Abstract
Shed hair from domestic animals readily adheres to clothing and other contact items, providing a source of transfer evidence for criminal investigations. Mitochondrial DNA is often the only option for DNA analysis of shed hair. Human mitochondrial DNA analysis has been accepted in the US court system since 1996. The murder trial of the State of Missouri versus Henry L. Polk, Jr. represents the first legal proceeding where cat mitochondrial DNA analysis was introduced into evidence. The mitochondrial DNA evidence was initially considered inadmissible due to concerns about the cat dataset and the scientific acceptance of the marker. Those concerns were subsequently addressed, and the evidence was deemed admissible. This report reviews the case in regards to the cat biological evidence and its ultimate admission as generally accepted and reliable. Expansion and saturation analysis of the cat mitochondrial DNA control region dataset supported the initial interpretation of the evidence.
Keywords: Forensic science, admissibility, Control region, feline, Felis silvestris catus, hair
A majority of households in the USA have domesticated pets, generally cats and/or dogs [1, 2]. Shed hairs from companion animals are abundant in pet-owning homes and easily adhere to clothing, personal effects and other items, potentially providing transfer evidence for criminal investigations [3–5]. Short tandem repeat (STR) analyses of cat hair have previously provided corroborative evidence in a murder trial [6], although the amount of nuclear DNA can sometimes be inadequate for STR profiling of hairs. Mitochondrial DNA (mtDNA) typing, which has recognized deficiencies for individual identification and exclusionary power, is often the only means to develop a DNA profile of the hair donor [7–13]. Although cat mtDNA typing has been previously used in a forensic investigation [14, 15], the defendant pled guilty in that earlier case and the cat mtDNA evidence was not subjected to legal scrutiny.
This case report presents the first analysis of mtDNA control region (CR) profiling of domestic cat hair used as corroborative evidence in a judicial proceeding. Prior to the initiation of the judicial process of the case, the complete domestic cat mtDNA sequence had been defined [16] and variation of the cat mtDNA control region (CR) had been evaluated in several studies [14, 15, 17, 18]. A hyper-variable repeat region within the cat mtDNA CR had been found to be too variable, thereby raising concerns of heteroplasmy and the regions use in forensic applications [15, 19, 20]. However, the remaining areas of the cat mtDNA CR do display sufficient variation for exclusion purposes and are appropriate regions for analysis in forensic investigations [21, 22]. In addition to the cat mtDNA CR sequence and dataset, although subjective, microscopic characterization of fur [5, 23–26] can tentatively support inclusion of cat hair in forensic investigations.
This report focuses on the available cat mtDNA datasets and their expansion during the course of the investigation and murder trial for the State of Missouri v. Henry L. Polk, Jr. (7CR104003803-01). Dataset sample size, representation, and sub-structuring are evaluated in regards to their relevance to the cat mtDNA testimonies. The mitotype sequences and definitions were published after the trial [21].
Background
On March 8, 2004, the Kansas City Police Department (KCPD) investigated a homicide crime scene in Clay County, Missouri. The victim was found severely beaten with his throat violently lacerated to near decapitation. The linings of the victim’s jean pockets had been turned inside out. Hinge lifts were taken of each pocket lining at the scene by KCPD crime scene investigators. The hinge lifts were examined by the KCPD Crime Laboratory, leading to the identification of a single hair. Morphological examination by light microscopy suggested the hair was from a domestic cat due to the banded coloration and the observation of frayed fibrils at the root end, both traits being more consistent with cat hairs than human or dog hairs [26–28].
A suspect was residing in a household with multiple cats (N > 10). Hair standards were collected from nine available cats in the suspect’s household. Hairs from four cats were determined to be microscopically similar to the evidentiary hair based on hair length and pigment size, density, and distribution. The hair pigmentation and density suggested that the cat had light orange coloration, which is consistent with all orange cats, tortoiseshell, or calico cats. The evidence hair was dissimilar to hair standards from the two cats belonging to the victim. The four similar standard hairs from cats living with the suspect and the evidence hair were submitted to QuestGen Forensics (now Zoogen Services) in Davis, California, for DNA analysis.
STR amplifications were unsuccessful; therefore, mtDNA control region (CR) sequence was generated from the hair DNA. QuestGen Forensics issued a report in January, 2005, stating that the mtDNA sequence of the evidentiary hair was identical to the sequence of two of the four standard hairs collected from cats at the suspect’s residence concluding that two of four suspect cats could not be excluded as contributors of the evidentiary hair. The mtDNA profile (mitotype) of the evidence and the two matching standards from the suspect’s cats did not match to any profiles in the QuestGen Forensics cat mtDNA dataset (N = 180 cats), indicating the evidence hair mtDNA mitotype was rare and defined as novel (unique) mitotype Uc1. The mitotype of the non - matching standards from the suspect’s cat’s hair were both determined to be mitotype C. The CR mitotype from the evidence, mitotype Uc1, is distinct from the next most similar mitotype, C1, by a single mutation, and two mutations from major mitotype C. The chance of a random match for the evidentiary hair was considered as 1/180 (0.0055) with the caveat that the true population frequency could not be accurately stated and could be lower.
In the fall of 2007, a hearing was conducted to determine if expert testimony with regard to DNA testing and analysis of cat hair evidence would be admitted under the Frye standard. On September 07, 2007, an order was issued stating: “The small number of cats which contributed to the datasets used for the testing and analysis raised doubt when the evidence was presented. No evidence of acceptance in any scientific community of this procedure was received. It is found and concluded that the procedure utilized by the witness, that of using the small data bases of cats in forming her opinions, has not gained acceptance in any scientific community. When compared to data bases used in human DNA testing and analysis, the sparcity of the data bases that are the foundation of the opinions offered in the Frye hearing supports this finding and conclusion.” With these judgments, the court concluded “No evidence with regard to the testing and DNA analysis of cat hair will be admitted in the trial of this case”.
In response to this order, prosecuting attorneys extended the investigation of the cat hair mtDNA. Cat hair standards from the two cats at the victim’s home were sent to QuestGen Forensics for comparative analyses and were also excluded as the source of the evidence hair by mtDNA mitotype comparison, which were mitotypes B and C.
In November 2007, all DNA extracts of the cat hairs from the case were transferred to the Veterinary Genetics Laboratory (VGL) Forensics Unit at University of California -Davis for confirmatory DNA testing and for comparison to a second cat mtDNA dataset (N = 129). The mtDNA CR sequences were confirmed as accurate, the same interpretations between the evidence hair and the hair standards from the suspect’s and victim’s cats were substantiated and the evidence hair mitotype remained unique as compared to the second independent dataset.
