Abstract
Introduction
the use of Next Generation Sequencing (NGS) in the diagnosis of rare genetic pathologies is becoming ever more widespread in clinical practice. The following study reports the first case of preimplantation diagnosis through NGS of a form of LAMA2-related muscular dystrophy.
Case report
a couple came to our Reproductive Medicine Centre for a preconceptional genetic consultation and for advice regarding secondary infertility. The couple already had a 3-year-old child who was suffering from a form of muscular dystrophy that has yet to be genetically defined. The disease had been diagnosed at the age of 6 months. A blood sample was taken from both parents and the child in order to analyze the DNA through the Illumina NextSeq 500 platform and an enrichment protocol, Trusight One Sequencing Panel, created by Illumina for the simultaneous sequencing of the exon regions of 4,813 clinically relevant genes. This led to the identification of 2 point mutations in the LAMA2 gene, each inherited by a parent. The couple then underwent a cycle of IVF (in vitro fertilization). A preimplantation genetic diagnosis was carried out on the embryos obtained after setting up a protocol for the analysis of a point mutation in the LAMA2 gene, (this mutation has yet to be described in literature) and the normal embryos together with the recessive LAMA2-related muscular dystrophy related carriers were transferred. There were no complications during pregnancy, which terminated with a cesarean section at 39 weeks and the birth of healthy 3430-gram baby.
Conclusions
given its robustness, reliability and reproducibility, NGS could also be useful in prenatal diagnosis. This technique could guarantee an ample and quick analysis of the genes involved in development, making it possible to organize medical interventions during pregnancy and after birth.
Keywords: Next Generation Sequencing, preimplantation genetic diagnosis, Lama2 related muscular dystrophy
Introduction
In recent years, a new technology, Next Generation Sequencing (NGS), has become an important instrument not only for the discovery of disease-provoking genes in research but also in clinical diagnostics.
In a recent study, it was demonstrated that the diagnostic result of the sequencing of the entire exome through NGS, for patients suffering from Mendelian diseases, was 25% (1). This figure suggests in what way NGS can be used to integrate conventional prenatal techniques to improve efficiency.
To date, one of the problems associated with Mendelian disorders regards the fact it is difficult to obtain a quick and certain diagnosis. In numerous cases, in fact, an efficient diagnosis can lead to a favourable prognosis of the pathology and make any possible medical intervention quicker. Through the use of a NGS platform, with one experiment, it is possible to identify pathological mutations in an ample number of selected genes that are potentially associated with the disorder being tested. We describe a case regarding the diagnosis of congenital muscular dystrophy, where a couple together with their child were analysed through NGS. The identification of the mutation that caused the pathology made it possible to perform a subsequent preimplantation genetic diagnosis (PGD) in the IVF cycle (in vitro fertilization) that the couple was about to undergo.
Case report
Family medical history
A couple of parents, (the father, 34 and the mother, 26 years of age) in good health and with no family history of well-known genetic diseases, came to our centre for preconception genetic counseling before undergoing a planned cycle of IVF due to a problem of secondary infertility following a tubal pregnancy. The couple already had a 3-year-old child, who was born at the 39th week of pregnancy through a cesarean section due to a breech delivery (3840 gr). In the previous pregnancy, there had been no physiological changes. The child, from the age of 6 months, started to develop progressively ever more serious symptoms such as muscle weakness, hypotonia, limited range of spontaneous movement and deformity, difficulty in swallowing and speech. A study of the family medical history had already revealed a form of muscular dystrophy of unknown genetic origin, given that the more common forms of Duchenne and amyotrophic dystrophy had been excluded.
At the end of the counseling, blood samples were taken from the child and the two parents.
Materials and Methods
DNA EXTRACTION
The DNA was extracted from the blood samples of the family using a QIAamp DNA Blood Mini Kit (Qiagen) following the manufacturer’s instructions.
ANALYSIS THROUGH NEXT GENERATION SEQUENCING (NGS)
Targeted resequencing
The targeted resequencing was performed using an illumina kit; a Trusight one sequencing panel on a NEXTSEQ500 platform. This kit makes it possible to perform enrichment and final analysis of a panel of approximately 5000 genes (http://www.illumina.com/products/trusight-one-sequencing-panel.ilmn).
