Abstract
The amalgamation of the research efforts of biologists, chemists and geneticists led by scientists at the Department of Zoology, University of Delhi has resulted in the development of a novel rifamycin derivative; 24-desmethylrifampicin, which is highly effective against multi-drug resistant (MDR) strains of Mycobacterium tuberculosis. The production of rifamycin analogue was facilitated by genetic-synthetic strategies that have opened an interdisciplinary route for the development of more such rifamycin analogues aiming at a better therapeutic potential. The results of this painstaking effort of nearly 25 years of a team of students and scientists led by Professor Rup Lal have been recently published in the Journal of Biological Chemistry (www.jbc.org/content/289/30/21142.long). This strategy can now find applications for developing newer rifamycin analogues that can be harnessed to overcome the problem of MDR, extensively drug resistant (XDR) and totally drug resistant (TDR) M. tuberculosis.
Keywords: 24-desmethylrifamycin, Rifampicin, Tuberculosis, Multidrug resistant Mycobacterium tuberculosis
Introduction
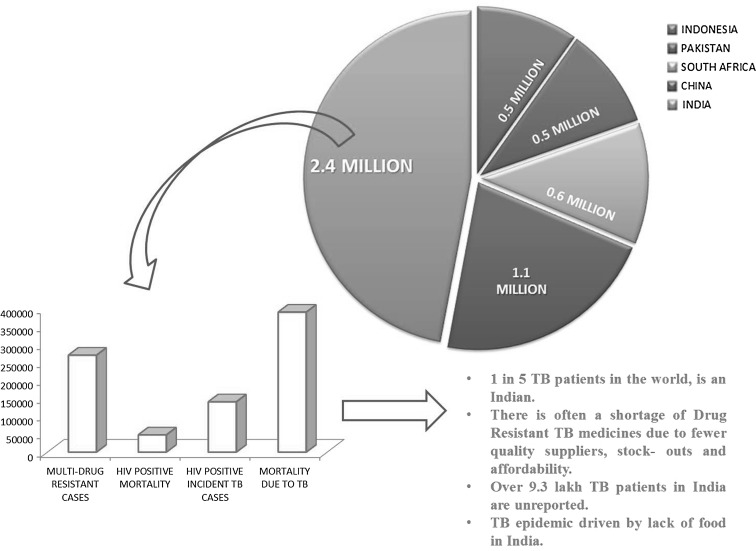
The emergence of rapidly evolving multi-drug resistant (MDR) and extensively drug resistant (XDR) pathogens has fuelled the need to find/generate novel drugs [1, 2]. Of all diseases affecting human beings, Tuberculosis (TB) has been one of the major threats and could be a potential pandemic. TB caused by Mycobacterium tuberculosis, is the second most infectious killer disease, next only to HIV infection. Of around nine million people affected world-wide by this deadly disease, approximately two to three million patients are in India (Fig. 1) [3]. The problem of drug resistant forms of TB is making the disease virtually incurable. The most common treatment of TB is through rifampicin, a drug which is used in combination with isoniazid and ethambutol. Rifampicin is a semisynthetic derivative of rifamycin B (Fig. 2) produced by an actinobacterium Amycolatopsis mediterranei [4]. In general, rifamycins bind to the β-subunit of the RNA polymerase, and inhibits mRNA transcription [5, 6]. Introduction of rifampicin in late 1960s led to considerable fall in the mortality rate due to TB and since then it has been used as the first line of drug. As MDR and TDR strains of M. tuberculosis have mutations in their rpoB, hence they are resistant to rifampicin and the disease becomes untreatable. Although novel antibiotics have been developed during the last 6–7 decades, however, bacteria have developed resistance to almost all of them. The big question is: Are we going to be mute spectators of this battle between bacteria and antibacterials? Is there an end in sight to this never ending battle? [7–10].
Fig. 1.
