Abstract
Acinetobacter baumannii is an opportunistic pathogen that exists in hospital environments. The emergence of multidrug resistant A. baumannii (MDRAB) has been reported worldwide. It is necessary to find a novel and effective treatment for MDRAB infection. In this study, three bacteriophages, designated as ØABP-01, ØABP-02 and ØABP-04 were selected for analysis. Transmission electron microscopy showed that bacteriophage ØABP-01 belonged to the Podoviridae family and bacteriophage ØABP-02 and ØABP-04 are classified into the family Myoviridae. ØABP-01 had the widest host range. ØABP-01, ØABP-02 and ØABP-04 exhibited a latent period of 15, 20 and 20 min. The burst sizes of the three bacteriophages were 110, 120 and 150 PFU/cell. DNA restriction analysis using EcoRI, HindIII, PstI, SphI, BamHI and SmaI showed different DNA fragment patterns between the three bacteriophages. ØABP-01 and ØABP-04 was positive for the endolysin gene as determined by PCR. In conclusion, bacteriophage ØABP-01 showed broad host-specificity, good lytic activity and a short latency period, making it an appropriate candidate for studying the control and diagnosis associated with MDRAB infections.
Keywords: Acinetobacter baumannii, Bacteriophage, Endolysin
Introduction
Acinetobacter baumannii is a Gram negative coccobacillus which is ubiquitous in the hospital setting. A. baumannii causes a diverse range of infections such as ventilator-associated pneumonia, skin and soft-tissue infections, urinary tract infection, wound and blood stream infection. It is an opportunistic pathogen that is a major cause of nosocomial infection which has a high mortality rate [1]. Emergence of multidrug resistant A. baumannii (MDRAB) has been reported worldwide [2–4]. The increased incidence of antibiotic resistance has led to the search for an alternative antimicrobial treatment. Phage therapy is one potential candidate for the treatment of multidrug resistant bacteria [5]. Clinical trials of bacteriophages and their derivatives as potential alternative agents for controlling multi drug resistance infection have been described in various bacterial pathogens [6–9]. Bacteriophages are able to replicate in the host cell and produce endolysin to lyse the host cell. Endolysin is a phage peptidoglycan hydrolases produced in the lytic cycle that degrades the bacterial cell wall [10, 11]. Phage endolysins have been studied in a variety of pathogenic bacteria including A. baumannii for theirs antibacterial activity [11–13]. In the past three years, A. baumannii bacteriophages and endolysin have been isolated and characterized [14–24]. However, there are geographic differences in A. baumannii host strains. Depending on their host specificity, bacteriophages and phage endolysin that have been isolated in one place may not be effective in another area. Thus, the aim of this study was to isolate and characterize the A. baumannii bacteriophages from the waste water treatment plant in two hospitals in Thailand.
Materials and Methods
Bacterial Strains and Media
Eleven clinical MDRAB isolates obtained from Buddhachinaraj hospital, Phitsanulok, Thailand were used for screening of A. baumannii bacteriophages [25]. All A. baumannii were grown in Luria–Bertani broth (LB) or Luria–Bertani Agar (LBA).
Bacteriophage Isolation
Phages were isolated from the inlets and outlets of wastewater treatment plants in two hospitals in Phitsanulok province. Samples were collected three times at monthly intervals from October–December 2010. Samples were centrifuged and filtered. Then, 5 ml each of the filtered supernatants were mixed with 5 ml double strength broth containing overnight culture A. baumannii. After 48 h growth at 25 °C, the culture was centrifuged and filtered through a membrane filter (0.22 µm pore size, Sartorius Stedim biotech, Germany). The presence of lytic phage in the filtrate was examined by using the double layer method [26].
