Abstract
Aim:
To investigate the metabolite changes caused by simvastatin or fenofibrate intervention in diet-induced hyperlipidemia rats using a GC-MS-based metabolomic profiling approach.
Methods:
SD rats were fed with high-lipid diet for 4 weeks to induce hyperlipidemia, then the rats were fed with normal diet, and orally administered with simvastatin (10 mg·kg−1·d−1) or fenofibrate (150 mg·kg−1·d−1) for 2 weeks. Blood samples were collected once a week, and potential biomarkers were examined using commercial assay kits and a metabolomic approach. The metabolomics data were analyzed using a multivariate statistical technique and a principal component analysis (PCA).
Results:
Oral administration of simvastatin or fenofibrate significantly decreased the plasma levels of total cholesterol (TC) and low-density lipoprotein (LDL) cholesterol and increased the plasma level of high-density lipoprotein (HDL) cholesterol in the hyperlipidemia rats. Plasma samples were scattered in the PCA scores plots in response to the diet and to the drugs administered. The main metabolites changed in the hyperlipidemia rats were cholesterol, creatinine, linoleic acid, β-hydroxybutyric acid, tyrosine, isoleucine and ornithine. The plasma level of creatinine was significantly lower in the simvastatin-treated rats than in the fenofibrate-treated rats. The plasma tyrosine concentration was declined following intake of high-lipid diet, which was reversed by fenobrate, but not by simvastatin.
Conclusion:
A series of potential biomarkers including tyrosine, creatinine, linoleic acid, β-hydroxybutyric acid and ornithine have been identified by metabolomic profiling, which may be used to identify the metabolic changes during hyperlipidemia progression.
Keywords: simvastatin, fenofibrate, hyperlipidemia, metabolomics, biomarker, creatinine, LDL cholesterol, GC-MS
Introduction
Hyperlipidemia, a metabolic disorder disease that involves abnormally increased levels of lipids (such as cholesterol) and lipid proteins in the blood, is one of the main risk factors that causes arteriosclerosis, cerebral stroke, coronary heart disease, myocardial infarction and renal failure in Chinese people1,2,3. Fibrates (ie, fenofibrate) and statins (ie, simvastatin) are currently the most commonly used drugs for the treatment of hyperlipidemia. Fibrates are activators of PPARα, which controls cholesterol levels by transcriptionally inducing the synthesis of the major apolipoproteins apoA-I and apoA-II2,3,4. Statins are also widely used to lower cholesterol levels because of their restraint effects on the 3-hydroxyl-3-methyl coenzyme A (HMG-CoA) reductase, which is responsible for the rate-limiting step in cholesterol biosynthesis3,4. Although the pharmacological mechanisms of fibrates and statins have been extensively studied, their lipid-regulating effects and adverse drug reactions in hyperlipidemia patients during long-term treatment remain unclear. Because hyperlipidemia is a systemic disease that affects the metabolism of many endogenous small-molecule metabolites, understanding the intervention of fibrates or statins within the body's metabolomic system is important. To date, conventional pharmacological methods could not effectively illuminate these changes. In recent years, the development of new methodologies for studying pharmacological efficacies, mechanisms and metabolomics has become a hotspot for pharmacology and pharmacodynamics research5.
Metabolomics is an important subject in the field of systemic biology, which includes metabolomics, genomics, transcriptomics and proteomics, and provides a wealth of information regarding the biochemical framework in biological samples. In recent years, rapid analytical platforms, including GC/TOF-MS, LC-MS/MS and NMR spectroscopy, have been applied to systematically detecting, characterizing and quantifying metabolites and the related metabolic pathways at the cellular, tissue, or organ levels. Among these technologies, GC-MS is a highly suitable technique for metabolomics analysis due to its high sensitivity and efficient separation in the gas state. When it is equipped with extensive databases, it can identify or verify various small-molecule metabolites in metabolomics studies6,7. Metabolomics has been widely used in discovering biomarkers and in monitoring therapeutic efficacy and pathogenesis in early detection and clinical diagnosis for many diseases, including tumors8,9, heart disease10,11 and diabetes mellitus12,13. Moreover, small changes in the activities of individual enzymes, genes and proteins or other biomarkers can be amplified in metabolomic analysis. In previous studies, we have successfully applied GC-TOF/MS- or GC-MS-based metabolomic approaches to study hyperlipidemia and osteoporosis by analyzing the serum or urine of rats or humans14,15,16.
Based on global and targeted metabolomic profiling, several endogenous metabolites that serve as safety biomarkers during treatment with atorvastatin (a statin) in hyperlipidemic SD rats have been developed17. Strauss and Vassallo et al revealed an increased toxicity in healthy rats when fibrates and statins were co-administered using a metabolomic approach18,19. However, little information on the metabolomic study of hyperlipidemia in a hyperlipidemia model has been reported in the literature.
The aim of this study was to investigate the lipid-regulating effects and adverse reactions of simvastatin or fenofibrate in diet-induced hyperlipidemia rats using a GC/MS-based metabolomic approach. The traditional pharmacology methods monitoring the dynamic circumstances in the pathological state were used to provide complementary information such as the total cholesterol (TC), triglyceride (TG) and high-density lipoprotein cholesterol (HDL-C) levels. We also aimed to illuminate the differences in the lipid-regulating effects and metabolic pathways between the simvastatin and fenofibrate treatment of hyperlipidemia rats using GC/MS-based metabolic profiling. This study may provide the evidence needed to allow for a clinical, rational administration treatment of hyperlipidemia patients with fibrates and statins.
Materials and methods
Chemicals and reagents
Simvastatin and fenofibrate were obtained from the Zhejiang Jiangbei Pharmaceutical Co, Ltd (Taizhou, China) and the Nhwa Pharma Corporation (Xuzhou, China), respectively. Commercial assay kits for total cholesterol (TC), triglycerides (TG), low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol (HDL-C) were purchased from Shanghai Rongsheng Biotechnology Co, Ltd (Shanghai, China). The [2H6]- salicylic acid (97%) that was used as an internal standard (IS) was purchased from the Cambridge Isotope Laboratories Inc (Andover, MA, USA). Alkane series (C8–C40), N-methyl-N-trimethylsilyl-trifluoroacetamide (MSTFA) and trimethyl chlorosilane (TMCS) were obtained from Fluka (Buchs, Switzerland). Methoxyamine was obtained from Supelco (Bellefonte, PA, USA). The HPLC-grade methanol, pyridine and n-heptane were purchased from the Sigma-Aldrich Corporation (St Louis, MO, USA). Deionized water was produced by a Milli-Q Reagent Water System (Millipore, MA, USA).
