Abstract
Hepatic stellate cells (HSCs) generate matrix, which in turn may also regulate HSCs function during liver fibrosis. We hypothesized that HSCs may endocytose matrix proteins to sense and respond to changes in microenvironment. Primary human HSCs, LX2, or mouse embryonic fibroblasts (MEFs) [wild-type; c-abl−/−; or Yes, Src, and Fyn knockout mice (YSF−/−)] were incubated with fluorescent-labeled collagen or gelatin. Fluorescence-activated cell sorting analysis and confocal microscopy were used for measuring cellular internalization of matrix proteins. Targeted PCR array and quantitative real-time PCR were used to evaluate gene expression changes. HSCs and LX2 cells endocytose collagens in a concentration- and time-dependent manner. Endocytosed collagen colocalized with Dextran 10K, a marker of macropinocytosis, and 5-ethylisopropyl amiloride, an inhibitor of macropinocytosis, reduced collagen internalization by 46%. Cytochalasin D and ML7 blocked collagen internalization by 47% and 45%, respectively, indicating that actin and myosin are critical for collagen endocytosis. Wortmannin and AKT inhibitor blocked collagen internalization by 70% and 89%, respectively, indicating that matrix macropinocytosis requires phosphoinositide-3-kinase (PI3K)/AKT signaling. Overexpression of dominant-negative dynamin-2 K44A blocked matrix internalization by 77%, indicating a role for dynamin-2 in matrix macropinocytosis. Whereas c-abl−/− MEF showed impaired matrix endocytosis, YSF−/− MEF surprisingly showed increased matrix endocytosis. It was also associated with complex gene regulations that related with matrix dynamics, including increased matrix metalloproteinase 9 (MMP-9) mRNA levels and zymographic activity. HSCs endocytose matrix proteins through macropinocytosis that requires a signaling network composed of PI3K/AKT, dynamin-2, and c-abl. Interaction with extracellular matrix regulates matrix dynamics through modulating multiple gene expressions including MMP-9.
Keywords: hepatic stellate cells, macropinocytosis, collagen, matrix metalloproteinase
hepatic stellate cells (HSCs) are pivotal for extracellular matrix homeostasis in liver. Quiescent HSCs contain numerous vitamin A lipid droplets and are located in the space of Disse. Upon liver injury, they are transformed to active myofibroblast-like cells, which are characterized by increased proliferation, α-smooth muscle actin expression, and robust collagen production (3–5, 13, 19, 26, 31). However, matrix also reciprocally regulates HSC function, including adhesion and migration through mechanical stiffness, integrin activation, and other pathways (2, 15, 20, 39, 57). However, the mechanisms by which HSCs sense their matrix surroundings are incompletely understood.
Collagen production and removal is a dynamic balance that determines tissue architecture. Disruption of this homeostasis can lead to tissue destruction or fibrosis. In liver, HSCs are important not only for collagen production that contributes to liver cirrhosis, but also for collagen degradation during cirrhosis resolution. Collagen has been reported to be internalized by macrophages and fibroblasts (1, 30) and recently by HSCs (36, 37). However, the mechanisms and relevance of this process remain undefined and were the focus of this investigation. Here we show that HSCs endocytose collagen and that this ultimately regulates production of the matrix-degrading protease matrix metalloproteinase 9 (MMP-9), implicating a feedback loop, whereby matrix sensing by HSCs regulates subsequent matrix degradation maneuvers. Furthermore, we provide mechanistic insight into the sensing process by showing that matrix endocytosis occurs through a macropinocytosis pathway that requires actin, dynamin, and c-abl. These studies add to our understanding of HSCs and their dynamic interaction with the matrix microenvironment.
MATERIALS AND METHODS
Reagents.