Phage Enrichment and Purification
Single plaques was cut out from the soft agar layer, diluted in liquid broth, centrifuged at 12,000 rpm and supernatant was used for phage propagation. Phage was propagated using liquid culture of A. baumannii hosts and isolated phages from supernatant. Hundred millilitre of bacterial host cells (OD600 of 0.3) was added with isolated phages at a multiplicity of infection (MOI) of 0.5 and incubated at 37 °C until complete lysis. After that, chloroform was added and bacterial debris was pelleted by centrifugation at 4,000 g for 10 min. The supernatants were passed through a membrane filter (0.22 µm pore size, Sartorius Stedim biotech, Germany)). Three repeated rounds of complete lysis were performed and the filtrates were subjected to the double layer method [26].
Host Range Analysis
Host range analysis was determined by spot test using 11 A. baumannii clinical isolates [25], A. baumannii ATCC19606 and 14 strains of different bacterial species (Enterococcus faecalis, Klebsiella pneumoniae, Proteus mirabilis, Proteus vulgaris, Pseudomonas fluorescens, Salmonella typhi, Salmonella typhimurium, Shigella flexneri, Vibrio parahaemolyticus, Bacillus cereus, Bacillus subtilis, Enterobacter aerogenes, Escherichia coli and Pseudomonas aeruginosa). 100 ul of overnight bacterial host cultures (108–109 CFU/ml) were added to 2.5 ml of 0.7 % soft agar at 45 °C. This mixture was poured onto 2 % solid agar plate to make double layer agar plates. After solidification, 5 µl aliquots of phage suspension (1.0 × 108 PFU) were spotted on the lawn of host bacteria. Plates were dried and incubated at 37 °C for 24 h. Clearance zone indicating lysis at the spot of phage inoculation implied that the host was sensitive.
One Step Growth
Cells of A. baumannii were harvested by centrifugation and then resuspended in fresh LB broth in a concentration of 1 × 109 CFU/ml. Phages were added at an MOI of 0.001 and allowed to absorb for 30 min at 4 °C. The mixtures were then centrifuged at 12,000×g for 10 min and the pellet was resuspended in 20 ml of LB broth. Samples were taken every 5 min over a period of 60 min and immediately assayed for plaque titre by the method described earlier. Latent period, burst time and burst size were calculated from the one-step growth curve. Each of the above experiments was repeated three times with triplicate samples.
Morphology of Phage
A drop of phage suspension (1012 PFU/ml) was applied to the surface of a formvar-coated grid and negatively stained with 0.5 % uranyl acetate for 3–5 min. After drying, the preparations were observed in a transmission electron microscope (Philips, Oregon, USA).
Analysis of Bacteriophage Nucleic Acid
Bacteriophage nucleic acid was extracted as described by Kutter and Sulakvelidze [27]. Phage lysate was treated with DNase I and purified using PEG8000. Purified phage particles were treated with SDS (10 %) at 65 °C for 15 min. An equal volume of phenol–chloroform (1:1) was added to remove proteinaceous materials. The extraction was repeated twice, and the nucleic acids were precipitated with 0.1 volume of 3 M sodium acetate and 1 volumes of isopropanol. Phage DNA was resuspended in TE buffer. Precipitation was repeated. The final pellet was washed twice with 70 % ethanol, air dried, and then resuspended in TE buffer. One μg samples of phage DNA was cut with restriction enzymes EcoRI, HindIII, PstI, SphI, BamHI and SmaI and analyzed by electrophoresis in 1 % agarose gel containing 0.5 ug/ml ethidium bromide. Pulsed-field gel electrophoresis was performed as described previously [28].
Detection of Endolysin Gene in Bacteriophage Genome
The sequence of endolysin was obtained from the GenBank Database of the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Primers specific for the endolysin gene (forward: 5′ATGATTCTGACTAAAGACGGATTTAGTATT3′ and reverse: 5′CTATAAGCTCCGTAGAGCACGTTC3′) were designed using Biology Workbench (http://workbench.sdsc.edu/). PCR was performed in a DNA thermal cycler using phage DNA ØABP-01, ØABP-02 or ØABP-04 genomic DNA as a template. A PCR was carried out: 2 min at 94 °C, 30 cycles of 20 s at 94 °C, 20 s at 63 °C, and 30 s at 72 °C, followed by 5 min at 72 °C. PCR products were analyzed by electrophoresis in 1 % agarose gel containing 0.5 µg/ml ethidium bromide and sequenced using Applied Biosystems. Phylogenetic analysis was constructed using Molecular Evolutionary Genetics Analysis software (MEGA version 5.05).