Animals, diets, and sample preparation
Thirty-two Sprague-Dawley (SD) male rats (age: 4 months) with a body weight of 200±10.0 g were obtained from the Sino-British Sippr/BK Lab Animal Ltd (Shanghai, China). The rats were acclimatized under controlled conditions (room temperature 20±1 °C, relative humidity 50%±5% and a 12 h light/12 h dark cycle) with standard solid food for six days prior to the experiments. This study was conducted in conformity with the Guidelines for Animal Experiments of Nanjing University of Technology (Nanjing, China).
The 32 rats were randomly assigned to four groups (n=8). The rats in the normal control group were fed with a normal diet from weeks 1–6. The rats in the hyperlipidemia model control group were fed with a high-lipid diet enriched with 1% (w/w) cholesterol, 10% (w/w) lard, 0.2% propylthiouracil, 5% egg yolk and 1% sodium tauroglycocholate14 during the first four weeks, followed by a normal diet in weeks 5–6 without treatment. Both the simvastatin and fenofibrate treatment groups were fed with a high-lipid diet in weeks 1−4 and a normal diet with simvastatin (10 mg/kg daily) and fenofibrate (150 mg/kg daily), respectively, in weeks 5 and 6. The diet and treatment plans for the four groups are listed in Table 1. Simvastatin and fenofibrate were orally administered as a suspension in water via gavage. Before and during the experiment, once a week for 6 weeks at 8:00 AM, 2 mL of blood was collected into tubes containing EDTA via retro-orbital bleeding from each rat under fasting conditions (but water ad libitum for 12 h). Plasma samples were obtained by centrifugation of the blood samples at 2000×g for 10 min at 4 °C; the samples were then divided into two equal parts in two new Eppendorf tubes. One portion was used for the detection of TC, TG, LDL-C and HDL-C levels using the commercial assay kits, and the other portion was used for the metabolomic analysis. All of the plasma samples were stored at −80 °C until analysis.
Table 1. Diet and treatment plans for all the rat groups. n=8.
| Group | Week 1–4 | Week 5–6 |
|---|---|---|
| Normal control | Normal diet | Normal diet |
| Model control | High-lipid diet | Normal diet |
| Simvastatin-treatment | High-lipid diet | Normal diet & Simvastatin (10 mg·kg−1·d−1) |
| Fenofibrate-treatment | High-lipid diet | Normal diet & Fenofibrate (150 mg·kg−1·d−1) |
Determination of TC, TG, LDL-C, and HDL-C
The levels of TC, TG, LDL-C and HDL-C in the rat plasma were determined using enzyme methods with commercial kits according to the manufacturers' protocols. The results obtained were compared with those determined in the metabolomics study.
Metabolomic studies
Plasma preparation
According to previous reports14,20, 400 μL of methanol containing 10 μL of an internal standard (IS) ([2H6]-salicylic acid, 1 mg/mL) was added to each plasma sample (100 μL) for one-step protein precipitation. The samples were vortex-mixed for 5 min and centrifuged at 12000×g for 10 min at 4 °C. The supernatant (100 μL) was transferred to a 1-mL GC vial and evaporated to dryness under vacuum in a SpeedVac concentrator. Subsequently, 50 μL of methoxyamine in pyridine (15 μg/mL) was added to dissolve each dried residue by vigorously vortexing for 10 min. The redissolved solution was trimethylsilylated for an additional 1 h by adding 50 μL of MSTFA with 1% TMCS as a catalyst after the methoxamination reaction proceeded for 16 h at room temperature. Then, 40 μL of heptane was added to each GC vial, and the vial was vortexed for 10 min before 1 μL of the derivative sample was taken for GC-MS analysis using a Finigan TRACE DSQ gas chromatograph (Thermo Finigan, San Jose, CA, USA) in splitless mode, as described below.
GC/MS analysis
The chromatographic separation was performed on a fused silica capillary column (30 m×0.25 mm ID) that was chemically bonded with a 0.25 μm DB1-MS stationary phase (J&W scientific, Folsom, CA, USA). Helium served as the carrier gas through the column. The GC/MS was operated under the following conditions: the injector temperature was at 270 °C, and the septum purge was turned on 1 min after injection and set to a flow rate of 20 mL/min. The gas flow rate through the column was 1 mL/min. The gradient column temperature program underwent the following steps: (1) 70 °C for 2 min; (2) 70 °C to 240 °C at a rate of 20 °C/min; (3) held at 240 °C for 1 min; and (4) elevated to 320 °C at a rate of 20 °C/min and held at 320 °C for 1 min. The transfer line temperature was maintained at 270 °C, and the ion source temperature was maintained at 200 °C. The ion source voltage was a 70 eV electron beam at a current of 2.0 mA. After a solvent delay of 4 min, the acceleration voltage was turned on, and the mass spectra were acquired in scan mode over the range from 50 to 650 m/z at an interval of 2 s14.
Data and statistical analysis
The mass spectrometry (MS)-based platform involves GC-single quad MS profiling followed by data analysis with the Xcalibur software (Thermo Finigan, San Jose, CA, USA), as reported previously21. The retention times were corrected, and all of the peaks were aligned using the internal standards to minimize batch errors. Peaks with intensities higher than 30-fold of the ratio of the signal-to noise (S/N) were recorded. The retention index for each peak was obtained by comparing its retention time with those of the alkane series (C8-C40). All endogenous compounds were identified and assigned by the AMDIS (Automatic Mass Spectral Deconvolution and Identification System) according to the MS spectra and retention index. The endogenous compounds were validated with the authentic reference standards listed in the NIST 2.0 (2005), the Wiley library and the in-house spectra library constructed in the Key Laboratory of Drug Metabolism and Pharmacokinetics, China Pharmaceutical University. In addition, potential metabolites and their physiological roles were searched on the Human Metabolome Database (http://www.hmdb.ca)22.
The multivariate statistical analysis (MVSA) technique and principal component analysis (PCA) were performed for the metabolomic data using the SIMCA-P v11 software (Umetrics, Umea, Sweden)23,24. The data were analyzed using a one-way analysis of variance (ANOVA) followed by a least significant difference (LSD) test, with statistical significance set at P<0.01 or P<0.05.