An overview of the current scenario of the TB pandemic highlighting the worsening situation in India. The pie chart represents the top five TB burdened countries with 2.4 million cases in India (adapted from WHO Tuberculosis Report-2013)
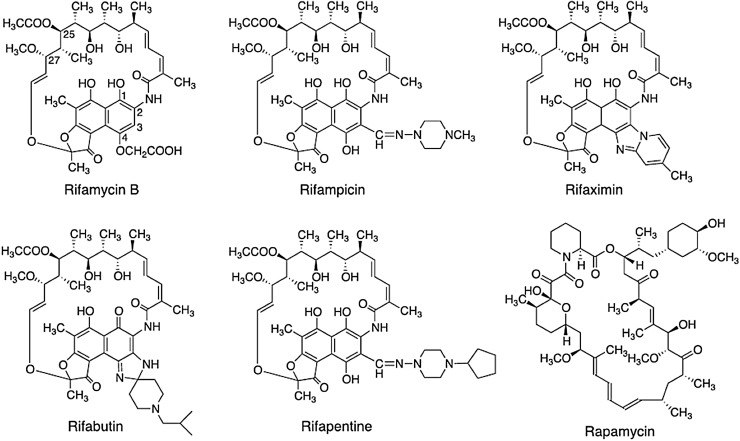
Fig. 2.
Chemical structures of the semi-synthetic derivatives of Rifamycin B
The appearance of the resistant forms of pathogens like M. tuberculosis is an evolutionary directed process; therefore any solution to the problem should hypothetically keep pace with the emerging drug resistant bacteria. Keeping in view the constant co-evolution of resistant pathogenic strains with the available antibiotics, the existing drugs are being rendered ineffective, fast.
Recent Developments in Drug Discovery for TB
A reasonable amount of effort has been put up to combat the problem posed by MDR strains of M. tuberculosis. However, the efforts are not commensurate with the rate at which MDR strains are appearing and thus are unable to keep pace. It is pertinent to mention here, that hypothetically if the resistant pathogenic strains are multiplying geometrically, the drugs that are needed to combat them are being discovered even slower than the arithmetic progression. Hence, drug discovery has to be dynamic and continuous effort. In this context, the burden due to TB should be of prime concern to most of the developing countries, especially India (Fig. 1).
This urgent necessity to have a long term solution to the problem is reflected by several institutes and laboratories across the world working on finding the solution to the disease but nothing significant has come out as yet. However, there has been development in the drug discovery for treating TB, which comprise of a range of antimycobacterial drugs such as: diarylquinolines, mycobacterial gyrase inhibitors, pyrazinamide analogs, spectinamides, etc. and many of them have also undergone successful clinical trials (Table 1). In the last 40 years, a new TB drug with a novel mechanism of action—bedaquiline—is now available, and was granted accelerated approval by the United States Food and Drug Administration (USFDA) in December 2012. But this drug, currently under clinical trials is also not free from limitations [11].
Table 1.
Recent developments in anti-tuberculosis drug discovery
| Drug-discovery | Drugs under pre-clinical trials | Drugs under clinical development |
|---|---|---|
| Diarylquinolines | Bedaquiline (TMC207) | |
| InhA inhibitors | SQ-109 | |
| LeusRS inhibitors | Q201 | |
| Mycobacterial gyrase | SPR-10199 | Novel regimens |
| Inhibitors | SQ609 | |
| Pyrazinamide analogues | CPZEN-45 | |
| Fluroquinolone | DC-159a | Moxifloxacin |
| Rifamycins | Rifapentine | |
| Nitroimidazole | PA-824 |
Thereby, keeping in view the limitations associated with the new drug available as well as the emergence of MDR strains of M. tuberculosis, we choose to generate anti-TB drug by modifying the polyketide backbone of rifamycin. Due to the availability of only few synthetic forms of rifamycin (rifampicin, rifapentine, rifabutin, rifamixin, rifamycin S, rifamycin SV) since 1960s till date, it clearly reflects the difficulties inherent to discovering anti-TB drugs. The difficulties include the recalcitrant nature of the parent drug rifamycin B to further undergo chemical modifications at C3 and C4 position of ansa chain of the molecule (Fig. 2). This has compelled the scientists to look for alternative strategies to produce rifamycin analogs to overcome the problem posed by the infections of M. tuberculosis and its MDR strains in particular using combinatorial biosynthetic approach.