Nucleotide Sequence Accession Number
The nucleotide sequences of two endolysin genes have been deposited to the GenBank Database under accession no. KF548002 and KF548003.
Results and Discussion
Phage Isolation, Host Range Analysis and Morphology
In this study, we used previously characterized A. baumannii strains to isolate bacteriophage from waste water treatment plants [25]. Fifty-four isolates of A. baumannii bacteriophages were collected from the inlets and outlets of the two treatment plants. Most of the bacteriophages isolated were from the inlets of hospital treatment plants (76 %). We determined the host range of 54 phage isolates using spot tests with A. baumannii and 14 bacterial species. No clear zone was observed against all 14 bacterial species tested bacteria. ØABP-01, ØABP-02 and ØABP-04 with high lytic activity on a broad range of A. baumannii were selected for further characterization. ØABP-01 was able to lyse all A. baumannii tested (n = 12). ØABP-02 and ØABP-04 lysed 50 % of A. baumannii tested (n = 6). Only ØABP-01 and ØABP-02 could completely lyse A. baumannii ATCC 19606. All 3 phages showed different plaque characteristics (Fig. 1). Plaques of ØABP-01 showed large clear zones (3–8 mm). ØABP-02 showed plaques with small clear zones inside (1–2 mm) and large opaque zones surrounding, whereas ØABP-04 showed plaques with clear zones inside and an opaque zone surrounding. The variation in plaque characteristics could be related to different types of bacteriophage. Morphology of the three bacteriophages was observed in a transmission electron microscope as shown in Fig. 2. ØABP-01 was found to have an icosahedral head (78 nm) with a short tail (9 nm), belonging to the Podoviridae family. ØABP-02 has an icosahedral head (80–85 nm) with a long tail (67–125 nm) and ØABP-04 has an icosahedral head (72 nm) with a long tail (110 nm). Both were classified into the Myoviridae family. All three bacteriophages were tailed bacteriophages and identified as members of order Caudovirales. To date, more than 96 % of isolated bacteriophages belong to this order [29]. ØABP-01 was classified in the Podoviridae family which is in accordance with previous studies in bacteriophages infecting A. baumannii phiAB1, AB2 and AB7-IBB2 [14, 21, 23]. Bacteriophage ØABP-02 and ØABP-04 were classified into the family Myoviridae. This family was also reported in bacteriophages ABp53, ZZ1 and AP22 [16, 18, 19].
Fig. 1.
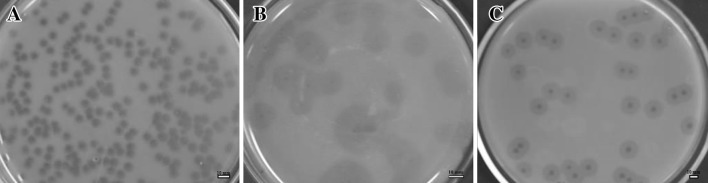
The appearance of plaques on a bacterial lawn formed by three A. baumannii bacteriophages. a ØABP-01 plaque characteristic showed large clear zone. b ØABP-02 plaque characteristic showed small clear zone inside with large opaque zone surround. c ØABP-04 plaque characteristic showed clear zone inside with an opaque zone surround
Fig. 2.