Results
Assay kit results
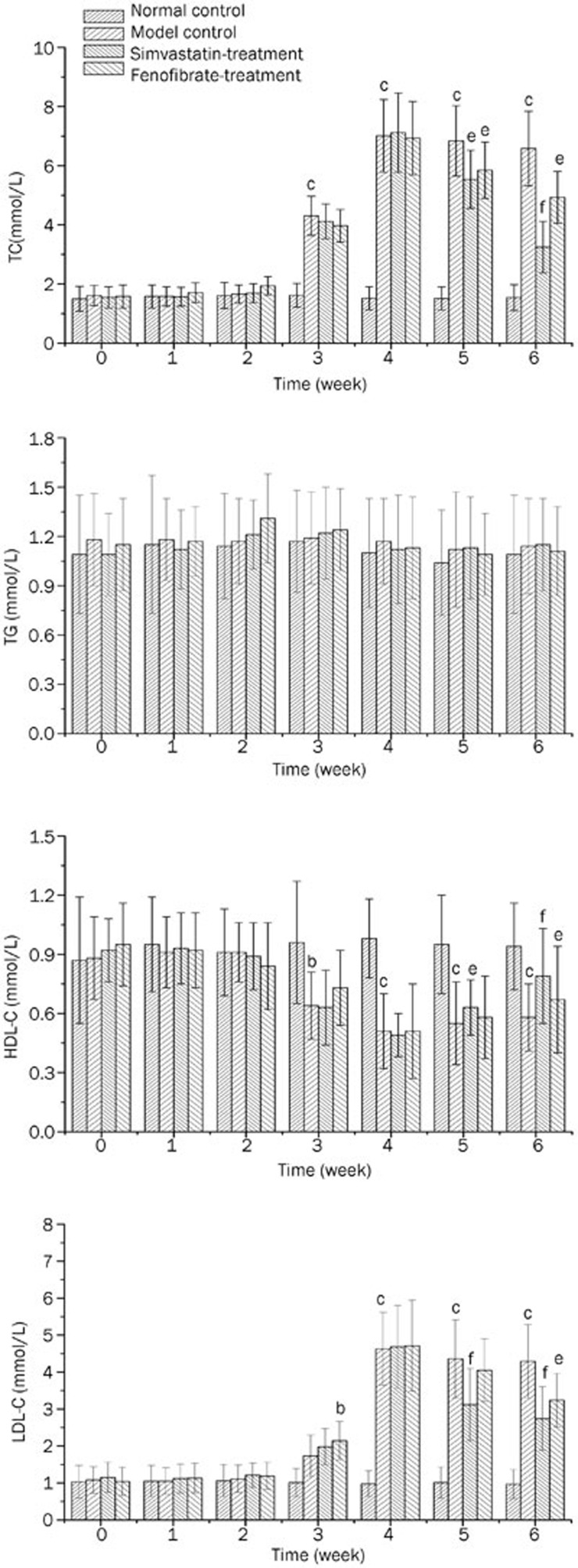
Varying trends in the plasma lipid levels were observed in the normal control, model control, simvastatin treatment and fenofibrate treatment groups (Figure 1). The TC, TG, LDL-C and HDL-C levels were observed to be stable in the normal control group during the six weeks of the experiment. Compared with the normal control rats, significant increases in both the TC (P<0.01) and LDL-C levels (P<0.05) were observed in the model control rats from week 3 post-diet induction (Figures 1A, 1D). The level of HDL-C in the plasma gradually decreased in the model control rats (P<0.05) starting in the 3rd week (Figure 1C).
Figure 1.
The plasma lipid levels of the rats from the four groups during the experiment. (A) TC levels; (B) TG levels; (C) HDL-C levels and (D) LDL-C levels. bP<0.05, cP<0.01 vs normal control group. eP<0.05, fP<0.01 vs model control group.
The results clearly showed that the oral administration of simvastatin or fenofibrate to the rats with lipid metabolic disorders resulted in the opposite effects, with a significant decrease in the TC (P<0.01 for simvastatin) and LDL-C levels (P<0.01/0.05 for simvastatin/fenofibrate) and an increase in the HDL-C levels (P<0.05 for simvastatin) in the 5th and 6th weeks, which represented the first and second weeks after treatment with the drugs.
Metabolomic profiling by GC/MS
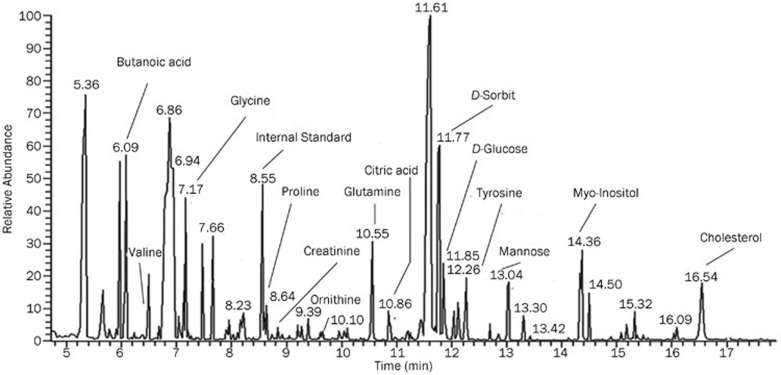
A typical GC-MS (total ion current) chromatogram with some chromatographic peaks of representative metabolites is shown in Figure 2. All of the data from the normal control (K), model control (M), fenofibrate treatment (B) and simvastatin treatment (T) groups during the 6-week experiment were processed using PCA to calculate the basic model and an overview of the data.
Figure 2.
A typical GC/MS chromatogram of the plasma samples obtained from a hyperlipidemic rat.