Oregon-green 488-conjugated gelatin, Fluorescein Conjugate DQ type I Collagen (DQ-collagen I), LysoTracker, and Dextran 10K were from Invitrogen (Grand Island, NY). Cysteine protease inhibitor E64D was from Calbiochem (San Diego, CA). Dynasore, Cytochalasin D, blebbistatin, methyl-β-cyclodextrin (MβCD), Filipin, nystatin, 5-ethylisopropyl amiloride (EIPA), PDGF-BB, and fibroblast growth factor were obtained from Sigma-Aldrich (Dorset, UK). VEGF and transforming growth factor (TGF)-β were from R&D Systems (Minneapolis, MN). Lysosomal-associated membrane protein-1 (LAMP-1) and CD63 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Early endosome antigen (EEA) antibody was from Cell Signaling Technology (Danvers, MA).
Cell culture.
Primary human HSCs were purchased from ScienCell Research Laboratories (Carlsbad, CA) and used at low passage (P3–5; catalog no. 5300). These cells have been well characterized and used widely in prior publications (10, 32, 46). Wild-type mouse embryonic fibroblast (MEF) cells, MEF cells isolated form c-abl and Arg double-knockout mice (c-abl−/− MEF), and MEF cells isolated from Yes, Src, and Fyn knockout mice (YSF−/− MEF) were kindly provided by Drs. E. Leof and M. McNiven, respectively (Mayo Clinic, Rochester, MN) (9, 58). Immortalized human-derived LX2 cells (54) are immortalized HSCs with a myofibroblast-like phenotype that were used in some experiments along with human HSCs. Cells were cultured in DMEM or specialized HSC medium, supplemented with 10% fetal bovine serum, 1 mmol/l l-glutamine, and 100 IU/ml streptomycin/penicillin.
Collagen endocytosis.
HSCs or LX2 cells were incubated with indicated concentrations of Oregon Green-labeled gelatin, DQ-collagen I, or Dextran 10K for indicated time in the absence or presence of 20 μm E64D. Cells were extensively washed and fixed with 4% paraformaldehyde and stained with LysoTracker. For subcellular localization analysis, cells were stained with EEA to mark early endosomes, LAMP-1 for late endosomes, and CD63 for multivesicular body marking. Images were taken with a Zeiss LSM 510 microscope.
For fluorescence-activated cell sorting (FACS), HSCs or LX2 cells were plated in six-well dishes, starved overnight with serum-free medium, and treated with indicated compounds for 30 min before incubation with fluorescent-labeled collagen or gelatin. Cells were extensively washed, trypsinized, and fixed with 2% formaldehyde and subjected to FACS analysis.
mRNA extraction, PCR array, real-time quantitative PCR.
To quantify the steady-state concentration of mRNAs, total RNA was extracted using RNeasy Mini kit (Qiagen, Crawley, West Sussex, UK) according to the manufacturer's instructions, quantified using a Spectrophotometer, and reverse transcribed. Real-time quantitative PCR was performed in an Applied Biosystems 7500 Real-time PCR System (Foster City, CA) to quantify the steady-state concentration of RNA using a SYBR Green PCR Kit. Primers are detailed in Table 1. The reaction contained 3.6–7.3 ng RNA and 0.5 μM primers. The relative gene expression (fold of GAPDH) of each gene was calculated by using ΔCT method. RT2 Profiler PCR Array (Venlo, Netherlands) was performed per the manufacturer's instruction.
Table 1.
Primers for RT-PCR analysis
| Gene | Primer |
|---|---|
| GAPDH forward | CCAGGGCTGCTTTTAACTCT |
| GAPDH reverse | GGACTCCACGACGTACTCA |
| MMP9 forward | GACGTCTTCCAGTACCGA |
| MMP9 reverse | CTCAGGGCACTGCAGGAT |
MMP9, matrix metalloproteinase 9.
Western blotting and zymography.
Conditioned medium was collected and centrifuged at 270 g for 3 min to remove cellular debris. Cells were lysed in RIPA buffer. Protein concentration in lysates was measured and used to normalize protein loading of gels. Cell extracts and conditioned medium were diluted fourfold in lysis buffer and reduced with 5% β-mercaptoethanol and then fractionated by PAGE and analyzed by Western blotting. Detection was performed using enhanced chemiluminescence.
