in studies included in the meta-analysis.
| Author, year | Ethnicity | Quality scorea | Gender (M/F) | Control | Case | P HWE | |||||||||
| Control | Case | AA | AG | GG | A | G | AA | AG | GG | A | G | ||||
| Wang 2010 [23] | United States(Mixed) | 8 | 0/102 | 0/46 | 48 | 44 | 9 | 140 | 62 | 14 | 24 | 8 | 52 | 40 | 0.81 |
| Lu 2009 [28] | China(Asian) | 7 | 0/157 | 0/221 | 112 | 38 | 7 | 262 | 52 | 138 | 73 | 10 | 349 | 93 | 0.12 |
| Li 2008 [29] | China(Asian) | 6 | 0/200 | 0/70 | 144 | 46 | 10 | 334 | 66 | 46 | 19 | 5 | 111 | 29 | 0.02 |
| Chen 2008 [30] | China(Asian) | 5 | 36/46 | 6/76 | 45 | 29 | 8 | 119 | 45 | 48 | 31 | 3 | 127 | 37 | 0.31 |
| Johansson 2005 [31] | Sweden(Caucasian) | 9 | 180/490 | 40/220 | 332 | 281 | 57 | 945 | 395 | 145 | 94 | 21 | 384 | 136 | 0.82 |
| Lee 2004 [19] | Korean(Asian) | 7 | 0/268 | 0/137 | 192 | 62 | 14 | 446 | 90 | 89 | 38 | 10 | 216 | 58 | <0.01 |
The quality score was determinded by using the Newcastle-Ottawa quality assessment scale.
ESR1: estrogen receptor 1 gene; M: Male; F: Female; HWE: Hardy-Weinberg equilibrium.
Quantitative synthesis
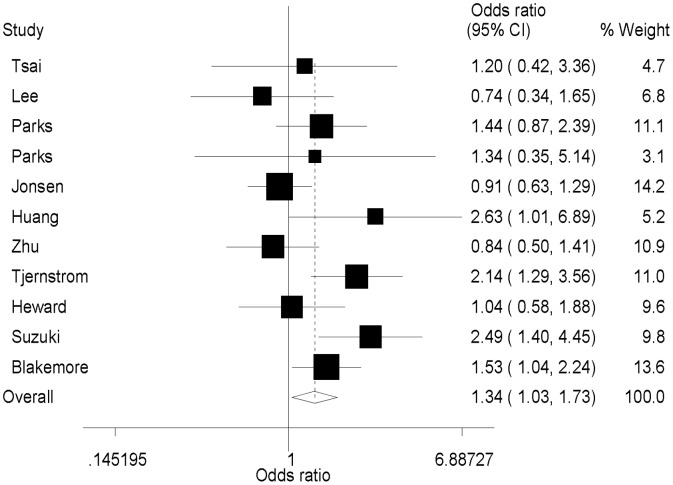
Eleven studies including 1171 SLE patients and 1834 controls assessed the relationship between IL1-RN VNTR polymorphisms and SLE susceptibility. IL1-RN VNTR polymorphism showed no significant association with SLE susceptibility in a dominant model (22/2L vs. LL: OR = 1.11, 95% CI = 0.87−1.40, P = 0.40), recessive model (22 vs. LL/2L: OR = 1.32, 95% CI = 0.88−1.97, P = 0.17) or additive model (22 vs. LL: OR = 1.32, 95% CI = 0.88−1.98, P = 0.19, Table 4). However, a significant association was observed in an allelic contrast model (2 vs. L: OR = 1.34, 95% CI = 1.03–1.73, P = 0.03, Figure 2). After excluding the studies in which the genotype distributions in the control groups deviated from HWE, the IL1-RN VNTR polymorphism showed no significant association with SLE susceptibility in all genetic models (Table S1). We performed subgroup analyses in Asian and Caucasian populations and found no significant differences in either ethnic subgroup (Table S1).