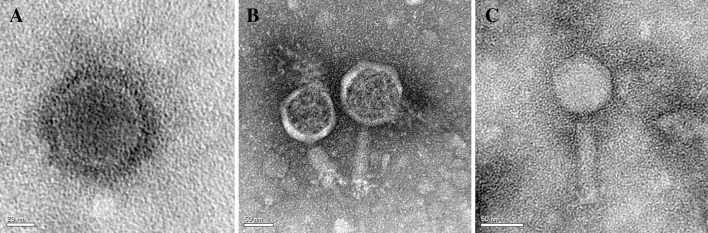
Electron micrograph of A. baumannii bacteriophages. a ØABP-01, b ØABP-02 and c ØABP-04
Phage Physiology
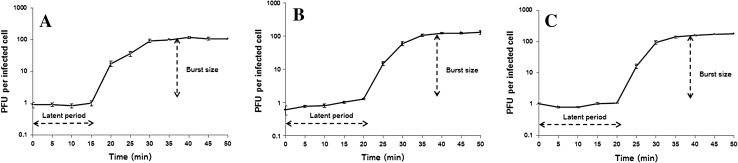
The growth cycle of all three bacteriophages was characterized by one-step growth. Burst size and latent period of three bacteriophages are shown in Fig. 3. The latent periods of ØABP-01, ØABP-02 and ØABP-04 were 15, 20 and 20 min and the burst sizes of ØABP-01, ØABP-02 and ØABP-04 were 110 ± 5.56, 120 ± 17.67 and 150 ± 10.96 PFU/cell. ØABP-01 had a shorter latent period and displayed good lytic activity compared to other bacteriophage isolates. Previous reports of bacteriophages infecting A. baumannii showed latent periods ranging from 9 to 25 min and burst size from 22 to 409 PFU/infected cell [14–16, 18–22].
Fig. 3.
One step growth of ØABP-01 (a), ØABP-02 (b) and ØABP-04 (c) bacteriophages against A. baumannii host strains. All data are shown as mean of three independent experiments. Error bar represent the standard error of mean
Phage Nucleic Acid Analysis
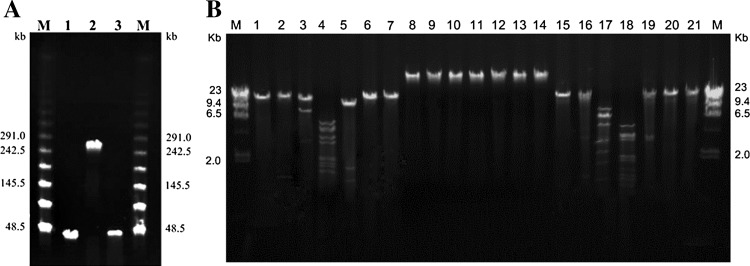
Analysis of all bacteriophage nucleic acids by digestion with DNaseI revealed that all phage nucleic acids were sensitive to DNase (data not shown). To date, most of A. baumannii bacteriophage isolates are DNA phages [14, 15, 16, 18, 19, 22]. PFGE analysis revealed that phage DNA ØABP-01 and ØABP-04 have genome sizes of approximately 30 kb and ØABP-02 showed a larger genome size of approximately 240 kb (Fig. 5a). Restriction analysis of ØABP-01 and ØABP-04 genomes showed they could be cut with restriction enzymes BamHI, EcoRI, HindIII and SphI (Fig. 4b). All six restriction enzymes used in this study appeared unable to cut the ØABP-02 genome (Fig. 4b).
Fig. 5.