MMP-9 activity in conditioned medium was evaluated by zymography as described (11). Briefly, 7.5% polyacrylamide gels containing 2 mg/ml gelatin were subjected to electrophoresis under nonreducing conditions. Following electrophoresis, SDS was removed by washing in 2.5% Triton X-100, and gels were incubated at 37°C for 18 h in 50 mM Tris·HCl, pH 8.0, 50 mM NaCl, 10 mM Ca2Cl, and 0.05% Triton X-100. Gels were then stained in 0.2% Coomassie Brilliant Blue. Gelatinase activity was detected as clear bands on a dark background. Densitometric analysis of bands was performed.
Gene ontology analysis.
We used the software package The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (17) for gene ontology analysis. In brief, the GeneBank IDs of the genes from PCR array were inputted into the tools for gene ontology analysis of biological process and cellular components. The results were ranked based on adjusted P values using Benjamini-Hochberg method for multiple-comparison corrections. The count represents the number of genes involved in that function group, with the percentage of genes in the input genes that are involved in that function.
Statistical analysis.
Results are expressed as means ± SE. Significance was established using the Student's t-test and ANOVA when appropriate. Differences were considered significant when P < 0.05.
RESULTS
HSCs and LX2 cells internalize collagen.
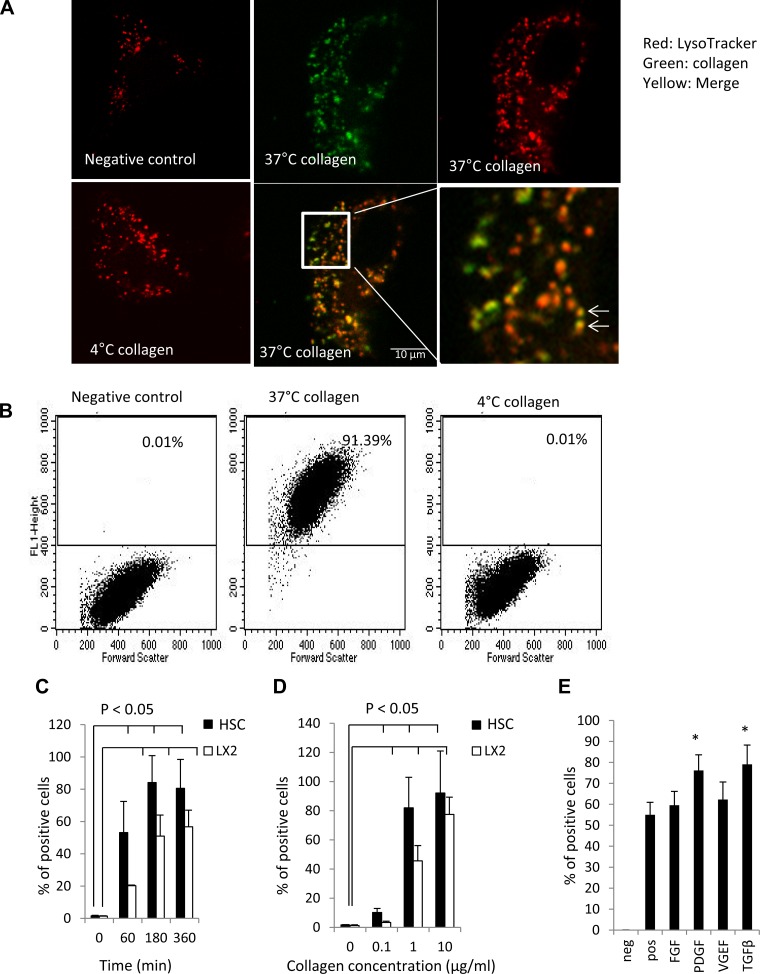
To test the hypothesis that collagen internalization could regulate extracellular matrix dynamics, we initially investigated whether primary HSCs can internalize collagen. Collagen I was chosen for this study because it is pathologically increased in liver cirrhosis (38). When DQ-collagen I (1 μg/ml) was incubated with HSCs for 3 h at 37°C to allow visualization of internalized molecules, a pronounced intracellular vesicular accumulation of fluorescent signal was observed (Fig. 1A). As seen in confocal microscopy images in Fig. 1A, these collagen-containing vesicles (green) colocalized with LysoTracker (red), a marker for lysosomes. These data indicate that the collagen is internalized and is routed to lysosomes. As endocytosis is susceptible to temperature changes, we tested specificity of effect by performing studies at 4°C, a temperature broadly used to block endocytosis. As shown in Fig. 1A, the intracellular collagen-containing vesicles were significantly decreased at 4°C compared with 37°C. The results were further confirmed by using FACS to quantify collagen internalization. At 37°C, 91% of cells contained intracellular collagen; however, only 0.01% cells were positive at 4°C, suggesting that collagen internalization occurs through endocytosis (Fig. 1B). To explore temporal kinetics of collagen endocytosis, we incubated HSCs or LX2 cells with 0.5 μg/ml collagen for varying durations. As shown in Fig. 1C, HSC collagen endocytosis increased with incubation