In addition, in mid-December, 2007, an independent feline genetics research laboratory at the University of California - Davis (L.A. Lyons) was contacted for information regarding cat mtDNA datasets and general expert witness testimony. Independently and without knowledge of the criminal investigation, the feline genetics research laboratory had generated mtDNA CR sequence that overlapped the region sequenced by QuestGen Forensics and the UC Davis VGL Forensics Unit. This data had been generated from a wide variety of cats in an effort to initiate the development of domestic cat mtDNA CR data for forensic profiling and studies regarding domestication, cat population dynamics, and population structuring [21, 22]. This dataset included 375 independent cats and the mtDNA sequence of the evidence remained unique within that dataset.
In January, 2008, a second admissibility hearing was conducted where representatives from all three laboratories, as well as additional expert witnesses, testified in regards to the cat DNA evidence. On May 29, 2008, the order was issued to accept mtDNA evidence stating: “The state’s motion to reconsider the order entered September 7, 2007, excluding DNA evidence pertaining to cat hair is sustained. That order is hereby withdrawn. Further evidence offered by the state, in combination with the evidence received before the September 7, 2007, order was entered, allows the court to find and conclude as follows: 1. The state has committed not to offer nuclear DNA evidence. Rulings hereinafter pertain only to mitochondrial DNA. 2. The polymerase chain reaction testing method (PCR) is generally accepted in the scientific community with regard to the extraction and analysis of animal DNA. 3. Mitochondrial DNA testing methods and procedures are generally accepted within the scientific community. 4. The state has committed to limit the DNA expert opinions offered as evidence to opinions based on the mitochondrial DNA extracted from the actual data base of tested cats. So limited, opinions are admissible under Frye standards as to whether or not cat hair from an unknown cat that has been subjected to PCR testing methods is either, (1) hair from a specific cat among the tested cats that make up the data base, or hair from a cat within the maternal line of that cat, or (2) is not hair from a cat among the number of tested cats that make up the data base or within the maternal line of any of the tested cats that make up the data base.”
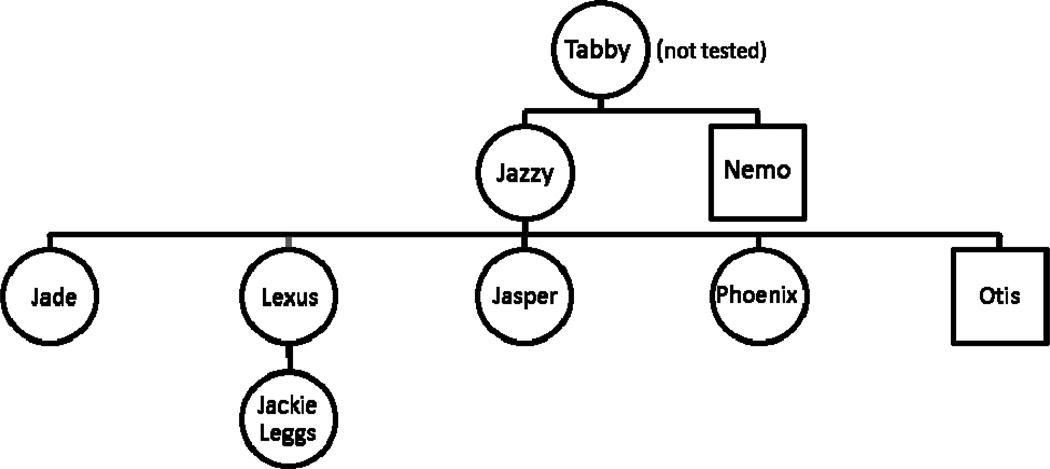
To further evaluate and support the rareness of the evidence hair’s unique mitotype after the January, 2008 admissibility hearing, buccal swabs were collected from eleven cats (MV1 – MV11, Table 1) reportedly living in the former household of the suspect at the time of the 2004 murder. Hair from three of the cats had previously been collected as part of the original criminal investigation, as reported by the owner, since the cats matched the determined coloration of the evidence hair. To examine possible genetic differentiation in randomly bred cats from different regions of the USA, buccal swabs from 23 randomly bred cats representing Clay County, Missouri, were collected by Dr. Lyons and the Clay County Sherriff’s Department. The buccal swab collection included four cats from the Liberty Animal Shelter, fifteen cats from a private practice animal clinic, and four cats that had been acquired from the area by a private owner (Table 2). The swab samples were genetically profiled using STR markers and mtDNA CR sequences by the UC Davis VGL Forensics Unit and independently analyzed by the feline genetics research laboratory using the mtDNA CR sequence [31, 32]. Two of the 23 Missouri cats were a mother-daughter pair, thus only one cat was included in the dataset and subsequent mtDNA database. Eight of eleven cats from the suspect’s household were reported to be related, consequently only four were considered independent cats to contribute to the overall mtDNA CR dataset (FCF118 - 9, 11, 12) (Fig. 1). STR analyses confirmed relationships of the cats. However, one kitten was not from its purported queen, but qualified to another queen in the lineage.
TABLE 1.
State of Missouri v. Henry L. Polk, Jr. cat DNA evidence sample identification.
| Item # | Sample Type | Mitotype | Identification |
|---|---|---|---|
| Evidence, victim’s left pocket, primarily brown with 2 white | |||
| 21-18 | Hair shaft | Uc1 | bands & white portion near root |
| 21-18 | Root end | Uc1 | Evidence |
| 29-2 | Hair | Uc1 | Reference, provided on two slides |
| 29-3 | Hair | Uc1 | Reference, Brown/black striped, real old cat |
| 29-8 | Hair | C | Reference, Brown/black outdoor cat |
| 29-9 | Hair | C | Reference, Brown/black & white outdoor cat |
| 28-1 | Hair | C | Elimination, Victim’s cat |
| 28-2 | Hair | B | Elimination, Victim’s cat |
| MV1 | Buccal | Uc1 | Lexus |
| MV2 | Buccal | Uc1 | Jade |
| MV3 | Buccal | Uc1 | Jazzy |
| MV4 | Buccal | Uc1 | Jackie Legs |
| MV5 | Buccal | Uc1 | Otis |
| MV6 | Buccal | C | Mitzi |
| MV7 | Buccal | Uc1 | Phoenix |
| MV8 | Buccal | C | Luckie |
| MV9 | Buccal | B | Taz |
| MV10 | Buccal | Uc1 | Jasper |
| MV11 | Buccal | Uc1 | Nemo |
TABLE 2.