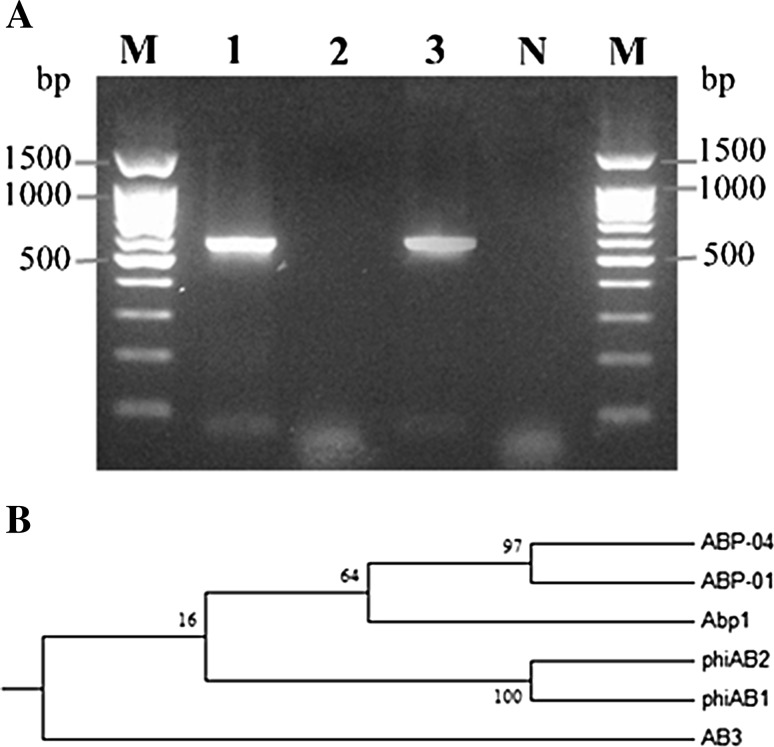
a Amplification of endolysin gene from A. baumannii bacteriophages detected by 1 % agarose gel electrophoresis. Lane M DNA marker, Lane 1 ØABP-01, Lane 2 ØABP-02, Lane 3 ØABP-04, Lane 4 Negative control. b Phylogenetic tree analysis of endolysin gene of phage ØABP-01, ØABP-04, phiAB, phiAB2, Abp1 and AB3. Phylogenetic analysis was conducted in MEGA version 5.05. Values on branches represent maximum likelihood support values
Fig. 4.
Nucleic acid analysis of A. baumannii bacteriophages. a Pulsed-field gel electrophoresis (PFGE) of undigested phages (4.5 V/cm, 30 h). Lambda ladder PFG marker (lane M); ØABP-01 (lane 1); ØABP-02 (lane 2); ØABP-04 (lane 3). b Restriction pattern of A. baumannii bacteriophages. Lambda DNA marker/HindIII (lane M); DNA of ØABP-01 uncut (lane 1); DNA of ØABP-01 cut with BamHI (lane 2), EcoRI (lane 3), HindIII (lane 4), SphI (lane 5), PstI (lane 6), SmaI (lane 7), DNA of ØABP-02 uncut (lane 8); DNA of ØABP-02 cut with BamHI (lane 9), EcoRI (lane 10), HindIII (lane 11), SphI (lane 12), PstI (lane 13), SmaI (lane 14),DNA of ØABP-04 uncut (lane 15); DNA of ØABP-04 cut with BamHI (lane 16), EcoRI (lane 17), HindIII (lane 18), SphI (lane 19), PstI (lane 20), SmaI (lane 21)
Detection of Endolysin Gene
The endolysin gene was detected in most A. baumannii bacteriophages isolated from different regions [13, 23, 25]. PCR was utilized to investigate the presence of the endolysin genes as shown in Fig. 5a. The amplified product of the endolysin gene (lys) could be detected from ØABP-01 to ØABP-04 genomes. Most double-stranded DNA phages accomplish host cell lysis through the holin-endolysin system. The absence of the endolysin gene in ØABP-02 may be due to different mechanisms to exit the host cell. Some DNA phage can exit the host cells by inhibition of specific host enzymes and impairing peptidoglycan biosynthesis [30]. The mechanism of ØABP-02 host lysis needs further investigation. Analysis of ABP-01 and ABP-04 endolysin sequences showed 558 and 576 bp fragment of the endolysin genes. Open Reading Frame analysis using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) revealed the endolysin protein of ABP-01 and ABP-04 consisting of 185 and 191 amino acid residues. Similarity of bacteriophage endolysin genes is essential for structural analysis which contributes to the potential of using endolysin as an antimicrobial agent. Sequencing the endolysin gene from ØABP-01 yields 94 % sequence identity to the endolysin gene (lys) phage from Abp1(Accession: JX658790.1), phiAB1(Accession: HQ186308.1) and phiAB2 (Accession: HM755898.1) and 93 % sequence identity to Acinetobacter phage AB3 (Accession: KC311669.1) obtained from GenBank. In addition, the sequence of the fragment of the endolysin gene from ØABP-04 yield 93 % identity to Abp1(Accession: JX658790.1), phiAB1(Acession: HQ186308.1) and phiAB2 (Accession: HM755898.1) and 92 % identity to Acinetobacter phage AB3(Accession: KC311669.1). A phylogenetic tree was constructed based on nucleotide sequences of known phage endolysin genes (Fig. 5b). Sequencing and phylogenetic relationships of the endolysin genes showed that ØABP-01 and ØABP-04 are closely related to phage Abp1, phiAB1 and phiAB2 isolated from China and Taiwan [13, 23, 24]. Despite the different origins of the host strains and the origin of the isolates, this implies that ØABP-01 and ØABP-04 may have common ancestral origins with other phages.