Library preparation and sequencing
A Trusight one sequencing panel contains all the reagents necessary for the amplification, amplicon enrichment, indexing of the samples and the use of NextSeq 500 without needing any external reagents. Each procedure was realized following the manufacturer’s instructions. For the quantification and validation of the genomic library, the Qubit® 2.0 Fluorometer system (Life techonologies) and 2100 Bioanalyzer Instruments (Agilent Technologies) were used.
Panel of muscular dystrophy pathologies
Considering the family medical history, a panel of genes was compiled comprising approximately 50 genes, which have been reported in literature to be linked to muscular dystrophy (Tab. 1). For analyses, this panel was inserted into the Variant Studio Software (Illumina) so restricting the analysis to genes of interest.
Table 1.
Panel of genes comprising approximately 50 genes, which have been reported in literature to be linked to muscular dystrophy.
| ACTA1 |
| AMPD1 |
| ANO5 |
| CAPN3 |
| COL6A1 |
| COL6A2 |
| COL6A3 |
| DAG1 |
| DES |
| DMD |
| DAG1 |
| DYSF |
| EMD |
| FKRP |
| FKTN |
| LAMA2 |
| LARGE |
| LMNA |
| MYOT |
| NEB |
| PEX1 |
| PEX12 |
| PEX14 |
| PEX2 |
| PEX26 |
| PEX5 |
| PEX6 |
| PMM2 |
| POMGNT1 |
| POMT1 |
| POMT2 |
| RYR1 |
| RYR2 |
| SEPN1 |
| SGCA |
| SGCB |
| SGCD |
| SGCE |
| SGCG |
| SIL1 |
| TCAP |
| TNNI2 |
| TNNT1 |
| TPM2 |
| TPM3 |
| TRIM32 |
| TTN |
Analysis of data
The NEXTSEQ500 system provides fully integrated on-instrument data analysis software. Basespace Reporter software performs secondary analyses on the base calls and Phred-like quality score (Qscore) generated by Real Time Analysis software (RTA) during the sequencing run. The Trusight one sequencing panel workflow in NEXTSEQ500 Reporter evaluates short regions of amplified DNA (amplicons) for variants through the alignment of reads against a “manifest file” specified while starting the sequencing run. The manifest file is provided by Illumina and contains all the information on the custom assay. The workflow requires the reference genome specified in the manifest file (Homo sapiens, hg19, build 37.2). The reference genome provides variant annotations and sets the chromosome sizes in the BAM file output. The Trusight one sequencing panel workflow performs demultiplexing of indexed reads, generates FASTQ files, aligns reads to a reference, identifies variants, and writes output files for the Alignment folder. SNPs and short indels are identified using the Genome Analysis Toolkit (GATK), by default. GATK calls raw variants for each sample, analyzes variants against known variants, and then calculates a false discovery rate for each variant. Variants are flagged as homozygous (1/1) or heterozygous (0/1) in the Variant Call File sample column. Because a SNP database dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP) is available in the Annotation subfolder of the reference genome folder, any known SNPs or indels are flagged in the VCF output file. A reference gene database is available in the Annotation subfolder of the reference genome folder and any SNPs or indels that occur within known genes are annotated.
Each single variant reported in the VCF output file was evaluated for the coverage and the Qscore and visualized via an Integrative Genome Viewer (IGV). Based on the guidelines of the American College of Medical Genetics and Genomics, all regions that were sequenced with a sequencing depth <30 were considered to be unsuitable for analysis. Furthermore, we established a minimum threshold in the Q-score of 30 (base call accuracy of 99.9%). For the variant calling and report, we used Variant Studio software (Illumina).
SANGER VALIDATION
The mutations identified as pathologies were confirmed using the Sanger method following the standard protocol (BigDye® Terminator v3.1 Cycle Sequencing Kit, Applied Biosystems®).
DESIGN OF PGD PRIMERS
The PGD primers were created using a Primer Express (Applied Biosystems®). A nested PCR was performed followed by minisequencing reactions (SNaP-shot® Multiplex Kit, Applied Biosystems®) in order to reveal the mutations in the LAMA2 gene in the blastomeres obtained from embryo biopsies (Tab. 2). Semi-nested PCRs were carried out for the amplification of STRs near the LAMA2 gene (D6S435, D6S407, D6S1690, D6S1620, D6S1705) which were used for the analysis of family linkage. For the PCR, the programme used was 94°C for 10min, 35 cycles at 94°C for 1min, 55°C for 1min and 72°C for 1min, with a final extension at 72°C for 10min for the external PCR and 65°C for 20min for the internal PCR.
Table 2.