Random-bred cat and cat breed mitotype frequencies for mtDNA CR.
| Mitotypes Frequencies (percentage of total) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Common | Minor | Rare ≤1.0 |
Unique | |||||||||||||
| Group | Location | Population | A | B | C | D | E | F | G | H | I | J | K | L | ||
| QuestGen (168)* |
Orange Co, CA | DSH (n = 99) | 19.2 | 25.3 | 26.3 | 8.1 | 2.0 | 4.0 | 3.0 | 1.0 | 0.0 | 0.0 | 6.1 | 0.0 | 5.1 | 0.0 |
| Various, USA | Breeds (n = 69) | 36.2 | 14.5 | 20.3 | 7.2 | 5.8 | 0.0 | 0.0 | 1.4 | 7.2 | 1.4 | 0.0 | 1.4 | 4.3 | 0.0 | |
| UC Davis VGL (78) |
Yolo Co, CA | DSH (n = 50) | 18.0 | 30.0 | 12.0 | 6.0 | 0.0 | 2.0 | 0.0 | 6.0 | 0.0 | 0.0 | 8.0 | 2.0 | 6.0 | 10.0 |
| Tompkins Co, NY | DSH (n = 28) | 39.3 | 21.4 | 25.0 | 3.6 | 7.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.6 | 0.0 | |
| Feline Genetics (196)† |
Yolo Co, CA | DSH (n = 15) | 13.3 | 13.3 | 13.3 | 20.0 | 0.0 | 6.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.7 | 6.7 | 20.0 |
| Tompkins Co, NY | DSH (n = 49) | 36.7 | 24.5 | 6.1 | 4.1 | 4.1 | 0.0 | 4.1 | 0.0 | 0.0 | 0.0 | 6.1 | 2.0 | 10.2 | 2.0 | |
| Maui Co, HI | DSH (n = 58) | 36.2 | 29.3 | 12.1 | 1.7 | 0.0 | 3.4 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 13.8 | |
| Brazos Co, TX | DSH (n = 24) | 16.7 | 37.5 | 20.8 | 4.2 | 0.0 | 0.0 | 0.0 | 16.7 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 0.0 | |
| Korea | DSH (n = 9) | 0.0 | 11.1 | 0.0 | 0.0 | 77.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 11.1 | |
| Israel | DSH (n = 9) | 0.0 | 11.1 | 11.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 33.3 | 0.0 | 44.4 | |
| Various, USA | Breeds (n = 32) | 43.8 | 21.9 | 9.4 | 12.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.1 | 9.4 | |
| Clay Co, MO‡ | DSH (n = 24) | 16.7 | 29.2 | 16.7 | 0.0 | 8.3 | 8.3 | 0.0 | 8.3 | 4.2 | 0.0 | 0.0 | 4.2 | 0.0 | 4.2 | |
| DSH Subtotal | DSH (242)§ | 26.9 | 26.0 | 12.8 | 4.5 | 4.5 | 1.7 | 1.7 | 2.9 | 0.0 | 0.0 | 2.9 | 2.5 | 4.5 | 9.1 | |
| Breeds Subtotal | Breeds (101) | 38.6 | 16.8 | 16.8 | 8.9 | 4.0 | 0.0 | 0.0 | 1.0 | 5.0 | 1.0 | 0.0 | 1.0 | 4.0 | 3.0 | |
| Total | DSH+Breeds (343) | 30.3 | 23.3 | 14.0 | 5.8 | 4.4 | 1.2 | 1.2 | 2.3 | 1.5 | .3 | 2.0 | 2.0 | 4.4 | 7.3 | |
| USA Subtotal | DSH (347) | 25.4 | 26.8 | 17.3 | 5.5 | 2.3 | 2.9 | 2.0 | 2.9 | 0.3 | 0.0 | 3.7 | 1.2 | 4.6 | 5.2 | |
| USA Total | DSH+Breeds (448) | 28.3 | 24.6 | 17.2 | 6.3 | 2.7 | 2.2 | 1.6 | 2.5 | 1.3 | 0.2 | 2.9 | 1.1 | 4.5 | 4.7 | |
To allow comparisons between laboratories, trimming of the 180 cat sequences resulted in 99 of 110 samples with complete overlapping mtDNA sequence in random-bred cats and 69 of 70 in breeds.
Clay County dataset includes the evidence sample.
Orange County pedigreed cats and Clay County are not included. DSH implies domestic shorthair.
FIG. 1. Relationship of cats obtained from the suspect’s home.
The source of the novel mitotype—Tabby—was deceased at the time of sampling. MtDNA analysis supported the relationships of the cats as reported by the owner. The mitotype of the related cats matched the hair evidence identified from the hinge lifts of the victim’s pockets and is a unique mitotype in the feline mtDNA CR dataset. Any of the related cats could be the contributor of the evidence hair, however, color, banding pattern and length narrowed the selection to four cats. The names of those four cats were not noted at the time of the evidence collection. Circles represent females and squares represent males. Three additional cats had different mitotypes, see Table 1.
On August 03, 2009, all three laboratories provided DNA testimony at trial. Considering all datasets combined (N = 448), the mitotype of the evidence only matched the mitotypes of cats living in the suspect’s home. The remaining cats from Clay County, Missouri, did not have mitotype Uc1. STR analyses supported the reported relationships, confirming that all mtDNA CR Uc1 mitotypes were due to shared maternal lineage of cats in the household.
The suspect was found guilty of first-degree murder with a later sentencing on October 01, 2009, with additional counts resulting in life in prison without parole plus 120 years. On September 13, 2011, in the Missouri Court of Appeals Western District, the case WD71598 State of Missouri v. Henry Lee Polk, Jr. was affirmed pursuant to rule 30.25(b) and issued per curiam. In the minutes of the Supreme Court of the State of Missouri for December 20, 2011, SC92139 – State of Missouri, Respondent, v. Henry Lee Polk, Jr., Appellant – appellant’s application for transfer from the Missouri Court of Appeals No. WD71598 was denied.
Materials and Methods
Mitochondrial DNA Analysis
In November, 2004, QuestGen Forensics received mounted hairs and envelopes of pulled hair from four reference cats at the suspect’s home (Items 29-2, 29-3, 29-8, and 29-9) along with an envelope containing the evidence hair (Item 21-18). All analyses of the evidence hair (21-18) were completed before the envelopes containing the reference cat hairs were opened. The root end of the evidence hair was cut from the shaft. The shaft portion of the hair was decontaminated of surface DNA prior to DNA isolation [29]. DNA was independently purified from the root end and shaft portion of the hair. Nuclear DNA amplification of feline STR markers from the root end extraction was unsuccessful for the evidence hair; thus mtDNA profiling was performed. Amplification of feline mtDNA CR (GenBank Acc. U20753 bases 16760-221) using a semi-nested amplification of two overlapping sections yielded amplicons from both the root-end DNA extract and the shaft DNA extract as previously described [15, 17].
Following the same SOPs and methodology, hair samples from the four known cats from the suspect’s residence (Items 29-2, 29-3, 29-8, and 29-9) were amplified for the same regions of the feline mtDNA CR. Analysis of the hairs from the two cats owned by the victim (Items 28-1 and 28-2) was performed under the same protocols once they were submitted in October, 2007. MtDNA sequence alignment was performed using the Staden software alignment package [30].