Table 4. Main results of meta-analysis of the association of IL1-RN VNTR, ESR1 PvuII and ESR1 XbaI polymorphisms with SLE susceptibility.
| Gene and Genetic models | Number of study | P heterogeneity | I2(%) | Type of effect model | ORs (95% CI) | P |
| IL1-RN VNTR | ||||||
| Dominant (22/2L vs. LL) | 8 | 0.19 | 29.3 | Fixed | 1.11 (0.87–1.40) | 0.40 |
| Recessive (22 vs. LL/2L) | 7 | 0.50 | 0 | Fixed | 1.32 (0.88–1.97) | 0.17 |
| Additive (22 vs. LL) | 7 | 0.46 | 0 | Fixed | 1.32 (0.88–1.98) | 0.19 |
| Allelic contrast (2 vs. L) | 11 | 0.02 | 51.4 | Random | 1.34 (1.03–1.73) | 0.03 |
| ESR1 PvuII | ||||||
| Dominant (CC/CT vs. TT) | 7 | 0.10 | 43.3 | Fixed | 1.25 (1.06–1.47) | 0.01 |
| Recessive (CC vs. TT/CT) | 7 | 0.22 | 26.8 | Fixed | 0.96 (0.79–1.17) | 0.71 |
| Additive (CC vs. TT) | 7 | 0.11 | 42.5 | Fixed | 1.10 (0.88–1.38) | 0.41 |
| Allelic contrast (C vs. T) | 7 | 0.00 | 85.5 | Random | 1.28 (0.95–1.74) | 0.11 |
| ESR1 XbaI | ||||||
| Dominant(GG/GA vs. AA) | 6 | 0.03 | 58.8 | Random | 1.19 (0.88–1.62) | 0.27 |
| Recessive(GG vs. AA/AG) | 6 | 0.39 | 6.1 | Fixed | 1.08 (0.77–1.51) | 0.67 |
| Additive(GG vs. AA) | 6 | 0.17 | 35.1 | Fixed | 1.09 (0.77–1.54) | 0.64 |
| Allelic contrast(G vs. A) | 6 | 0.03 | 60.4 | Random | 1.15 (0.89–1.49) | 0.27 |
IL1-RN: Interleukin-1 receptor antagonist gene; VNTR: variable number of tandem repeats; ESR1: estrogen receptor 1 gene; OR: odds ratio; CI: confidence interval.
Figure 2. Forest plot of the association between SLE susceptibility and IL1-RN VNTR polymorphism (2 versus L).
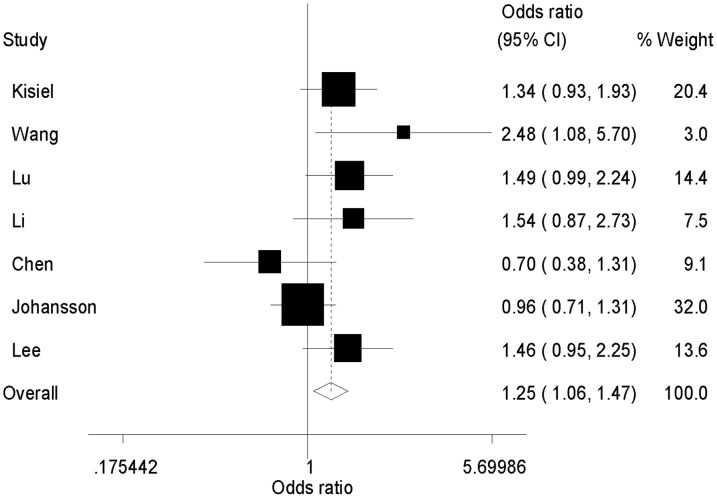
Seven studies compared the ESR1 PvuII polymorphism in SLE patients and controls. Individuals carrying variant genotypes had an increased risk of SLE in the dominant model (CC/CT vs. TT: OR = 1.25, 95% CI = 1.06–1.47, P = 0.01, Figure 3) but not in other genetic models (CC vs. TT/CT: OR = 0.96, 95% CI = 0.79–1.17, P = 0.71; CC vs. TT: OR = 1.10, 95% CI = 0.88–1.38, P = 0.41; C vs. T: OR = 1.28, 95% CI = 0.95–1.74, P = 0.11, Table 4). After excluding the study in which the genotype distribution in the control group deviated from the HWE, ESR1 PvuII polymorphism was significantly associated with SLE susceptibility in both dominant and allelic contrast models (Table S2). When grouped by ethnicity, a significant association was still observed in the dominant model in the Asian group (CC/CT vs. TT: OR = 1.33, 95% CI = 1.04–1.69, P = 0.02, Table S2) but not in the Caucasian group (CC/CT vs. TT: OR = 1.11, 95% CI = 0.88–1.40, P = 0.38, Table S2).