Mutations in the LAMA2 gene in the blastomeres obtained from embryo biopsies.
| Lama2 ex13 external forward | AGCCCATCTCCTATCCCGTA |
| Lama2 ex13 external reverse | GTCAGTGTCAAACCAGGCAG |
| Lama2 ex13 internal forward | CCAGCAGTAGGAGGACAGTT |
| Lama2 ex13 internal reverse | CGGGTGAACATGCTCTCAGT |
| Lama2 Q622X C>T (forward) | AGAAGATACAGAACGTGTTCTC |
| Lama2 ex36 external forward | CAGGAACACTCACGGCAAAA |
| Lama2 ex36 external reverse | CACACCAACTCATCTTCAGCA |
| Lama2 ex36 internal forward | TTACCCCCTGCAGCTGTAAA |
| Lama2 ex36 internal reverse | GCCCTTCCAAATTTCTCTCA |
| Lama2 R1706X C>T (reverse) | TTTCTCTCAAAGGCCTCGTCTC |
The fragments were analysed using an Applied Biosystems 3130 Genetic Analyzer and the GeneMapper software.
Results
Identification of mutations
The DNA of the affected child and the two parents was processed during the same analysis. For the identifications, the variants recorded in literature and registered as pathogenetic, plus all the stop and frameshift mutations were taken into consideration. All the mutations identified that were not recorded in literature were examined considering their predictive effect through a Polyphen, Sift and Mutation Taster and their heredity. If the mutation identified in the child derives from only one of the parents (who showed no clinical signs), the penetration value for the disorder associated to the gene was analysed. For every mutation, the depth of the reads was taken into consideration, which under our conditions (platform and sequencing system used), ranged between 120X and 200X, together with the sequence alignment.
The variants identified (Tab. 3) included two stop mutations in the Lama2 gene, p.Gln622Ter and p.Arg1706Ter, which were inherited by a parent and, therefore, present in trans in the affected child. Regarding the other mutations, we did not identify anything pathogenetic given the absence of mutations described and given the direct transmission from one of the parents.
Table 3.
The variants identified in the Lama2 gene.
| Gene | Variant | Chr | Type | Genotype | Exonic | Consequence | PolyPhen | HGVSc HGVSp |
dbSNP ID | Inherited From |
|---|---|---|---|---|---|---|---|---|---|---|
| AMPD1 | T>T/A | 1 | snv | het | yes | missense_variant | probably_damaging (1) |
NM_000036.2:c.959A>T NP_000027.2:p.Lys320Ile |
rs34526199 | FATHER |
| COL6A1 | G>G/A | 21 | snv | het | yes | missense_variant | possibly_damaging (0.817) |
NM_001848.2:c.2549G>A NP_001839.2:p.Arg850His |
rs1053312 | MOTHER |
| COL6A1 | C>C/T | 21 | snv | het | yes | missense_variant | probably_damaging (0.909) |
NM_001848.2:c.2594C>T NP_001839.2:p.Thr865Met |
MOTHER | |
| COL6A2 | G>G/A | 21 | snv | het | yes | missense_variant | probably_damaging (0.987) |
NM_001849.3:c.2039G>A NP_001840.3:p.Arg680His |
rs1042917 | FATHER |
| DAG1 | C>C/T | 3 | snv | het | yes | stop_gained |
NM_001177643.2:c.1627C>T NP_001171114.1:p.Gln543Ter |
MOTHER | ||
| DMD | C>C/A | X | snv | het | yes | stop_gained |
NM_004006.2:c.10759G>T NP_003997.1: p.Glu3587Ter |
MOTHER | ||
| DMD | C>C/A | X | snv | het | yes | missense_variant | probably_damaging (0.941) |