In November 2007, the eight cat hair DNA extracts (evidence hair 21-18 shaft and 21-18 root; knowns 29-2, 29-3, 29-8, 29-9; and elimination samples 28-1, and 28-2) and three extraction negative controls were transferred from QuestGen Forensics to the VGL Forensics Unit for confirmatory analysis with identities masked. Aliquots of the DNA isolates from QuestGen Forensics were amplified by PCR for two regions, HVI (HV1A) and HVII (HV1B) [16, 19], of the feline mtDNA CR using primers: F16268-CCACTATCAGCACCCAAAGC, R16567-CATGCTTAATATTCATGGGGACT and F16796-CAGTCTTCTATGGACCTC, R240-GTCCTGTGGAACAATAGG. DNA sequences were generated using Big Dye Terminator v3.1 and sequencing products were electrophoretically separated on an AB3730 DNA Analyzer (Applied Biosystems). The portions of the mtDNA sequence that overlapped with the QuestGen data, part of the HVII, were identified and compared by sequence alignment to each other and the UC Davis VGL Forensics cat mtDNA dataset using Sequencher® v. 4.8 (GeneCodes Corporation, Ann Arbor, MI).
STR Analysis
Twenty-two autosomal STR markers and the SRY gene were amplified in the DNA from the buccal swabs of the cats collected at the suspect’s residence. The markers included the International Society for Animal Genetics (ISAG) recommended markers for parentage analysis of domestic cats [31] and additional feline-derived STR markers; FCA005, FCA026, FCA008, FCA126, FCA132, FCA201, FCA224, FCA023, FCA290, FCA043, FCA058, FCA77, and FCA090 [6, 32, 33].
Cat mtDNA Dataset
The QuestGen Forensics cat mtDNA CR dataset (N = 180; n = 158 after sequence trimming for comparison) consisted of 110 random-bred cats from animal shelters in Orange County, (Southern) California, and 70 cats of various known breeds. Some of the animal shelter cats may have been “hobby-bred” pedigreed cats but most were of random-bred heritage. The UCD VGL Forensic Unit cat mtDNA dataset (N = 129; n = 78 after removal of cats that were duplicated in the feline genetics research laboratory dataset) included random-bred cats from eastern and western USA and twelve cats from miscellaneous breeds. Additional mtDNA CR profiles (N = 196) from 164 random-bred cats representing other locations of the USA and non-USA cats and 32 cats representing breeds were generated by the feline genetics research laboratory [31, 32]. Cats that represented fancy breeds (n = 101) were combined to produce breed-specific mitotype frequencies for comparison to randomly bred cats (Tables 2 and 3).
Table 3.
Cats breeds represented in the mtDNA CR dataset*
| Breed | No. | Breed | No. |
|---|---|---|---|
| Abyssinian | 6 | Norwegian Forest Cat | 5 |
| American Shorthair | 4 | Persian | 6 |
| Birman | 9 | Russian Blue | 6 |
| British Shorthair | 10 | Scottish Fold | 1 |
| Burmese | 2 | Siamese | 8 |
| Chartreux | 4 | Siberian | 4 |
| Egyptian Mau | 3 | Singapura | 2 |
| Havana Brown | 1 | Sokoke | 2 |
| Japanese Bobtail | 5 | Turkish Angora | 5 |
| Korat | 11 | Turkish Van | 5 |
| Maine Coon | 5 | Total | 101 |
Includes data generated by both QuestGen (n = 69) and the UC Davis research laboratory (n = 32).
The overall composition of the ~408 bp cat mtDNA CR dataset was 448 cats, consisting of 347 randomly bred cats (Table 2) and 101 individuals representing breeds (Table 3), excluding cats from Clay County, Missouri. After the adjudication of the trial, all sequences from the different datasets were compared to a consensus “Sylvester” cat CR mtDNA reference sequence to define the DNA variants and establish mitotype nomenclature [21, 22] and to estimate mitotype frequencies.
Results of the Combined Cat mtDNA CR dataset
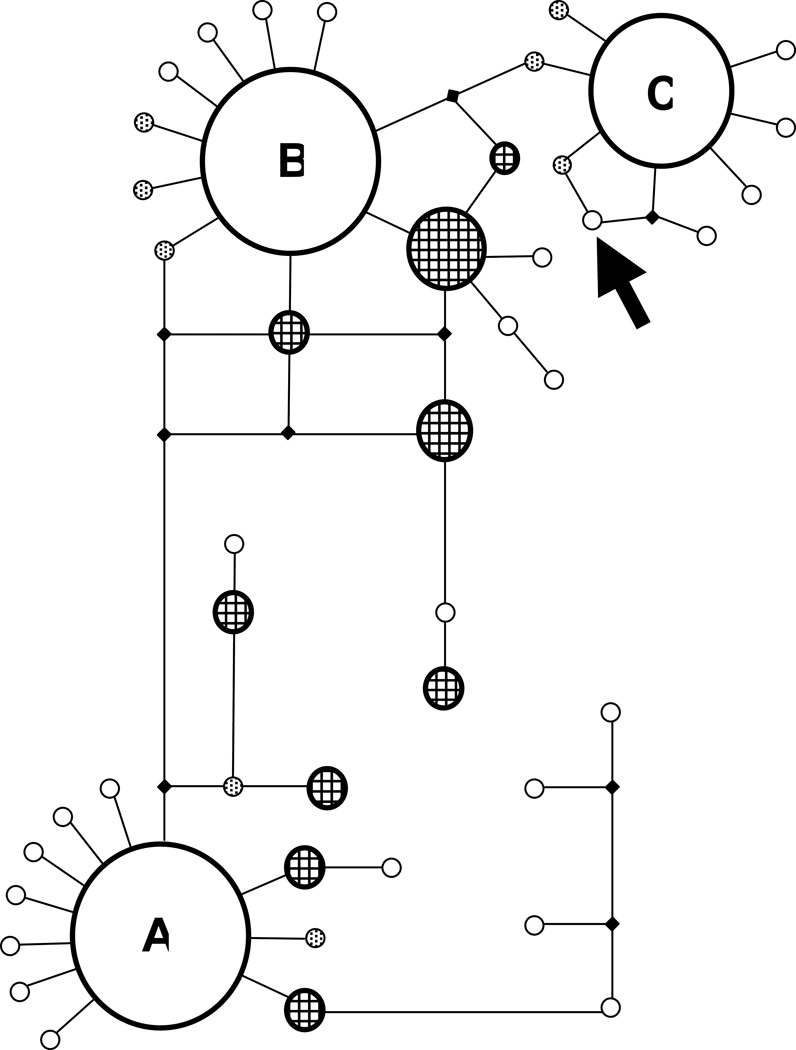
The sizes of the datasets for the three laboratories vary slightly from the reports provided at the hearings for these mitotype analyses. Only sequences that had complete overlap across the region to define the mitotypes were included and duplicate cats between the laboratories were excluded. The frequencies of the mtDNA CR mitotypes in the three datasets are presented in Table 2. Regardless of the individual dataset, the composition of the mtDNA mitotypes is comparable. The DNA variants of the mitotypes are published [21]. Three mitotypes (A, B, C) are common, representing 67.6% of cats (Fig. 2, Table 2). An additional 20.6% of cats are represented by an additional nine minor mitotypes D – L. Rare mitotypes, implying those with less than 1% of the total sample set (n ≤ 3), comprise 4.4%. Unique sequences (i.e., found in only one cat in the dataset) account for 7.3% of cats. The probability of a cat having an uncommon mtDNA CR mitotype is ~11% in the USA. The cats representing breeds from the USA and cats from Clay County, Missouri, had a similar mitotype distribution to other regions of the USA. The mitotype of the evidence hair and the maternally related cats was not found in the Missouri dataset or the combined USA datasets. Considering the likelihood ratio from the combined UC Davis datasets of non-overlapping cats (N = 376), 1/376 = 0.00266, implies a 0.27% chance of finding an unrelated cat with the same mitotype. Applying a 95% confidence interval, where 1-α1/N (α = 0.05, N = 376), the upper bound probability of this mitotype occurring in a random population of unrelated cats in 0.79%.