Figure 3. Forest plot of the association between SLE susceptibility and ESR1 PvuII polymorphism (CC/CT versus TT).
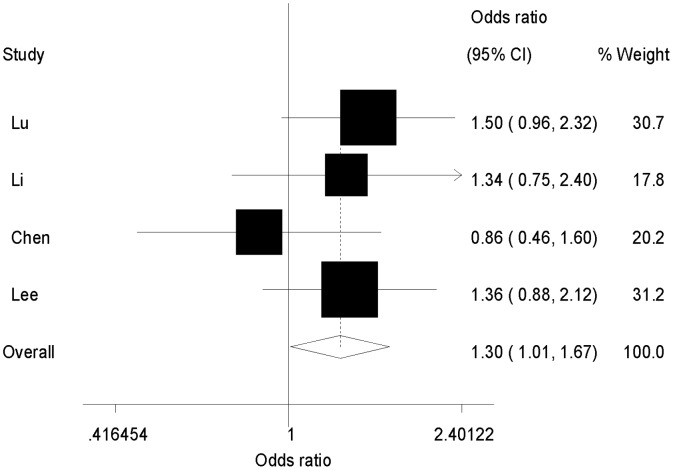
Six studies investigated the association between ESR1 XbaI polymorphism and SLE susceptibility. No significant relationships were identified for any of the genetic models in the whole study set (GG/GA vs. AA: OR = 1.19, 95% CI = 0.88–1.62, P = 0.27; GG vs. AA/GA: OR = 1.08, 95% CI = 0.77–1.51, P = 0.67; GG vs. AA: OR = 1.09, 95% CI = 0.77–1.54, P = 0.64; G vs. A: OR = 1.15, 95% CI = 0.89–1.49, P = 0.27, Table 4). Exclusion of the two studies in which the genotype distributions in the control groups deviated from the HWE had no significant effect on the results (Table S3). However, stratified analysis by ethnicity demonstrated a marginally significant association in the dominant model (GG/GA vs. AA: OR = 1.30, 95% CI = 1.01–1.67, P = 0.04, Figure 4, Table S3) in individuals of Asian descent.
Figure 4. Forest plot of the association between SLE susceptibility and ESR1 XbaI polymorphism in Asian descent (GG/GA versus AA).
Tests of heterogeneity
For IL1-RN VNTR, ESR1 PvuII, and ESR1 XbaI, heterogeneity between studies was observed in the overall analysis of the allelic contrast model (P heterogeneity = 0.02, 0.00, 0.03, respectively, Table 4). In addition, heterogeneity was also found for ESR1 XbaI in the dominant model (P heterogeneity = 0.03). Ethnicity was assessed as a potential source of heterogeneity. Ethnicity (χ2 = 11.49, df = 2, P = 0.003) contributed to the heterogeneity for ESR1 PvuII. Ethnicity also contributed to the heterogeneity for the dominant model (χ2 = 10.00, df = 2, P = 0.007) and the allelic contrast model (χ2 = 8.83, df = 2, P = 0.010) for ESR1 XbaI.
Sensitivity analysis
Sensitivity analysis revealed that heterogeneity decreased after some studies were removed: Jonsen et al. 2004 [21] for IL1-RN VNTR (2 vs. L: P heterogeneity = 0.08, I2 = 41.8%); Johansson et al. 2005 [31] for ESR1 XbaI (G vs. A: P heterogeneity = 0.25, I2 = 26.4%); and Johansson et al. 2005 [31] for ESR1 XbaI (GG/GA vs. AA: P heterogeneity = 0.48, I2 = 0.0%). The results of the association between ESR1 PvuII and SLE susceptibility were not substantially altered.