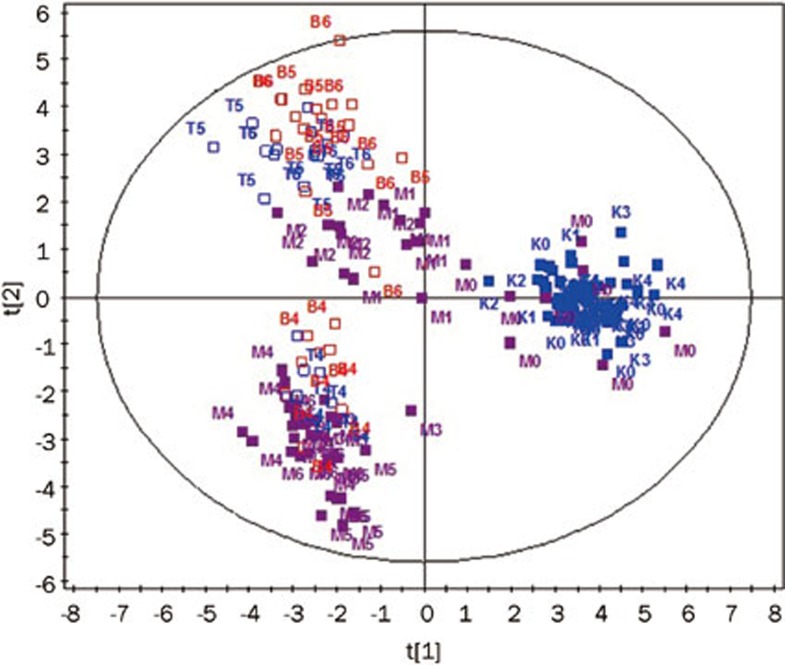
A full-scale map of the PCA scores plot depicting the entire metabolomic profiling of the four groups is shown in Figure 3. Samples from different groups at the same period or the same group in different periods were scattered clearly for the first two components. According to the statistical parameters from the PCA model, these two components explained the R2 value of 52.7% for all of the GC-MS response variables and predicted a Q2 value of 53.1% for all of the observation/sample variables (R2 is the percentage of all of the GC-MS response variables explained by the model; Q2 is a measurement of the predictive ability of the model)15,23. The scores plot shows that the samples from the model control, the fenofibrate treatment and the simvastatin treatment groups changed in a time-dependent manner, whereas the samples from the normal control group were clustered in the same area throughout the experiment.
Figure 3.
The PCA scores plots of the four groups. The letters K, M, T and B denote the following different treatment groups: K (blue) the normal control group; M (purple) the model control group; T (blue hollow) the simvastatin treatment group and B (red) the fenofibrate treatment group. The numbers 1–6 represent the following different time points during the experiment: (0) before the experiment and (1–6) for the weeks during the experiment. For instance, T4 indicates the sample in the simvastatin treatment group at the 4th week after the experiment had begun.
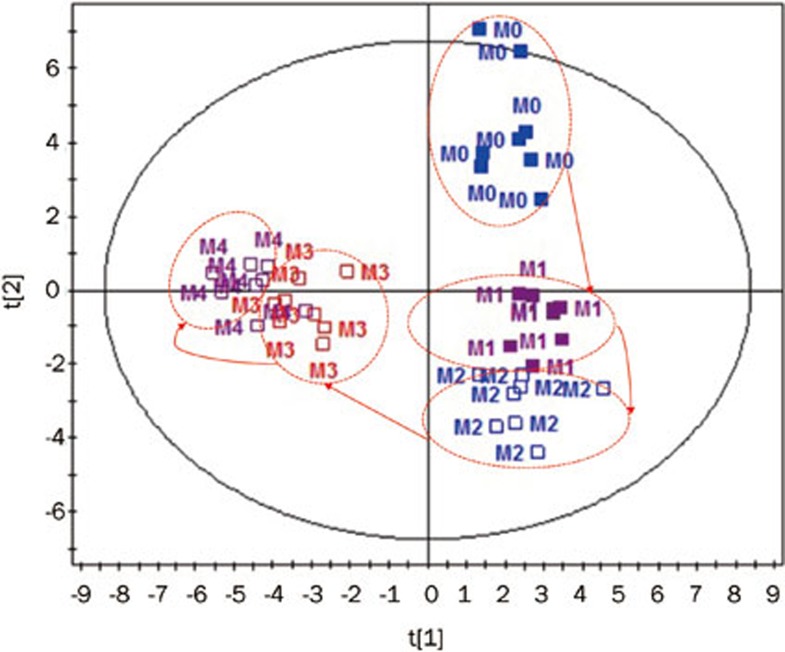
The two principal components analysis model for the model control group (M0–M4) (Figure 4) showed the metabolomic movement during the 4-week high-lipid diet induction, which could be classified clearly, with an R2 of 62.5% and a Q2 of 75.5%. Interestingly, a distinct segregation between the pre-experiment and the diet induction was observed starting in the 2nd week (M2). Significant differences were not found in the classical pathology biomarkers, including TC, TG, LDL-C and HDL-C, in the 2nd week (Figure 1). This possibly indicates that the fingerprint of the metabolome is more sensitive than the classic biochemical indicators.
Figure 4.
The PCA scores plots of the model control group (M) from weeks 1–4.
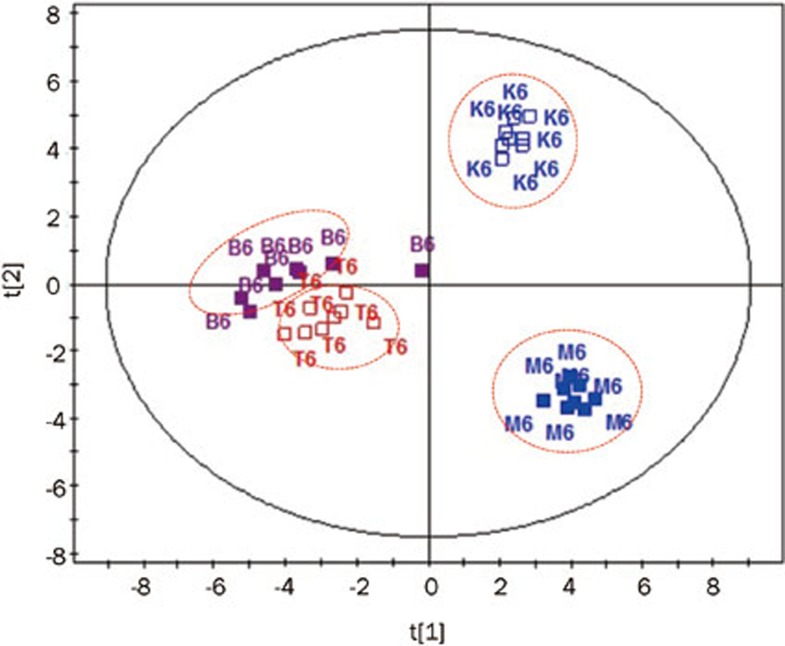
The metabolic patterns changed after treatment with either fenofibrate or simvastatin for two weeks (Figure 5). Interestingly, the fenofibrate treatment (B6) and simvastatin treatment (T6) samples gathered in two different regions, indicating that the rats had different metabolic responses to fenofibrate and simvastatin.