Conclusion
Three bacteriophages classified as Podoviridae and Myoviridae family members were characterized. Bacteriophages in this study are quite different from those of other reports in genome size, morphology, and one-step growth curve. ØABP-01 has shown good specificity in host range and good lytic activity against A. baumannii is a valuable candidate for further study.
Acknowledgments
The authors thank Prof. Gavin Reynolds and staff from Naresuan University Language Center for editing this manuscript. We are grateful to staff from Buddhachinaraj hospital and Bang Rakam hospital, Phitsanulok for their valuable cooperation during phage sample collection. This work was supported by grant from Naresuan University Research fund (R2555C085).
References
- 1.Choi WS, Kim SH, Jeon EG, Son MH, Yoon KY, et al. Nosocomial outbreak of carbapenem-resistant Acinetobacter baumannii in intensive care units and successful outbreak control program. J Korean Med Sci. 2010;25:999–1004. doi: 10.3346/jkms.2010.25.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 4.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu TK, Koeris MS. The next generation of bacteriophage therapy. Curr Opin Microbiol. 2011;14:524–531. doi: 10.1016/j.mib.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 6.Soothill JS. Treatment of experimental infections of mice with bacteriophages. J Med Microbiol. 1992;37:226–258. doi: 10.1099/00222615-37-4-258. [DOI] [PubMed] [Google Scholar]
- 7.Wills QF, Kerrigan C, Soothill JS. Experimental bacteriophage protection against Staphylococcus aureus abscesses in a rabbit model. Antimicrob Agents Chemother. 1992;49:1220–1221. doi: 10.1128/AAC.49.3.1220-1221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McVay CS, Velásquez M, Fralick JA. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob Agents Chemother. 2007;51:1934–1938. doi: 10.1128/AAC.01028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morello E, Saussereau E, Maura D, Huerre M, Lhousseine T, Laurent D. Pulmonary bacteriophage therapy on Pseudomonas aeruginosa cystic fibrosis strains: first steps towards treatment and prevention. PLoS One. 2011;6:1–9. doi: 10.1371/journal.pone.0016963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borysowski J, Weber-Dabrowska B, Górski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med. 2006;231:366–377. doi: 10.1177/153537020623100402. [DOI] [PubMed] [Google Scholar]
- 11.Fischetti VA. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermoso JA, García JL, García P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Lai MJ, Lin NT, Hu A, Soo PC, Chen LK, Chen LH, Chang KC. Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl Microbiol Biotechnol. 2011;90:529–539. doi: 10.1007/s00253-011-3104-y. [DOI] [PubMed] [Google Scholar]
- 14.Lin NT, Chiou PY, Chang KC, Chen LK, Lai MJ. Isolation and characterization of phi AB2: a novel bacteriophage of Acinetobacter baumannii. Res Microbiol. 2010;161:308–314. doi: 10.1016/j.resmic.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Liang L, Lin S, Jia S. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 2010;10:131. doi: 10.1186/1471-2180-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CN, Tseng TT, Lin JW, Fu YC, Weng SF, Tseng YH. Lytic myophage Abp53 encodes several proteins similar to those encoded by host Acinetobacter baumannii and phage phiKO2. Appl Environ Microbiol. 