Publication bias
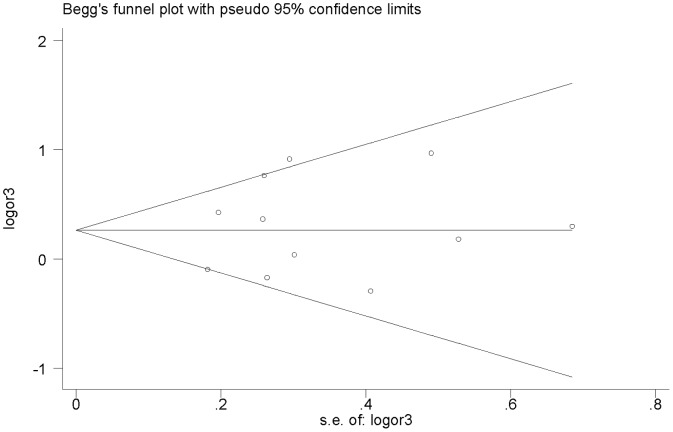
No publication bias was detected among studies regarding the association between the IL1-RN VNTR polymorphism and SLE (P = 0.83 for 2 vs. L, Figure 5). Similarly, the results of Egger’s and Begg’s tests showed no publication bias for the ESR1 PvuII or ESR1 XbaI polymorphisms in all models.
Figure 5. Begg’s funnel plot for publication bias test. IL1-RN VNTR: 2 versus L.
Discussion
Studies of gene polymorphisms potentially related to SLE have recently attracted growing attention. In the present study, we performed a meta-analysis of the associations between IL1-RN VNTR, ESR1 PvuII, and ESR1 XbaI polymorphisms and SLE susceptibility. The analysis indicated an association between the 2 allele of the VNTR polymorphism in intron 2 of IL1-RN and increased SLE susceptibility (2 vs. L: OR = 1.34, 95% CI = 1.03–1.73, P = 0.03). There was also an association between ESR1 PvuII and SLE in the dominant model (CC/CT vs. TT: OR = 1.25, 95% CI = 1.06–1.47, P = 0.01), which was pronounced among Asian individuals (CC/CT vs. TT: OR = 1.33, 95% CI = 1.04–1.69, P = 0.02). There was no significant association between the ESR1 XbaI polymorphism and SLE susceptibility in the overall analysis, but the GG/GA genotype was associated with SLE susceptibility in Asians (GG/GA vs. AA: OR = 1.30, 95% CI = 1.01–1.67, P = 0.04).
IL1 is a potent pro-inflammatory cytokine in acute and chronic inflammation in SLE [37]. IL1RA is a natural antagonist of IL1 and its anti-inflammatory activity is mediated through several different pathways [38] and investigations found decreased production of IL1RA in active SLE [39]. The IL1-RN VNTR 2 allele was associated with increased production of IL1β in vitro [40], [41] and the concentration of IL1RA was shown to be correlated with IL1β [42]. Also, this meta-analysis identified carriage of the 2 allele as a risk factor for SLE susceptibility (2 vs. L: OR = 1.34, 95% CI = 1.03–1.73, P = 0.03). In support of this, the IL1-RN VNTR contains three potential protein-binding sites: an acute phase response element, an interferon α and an interferon β silencer B [43]. The 2 allele of IL1-RN VNTR only has 2 repeats. This could affect mRNA length and subsequent protein processing and stability [44], which could in turn affect the production of IL1RA.
Our study also showed that the ESR1 PvuII CC/CT and ESR1 XbaI GG/GA genotypes could increase susceptibility to SLE. Estrogen can affect both innate and adaptive immune responses in mice [45] and SLE patients [46] through different pathways [47]–[49], and estrogen receptors are expressed in most immunocompetent cells [50]. Some researchers determined that IL1β levels were higher during the luteal period compared with the follicular period of the female reproductive cycle, which was consistent with the results of in vivo [51]–[53] and in vitro tests [18]. The polymorphisms PvuII and XbaI are located in intron 1 of ESR1 but are still able to affect the gene, and thus affect estrogen concentrations. The C allele of PvuII can produce a binding site for the B-myb transcription factor, which could enhance the ability to up-regulate downstream receptor structures compared with the T allele [54]. In the present study, SLE susceptibility was associated with the ESR1 PvuII C allele but not with XbaI in overall analysis. However, we could not rule out the possibility of an association between the ESR1 XbaI polymorphism and SLE susceptibility because PvuII and XbaI are tightly linked [55] and it is difficult to identify which one has a role to play.
Given its multifactorial nature, it is likely that the pathogenesis of SLE may be modulated by age, gender, ethnicity, environmental factors and other variables. We therefore carried out subgroup analysis based on ethnicity. Associations between SLE susceptibility and the ESR1 PvuII C allele and the ESR1 XbaI G allele were found in individuals of Asian descent. This may be attributable to genetic heterogeneity among different populations. Consistent with this, ethnicity contributed to the heterogeneity for ESR1 PvuII and ESR1 XbaI. Moreover, sensitivity analysis revealed that the heterogeneity was reduced by removing Johansson et al.’s [31] study, which was the only study in the meta-analysis of the association between ESR1 XbaI polymorphism and SLE susceptibility that was based on Caucasians. It is also possible that differences in lifestyle and environmental factors between different populations may interact with genes to affect the pathogenesis of SLE.