NM_004006.2:c.8163G>T NP_003997.1: p.Lys2721Asn |
MOTHER | |
| LAMA2 | C>C/T | 6 | snv | het | yes | stop_gained |
NM_000426.3:c.1864C>T NP_000417.2: p.Gln622Ter |
FATHER | ||
| LAMA2 | C>C/T | 6 | snv | het | yes | stop_gained |
NM_000426.3:c.5116C>T NP_000417.2: p.Arg1706Ter |
MOTHER | ||
| LAMA2 | C>T/T | 6 | snv | hom | yes | missense_variant | possibly_damaging (0.886) |
NM_000426.3: c.7760C>T NP_000417.2: p.Ala2587Val |
rs6569605, rs2229848 | FATHER |
| LARGE | C>C/A | 22 | snv | het | yes | missense_variant | probably_damaging (0.997) |
NM_004737.4: c.1447G>T NP_004728.1: p.Asp483Tyr |
MOTHER | |
| NEB | C>C/T | 2 | snv | het | yes | missense_variant | probably_damaging (0.999) |
NM_001164507.1:c.21856G>A NP_001157979.1:p.Asp7286Asn |
rs35625617 | MOTHER |
| NEB | C>C/G | 2 | snv | het | yes | missense_variant | probably_damaging (0.997) |
NM_001164507.1:c.7839G>C NP_001157979.1:p.Lys2613Asn |
rs13013209 | FATHER |
| NEB | C>T/T | 2 | snv | hom | yes | missense_variant | probably_damaging (0.937) |
NM_001164507.1:c.4471G>A NP_001157979.1:p.Val1491Met |
rs7426114 | FATHER |
| NEB | T>A/A | 2 | snv | hom | yes | missense_variant | probably_damaging (0.997) |
NM_001164507.1:c.3081A>T NP_001157979.1:p.Lys1027Asn |
rs6735208 | FATHER |
| PEX5 | G>G/C | 12 | snv | het | yes | missense_variant | possibly_damaging (0.744) |
NM_001131023.1:c.178G>C NP_001124495.1:p.Ala60Pro |
MOTHER | |
| SEPN1 | G>A/A | 1 | snv | hom | yes | missense_variant |
NM_020451.2: c.425G>A NP_065184.2: p.Cys142Tyr |
rs7349185 | MOTHER | |
| SEPN1 | C>A/A | 1 | snv | hom | yes | missense_variant |
NM_020451.2: c.1506C>A NP_065184.2: p.Asn502Lys |
rs2294228 | MOTHER | |
| TTN | C>C/T | 2 | snv | het | yes | missense_variant |
NM_001267550.1:c.33287G>A NP_001254479.1:p.Arg11096His |
rs36051007 | MOTHER | |
| TTN | C>C/T | 2 | snv | het | yes | missense_variant |
NM_001267550.1:c.31864G>A NP_001254479.1:p.Gly10622Arg |
rs2244492 | FATHER | |
| TTN | C>T/T | 2 | snv | hom | yes | missense_variant |
NM_001267550.1:c.10726G>A NP_001254479.1:p.Ala3576Thr |
rs6433728 | FATHER | |
| TTN | C>T/T | 2 | snv | hom | yes | missense_variant |
NM_001267550.1:c.10256G>A NP_001254479.1:p.Ser3419Asn |
rs2291310 | FATHER | |
| TTN | C>T/T | 2 | snv | hom | yes | missense_variant |
NM_001267550.1:c.9781G>A NP_001254479.1:p.Val3261Met |
rs2291311 | FATHER | |
| TTN | G>A/A | 2 | snv | hom | yes | missense_variant |
NM_001267550.1:c.3884C>T NP_001254479.1:p.Ser1295Leu |
rs1552280 | MOTHER | |
| TTN | T>C/C | 2 | snv | hom | yes | missense_variant |
NM_001267550.1:c.3601A>G NP_001254479.1:p.Lys1201Glu |
rs10497520 | MOTHER | |
| TTN-AS1, TTN | A>A/G | 2 | snv | het | yes | missense_variant |
NM_001267550.1:c.78674T>C NP_001254479.1:p.Ile26225Thr |
rs12463674 | FATHER | |
| TTN-AS1, TTN | C>G/G | 2 | snv | hom | yes | missense_variant |
NM_001267550.1:c.67246G>C NP_001254479.1:p.Ala22416Pro |
rs4145333 | FATHER |
The clinical conditions associated with LAMA2-related muscular dystrophy overlap perfectly with the family medical history of the affected child.