FIG. 2. Minimal spanning network of cat mtDNA CR mitotypes.
Circle sizes represent relative frequency of each mitotype. Major mitotypes A, B and C are indicated. Minor mitotypes (n = 9) are indicated with crosshatches, stippled circles indicate rare mitotypes (n = 8) and open circles are unique mitotypes (n = 26). The arrow indicates the evidence sample – a novel mtDNA mitotype. The CR mitotype from the evidence is distinct from the next most similar mitotype, C1, by a single mutation and two mutations from major mitotype C. Cats represent USA random bred and breeds.
Discussion
Transferred pet hair is perceived as a nuisance, but has proven to be a benefit to the forensic science community as crime scene evidence. In the case of the State of Missouri v. Henry L. Polk, Jr., cat hair provided corroborative evidence implicating the suspect. Transferred cat hair from the victim’s pocket was excluded as originating from the victim’s cats, but could not be excluded from cats living with the suspect. When evidence provides exclusionary information, dataset size is of limited concern. Indeed, when considering exclusion, published forensic cases have used as few as four animals [34].
A variety of issues can lead to the inadmissibility of evidence at trial, notably issues that raise doubts about the general acceptance of the technique or the validity of the underlying statistics [35, 36]. Although non-human DNA evidence is increasingly used in forensic investigations, in the case of the State of Missouri v. Henry L. Polk, Jr., the admissibility of the mitochondrial DNA was questioned—particularly because the evidence supported inclusion, not exclusion. In September 2007, the court order issued (Case No: 7CR104003803-01, 7CR105000217-01) brought into question “the size of data bases required for animal DNA profiling and the documentation of DNA markers”. This order cites an appeal from the State of Washington (Washington v. Leuluaialii) that overruled the admission of STR canine DNA evidence in a case alleging animal cruelty and two counts of aggravated murder [37]. This concern was raised even though canine evidence using the same dataset had been accepted subsequent to the Washington appeal in several states including; Georgia, Nevada, Arizona, Michigan, Iowa, South Carolina, Illinois, California, Wisconsin, Indiana, and Delaware [38–41]. Although the Washington trial court’s admission of canine evidence was ruled harmless and the convictions upheld, this appeal highlights the concern over the statistical significance and appropriate composition of animal DNA datasets for consideration in criminal proceedings.
The primary concern expressed in the trial judge’s initial ruling in the Polk case was that the mtDNA sequence comparisons could not be considered generally accepted due to the small number of cats in the dataset. This ruling inherently implied that a limited database may not address concerns regarding sub-structuring and sampling saturation, which was unknown for cats at the time. However, no standard size for a dataset had been a priori determined to be “adequate” for any population or species, although the concerns have been scientifically examined [42–44]. In addition, the 2007 Frye hearing ruling was based on the previous ruling that pertained to a canine STR dataset that involved the same DNA laboratory service, not the published scientific studies on sub-structuring and mtDNA mitotype saturation points. At the time, many canine datasets with known and unknown breed compositions of ~ 100 individuals had been developed for the dog mtDNA CR [45–48], and the initial cat dataset was of comparable size with 180 cats. Expert testimony could have been acquired concerning cat population dynamics and sub-structuring at the time. Although based on STR data, the work of Lipinski et al. [49] and Menotti-Raymond et al. [50] became publicly available on December 03, 2007, implying that data had already been peer-reviewed and commentary on cat population dynamics could have been obtained. Data was also available as to the composition of pet cats in regards to representing a breed or a randomly bred individual. Only ~10 – 15% of the feline patients at highly specialized referral clinics, such as the UC Davis Veterinary Medical Teaching Hospital, are represented by pedigreed cats [51]. This suggests a vast majority of cats are randomly bred in the USA, which is consistent with other surveys of pet ownership [1, 2]. The cats in question for this case were randomly bred, thus, the representative database needed to be comprised of non-pedigreed cats, which was considered in the mitotype comparisons.
Population stratification or sub-structuring has been a concern and debated in statistical analyses in human DNA profiling [52]. Knowledge of the population dynamics specific to a species helps anticipate whether a smaller versus a larger dataset would be required and if the level of population stratification is a concern. Breeds may have significant founder effects, thus are always subject to sub-structuring. DNA mutation rate estimates of mtDNA CR DNA are also different between species [52–58], leading to higher or lower exclusionary power for a given mitochondrial region. The dog dataset was anticipated to require a smaller sampling compared to humans since the domestic dog mtDNA CR contains fewer polymorphisms [59] and mitotypes and provides less power of exclusion than humans [60]. As noted in earlier studies on mtDNA mitotype saturation [42–44, 61], different animal populations, including humans, have different population dynamics. Population structures are particularly evidenced by different but acceptable levels of inbreeding and outcrossing as well as different migration rates. The less polymorphic right domain of the dog mtDNA CR was predicted to require a 350 – 450 sample size to reach a 1 – 5% cut-off level of mitotype or polymorphic site saturation, which is less than half the sample size required for humans in a comparable region [44].
Cats have a far more recent evolutionary history compared to dogs and less overall mtDNA mitotype diversity [49, 62]. Thus, at the time of the Frye hearing, by logical assumption the sub-structuring of random-bred cat populations should have been anticipated to be minimal. Therefore, sampling in any state in the USA should be fairly representative of all USA cats. STR analyses show insignificant sub-structuring of random-bred cats in the USA [49], although cat populations have not been examined as widely as human populations. Additionally, linkage disequilibrium estimates in cats suggest less intense selection as compared to dogs [63, 64], and recent publications of the cat mtDNA CR datasets support these findings [21, 22]. Indeed, cat breeds seem to have been founded by individuals with multiple mtDNA types [21]. However, these aspects of cats had not been all peer-reviewed and published at the time of the hearing. Although they were available via expert opinion, the conservative approach taken by the judge seems warranted.