This meta-analysis had some inevitable limitations. First, three studies on the IL1-RN VNTR polymorphism did not provide genotype data, and the data used to analyze the various genetic models were thus not completely consistent. This may lead to misinterpretation of the association between the IL1-RN VNTR polymorphism and SLE susceptibility. Second, although all eligible studies were included in our study, the small sample size and low statistical power (Table S4, S5 and S6) associated with the low incidence of SLE means that there is a possibility of false negative results. We expect more participants being tested in the future to draw a more reliable conclusion. Third, deviation of genotype distributions from HWE in the control populations in some studies may reflect genotyping errors or control selection bias. The results relating to IL1-RN VNTR and ESR1 PvuII changed when these studies were excluded, suggesting that these results should be interpreted with caution. Fourth, as mentioned above, various factors affect the pathology of SLE; the lack of individual data meant that we only pooled the data based on unadjusted information. Finally, the quality scores were not high for some studies and these studies may have distorted the results.
In conclusion, this meta-analysis indicated that the IL1-RN VNTR 2 allele and the ESR1 PvuII CC/CT and ESR1 XbaI GG/GA genotypes may increase the susceptibility to SLE, especially in individuals of Asian descent. However, this conclusion should be interpreted cautiously because of the low statistical power and considerable heterogeneity. Also, further functional studies are needed to investigate the functions of these alleles. Well-designed, large studies in different ethnic groups and with more detailed information on age, sex, and age of onset of the disease are needed to validate our results. Studies of gene-environment interactions in relation to IL1-RN-ESR1 should also be performed to confirm our preliminary findings.
Supporting Information
Meta analysis of the association of IL1-RN VNTR polymorphism with SLE susceptibility.
(XLS)
Meta analysis of the association of ESR1 Pvu II polymorphism with SLE susceptibility.
(XLS)
Meta analysis of the association of ESR1 Xba I polymorphism with SLE susceptibility.
(XLS)
The statistical power of IL1-RN VNTR 2 allele in studied included in the meta-analysis.
(XLS)
The statistical power of ESR1 Pvu II CC/CT genotypes in studied included in the meta-analysis.
(XLS)
The statistical power of ESR1 Xba I GG/GA genotypes in studied included in the meta-analysis.
(XLS)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the paper and its Supporting Information files.
Funding Statement
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81273146, 81102165). (http://www.nsfc.gov.cn) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tsokos GC (2011) Systemic lupus erythematosus. N Engl J Med 365: 2110–2121. [DOI] [PubMed] [Google Scholar]
- 2. D'Cruz DP, Khamashta MA, Hughes GR (2007) Systemic lupus erythematosus. Lancet 369: 587–596. [DOI] [PubMed] [Google Scholar]
- 3. Yap DY, Lai KN (2013) The role of cytokines in the pathogenesis of systemic lupus erythematosus - from bench to bedside. Nephrology (Carlton) 18: 243–255. [DOI] [PubMed] [Google Scholar]
- 4. Boswell JM, Yui MA, Burt DW, Kelley VE (1988) Increased tumor necrosis factor and IL-1 beta gene expression in the kidneys of mice with lupus nephritis. J Immunol 141: 3050–3054. [PubMed] [Google Scholar]
- 5. Segal R, Bermas BL, Dayan M, Kalush F, Shearer GM, et al. (1997) Kinetics of cytokine production in experimental systemic lupus erythematosus: involvement of T helper cell 1/T helper cell 2-type cytokines in disease. J Immunol 158: 3009–3016. [PubMed] [Google Scholar]
- 6. Smith DE, Renshaw BR, Ketchem RR, Kubin M, Garka KE, et al. (2000) Four new members expand the interleukin-1 superfamily. J Biol Chem 275: 1169–1175. [DOI] [PubMed] [Google Scholar]