Sanger confirmation
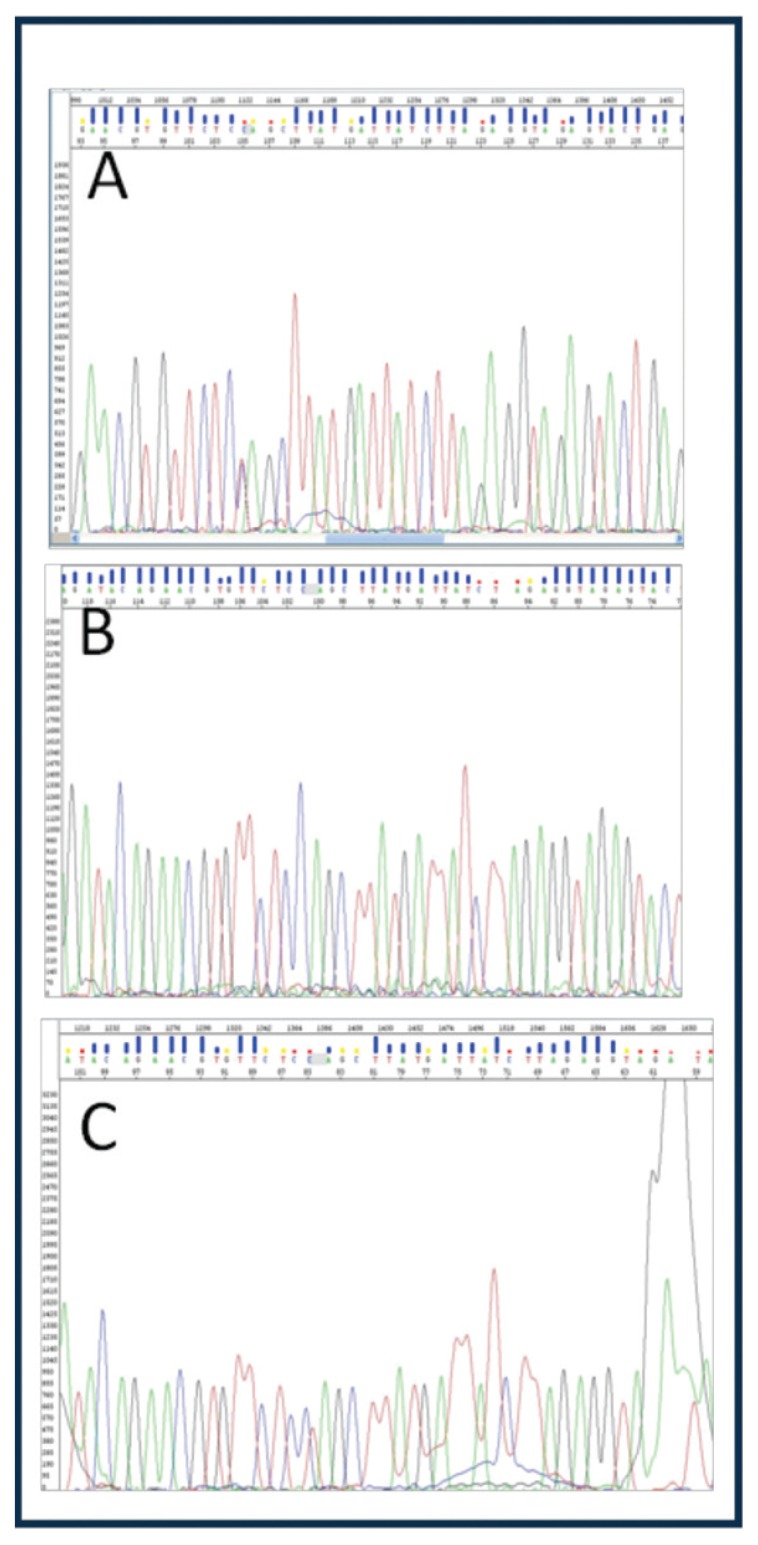
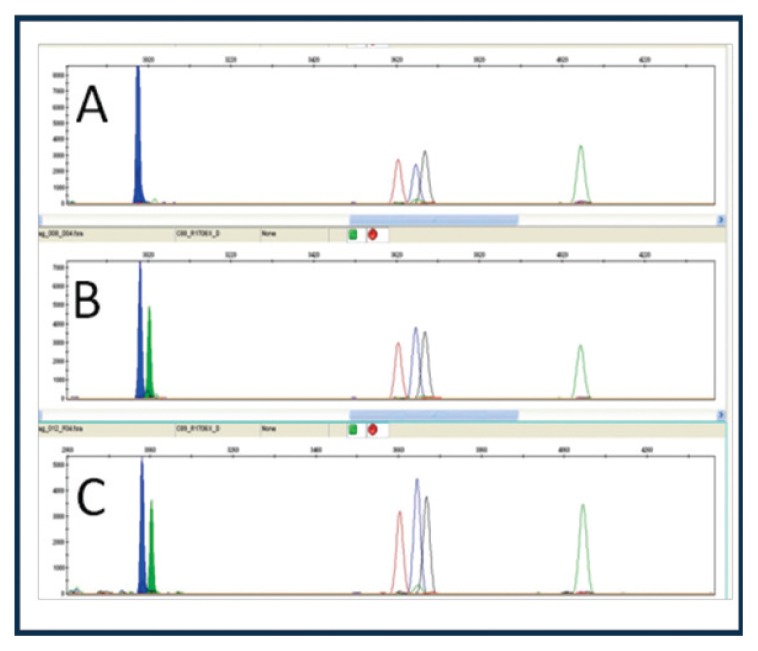
The p.Gln622Ter and p.Arg1706Ter mutations of the Lama2 gene were confirmed using the Sanger method. Both mutations were confirmed in the sample analysed. The affected child was shown to be heterozygous for the two mutations (Figs. 1, 2).