time; 53% of HSC cells were positive at 60 min, and 81% were positive at 360 min. The extracellular concentration of collagen also regulated collagen internalization; 30% of HSC cells incubated with 0.1 μg/ml gelatin for 3 h internalized collagen, which increased to 81% with 1.0 μg/ml and plateaued with subsequent increases in collagen concentration (Fig. 1D). Although LX2 cells showed slower endocytosis kinetics, they were able to internalize collagen as well (Fig. 1, C and D). Experiments using human and mouse pancreatic stellate cells showed similar results to that of LX2 and HSCs, suggesting that collagen internalization is a general phenomenon of stellate cells (data not shown). Additionally, experiments carried out using Oregon-green 488 gelatin (1 μg/ml; degradation product of full-length collagen) showed similar concentration- and temperature-dependent internalization, indicating that both collagen and its degradation products can be effectively internalized. Finally, we evaluated effects of HSC growth factor activation on collagen endocytosis. Both TGF-β and PDGF-BB significantly, albeit modestly, increased collagen internalization compared with vehicle-treated HSCs (Fig. 1E).
Fig. 1.
Human hepatic stellate cells (HSCs) internalize collagen in a time- and concentration-dependent manner. A: human HSCs were incubated with DQ collagen I (1 μg/ml, green) at indicated temperature for 3 h. LysoTracker (red) was added in the last 15 min of incubation. Pictures were taken using a LSM 520 confocal microscopy. In negative control (cells incubated with collagen without fluorescent labeling) or HSCs incubated with DQ collagen I (1 μg/ml) at 4°C for 3 h, LysoTracker (red) was shown scattered in the cytoplasm, but there was no collagen internalization as quantified by confocal microscopy. However, in HSCs incubated with DQ collagen I (1 μg/ml) at 37°C for 3 h, internalized collagen (green) colocalized with Lysotracker (red), as shown in the merged image as yellow. High-zoom colocalization image is also shown with colocalization, highlighted with arrows. B: human HSCs were incubated with DQ-collagen I (1 μg/ml) at indicated temperature for 3 h and then subjected to fluorescence-activated cell sorting (FACS) analysis. C: LX2 cells and human HSCs were incubated with DQ-collagen I (0.5 μg/ml) for indicated time and then subjected to FACS analysis (n = 3 independent experiments; P < 0.05 compared with time zero). D: human HSCs and LX2 were incubated with DQ-collagen I at indicated concentration at 37°C for 3 h and then subjected to FACS analysis (n = 3; P < 0.05 compared with time zero). E: HSC were preincubated with transforming growth factor (TGF)-β1 (5 ng/ml), PDGF-BB (100 ng/ml), FGF (10 ng/ml), or VEGF (5 ng/ml) for 30 min at 37°C before incubation with DQ collagen I (0.5 μg/ml) for 3 h at 37°C and then subjected to FACS analysis (n = 3; *P < 0.05 compared with control).