Here, we describe nearly 25 years of research efforts at the Department of Zoology, University of Delhi which led to the development of a proof of concept for the production of rifamycin analogs by manipulating the rifamycin polyketide synthase gene cluster in A. mediterranei that produces rifamycin B. The production of a rifamycin analogue: 24-desmethylrifamycin B and its semisynthetic derivatives: 24-desmethylrifampicin and 24-desmethylrifamycin S, were found to be effective against MDR strains of M. tuberculosis.
Modification of Rifamycin Polyketide Backbone: Generating Rifamycin Analogue
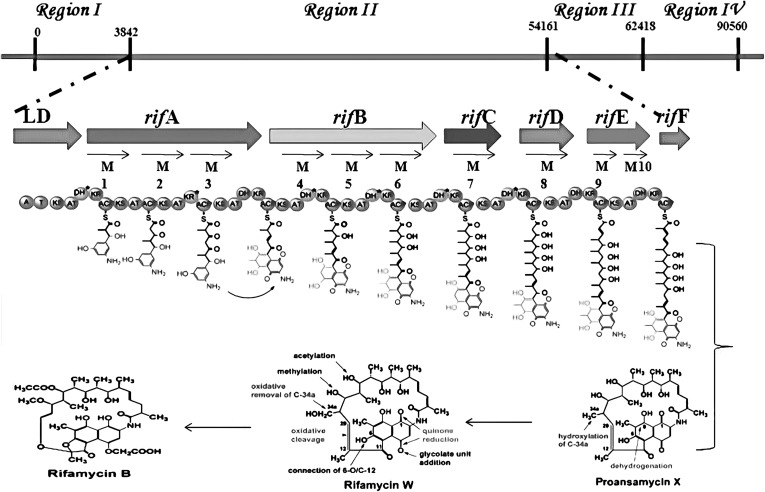
Till date, only six semisynthetic clinically effective derivatives (Fig. 2) of rifamycin B could be produced by the chemical modifications of the naphthaquinone ring of rifamycin B. Rifampicin, being one of them is majorly used in combating TB. Other than the already implemented chemical modifications, no further alterations were possible to produce effective rifamycin analogues. Also, other methods of modification are generally not applicable to this molecule or the producer organism. However, with the availability of rifamycin polyketide synthase gene cluster (rifPKS) in 1998 [12] (Fig. 3) and with the development of cloning vectors and transformation system for the rifamycin producer strain A. mediterranei by our group here at the University of Delhi [13–18], the possibilities of manipulating the rifPKS to produce more effective analogues of rifamycin B were raised [16].
Fig. 3.
Biosynthetic organization and polyketide chain extension of rifamycin B biosynthesis. The synthesis of the hypothetical intermediate by rifamycin PKS of region II during the biosynthesis of rifamycin B. The chain is assembled of starter unit (AHBA), with two acetate and eight propionate extender units. Rif F, translationally coupled to Rif E, encodes amide synthase and thus displaces the thioesterase linkage of the assembled polyketide chain. Proansamycin X then undergoes post PKS modifications to form rifamycin B
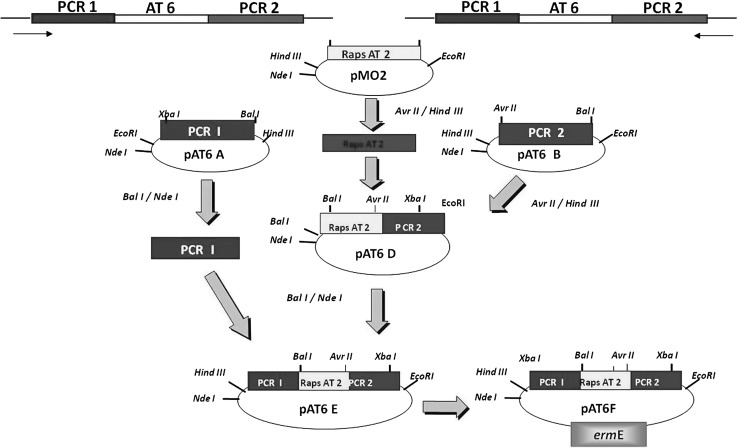
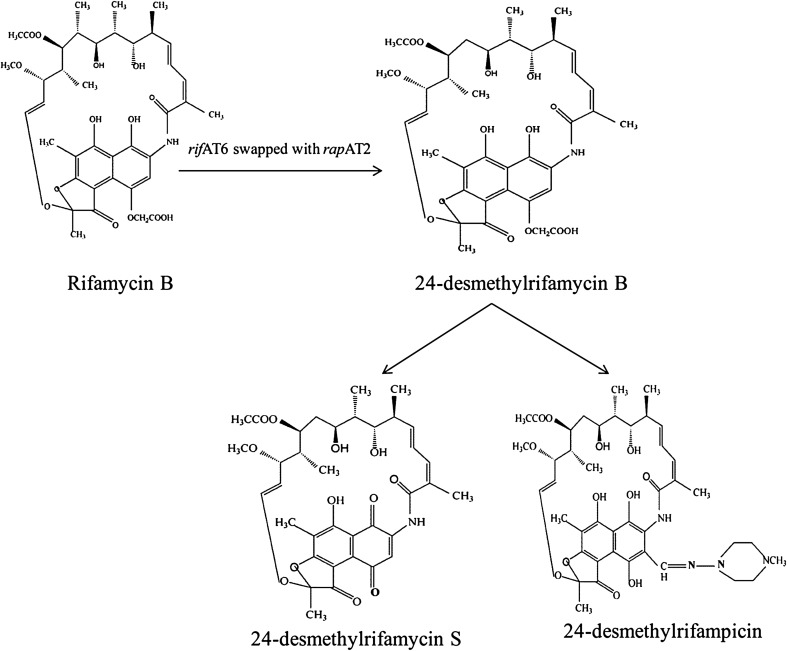