Figure 5.
The PCA scores plots of the samples from the normal control (K), model control (M), simvastatin treatment (T) and fenofibrate treatment group (B) at the 6th week.
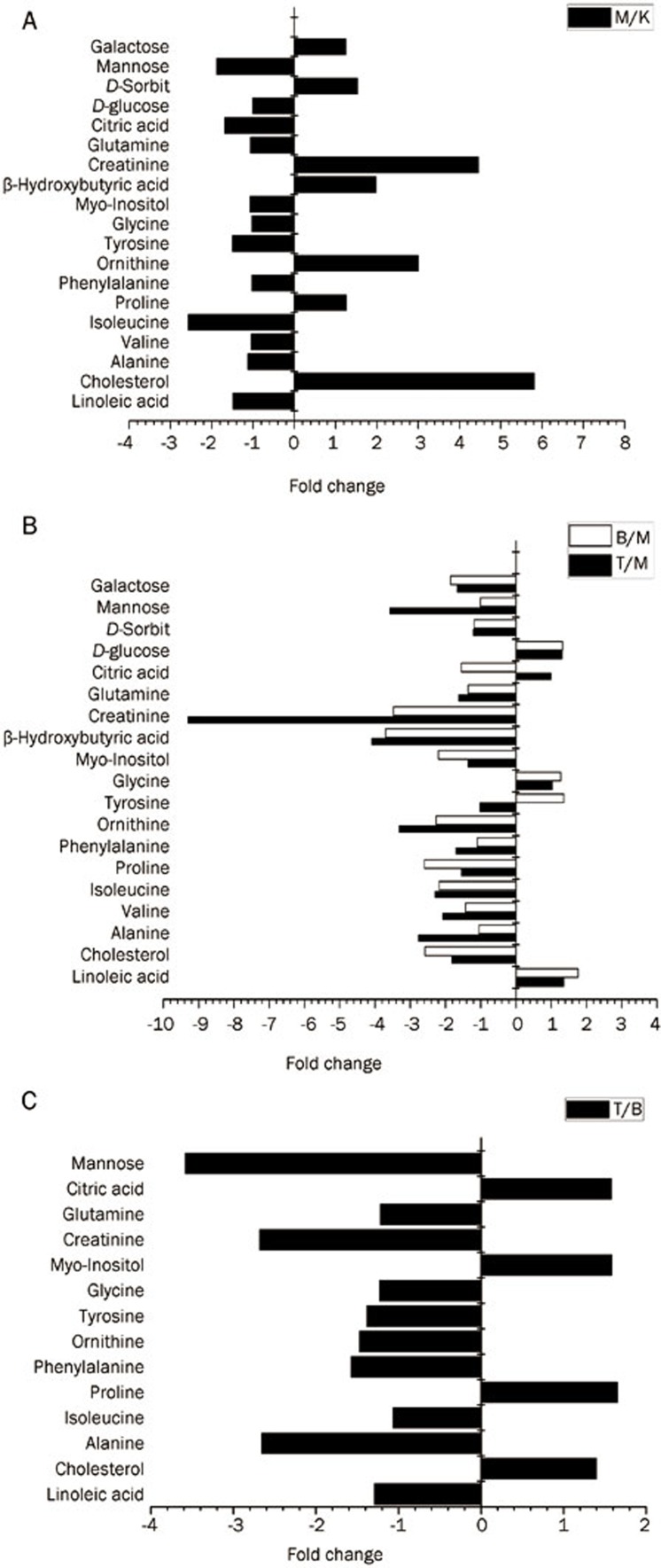
The major metabolites were identified and included a number of fatty acids, amino acids, carbohydrates and other metabolites. These potential biomarkers were further analyzed by ANOVA, and the relative fold change was calculated. Differences in the metabolites in model control, the fenofibrate treatment and the simvastatin treatment groups compared with the normal control group in the 6th week are listed in Figure 6 and Table 2. The fold changes between the model control group and the normal control group suggest that there is a significant increase in the levels of endogenous metabolites, including cholesterol, ornithine, β-hydroxybutyric acid, creatinine, galactose and D-sorbit, and a significant decrease in the levels of linoleate, isoleucine, tyrosine, citrate and mannose in diet-induced hyperlipidemia rats compared with the normal group. The level of cholesterol, a classical biomarker of hyperlipidemia, in both treatment groups was significantly lower than that observed in model control group. Furthermore, metabolite differences existed between the simvastatin and the fenofibrate treatment in 6th week according to the fold change and P value (P<0.05). The levels of linoleic acid, alanine, valine, phenylalanine, ornithine, tyrosine, mannose, glycine, glutamine and creatinine were lower in the plasma of the simvastatin treatment group, and the levels of cholesterol, proline, myo-inositol and critic acid were higher in the fenofibrate-treated animals.
Figure 6.
The relative fold changes of plasma metabolite levels at 6th week. Different groups of rats are represented using the following notation: (A) the model control group (M) versus the normal control group (K); (B) the simvastatin treatment group (T) or fenofibrate treatment group (B) versus the model control group (M); (C) the simvastatin treatment group (T) versus the fenofibrate treatment group (B).
Table 2. Significant alterations of endogenous metabolites detected by GC-MS. bP<0.05, cP<0.01 vs normal control group. eP<0.05, fP<0.01 vs model control group. hP<0.05, iP<0.01 vs fenofibrate-treatment group.