Recently, a saturation analysis for the cat mtDNA CR dataset has been conducted (companion paper, Grahn et al.). This study considered randomly bred cat datasets from around the world, ranging from 64 – 514 individual cats per population, including USA populations and sub-populations. These datasets included the cat mtDNA CR sequences considered in the criminal proceedings. The USA population of cats (n = 514) required 50 and 110 cats to meet the 5% and 1% mtDNA CR saturation cut-off values. Therefore, retrospectively, the initial dataset used for comparison in this criminal case (n = 180), even when not considering the inclusion of the 70 pedigreed cats, was a sufficient dataset for consideration of the cat mtDNA CR mitotype frequencies. As predicted, these estimates for saturation are lower than the dog mtDNA CR.
To further consider the potential of local population sub-structuring and the analysis of cats that could have been the contributor versus a general dataset, additional samples from random-bred cats within the area of the crime scene in Missouri (n = 24) and additional cats associated with the evidence were collected. The descriptive colorations of cat hairs are subjective and can appear different due to lighting and seasonal variation. No standard for hair coloration terminology has been defined for the cat. Thus, the DNA analysis was extended to all cats in the household to alleviate concerns of subjective errors in the collection of the reference hairs based on coloration. The novel mitotype found in hairs 21-18, 29-2, and 29-3 was also observed in eight of the eleven cats from the suspect’s home, a finding consistent with their reported descent from a single female. Genotyping of the eight cats confirmed that maternal lineage. The novel mitotype of the evidence was not observed amongst the 23 random-bred cats from Missouri. The Missouri cats reflected a mitotype distribution that is comparable to random-bred cats in both datasets. These results further support the rarity of the evidence hair.
Current domestic cat mtDNA data does not support significant sub-structuring within domestic cat populations within the US [21]. However, cats have not been examined to the extent of human populations and less dramatic sub-structuring likely occurs for cats due to founder effects. The distribution of mtDNA CR mitotype frequencies was similar in cats from all regions of the USA including California, Hawaii, Missouri, and New York. Expansion of the cat mtDNA CR dataset from 180 cats to > 500 cats and examination of local populations had no effect on the accuracy of the DNA data interpretation, reaffirming the original testimony. Fortunately, the implicating mitotype in this case was rare. Therefore, the issue of population sub-structuring was not a significant concern. Saturation studies now suggest the USA cat mtDNA CR dataset is sufficient for forensic interpretations. To date, over 1,300 DNA sequences from the same mtDNA region have been generated from random-bred cats from around the world and fancy breed cats [21]. The dataset is available to the forensic community and the consensus “Sylvester” reference sequence is published for standardization of future cat mtDNA CR studies [21, 22]. The use of cat mitochondrial DNA analysis has been accepted in court and has been shown to be reliable and of value in the forensic analysis of shed cat hair. However, when considering mtDNA, especially when multiple suspect cats are in the same household and mitotypes are not unique, multiple comparison tests, such as Bonferroni corrections or Bayes factor of calculation, should be considered.
Highlights.
First cat mtDNA control region data supports murder conviction
Shed cat hair can be valuable trace evidence in criminal investigations
Cat genetic databases are sufficient and available for evidence admissibility
STR and mtDNA genetic data can be obtained from shed cat hairs
Morphological analysis of hair, including color, can predict phenotype of the contributing cat.
Acknowledgements
We appreciate comments to the early development of the manuscript by Christy R. Tarditi, B.S., and technical support of Elyse Hammer, B.S. Funding support for the background dataset sets was provided to Dr. Lyons from the National Institutes of Health, NIH-NCRR R24 RR016094, and to Dr. Lyons and the †VGL by the University of California, Davis, Forensic Science Graduate Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have no conflicts of interest to declare.
References
- 1.APPMA. National Pet Owner’s Survey. Greenwich, CT: American Pet Product Manufacturing Association; 2006. [Google Scholar]
- 2.AVMA. US Pet Ownership and Demographics Sourcebook. Schaumburg, IL: American Veterinary Medical Association; 2007. [Google Scholar]
- 3.Dachs J, McNaught IJ, Robertson J. The persistence of human scalp hair on clothing fabrics. Forensic Sci. Int. 2003;138:27–36. doi: 10.1016/j.forsciint.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 4.D’Andrea F, Frides F, Coquoz R. Preliminary Experiments on the Transfer of Animal Hair During Simulated Criminal Behavior. Journal of Forensic Science. 1998;43:1257–1258. [Google Scholar]
- 5.Deedrick DW. Hairs, Fibers, Crime, and Evidence. FBI Forensic Science Communications. 2000;2 [Google Scholar]
- 6.Menotti-Raymond MA, David VA, O’Brien SJ. Pet cat hair implicates murder suspect. Nature. 1997;386:774. doi: 10.1038/386774a0. [DOI] [PubMed] [Google Scholar]
- 7.Allen M, Engstrom AS, Meyers S, Handt O, Saldeen T, von Haeseler A, Paabo S, Gyllensten U. Mitochondrial DNA sequencing of shed hairs and saliva on robbery caps: sensitivity and matching probabilities. Journal Forensic Science. 1998;43:453–464. [PubMed] [Google Scholar]
- 8.Bar W, Brinkmann B, Budowle B, Carracedo A, Gill P, Holland M, Lincoln PJ, Mayr W, Morling N, Olaisen B, Schneider PM, Tully G, Wilson M. DNA Commission of the International Society for Forensic Genetics: guidelines for mitochondrial DNA typing. International Journal Legal Medicine. 2000;113:193–196. doi: 10.1007/s004140000149. [DOI] [PubMed] [Google Scholar]
- 9.Bar W, Brinkmann B, Budowle B, Carracedo A, Gill P, Holland M, Lincoln PJ, Mayr W, Morling N, Olaisen B, Schneider PM, Tully G, Wilson M. Guidelines for mitochondrial DNA typing. DNA Commission of the International Society for Forensic Genetics. Vox Sang. 2000;79:121–125. doi: 10.1159/000031227. [DOI] [PubMed] [Google Scholar]
- 10.Carracedo A, Bar W, Lincoln P, Mayr W, Morling N, Olaisen B, Schneider P, Budowle B, Brinkmann B, Gill P, Holland M, Tully G, Wilson M. DNA commission of the international society for forensic genetics: guidelines for mitochondrial DNA typing. Forensic Sci. Int. 2000;110:79–85. doi: 10.1016/s0379-0738(00)00161-4. [DOI] [PubMed] [Google Scholar]