2011;77:6755–6762. doi: 10.1128/AEM.05116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon J, Kim JW, Yong D, Lee K, Chong Y. Complete genome sequence of the podoviral bacteriophage YMC/09/02/B1251 ABA BP, which causes the lysis of an OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolate from a septic patient. J Virol. 2012;86:12437–12438. doi: 10.1128/JVI.02132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, Li ZJ, Wang SW, Wang SM, Huang DH, et al. Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 2012;12:156. doi: 10.1186/1471-2180-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popova AV, Zhilenkov EL, Myakinina VP, Krasilnikova VM, Volozhantsev NV. Isolation and characterization of wide host range lytic bacteriophage AP22 infecting Acinetobacter baumannii. FEMS Microbiol Lett. 2012;332:40–46. doi: 10.1111/j.1574-6968.2012.02573.x. [DOI] [PubMed] [Google Scholar]
- 20.Shen GH, Wang JL, Wen FS, Chang KM, Kuo CF, et al. Isolation and characterization of φkm18p, a novel lytic phage with therapeutic potential against extensively drug resistant Acinetobacter baumannii. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0046537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thawal ND, Yele AB, Sahu PK, Chopade BA. Effect of a novel podophage AB7-IBB2 on Acinetobacter baumannii biofilm. Curr Microbiol. 2012;65:66–72. doi: 10.1007/s00284-012-0127-2. [DOI] [PubMed] [Google Scholar]
- 22.Yele AB, Thawal ND, Sahu PK, Chopade BA. Novel lytic bacteriophage AB7-IBB1 of Acinetobacter baumannii: isolation, characterization and its effect on biofilm. Arch Virol. 2012;157:1441–1450. doi: 10.1007/s00705-012-1320-0. [DOI] [PubMed] [Google Scholar]
- 23.Chang KC, Lin NT, Hu A, Lin YS, Chen LK, Lai MJ. Genomic analysis of bacteriophage ϕAB1, a ϕKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Genomics. 2012;97:249–255. doi: 10.1016/j.ygeno.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Huang G, Le S, Peng Y, Zhao Y, Yin S, Zhang L, Yao X, Tan Y, Li M, Hu F. Characterization and genome sequencing of phage Abp1, a new phiKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Curr Microbiol. 2013;66:535–543. doi: 10.1007/s00284-013-0308-7. [DOI] [PubMed] [Google Scholar]
- 25.Niumsup PR, Boonkerd N, Tansawai U, Tiloklurs M. Carbapenem-resistant Acinetobacter baumannii producing OXA-23 in Thailand. Jpn J Infect Dis. 2009;62:152–154. [PubMed] [Google Scholar]
- 26.Adams MH. Bacteriophages. NewYork: Inter science Publishers Inc; 1959. [Google Scholar]
- 27.Kutter E, Sulakvelidze A. Bacteriophages: biology and applications. USA: CRC Press; 2005. [Google Scholar]
- 28.Clokie MRJ, Kropinski AM. Bacteriophages: methods and protocols, volume 2: molecular and applied aspects. Totowa: Humana Press; 2009. [Google Scholar]
- 29.Ackermann HW. Frequency of morphological phage descriptions in the year 2000: brief review. Arch Virol. 2001;146:843–857. doi: 10.1007/s007050170120. [DOI] [PubMed] [Google Scholar]
- 30.Bernhardt TG, Wang IN, Struck DK, Young R. A protein antibiotic in the phage Qbeta virion: diversity in lysis targets. Science. 2001;292:2326–2329. doi: 10.1126/science.1058289. [DOI] [PubMed] [Google Scholar]