| Metabolites | Biological role | Fold change |
|||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Linoleic acid | An important role in the pathogenesis of essential hypertension | 0.67b | 1.36e | 1.76f | 0.77h |
| Cholesterol | Biomarker of hyperlipidemia | 5.82b | 0.54f | 0.39f | 1.40h |
| Alanine | An metabolic intermediate in both glycolysis and gluconeogenesis | 0.89 | 0.36f | 0.95 | 0.38i |
| Valine | 0.96 | 0.48f | 0.70e | 0.68h | |
| Isoleucine | Essential amino acids, stress, energy and muscle metabolism | 0.39c | 0.43e | 0.46e | 0.94 |
| Proline | Non-essential amino acid synthesized from glutamic acid | 1.26b | 0.64e | 0.38f | 1.65i |
| Phenylalanine | An essential amino acid and the precursor for the amino acid tyrosine | 0.97 | 0.58e | 0.91 | 0.64h |
| Ornithine | Amino acid produced in the urea cycle from arginine | 3.00c | 0.30f | 0.44f | 0.68h |
| Tyrosine | A precursor for epinephrine | 0.67c | 0.98 | 1.36e | 0.72h |
| Glycine | 0.97 | 1.03 | 1.27 | 0.81h | |
| Myo-Inositol | A crucial role in the phosphatidylinositol signaling pathway | 0.93 | 0.72e | 0.45f | 1.59h |
| β-Hydroxybutyric acid | A four-carbon fatty acid | 1.98 | 0.24f | 0.27f | 0.90 |
| Creatinine | An indicator of renal function; A breakdown product of creatine phosphate in muscle | 4.46c | 0.11f | 0.29f | 0.37i |
| Glutamine | A key molecule in cellular metabolism, γ-glutamyl cycle | 0.94 | 0.61f | 0.74e | 0.82h |
| Citric acid | Tricarboxylic acid cycle | 0.59c | 1.02 | 0.64f | 1.58h |
| D-glucose | Primary source of energy, available from glycogenolysis and gluconeogenesis or food | 0.99 | 1.31e | 1.32e | 0.99 |
| D-Sorbit | 1.53c | 0.81e | 0.84e | 0.96 | |
| Mannose | An important roles in protein quality control | 0.53c | 0.28f | 0.99 | 0.28i |
| Galactose | Component of glycerolipid and glycosphingolipid metabolism | 1.24 | 0.59f | 0.54f | 1.10 |
A: Ratio of abundance of metabolites in plasma samples between diet-induced hyperlipidemia group and normal control group at week 6.
B: Ratio of abundance of metabolites in plasma samples between simvastatin-treatment group and model control group at week 6.
C: Ratio of abundance of metabolites in plasma samples between fenofibrate-treatment group and model control group at week 6.
D: Ratio of abundance of metabolites in plasma samples between simvastatin-treatment group and fenofibrate-treatment group at week 6.
Discussion
Metabolomics is a global level tool that is employed in the prognosis or diagnosis of diseases by investigating the endogenous levels of small molecule metabolites in clinical practice5. In this study, metabolomics analysis was applied to systematically monitor and evaluate the development of animal models and the treatment effects of simvastatin and fenofibrate in hyperlipidemia rats. The results showed that the conventional biochemical indicators evaluated by commercial assay kits, ie, TC, TG, HDL-C and LDL-C, changed significantly in the 3rd week of the experiment. In contrast, metabolic data indicated that the model rats administered the high-lipid diet only for 1 week (M1) have been different from the normal rats (M0) in the process of developing hyperlipidemia (Figure 4). Thus, metabolic profiling is a more sensitive and effective approach than the conventional method (based on assay kits) for detecting the disease status and/or biological responses.
The changes in the plasma biochemical indexes showed that the rats had developed a metabolic disorder of lipids and/or lipid proteins in the plasma. This indicated that the diet-induced hyperlipidemia model had been successfully established in the male rats after a continuous four-week administration of a high-lipid diet, which was also verified by the metabolomics analysis in the scores plot.
Simvastatin can competitively inhibit HMG-coenzyme A reductase, an enzyme that catalyzes the rate-limiting step of cholesterol biosynthesis in liver cells25,26,27. It increases the levels of HDL, which has cardiovascular protective effects and reduces the susceptibility of lipoproteins to oxidation in humans25. Fenofibrate acts as a PPAR-α receptor agonist: it increases fatty acid β-oxidation and the excretion of cholesterol via bile28. Even if their mechanisms of action are different, both drugs are able to prevent and treat hyperlipidemia efficiently. Following the oral administration of 10 mg·kg−1·d−1 simvastatin or 150 mg·kg−1·d−1 fenofibrate for two weeks, rats with diet-induced hyperlipidemia showed a trend of reversal in TC, HDL-C and LDL-C levels. The metabolic profiles of the normal control, the model control, the fenofibrate treatment and the simvastatin treatment groups could be clearly distinguished from one another using a PCA score plot analysis (Figure 5). Numerous endogenous molecules, including fatty acids, cholesterol, amino acids and carbohydrates, in the model control group exhibited significant differences (P<0.05) compared with the normal control group. It is proposed that further treatment for 2 weeks may continue to improve the hyperlipidemia rats; however, there are no data to support this notion.
Many potential biomarkers (Table 2) significantly changed and normalized following treatment with simvastatin or fenofibrate. Interestingly, some of the biomarkers, such as cholesterol, β-hydroxybutyric acid, linoleic acid, creatinine and ornithine, changed with the same trend in both treatment groups, but other biomarkers, including tyrosine, varied in an opposite manner.
Cholesterol is an important biomarker of hyperlipidemia, and its level significantly decreased after two weeks of simvastatin or fenofibrate therapy. Endogenous cholesterol synthesis by acetyl-CoA should be lower, but the β-oxidation of fatty acids should be higher in the diet-induced hyperlipidemia rats because of the feedback regulation during excessive lipid metabolism. These processes should result in an accumulation of acetyl-CoA in vivo14. The excess of acetyl-CoA may have been converted to acetone bodies, as indicated by the high level of β-hydroxybutyric acid in the model control group. The lipid-lowering effect led to a decrease in the β-hydroxybutyric acid level in the plasma after simvastatin or fenofibrate therapy in our study. Linoleic acid (LA, 18:2n-6) is an essential fatty acid (EFA) in human nutrition and thus exclusively reflects the dietary intake29. LA also plays an important role in reducing the plasma TC and LDL-C levels30. As a polyunsaturated fatty acid (PUFA), LA is a precursor of prostaglandins (PGs) via the arachidonic acid (AA) pathway. PGs have many beneficial effects against cardiovascular risk, including hyperlipidemia and essential hypertension31,32. Ornithine, a central intermediate metabolite of the urea cycle, was notably increased in the rats induced with hyperlipidemia by the high-cholesterol diet. This trend was quickly inversed after treatment for 2 weeks with either simvastatin or fenofibrate, which may be caused by the dysfunction in lipid metabolism33,34. This finding indicates that ornithine might be a potential biomarker of the pathological process and lipid regulation in hyperlipidemia.