- 11.Holland MM, Parsons TJ. Mitochondrial DNA Sequence Analysis: Validation and Use for Forensic Casework. Forensic Sci. Rev. 1999;11:21–50. [PubMed] [Google Scholar]
- 12.Wilson MR, DiZinno JA, Polanskey D, Replogle J, Budowle B. Validation of mitochondrial DNA sequencing for forensic casework analysis. Int. J. Legal Med. 1995;108:68–74. doi: 10.1007/BF01369907. [DOI] [PubMed] [Google Scholar]
- 13.Wilson MR, Holland MM, Stoneking M, DiZinno JA, Budowle B. Guidelines for the use of mitochondrial DNA sequencing in forensic science. Crime Laboratory Digest. 1993;20:68–77. [Google Scholar]
- 14.Halverson JL, Basten C. Forensic DNA identification of animal-derived trace evidence: tools for linking victims and suspects. Croat. Med. J. 2005;46:598–605. [PubMed] [Google Scholar]
- 15.Halverson JL, Lyons LA. Forensic DNA Identification of Feline Hairs: Casework and a Mitochondrial Database. Proc. Am. Acad. Forensic Sci. 2004:B150. X. [Google Scholar]
- 16.Lopez JV, Cevario S, O’Brien SJ. Complete nucleotide sequences of the domestic cat (Felis catus) mitochondrial genome and a transposed mtDNA tandem repeat (Numt) in the nuclear genome. Genomics. 1996;33:229–246. doi: 10.1006/geno.1996.0188. [DOI] [PubMed] [Google Scholar]
- 17.Tamada T, Kurose N, Masuda R, Genetic diversity in domestic cats Felis catus of the Tsushima Islands. based on mitochondrial DNA cytochrome b and control region nucleotide sequences. Zoolog. Sci. 2005;22:627–633. doi: 10.2108/zsj.22.627. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, Yang ZH, Liang WB, Zhou B, Zhang L. [Species identification by multiplex amplifying mtDNA-HV I, HV II and cytb regions] Sichuan Da Xue Xue Bao Yi Xue Ban. 2006;37:787–789. [PubMed] [Google Scholar]
- 19.Eizirik E, Bonatto SL, Johnson WE, Crawshaw PG, Jr., Vie JC, Brousset DM, O’Brien SJ, Salzano FM. Phylogeographic patterns and evolution of the mitochondrial DNA control region in two neotropical cats (Mammalia, felidae) Journal Molecular Evolution. 1998;47:613–624. doi: 10.1007/pl00006418. [DOI] [PubMed] [Google Scholar]
- 20.Fridez F, Rochat S, Coquoz R. Individual identification of cats and dogs using mitochondrial DNA tandem repeats? Science Justice. 1999;39:167–171. doi: 10.1016/S1355-0306(99)72042-3. [DOI] [PubMed] [Google Scholar]
- 21.Grahn RA, Kurushima JD, Billings NC, Grahn JC, Halverson JL, Hammer E, Ho CK, Kun TJ, Levy JK, Lipinski MJ, Mwenda JM, Ozpinar H, Schuster RK, Shoorijeh SJ, Tarditi CR, Waly NE, Wictum EJ, Lyons LA. Feline non-repetitive mitochondrial DNA control region database for forensic evidence. Forensic Sci. Int. Genet. 2011;5:33–42. doi: 10.1016/j.fsigen.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarditi CR, Grahn RA, Evans JJ, Kurushima JD, Lyons LA. Mitochondrial DNA sequencing of cat hair: an informative forensic tool. J. Forensic Sci. 2011;56(Suppl. 1):S36–S46. doi: 10.1111/j.1556-4029.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bisbing RE. The Forensic Identification and Association of Human Hair. Forensic Science Handbook. 1982;1:185–221. [Google Scholar]
- 24.Harding H, Rogers G. In: Physiology and Growth of Human Hair. Robertson J, editor. London, Taylor and Francis: Forensic Examination of Hair; 1999. pp. 1–62. [Google Scholar]
- 25.Hendricks WH, Tarttelin MF, Moughan PJ. Seasonal Hair Growth in the Adult Domestic Cat (Felis catus) Comparative Biochemistry Physiology. 1996;116A:29–35. [Google Scholar]
- 26.Moore JE. A key to the Identification of Animal Hairs. Journal of Forensic Science Society. 1988;28:335–339. [Google Scholar]
- 27.Brunner H, J B. Coman, The Identification of Mammalian Hair. Melbourne: Inkata Press Proprietary Limited; 1974. [Google Scholar]
- 28.Hicks JW. Microscopy of Hairs: A Practical Guide and Manual. Washington D.C.: Federal Bureau of Investigation Laboratory; 1077. [Google Scholar]
- 29.Jehases E, Gilissen A, Cassiman J, Decorte R. Evaluation of a decontamination protocol for hair shafts for mtDNA sequencing. Forensic Journal International. 1998;94:65–71. doi: 10.1016/s0379-0738(98)00052-8. [DOI] [PubMed] [Google Scholar]
- 30.Staden R. The Staden sequence analysis package. Mol. Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 31.Lipinski MJ, Amigues Y, Blasi M, Broad TE, Cherbonnel C, Cho GJ, Corley S, Daftari P, Delattre DR, Dileanis S, Flynn JM, Grattapaglia D, Guthrie A, Harper C, Karttunen PL, Kimura H, Lewis GM, Longeri M, Meriaux JC, Morita M, Morrin-O’donnell R C, Niini T, Pedersen NC, Perrotta G, Polli M, Rittler S, Schubbert R, Strillacci MG, Van Haeringen H, Van Haeringen W, Lyons LA. An international parentage and identification panel for the domestic cat (Felis catus) Anim. Genet. 2007;38:371–377. doi: 10.1111/j.1365-2052.2007.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menotti-Raymond M, David VA, Lyons LA, Schaffer AA, Tomlin JF, Hutton MK, O’Brien SJ. A genetic linkage map of microsatellites in the domestic cat (Felis catus) Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- 33.Menotti-Raymond M, David VA, Stephens JC, Lyons LA, O’Brien SJ. Genetic individualization of domestic cats using feline STR loci for forensic applications. Journal Forensic Science. 1997;42:1039–1051. [PubMed] [Google Scholar]
- 34.Schneider PM, Seo Y, Rittner C. Forensic mtDNA hair analysis excludes a dog from having caused a traffic accident. International Journal Legal Medicine. 1999;112:315–316. doi: 10.1007/s004140050257. [DOI] [PubMed] [Google Scholar]
- 35.Page M, Taylor J, Blenkin M. Forensic identification science evidence since Daubert: Part I--A quantitative analysis of the exclusion of forensic identification science evidence. J. Forensic Sci. 56:1180–1184. doi: 10.1111/j.1556-4029.2011.01777.x. [DOI] [PubMed] [Google Scholar]
- 36.Page M, Taylor J, Blenkin M. Forensic identification science evidence since Daubert: Part II--judicial reasoning in decisions to exclude forensic identification evidence on grounds of reliability. J. Forensic Sci. 56:913–917. doi: 10.1111/j.1556-4029.2011.01776.x. [DOI] [PubMed] [Google Scholar]
- 37.118 Wash. App. 70, 77P.3d 1192.
- 38.The Court affirmed an order releasing cat hairs for DNA testing to the doctor who performed DNA testing on canine hairs in the defendants’ murder case. Illinois v. Michael Slover, Jr. et al, App. Ct. Illinois, 4th District, NO. 4-02-0892.
- 39.An expert testified that dog DNA from feces found on a pair of shoes and the feces from the ground at the scene of a triple murder were likely from the same animal. Stroud v. Indiana, Indiana Supreme Court, No. 71S00-0011-DP-00642.(May 25, 2004).