Even though, the co-linear nature of the rifamycin polyketide synthase gene cluster was decrypted in 1998 [12], still, genetically modifying it seemed to be a daunting task. However, by exploiting the modular nature of rifPKS biosynthetic gene cluster, we for the first time could manipulate and modify the rifamycin polyketide ansa chain by combinatorial biosynthetic approach. The efforts led to the swapping of acyltransferase (AT) domain of the sixth module (AT6) of rifamycin polyketide synthase (which adds propionate unit to the growing polyketide chain) with that of AT domain of the second module (AT2) of rapamycin PKS (rapPKS) (which adds acetate unit) in A. mediterranei S699. The purpose of swapping AT domain was to alter the substrate specificity in rifPKS gene cluster. This was facilitated by designing and constructing AT6 replacement cassette (pAT6F) (Fig. 4). The plasmid AT6F was electro-transformed into producer organism—A. mediterranei S699. The resulting mutant strain was generated using homologous recombination and was found to produce a derivative 24-desmethylrifamycin B (MW:740), which lacked a pendant methyl group at C-33 of the rifamycin B skeletal structure (MW:754) (Fig. 5). It was confirmed using NMR and LC–MS studies. The derivatives of 24-desmethylrifamycin B; 24-desmethylrifampicin & 24-desmethylrifamycin S showed much better antibacterial activities against MDR strains of M. tuberculosis in comparison to the available rifampicin (Fig. 5). The findings were also corroborated with free energy perturbation studies. The results have been published in the Journal of Biological Chemistry (www.jbc.org/content/289/30/21142.long).
Fig. 4.
Strategy for the construction of functional cassette pAT6F in the plasmid pIJ4026, which was electroporated into Amycolatopsis mediterranei to swap rifAT6 with rapAT2. The flanking regions immediately upstream (41862–43533 bp-PCRI) and downstream (44488–45989 bp-PCRII) of rifPKS of AT6 domain were amplified using primer pair I, II and primer pair III, IV, respectively to obtain PCR I and PCRII amplified products. PCRI and PCRII were cloned in pUC18 and named as pAT6A and pAT6B, respectively. The plasmid pMO2 was digested to release rapAT2, which was ligated with linearized pAT6B to form plasmid pAT6D. PCRI from pAT6A was digested and ligated into linearized pAT6D to form the plasmid pAT6E. The entire construct PCRI + rapAT2 + PCRII was digested and eluted from pAT8E and finally cloned in the plasmid pIJ4026, which confers erythromycin resistance to the host organism, A. mediterranei S699. This final construct pAT6F was transformed in the wild type strain (A. mediterranei S699)
Fig. 5.