Figure 1.
p.Gln622Ter mutation (c.1864C>T); A: heterozygous father, B: wild type mother, C: heterozygous child.
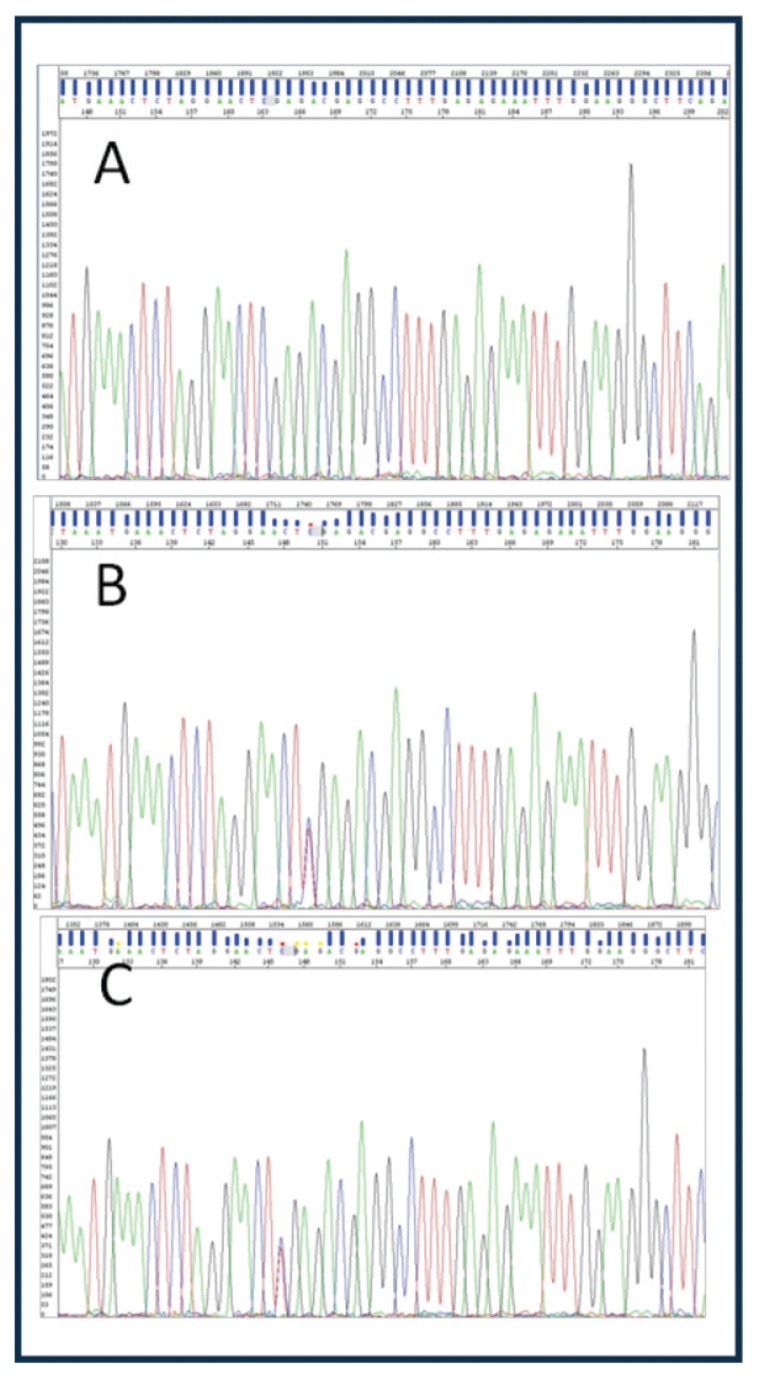
Figure 2.
p.Arg1706Ter mutation (c.5116C>T); A: wild type father, B: heterozygous mother, C: herterozygous child.