- 7. Dinarello CA (1998) Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int Rev Immunol 16: 457–499. [DOI] [PubMed] [Google Scholar]
- 8. Vamvakopoulos JE, Taylor CJ, Morris-Stiff GJ, Green C, Metcalfe S (2002) The interleukin-1 receptor antagonist gene: a single-copy variant of the intron 2 variable number tandem repeat (VNTR) polymorphism. Eur J Immunogenet 29: 337–340. [DOI] [PubMed] [Google Scholar]
- 9. Blakemore AI, Tarlow JK, Cork MJ, Gordon C, Emery P, et al. (1994) Interleukin-1 receptor antagonist gene polymorphism as a disease severity factor in systemic lupus erythematosus. Arthritis Rheum 37: 1380–1385. [DOI] [PubMed] [Google Scholar]
- 10. Tsai LJ, Lan JL, Lin CY, Hsiao SH, Tsai LM, et al. (2006) The different expression patterns of interleukin-1 receptor antagonist in systemic lupus erythematosus. Tissue Antigens 68: 493–501. [DOI] [PubMed] [Google Scholar]
- 11. Lee YH, Kim HJ, Rho YH, Choi SJ, Ji JD, et al. (2004) Interleukin-1 receptor antagonist gene polymorphism and rheumatoid arthritis. Rheumatol Int 24: 133–136. [DOI] [PubMed] [Google Scholar]
- 12. Parks CG, Cooper GS, Dooley MA, Treadwell EL, St Clair EW, et al. (2004) Systemic lupus erythematosus and genetic variation in the interleukin 1 gene cluster: a population based study in the southeastern United States. Ann Rheum Dis 63: 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ansar Ahmed S, Penhale WJ, Talal N (1985) Sex hormones, immune responses, and autoimmune diseases. Mechanisms of sex hormone action. Am J Pathol 121: 531–551. [PMC free article] [PubMed] [Google Scholar]
- 14. Tucker LB, Menon S, Schaller JG, Isenberg DA (1995) Adult- and childhood-onset systemic lupus erythematosus: a comparison of onset, clinical features, serology, and outcome. Br J Rheumatol 34: 866–872. [DOI] [PubMed] [Google Scholar]
- 15. Roubinian JR, Talal N, Greenspan JS, Goodman JR, Siiteri PK (1978) Effect of castration and sex hormone treatment on survival, anti-nucleic acid antibodies, and glomerulonephritis in NZB/NZW F1 mice. J Exp Med 147: 1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan Y, Shimizu I, Shen M, Aoyagi E, Takenaka H, et al. (2008) Effects of estradiol and progesterone on the proinflammatory cytokine production by mononuclear cells from patients with chronic hepatitis C. World J Gastroenterol. 14: 2200–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cunningham M, Gilkeson G (2011) Estrogen receptors in immunity and autoimmunity. Clin Rev Allergy Immunol 40: 66–73. [DOI] [PubMed] [Google Scholar]
- 18. Kim C, Cadet P (2010) Environmental toxin 4-nonylphenol and autoimmune diseases: using DNA microarray to examine genetic markers of cytokine expression. Arch Med Sci 6: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee YJ, Shin KS, Kang SW, Lee CK, Yoo B, et al. (2004) Association of the oestrogen receptor alpha gene polymorphisms with disease onset in systemic lupus erythematosus. Ann Rheum Dis 63: 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kisiel BM, Kosinska J, Wierzbowska M, Rutkowska-Sak L, Musiej-Nowakowska E, et al. (2011) Differential association of juvenile and adult systemic lupus erythematosus with genetic variants of oestrogen receptors alpha and beta. Lupus 20: 85–89. [DOI] [PubMed] [Google Scholar]
- 21. Jonsen A, Bengtsson AA, Sturfelt G, Truedsson L (2004) Analysis of HLA DR, HLA DQ, C4A, FcgammaRIIa, FcgammaRIIIa, MBL, and IL-1Ra allelic variants in Caucasian systemic lupus erythematosus patients suggests an effect of the combined FcgammaRIIa R/R and IL-1Ra 2/2 genotypes on disease susceptibility. Arthritis Res Ther. 6: R557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang CM, Wu MC, Wu JY, Tsai FJ (2002) Interleukin-1 receptor antagonist gene polymorphism in chinese patients with systemic lupus erythematosus. Clin Rheumatol 21: 255–257. [DOI] [PubMed] [Google Scholar]