Tyrosine is an essential amino acid that might also be a potential biomarker for hyperlipidemia14. Tyrosine is a precursor to catecholamines (epinephrine, norepinephrine and dopamine) and is known to promote lipid metabolism35. Epinephrine has been shown to cause an increase in endogenous lipid hydrolysis in muscle and adipose tissue36. Epinephrine intervention has also been shown to alter lipid distribution, lipid transport and phospholipid metabolism in the liver, aorta and plasma37. A decrease in the plasma tyrosine levels was observed in the hyperlipidemia rats (0.67-fold), possibly due to the excessive lipid metabolism in response to the high-lipid diet. The tyrosine levels in the plasma were further increased in the fenofibrate treatment group. In this study, the effects of fenofibrate on lipid metabolism regulation were verified by examining its effects on the epinephrine pathway and evaluating the tyrosine levels. Data from many studies in rodents and humans indicate that fibrates promote the hydrolysis of triglycerides, thereby enhancing the decomposition rate of many fatty acids and stimulating reverse cholesterol transport, which may relieve stress or substitute for the regulation of the lipid metabolism pathway by epinephrine38,39,40. In contrast to the fenofibrate treatment, no significant alterations in the tyrosine levels were observed in the simvastatin treatment group, possibly because simvastatins competitively inhibit the biosynthesis of cholesterol but do not intervene in lipid transport or lipolysis.
Creatinine is a product of creatine and phosphocreatine in muscle41. They both play a vital role in regenerating adenosine triphosphate (ATP) in skeletal muscle to energize muscle contraction through the creatine kinase (CK) reaction42. Creatinine transfers from the blood plasma to the kidneys before eventually being eliminated from the body by glomerular filtration and partial tubular excretion43,44. Serum creatinine is an important indicator of renal health because it is an easily measured byproduct of muscle metabolism44. The creatinine levels increased significantly by 4.46-fold in the diet-induced hyperlipidemia group compared with the normal animals. The accumulation of plasma creatinine may indicate a potential risk for or existing renal damage caused by hyperlipidemia41,45. The creatinine levels in the plasma significantly decreased following treatment with simvastatin (0.11-fold) or fenofibrate (0.29-fold). These results suggest, for the first time, that both simvastatin and fenofibrate could prevent renal damage caused by lipid disorders. However, further examination is required to confirm this hypothesis.
Conclusion
A metabolomic profiling approach based on a GC-MS assay has been successfully applied to monitor the pathological changes in diet-induced hyperlipidemia rats and the therapeutic processes associated with simvastatin or fenofibrate treatment. Metabolic profiling is a more sensitive and effective approach compared with the conventional methods (based on assay kits) that are used to detect disease status or biological responses. A series of potential biomarkers, including tyrosine, creatinine, LA, β-hydroxybutyric acid and ornithine, were identified by global metabolomics and may be used to identify the metabolic effects of hyperlipidemia during disease progression. Further studies of these potential biomarkers would provide more useful and valuable information for the rational clinical usage of statins or fibrates.
Author contribution
Qiu-yu XU and Yin-hui LIU were involved in all phases of this project. Qi ZHANG and Zi-mei WU designed the research project. Lei LIU and Di YAO contributed analytic tools. Guang-bo CUI and Jing-jing SUN performed the animal experiments in the project. Bo MA and Zhen-dong YANG analyzed the data.
Acknowledgments
This study was supported by a grant from the “National Natural Science Foundation of China (Grant No 81373478, 81302835)” and from the “Specialized Research Fund for the Clinical Medicine of Jiangsu Province (Grant No SBL201330086)”, and from the “Jiangsu Province Science Foundation for Youths (No BK20130951).”
References
- Jain KS, Kathiravan MK, Somani RS, Shishoo CJ. The biology and chemistry of hyperlipidemia. Bioorg Med Chem. 2007;15:4674–99. doi: 10.1016/j.bmc.2007.04.031. [DOI] [PubMed] [Google Scholar]
- Farnier M, Davignon J. Current and future treatment of hyperlipidemia: the role of statins. Am J Cardiol. 1998;82:3J–10J. doi: 10.1016/s0002-9149(98)00423-8. [DOI] [PubMed] [Google Scholar]
- Fazio S, Linton MF. The role of fibrates in managing hyperlipidemia: mechanisms of action and clinical efficacy. Curr Atheroscler Reports. 2004;6:148–57. doi: 10.1007/s11883-004-0104-8. [DOI] [PubMed] [Google Scholar]
- Staels B, Dallongeville J, Auwerx J, Schoonjans K, Leitersdorf E, Fruchart JC. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation. 1998;98:2088–93. doi: 10.1161/01.cir.98.19.2088. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- Kanani H, Chrysanthopoulos PK, Klapa MI. Standardizing GC-MS metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:191–201. doi: 10.1016/j.jchromb.2008.04.049. [DOI] [PubMed] [Google Scholar]
- Koek MM, van der Kloet FM, Kleemann R, Kooistra T, Verheij ER, Hankemeier T. Semi-automated non-target processing in GC x GC-MS metabolomics analysis: applicability for biomedical studies. Metabolomics. 2011;7:1–14. doi: 10.1007/s11306-010-0219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang Q, Yin P, Xing W, Wu Z, Chen S, et al. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal Bioanal Chem. 2012;403:203–13. doi: 10.1007/s00216-012-5782-4. [DOI] [PubMed] [Google Scholar]
- Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, et al. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011;71:6590–600. doi: 10.1158/0008-5472.CAN-11-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Shah YM, Morimura K, Krausz KW, Miyazaki M, Richardson TA, et al. Metabolomics reveals that hepatic stearoyl-CoA desaturase 1 downregulation exacerbates inflammation and acute colitis. Cell Metab. 2008;7:135–47. doi: 10.1016/j.cmet.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee EP, Gerszten RE. Metabolomics and cardiovascular biomarker discovery. Clin Chem. 2012;58:139–47. doi: 10.1373/clinchem.2011.169573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–26. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie RD, Beyan H. Metabolomics makes a mark: early changes associated with autoimmune diabetes. Diabetes. 2011;60:2688–90. doi: 10.2337/db11-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang GJ A JY, Wu D, Zhu LL, Ma B, et al. Application of GC/MS-based metabonomic profiling in studying the lipid-regulating effects of Ginkgo biloba extract on diet-induced hyperlipidemia in rats. Acta Pharmacol Sin. 2009;30:1674–87. doi: 10.1038/aps.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Zhang Q, Wang GJ A JY, Wu D, Liu Y, et al. GC-TOF/MS-based metabolomic profiling of estrogen deficiency-induced obesity in ovariectomized rats. Acta Pharmacol Sin. 2011;32:270–8. doi: 10.1038/aps.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Liu J, Zhang Q, Ying H A J, Sun J, et al. Metabolomic profiles delineate signature metabolic shifts during estrogen deficiency-induced bone loss in rat by GC-TOF/MS. PloS One. 2013;8:e54965. doi: 10.1371/journal.pone.0054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar BS, Lee YJ, Yi HJ, Chung BC, Jung BH. Discovery of safety biomarkers for atorvastatin in rat urine using mass spectrometry based metabolomics combined with global and targeted approach. Anal Chim Acta. 2010;661:47–59. doi: 10.1016/j.aca.2009.11.063. [DOI] [PubMed] [Google Scholar]
- Strauss V, Mellert W, Wiemer J, Leibold E, Kamp H, Walk T, et al. Increased toxicity when fibrates and statins are administered in combination--a metabolomics approach with rats. Toxicol Lett. 2012;211:187–200. doi: 10.1016/j.toxlet.2012.03.798. [DOI] [PubMed] [Google Scholar]
- Vassallo JD.Integration of transcriptomic and metabolomic profiling to identify mechanisms and biomarkers of statin-induced myopathy [dissertation]. Bethlehem (USA): Lehigh University; 2012
- A J, Trygg J, Gullberg J, Johansson AI, Jonsson P, Antti H, et al. Extraction and GC/MS analysis of the human blood plasma metabolome. Anal Chem. 2005;77:8086–94. doi: 10.1021/ac051211v. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang G, Du Y, Zhu L A J. GC/MS analysis of the rat urine for metabonomic research. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;854:20–5. doi: 10.1016/j.jchromb.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35 (Database issue):D521–6. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Enciso M, Tenenhaus M. Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Human Genet. 2003;112:581–92. doi: 10.1007/s00439-003-0921-9. [DOI] [PubMed] [Google Scholar]
- Lu Y A J, Wang G, Hao H, Huang Q, Yan B, et al. Gas chromatography/time-of-flight mass spectrometry based metabonomic approach to differentiating hypertension- and age-related metabolic variation in spontaneously hypertensive rats. Rapid Commun Mass Spectrom. 2008;22:2882–8. doi: 10.1002/rcm.3670. [DOI] [PubMed] [Google Scholar]
- Stancu C, Sima A. Statins: mechanism of action and effects. J Cell Mol Med. 2001;5:378–87. doi: 10.1111/j.1582-4934.2001.tb00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunwald E, Sacks FM, Pfeffer MA, Ridker PM. HMG CoA reduction in patients with average cholesterol concentrations. Clin Chem. 2011;57:1072–3. doi: 10.1373/clinchem.2011.164038. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd RA. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–21. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Jeong S, Nicol CJ, Lee H, Han M, Kim JJ, et al. Fenofibrate regulates obesity and lipid metabolism with sexual dimorphism. Exp Mol Med. 2002;34:481–8. doi: 10.1038/emm.2002.67. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Saitoh M, Moriuchi A, Nomura M, Okuyama H. Effect of dietary alpha-linolenate/linoleate balance on brain lipid compositions and learning ability of rats. J Lipid Res. 1987;28:144–51. [PubMed] [Google Scholar]
- Wijendran V, Hayes KC. Dietary n-6 and n-3 fatty acid balance and cardiovascular health. Annu Rev Nutr. 2004;24:597–615. doi: 10.1146/annurev.nutr.24.012003.132106. [DOI] [PubMed] [Google Scholar]
- Russo GL. Dietary n-6 and n-3 polyunsaturated fatty acids: from biochemistry to clinical implications in cardiovascular prevention. Biochem Pharmacol. 2009;77:937–46. doi: 10.1016/j.bcp.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Das UN. Essential fatty acids: biochemistry, physiology and pathology. Biotech J. 2006;1:420–39. doi: 10.1002/biot.200600012. [DOI] [PubMed] [Google Scholar]
- Gu S A J, Wang G, Zha W, Yan B, Zhang Y, et al. Metabonomic profiling of liver metabolites by gas chromatography-mass spectrometry and its application to characterizing hyperlipidemia. Biomed Chromatogr. 2010;24:245–52. doi: 10.1002/bmc.1279. [DOI] [PubMed] [Google Scholar]
- Sugino T, Shirai T, Kajimoto Y, Kajimoto O. L-ornithine supplementation attenuates physical fatigue in healthy volunteers by modulating lipid and amino acid metabolism. Nutr Res. 2008;28:738–43. doi: 10.1016/j.nutres.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Wyngaarden JB. Precursors of adrenal epinephrine and norepinephrine in vivo. Biochim Biophys Acta. 1956;20:48–52. doi: 10.1016/0006-3002(56)90261-x. [DOI] [PubMed] [Google Scholar]
- Peters SJ, Dyck DJ, Bonen A, Spriet LL. Effects of epinephrine on lipid metabolism in resting skeletal muscle. Am J Physiol. 1998;275:E300–9. doi: 10.1152/ajpendo.1998.275.2.E300. [DOI] [PubMed] [Google Scholar]
- Dury A. Effects of epinephrine on lipid partition and metabolism in the rabbit. Circ Res. 1957;5:47–53. doi: 10.1161/01.res.5.1.47. [DOI] [PubMed] [Google Scholar]
- Patterson AD, Slanar O, Krausz KW, Li F, Hofer CC, Perlik F, et al. Human urinary metabolomic profile of PPARalpha induced fatty acid beta-oxidation. J Proteome Res. 2009;8:4293–300. doi: 10.1021/pr9004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SM. Lipid metabolome-wide effects of the PPARgamma agonist rosiglitazone. J Lipid Res. 2002;43:1809–17. doi: 10.1194/jlr.m200169-jlr200. [DOI] [PubMed] [Google Scholar]
- Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–4. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80:1107–213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- Guerrero-Ontiveros ML, Wallimann T. Creatine supplementation in health and disease. Effects of chronic creatine ingestion in vivo: down-regulation of the expression of creatine transporter isoforms in skeletal muscle. Mol Cell Biochem. 1998;184:427–37. [PubMed] [Google Scholar]
- Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- Lassnigg A. Minimal changes of serum creatinine predict prognosis in patients after cardiothoracic surgery: a prospective cohort study. J Am Soc Nephrol. 2004;15:1597–605. doi: 10.1097/01.asn.0000130340.93930.dd. [DOI] [PubMed] [Google Scholar]
- Keane WF, Mulcahy WS, Kasiske BL, Kim Y, O'Donnell MP. Hyperlipidemia and progressive renal disease. Kidney Int Suppl. 1991;31:S41–8. [PubMed] [Google Scholar]