PGD protocol
Following the identification of mutations, the diagnostic protocol is then validated which is to be utilized in the preimplantation genetic diagnosis. To this aim, single lymphocyte cells are used which are obtained from the blood samples of the parents and child. This step is necessary in order to establish the amplification efficiency and the ADO ratio (allele drop out) for all the primers that will be used in the clinical study.
The minisequencing protocol is, therefore, used to diagnose the embryos associated with the linkage which makes it possible to exclude any possible aneuploidy and recombination events.
IVF AND A DAY-3 EMBRYO BIOPSY
The patient underwent a microflare ovarian stimulation protocol using gonadotrophin r-FSH and a GnRH antagonist. The stimulation was started during the second day of the spontaneous cycle and was continued for 10 days during which, on alternate days, sonographies were performed and hormonal doses were administered to evaluate the ovarian response. When 17β estradiol in the plasma reaches 2450 and there are 10 follicles with a diameter of > 16, in vitro maturation was induced with hcg and, after 35 hours, a pick up was carried out during the 13th day of the cycle. A total of 9 oocytes were obtained of which 8 were mature and on these an Intracytoplasmatic Sperm Injection (ICSI) was performed with the sperm of the partner. Eight embryos were obtained with a fertilization rate of 100%.
PGD was performed at day3 embryo stage. A hole was created in the pellucid zone by laser-assisted microdissection (Cronus 3 of the Research Instruments). A calcium/magnesium-free medium (Global, Guilford, CT, USA) was used to loosen the cell–cell adhesion allowing a less traumatic suction removal of the cell from the embryo. The biopsied cell, after washing, was then loaded into a PCR tube (Eppendorf) and labelled with the corresponding embryo number and sent to our internal genetic laboratory.
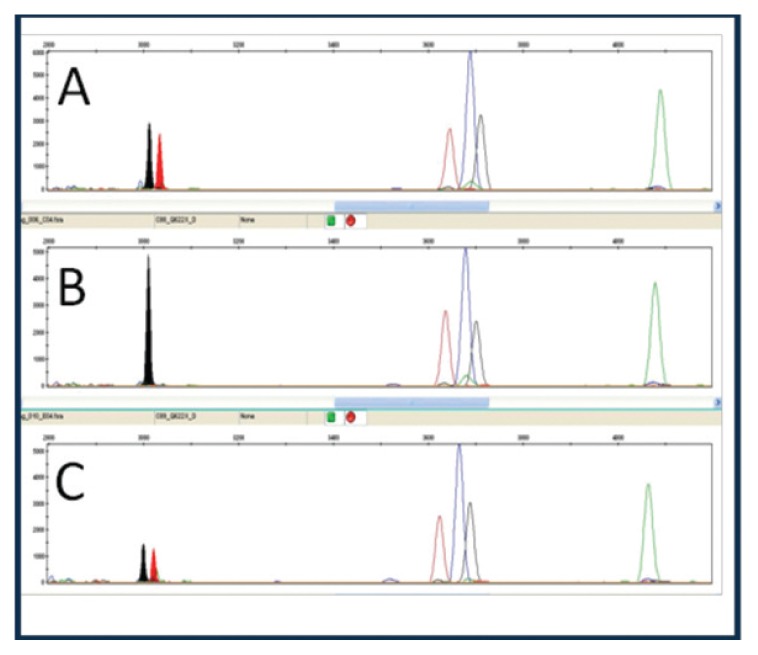
The mutations were analysed through minisequencing reactions (Figs. 3, 4) and the healthy haplotype and the defective haplotype of the parents were identified through STR linkage analysis and compared to the affected child (Tab. 4). Through the preimplantation genetic diagnosis of the embryos, it was possible to identify the affected embryos and carry out the transfer of the carrier and unaffected embryos.
Figure 3.
p.Gln622Ter mutation; A: heterozygous father, B: wild type mother, C: affected heterozygous child.
Figure 4.
p.Arg1706Ter mutation; A: wild type father, B: heterozygous mother, C: affected heterozygous child.
Table 4.