- 23. Zhu W, Xie HF, Shi W, Liu ZR (2000) Interleukin-1 receptor antagonist gene polymorphism in patients with systemic lupus erythematosus. Chin J Dermatol 33: 35. [Google Scholar]
- 24. Tjernstrom F, Hellmer G, Nived O, Truedsson L, Sturfelt G (1999) Synergetic effect between interleukin-1 receptor antagonist allele (IL1RN*2) and MHC class II (DR17,DQ2) in determining susceptibility to systemic lupus erythematosus. Lupus 8: 103–108. [DOI] [PubMed] [Google Scholar]
- 25. Heward J, Allahabadia A, Gordon C, Sheppard MC, Barnett AH, et al. (1999) The interleukin-1 receptor antagonist gene shows no allelic association with three autoimmune diseases. Thyroid 9: 627–628. [DOI] [PubMed] [Google Scholar]
- 26. Suzuki H, Matsui Y, Kashiwagi H (1997) Interleukin-1 receptor antagonist gene polymorphism in Japanese patients with systemic lupus erythematosus. Arthritis Rheum 40: 389–390. [DOI] [PubMed] [Google Scholar]
- 27. Wang J, Nuite M, McAlindon TE (2010) Association of estrogen and aromatase gene polymorphisms with systemic lupus erythematosus. Lupus 19: 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu ZM, Wang ZE, Wu CX, Wang CY, Zhang BC, et al. (2009) Association of Estrogen Receptor α Gene Polymorphisms with Cytokine Genes Expression in Systemic Lupus Erythematosus. Croatian Medical Journal 50: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li F, Che ZX, Liu YQ, Fu HL, Wang ZE, et al. (2008) Association between estrogen receptor α gene polymorphisms and systemic lupus erythematosus in Chinese women. Chin J Clinical Laboratory Science 26: 112–114. [Google Scholar]
- 30. Chen H, Fan Y, Men JL (2008) Relationship between estrogen receptor gene polymorphisms and SLE. Chin J Dermatol 41: 46–48. [Google Scholar]
- 31. Johansson M, Arlestig L, Moller B, Smedby T, Rantapaa-Dahlqvist S (2005) Oestrogen receptor {alpha} gene polymorphisms in systemic lupus erythematosus. Ann Rheum Dis 64: 1611–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, et al. (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25: 1271–1277. [DOI] [PubMed] [Google Scholar]
- 33. Hochberg MC (1997) Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40: 1725. [DOI] [PubMed] [Google Scholar]
- 34.Wells GA, Shea B, O’Connell D, Peterson J, Welch V, et al. Home>Our Research>Research Programs>Clinical Epidemiology>The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-analyses [Web page]. Ottawa, ON: Ottawa Hospital Research Institute; n.d. [Available online at: http://www.ohri.ca/programs/clinical_epidemiology/oxford. asp; Accessed 2012 February 10.
- 35. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rus V, Atamas SP, Shustova V, Luzina IG, Selaru F, et al. (2002) Expression of cytokine- and chemokine-related genes in peripheral blood mononuclear cells from lupus patients by cDNA array. Clin Immunol 102: 283–290. [DOI] [PubMed] [Google Scholar]
- 38. Garat C, Arend WP (2003) Intracellular IL-1Ra type 1 inhibits IL-1-induced IL-6 and IL-8 production in Caco-2 intestinal epithelial cells through inhibition of p38 mitogen-activated protein kinase and NF-kappaB pathways. Cytokine 23: 31–40. [DOI] [PubMed] [Google Scholar]
- 39. Hsieh SC, Tsai CY, Sun KH, Tsai YY, Tsai ST, et al. (1995) Defective spontaneous and bacterial lipopolysaccharide-stimulated production of interleukin-1 receptor antagonist by polymorphonuclear neutrophils of patients with active systemic lupus erythematosus. Br J Rheumatol 34: 107–112. [DOI] [PubMed] [Google Scholar]
- 40. Danis VA, Millington M, Hyland VJ, Grennan D (1995) Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1Ra) gene polymorphism. Clin Exp Immunol 99: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santtila S, Savinainen K, Hurme M (1998) Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol 47: 195–198. [DOI] [PubMed] [Google Scholar]
- 42. Vamvakopoulos J, Green C, Metcalfe S (2002) Genetic control of IL-1beta bioactivity through differential regulation of the IL-1 receptor antagonist. Eur J Immunol 32: 2988–2996. [DOI] [PubMed] [Google Scholar]