Chemical structures of Rifamycin B and its analogues 24-desemthylrifamycin B, 24-desmethylrifamycin S and 24-desmethylrifampicin (lacking methyl group)
Fighting Against TB Requires Strong Collaborations
R&D efforts of over two decades by researchers at the University of Delhi under the leadership of Prof. Lal bore fruit with the development of a new compound 24-desmethylrifampicin (Fig. 5) which is effective against MDR strains of M. tuberculosis. While rifPKS was manipulated and mutants were generated by team at the University of Delhi, the chemical structure of the analogue 24-desmethylrifamycin B and the derivative 24-desmethylrifampicin so produced was confirmed by the US collaborators. Further testing of the analogue against MDR strains of M. tuberculosis was done in association with Institute of Genomics and Integrative Biology (CSIR-IGIB), Delhi. The analogue 24-desmethylrifampicin was found to be better than the already existing rifampicin used against MDR strains [19]. Thus it can be hypothesised that, this drug could find application in pharmaceutical sector. In addition, these results are critical additions to the technological development and scientific advancements necessary for developing an anti-TB drug that effectively eliminates the threat of the deadly MDR strains of M. tuberculosis.
Stepping Stones for Generation of 24-Desmethylrifampicin
Looking at the ugly side of the disease over the years, where TB claims one millions lives per year globally with nearly two hundred people dying of it every hour and 20–25 % of these deaths are reported from India alone, it was urgent to take up this challenge to develop a drug that should be effective against MDR-TB.
The prerequisites for the generation of rifamycin analogues involved developing a cloning vector and transformation system to negotiate with the bacterium A. mediterranei that produces Rifamycin B. Thus, a work initiated in 1988 resulted in crossing the first hurdle by developing a system of cloning vectors and transformation system by our team [13, 15, 18].
In order to generate rifamycin analogue, the genetic manipulations were carried out in the rifamycin B producer actinobacterium A. mediterranei that harbours rifamycin polyketide synthase gene cluster. Applying the genetic-synthetic strategy, the mutant strains were generated that produced 24-desmethylrifamycin B (lacking one methyl group in comparison to rifamycin B) (Fig. 3). The derivatives of 24-desmethylrifamycin B: 24-desmethylrifampicin and 24-desmethylrifamycin S, were tested against MDR strains of M. tuberculosis, where the activity was found to be 10 times better than the existing commercially available rifampicin [19].
In conclusion, the task of genetically modifying the bacterium appeared very difficult and it was only through persistent efforts of several years that the new compound 24-desmethylrifampicn was developed in 2011. Even though, it is may appear to be small but is a significant milestone in the arena of antibiotic discovery. However, a lot still needs to be done. Foremost, this strategy can be used to create library of analogues and it is the harbinger of reinvigorated interest in the field of drug discovery. Developing more analogues on similar approaches could serve as a great service to the humanity. With the preliminary data available, it can be said that 24-desmethylrifampicin could prove to be a potential drug to fight against MDR-TB.
Acknowledgments
This work was supported by the Department of Biotechnology (DBT). AS, UM, RK & PS thank the University Grant Commission for providing fellowships.