Healthy haplotype and the defective haplotype of the parents compared to the affected child.
| STR | Affected child (bp) | Mother carrier (bp) | Father carrier (bp) | |||
|---|---|---|---|---|---|---|
| D6S435 | 190 | 204 | 190* | 202 | 204** | 220 |
| D6S407 | 210 | 192 | 210 | 218 | 192 | 186 |
| D6S1690 | 144 | 130 | 144 | 160 | 130 | 138 |
| D6S1620 | 184 | 174 | 184 | 188 | 174 | 162 |
| D6S1705 | 115 | 124 | 115 | 117 | 124 | 124 |
affected maternal haplotype
affected paternal haplotype
Discussion
This study analyses the first case report regarding the identification of two mutations associated with a Lama2-gene-related muscular dystrophy using Next Generation Sequencing (NGS).
The LAMA2 gene provides instructions for the alpha-2 subunit of laminin protein. This subunit, together with the beta-1 and gamma-1 subunits, forms the laminin 2 protein, also known as merosin or laminin-211. Laminin 2 and laminin 4 play a particularly important role in the skeletal muscles. LAMA2-related muscular dystrophy is a disorder that causes weakness and wasting (atrophy) of muscles used for movement (skeletal muscles). This condition generally appears in one of two ways: as a severe, early-onset type or a milder, late-onset form (2–4). Affected infants have severe muscle weakness, lack of muscle tone (hypotonia), little spontaneous movement, and joint deformities (contractures). Weakness of the muscles in the face and throat can result in feeding difficulties and an inability to grow and gain weight at the expected rate (failure to thrive). Hypotonia also affects the muscles used for breathing, which causes a weak cry and breathing problems that can lead to frequent, potentially life-threatening lung infections. As affected children grow, they often develop an abnormal, gradually worsening side-to-side curvature of the spine (scoliosis) and inward curvature of the back (lordosis). Children with early-onset LAMA2-related muscular dystrophy usually do not learn to walk unassisted. Speech problems may result from weakness of the facial muscles and tongue, but intelligence is usually normal.
We used a Next Generation Sequencing approach based on the use of a gene panel produced by Illumina including approximately 5000 genes for sequencing and filtering for analysis a pool of genes known to be involved in dystrophinopathies. Genetic mutations identification made it possible to validate a protocol for PGD that could be used for the analyses of the embryos obtained with IVF that the couple had undergone. The analyses of the embryos made it possible to transfer only embryos unaffected or carriers for Lama2-gene-related muscular dystrophy.
The advantage of this approach was found to be the robustness of the experimental design and the results obtained, the reproducibility and speed of execution, which is possible due to the fact it is not necessary to create a customized panel according to the pathology or the clinical indications. From the data obtained, it will be possible to extrapolate for analysis genes pool selected according to clinical indications or presumed pathology, excluding all the remaining genes from the analysis.
For these reasons, this approach opens up new scenarios in the diagnosis of rare pathologies, for which often the data available in literature is incomplete, and in prenatal diagnosis (5, 6).
In prenatal diagnosis this experimental approach could be used for the analysis of genes clearly associated to common and congenital Mendelian pathologies and/or are involved in fetal development. Furthermore, the mutations that are identified should be then confirmed through the Sanger sequencing method and a search must be carried out in the parents to identify the origin and to confirm the pathogenicity.
References
- 1.Yang Y, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim HJ, et al. Congenital muscular dystrophy type 1A with residual merosin expression. Korean J Pediatr. 2014;57(3):149–152. doi: 10.3345/kjp.2014.57.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Blasi C, et al. LAMA2 gene analysis in congenital muscular dystrophy: new mutations, prenatal diagnosis, and founder effect. Arch Neurol. 2005;62(10):1582–1586. doi: 10.1001/archneur.62.10.1582. [DOI] [PubMed] [Google Scholar]
- 4.Chan SH, et al. Limb girdle muscular dystrophy due to LAMA2 mutations: diagnostic difficulties due to associated peripheral neuropathy. Neuromuscul Disord. 2014 doi: 10.1016/j.nmd.2014.05.008. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 5.Jones D, et al. First trimester diagnosis of Meckel-Gruber Syndrome by fetal ultrasound, with molecular identification of CC2D2A mutations by Next-Generation Sequencing. Ultrasound Obstet Gynecol. 2014 doi: 10.1002/uog.13381. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 6.Carss KJ, et al. Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum Mol Genet. 2014;23(12):3269–3277. doi: 10.1093/hmg/ddu038. [DOI] [PMC free article] [PubMed] [Google Scholar]