- 43. Tarlow JK, Blakemore AI, Lennard A, Solari R, Hughes HN, et al. (1993) Polymorphism in human IL-1 receptor antagonist gene intron 2 is caused by variable numbers of an 86-bp tandem repeat. Hum Genet 91: 403–404. [DOI] [PubMed] [Google Scholar]
- 44. Korthagen NM, van Moorsel CH, Kazemier KM, Ruven HJ, Grutters JC (2012) IL1RN genetic variations and risk of IPF: a meta-analysis and mRNA expression study. Immunogenetics 64: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nilsson N, Carlsten H (1994) Estrogen induces suppression of natural killer cell cytotoxicity and augmentation of polyclonal B cell activation. Cell Immunol 158: 131–139. [DOI] [PubMed] [Google Scholar]
- 46. Kanda N, Tsuchida T, Tamaki K (1999) Estrogen enhancement of anti-double-stranded DNA antibody and immunoglobulin G production in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Arthritis Rheum 42: 328–337. [DOI] [PubMed] [Google Scholar]
- 47. Paharkova-Vatchkova V, Maldonado R, Kovats S (2004) Estrogen preferentially promotes the differentiation of CD11c+ CD11b(intermediate) dendritic cells from bone marrow precursors. J Immunol 172: 1426–1436. [DOI] [PubMed] [Google Scholar]
- 48. Suzuki T, Shimizu T, Yu HP, Hsieh YC, Choudhry MA, et al. (2007) Salutary effects of 17beta-estradiol on T-cell signaling and cytokine production after trauma-hemorrhage are mediated primarily via estrogen receptor-alpha. Am J Physiol Cell Physiol 292: C2103–2111. [DOI] [PubMed] [Google Scholar]
- 49. Cohen JH, Danel L, Cordier G, Saez S, Revillard JP (1983) Sex steroid receptors in peripheral T cells: absence of androgen receptors and restriction of estrogen receptors to OKT8-positive cells. J Immunol 131: 2767–2771. [PubMed] [Google Scholar]
- 50. Speirs V, Kerin MJ, Newton CJ, Walton DS, Green AR, et al. (1999) Evidence for transcriptional activation of ERalpha by IL-1beta in breast cancer cells. Int J Oncol 15: 1251–1254. [DOI] [PubMed] [Google Scholar]
- 51. Bouman A, Moes H, Heineman MJ, de Leij LF, Faas MM (2001) The immune response during the luteal phase of the ovarian cycle: increasing sensitivity of human monocytes to endotoxin. Fertil Steril 76: 555–559. [DOI] [PubMed] [Google Scholar]
- 52. Cannon JG, Dinarello CA (1985) Increased plasma interleukin-1 activity in women after ovulation. Science 227: 1247–1249. [DOI] [PubMed] [Google Scholar]
- 53. Polan ML, Loukides JA, Honig J (1994) Interleukin-1 in human ovarian cells and in peripheral blood monocytes increases during the luteal phase: evidence for a midcycle surge in the human. Am J Obstet Gynecol 170: 1000–1006 discussion 1006–1007. [DOI] [PubMed] [Google Scholar]
- 54. Herrington DM, Howard TD, Brosnihan KB, McDonnell DP, Li X, et al. (2002) Common estrogen receptor polymorphism augments effects of hormone replacement therapy on E-selectin but not C-reactive protein. Circulation 105: 1879–1882. [DOI] [PubMed] [Google Scholar]
- 55. Liu ZH, Cheng ZH, Gong RJ, Liu H, Liu D, et al. (2002) Sex differences in estrogen receptor gene polymorphism and its association with lupus nephritis in Chinese. Nephron 90: 174–180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Meta analysis of the association of IL1-RN VNTR polymorphism with SLE susceptibility.
(XLS)
Meta analysis of the association of ESR1 Pvu II polymorphism with SLE susceptibility.
(XLS)
Meta analysis of the association of ESR1 Xba I polymorphism with SLE susceptibility.
(XLS)
The statistical power of IL1-RN VNTR 2 allele in studied included in the meta-analysis.
(XLS)
The statistical power of ESR1 Pvu II CC/CT genotypes in studied included in the meta-analysis.
(XLS)
The statistical power of ESR1 Xba I GG/GA genotypes in studied included in the meta-analysis.
(XLS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the paper and its Supporting Information files.