References
- 1.Kalia VC, Rani A, Lal S, Cheema S, Raut CP. Combing databases reveals potential antibiotic producers. Expert Opin on Drug Discov. 2007;2:211–224. doi: 10.1517/17460441.2.2.211. [DOI] [PubMed] [Google Scholar]
- 2.Purohit HJ, Cheema S, Lal S, Raut CP, Kalia VC. In search of drug targets for Mycobacterium tuberculosis. Infect Disord Drug Targets. 2007;7:245–250. doi: 10.2174/187152607782110068. [DOI] [PubMed] [Google Scholar]
- 3.Global tuberculosis report 2013-www.who.in/tb/publications/global_report/en/
- 4.Maggi N, Pasqalucci CR, Ballotta R, Sensi P. Rifampicin: a new orally active rifamycin. Chemotherapea. 1966;11:285–292. doi: 10.1159/000220462. [DOI] [PubMed] [Google Scholar]
- 5.Wehrli W, Knusel F, Schmid K, Staehelin, M (1968) Interaction of rifamycin with bacterial RNA polymerase. Proc Natl Acad Sci USA 61: 667–673. (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC225211/?page=1) [DOI] [PMC free article] [PubMed]
- 6.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 7.Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Critical Rev Microbiol. 2011;37:121–140. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]
- 8.Kalia VC. Quorum sensing inhibitors: an over view. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Kalia VC. Microbes, antimicrobials and resistance: the battle goes on. Indian J Microbiol. 2014;54:1–2. doi: 10.1007/s12088-013-0443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalia VC, Wood TK, Kumar P. Evolution of resistance to quorum sensing inhibitors. Microb Ecol. 2014;68:13–23. doi: 10.1007/s00248-013-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diacon AH, Dawson R, Bidlingmaier-Groote FV, Symons G, Venter A, Donald PR, Niekerk CV, Everitt D, Winter H, Becker P, Mendel CM, Spigelman MK. 14-Day bactericidal activity of PA-824, bedaquiline, pyrazinamide and moxifloxacin combinations: a randomized trial. Lancet. 2012;380:986–993. doi: 10.1016/S0140-6736(12)61080-0. [DOI] [PubMed] [Google Scholar]
- 12.August PR, Tang L, Yoon YJ, Ning S, Müller R, Yu TW, Taylor M, Hoffmann D, Kim CG, Zhang X, Hutchinson CR, Floss HG. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Bio. 1998;5:69–79. doi: 10.1016/S1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 13.Lal R, Khanna R, Dhingra N, Khanna M, Lal S. Development of an improved cloning vector and transformation system in Amycolatopsis mediterranei (Nocardia mediterranei) J Antibiot (Tokyo) 1998;51:161–169. doi: 10.7164/antibiotics.51.161. [DOI] [PubMed] [Google Scholar]
- 14.Lal R (1999) Cloning vector and the process for the preparation thereof. US Patent US00598560A
- 15.Khanna M, Dua M, Lal R. Selection of suitable marker genes for the development of cloning vectors and electroporation in different strains of Amycolatopsis mediterranei. Microbiol Res. 1998;153:205–211. doi: 10.1016/S0944-5013(98)80002-5. [DOI] [PubMed] [Google Scholar]
- 16.Lal R, Kumari R, Kaur H, Khanna R, Dhingra N, Tuteja D. Regulation and manipulation of the gene clusters encoding type-I PKSs. Trends Biotechnol. 2000;18:264–274. doi: 10.1016/S0167-7799(00)01443-8. [DOI] [PubMed] [Google Scholar]
- 17.Tuteja D, Dua M, Khanna R, Dhingra N, Khanna M, Kaur H, Saxena D, Lal R. The importance of homologous recombination in the generation of large deletions in hybrid plasmids in Amycolatopsis mediterranei. Plasmid. 2000;43:1–11. doi: 10.1006/plas.1999.1426. [DOI] [PubMed] [Google Scholar]
- 18.Dhingra G, Kumari R, Bala S, Majumdar S, Malhotra S, Sharma P, Lal S, Cullum J, Lal R. Development of cloning vectors and transformation methods for Amycolatopsis. J Ind Microbiol Biotechnol. 2003;30:195–204. doi: 10.1007/s10295-003-0040-6. [DOI] [PubMed] [Google Scholar]
- 19.Nigam A, Almabruk KH, Saxena A, Jongtae Y, Mukherjee U, Kaur H, Kohli P, Kumari R, Singh P, Zakharov LN, Singh Y, Mahmud T, Lal R. Modification of rifamycin polyketide backbone leads to improved drug activity against rifampicin resistant Mycobacterium tuberculosis. J Biol Chem. 2014;289:21142–21152. doi: 10.1074/jbc.M114.572636. [DOI] [PMC free article] [PubMed] [Google Scholar]