- 40.MtDNA testing of dog hairs ruled not novel and admissible California v. Westerfield, Superior Court of California for San Diego County. (July 01, 2002).
- 41.MtDNA testing of dog hairs used in a murder case – no admissibility issue, Huck v. Florida , District of Appeals of Florida, 5 th District, No. 5D03-1906.(July 16, 2004).
- 42.Pereira L, Cunha C, Amorim A. Predicting sampling saturation of mtDNA haplotypes: an application to an enlarged Portuguese database. Int. J. Legal Med. 2004;118:132–136. doi: 10.1007/s00414-003-0424-1. [DOI] [PubMed] [Google Scholar]
- 43.Pfeiffer H, Forster P, Ortmann C, Brinkmann B. The results of an mtDNA study of 1,200 inhabitants of a German village in comparison to other Caucasian databases and its relevance for forensic casework. Int. J. Legal Med. 2001;114:169–172. doi: 10.1007/s004140000165. [DOI] [PubMed] [Google Scholar]
- 44.Webb K, Allard M. Assessment of minimum sample sizes required to adequately represent diversity reveals inadequacies in datasets of domestic dog mitochondrial DNA. Mitochondrial DNA. 2010;21:19–31. doi: 10.3109/19401730903532044. [DOI] [PubMed] [Google Scholar]
- 45.Angleby H, Savolainen P. Forensic informativity of domestic dog mtDNA control region sequences. Forensic Sci. Int. 2005;154:99–110. doi: 10.1016/j.forsciint.2004.09.132. [DOI] [PubMed] [Google Scholar]
- 46.Gundry RL, Allard MW, Moretti TR, Honeycutt RL, Wilson MR, Monson KL, Foran DR. Mitochondrial DNA analysis of the domestic dog: control region variation within and among breeds. J. Forensic Sci. 2007;52:562–572. doi: 10.1111/j.1556-4029.2007.00425.x. [DOI] [PubMed] [Google Scholar]
- 47.Himmelberger AL, Spear TF, Satkoski JA, George DA, Garnica WT, Malladi VS, Smith DG, Webb KM, Allard MW, Kanthaswamy S. Forensic utility of the mitochondrial hypervariable region 1 of domestic dogs, in conjunction with breed and geographic information. Journal Forensic Science. 2008;53:81–89. doi: 10.1111/j.1556-4029.2007.00615.x. [DOI] [PubMed] [Google Scholar]
- 48.Wetton JH, Higgs JE, Spriggs AC, Roney CA, Tsang CS, Foster AP. Mitochondrial profiling of dog hairs. Forensic Sci. Int. 2003;133:235–241. doi: 10.1016/s0379-0738(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 49.Lipinski MJ, Froenicke L, Baysac KC, Billings NC, Leutenegger CM, Levy AM, Longeri M, Niini T, Ozpinar H, Slater MR, Pedersen NC, Lyons LA. The ascent of cat breeds: Genetic evaluations of breeds and worldwide random-bred populations. Genomics. 2008;91:12–21. doi: 10.1016/j.ygeno.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menotti-Raymond M, David VA, Pflueger SM, Lindblad-Toh K, Wade CM, O’Brien S, Johnson WE. Patterns of molecular genetic variation among cat breeds. Genomics. 2008;91:1–11. doi: 10.1016/j.ygeno.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 51.Louwerens M, London CA, Pedersen NC, Lyons LA. Feline lymphoma in the post-feline leukemia virus era. J. Vet. Intern. Med. 2005;19:329–335. doi: 10.1892/0891-6640(2005)19[329:flitpl]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 52.Buckleton JS, Krawczak M, Weir BS. The interpretation of lineage markers in forensic DNA testing. Forensic Sci. Int. Genet. 2011;5:78–83. doi: 10.1016/j.fsigen.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoelzel AR, Lopez JV, Dover GA, O’Brien SJ. Rapid evolution of a heteroplasmic repetitive sequence in the mitochondrial DNA control region of carnivores. Journal Molecular Evolution. 1994;39:191–199. doi: 10.1007/BF00163807. [DOI] [PubMed] [Google Scholar]
- 54.Lopez JV, Culver M, Stephens JC, Johnson WE, O’Brien SJ. Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Mol. Biol. Evol. 1997;14:277–286. doi: 10.1093/oxfordjournals.molbev.a025763. [DOI] [PubMed] [Google Scholar]
- 55.Savolainen P, Lundeberg J. Forensic evidence based on mtDNA from dog and wolf hairs. Journal Forensic Science. 1999;44:77–81. [PubMed] [Google Scholar]
- 56.Savolainen P, Rosen B, Holmberg A, Leitner T, Uhlen M, Lundeberg J. Sequence analysis of domestic dog mitochondrial DNA for forensic use. Journal Forensic Science. 1997;42:593–600. [PubMed] [Google Scholar]
- 57.Sigurgardottir S, Helgason A, Gulcher JR, Stefansson K, Donnelly P. The mutation rate in the human mtDNA control region. Am. J. Hum. Genet. 2000;66:1599–1609. doi: 10.1086/302902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 59.Parsons TJ, Muniec DS, Sullivan K, Woodyatt N, Alliston-Greiner R, Wilson MR, Berry DL, Holland KA, Weedn VW, Gill P, Holland MM. A high observed substitution rate in the human mitochondrial DNA control region. Nat. Genet. 1997;15:363–368. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- 60.Carter RW. Mitochondrial diversity within modern human populations. Nucleic Acids Res. 2007;35:3039–3045. doi: 10.1093/nar/gkm207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfeiffer H, Brinkmann B, Huhne J, Rolf B, Morris AA, Steighner R, Holland MM, Forster P. Expanding the forensic German mitochondrial DNA control region database: genetic diversity as a function of sample size and microgeography. Int. J. Legal Med. 1999;112:291–298. doi: 10.1007/s004140050252. [DOI] [PubMed] [Google Scholar]
- 62.Driscoll CA, Menotti-Raymond M, Roca AL, Hupe K, Johnson WE, Geffen E, Harley EH, Delibes M, Pontier D, Kitchener AC, Yamaguchi N, O’Brien S J, Macdonald DW. The Near Eastern origin of cat domestication. Science. 2007;317:519–523. doi: 10.1126/science.1139518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, Schaffer AA, Agarwala R, Narfstrom K, Murphy WJ, Giger U, Roca AL, Antunes A, Menotti-Raymond M, Yuhki N, Pecon-Slattery J, Johnson WE, Bourque G, Tesler G, O’Brien SJ. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alhaddad H, Khan R, Grahn RA, Gandolfi B, Mullikin JC, Cole SA, Gruffydd-Jones TJ, Haggstrom J, Lohi H, Longeri M, Lyons LA. Extent of linkage disequilibrium in the domestic cat Felis silvestris catus, and its breeds. PLoS One. 2013;8:e53537. doi: 10.1371/journal.pone.0053537. [DOI] [PMC free article] [PubMed] [Google Scholar]