Collagen endocytosis occurs through macropinocytosis.
We next sought to discern the mechanism of collagen endocytosis by HSCs. A recent study suggested that macropinocytic endocytosis may be an important route for cell sensing (8). To test whether stellate cells use macropinocytosis as an endocytic pathway for collagen, we first performed studies with Dextran 10K, a classic marker for macropinocytosis. We treated cells with DQ-collagen I and Alexa Fluor 647 Dextran 10K together for 3 h. FACS analysis revealed that about 62% of cells cointernalized collagen and Dextran 10K (Fig. 2, A and B). We next evaluated the effect of EIPA, an ion exchange inhibitor that inhibits macropinocytosis (21), on collagen endocytosis. Cells pretreated with EIPA showed significantly reduced internalization of Dextran 10K as well as collagen (Fig. 2, A and B) as assessed by FACS. Additionally, under confocal microscopy, Dextran 10K colocalized with collagen and LysoTracker (Fig. 2C), indicating that HSCs internalize collagen through macropinocytosis. Finally, colocalization analysis showed internalized collagen colocalized with CD63, a marker of multivesicular bodies (Pearson's coefficient 0.56 ± 0.14) with lesser colocalization with EEA and LAMP-1 (Pearson's coefficient 0.15 ± 0.08), indicating that internalized collagen is eventually targeted for degradation (Fig. 2D), which is consistent with previous reports (22, 30).
Fig. 2.
Collagen endocytosis occurs through macropinocytosis. Human HSCs were pretreated with 5-ethylisopropyl amiloride (EIPA) (50 μM) for 30 min before incubation with Dextran 10K or DQ-collagen I (0.5 μg/ml) or both for 3 h, as labeled in the graph. A: representative FACS analysis is shown. B: quantification data of FACS are depicted (n = 3, *P < 0.05). C: human HSCs were incubated with DQ collagen I (1 μg/ml, green) and Dextran 10K (0.2 mg/ml) (blue) at 37°C for 3 h. Representative micrographs were taken using a LSM 520 confocal microscopy and showed internalized collagen (green) colocalized with Dextran 10K (blue) in the merged image. High-zoom colocalization image was also shown. D: LX2 cells were incubated with DQ-collagen I (1 μg/ml, green) for 3 h at 37°C. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton-X. Early endosome antigen (EEA), lysosomal-associated membrane protein-1 (LAMP-1), and CD63 (red) antibodies were used for fluorescence analysis.
Collagen macropinocytosis occurs through an actin-regulated signaling network that requires dynamin-2 and phosphoinositide-3-kinase/AKT.
We next sought to examine the signaling mechanism of collagen macropinocytosis in greater detail. The involvement of the large GTPase- and endocytosis-regulating protein, dynamin-2, in collagen internalization was investigated using a dominant-negative dynamin-2 construct containing a well-characterized K44A point mutation (7). HSCs were transiently transfected to express control adenovirus or K44A dynamin-2. Cells expressing K44A dynamin-2 showed significantly decreased collagen internalization by 77% (Fig. 3A). The data were further confirmed by pretreatment of HSCs or LX2 cells with Dynasore (20 μM), a pharmacological inhibitor of dynamin, which diminished HSC collagen endocytosis by 51% (Fig. 3A). Because macropinocytosis is an actin-dependent endocytic process, we next tested pharmacological inhibitors of actin cytoskeleton dynamics and myosin activity on collagen internalization by HSCs. Cells were preincubated with Cytochalasin D (0.5 μM) to inhibit actin polymerization, ML7 (1 μM) to inhibit myosin light chain kinase, or blebbistatin (50 μM) to block myosin II for 30 min before cells were incubated with DQ-collagen I for 3 h. FACS analysis showed that Cytochalasin D, ML7, and blebbistatin all decreased collagen internalization by 47%, 45%, and 39%, respectively (Fig. 3B). These data suggest that actin and myosin are important for collagen endocytosis in HSCs. Because dynamin regulates both clathrin- and caveolae-dependent endocytosis, we next explored which of these pathways mediated collagen endocytosis using a panel of pharmacological inhibitors. Cells were pretreated with inhibitors of caveolae-dependent endocytosis [5 mM MβCD (49), 5 μg/ml filipin (44), 30 μM nystatin (41), or clathrin-dependent endocytosis (0.45 M sucrose) (16)] for 30 min, and then collagen endocytosis was measured. Whereas sucrose inhibited endocytosis, inhibitors of caveolae pathway did not, indicating that collagen endocytosis occurs through a clathrin-dependent pathway (Fig. 3C). Phosphoinositide-3-kinase (PI3K) and the downstream effector protein AKT are important in regulating completion of macropinocytosis (43). We next tested their roles in collagen internalization by HSCs using pharmacological inhibitors. Wortmannin and AKT inhibitor blocked collagen internalization by 70% and 90%, respectively (Fig. 3D). Under the current concentration and treatment durations, we did not observe significant effects of the pharmacological inhibitors on cell viability based on DAPI uptake (Fig. 3E). These studies indicate that collagen macropinocytosis occurs through a dynamin-, PI3K/AKT-dependent actin-regulated pathway.
Fig. 3.
Collagen internalization is regulated by Dynamin-2, phosphoinositide-3-kinase/AKT, and actin cytoskeleton. Human HSCs were infected with K44A mutant Dynamin-2 adenovirus for 24 h or pretreated with Dynasore (A), blebbistatin, ML7, or Cytochalasin D (B), methyl-β-cyclodextrin (MβCD), filipin, nystatin, or 0.5 M sucrose (C), or wortmannin or AKT inhibitor (D) for 30 min before DQ-collagen I (0.5 μg/ml) incubation for 3 h. Cells were subjected to FACS analysis to measure matrix protein endocytosis (n = 4, *P < 0.05 compared with positive control). E: DAPI exclusion assay was used to identify the percentage of viable cells, which was not significantly different between control and compounds assessed.
C-abl, but not Src is critical for macropinocytosis.
Both c-abl and Src family nonreceptor tyrosine kinases are important for matrix dynamics and have been implicated in PI3K/AKT signaling. We therefore hypothesized that c-abl and Src may be important for HSC collagen endocytosis. We used MEF cells isolated from c-abl and Arg double-knockout mice (c-abl−/−) and MEF cells from YSF−/−. These double- and triple-knockout cells delete multiple family members with redundant functions. Compared with wild-type MEF, c-abl−/− MEF showed less collagen internalization at a range of collagen concentrations (Fig. 4A). Surprisingly, YSF−/− MEF cells demonstrated significantly enhanced collagen endocytosis compared with wild-type cells (Fig. 4A). To explore this unexpected result further, we measured c-abl activity using phospho-CrkII as a readout. Western blot data showed increased phospho-CrkII in YSF−/− MEF compared with WT MEF (Fig. 4B). These studies indicate that c-abl activity is upregulated in YSF−/− MEF and may account for the unanticipated increase in collagen endocytosis observed in these cells.
Fig. 4.
Knockdown of c-abl decreases but knockdown of Yes, Src, and Fyn (YSF) increases collagen endocytosis in mouse embryonic fibroblasts (MEF) cells. MEF cells isolated form wild-type (WT), c-abl−/− mice (c-abl−/−), and Yes−/−Src−/−Fyn−/− (YSF−/−) cells were starved overnight and then incubated with indicated concentration of DQ-collagen I (0.5 μg/ml) for 3 h before the cells were subjected to FACS analysis. A: FACS analysis comparing collagen uptake among WT, c-abl−/−, and YSF−/− MEF cells after incubation with indicated concentration of DQ-collagen I for 3 h. B: p-CRKII activity is shown in cells, including quantitation of densitometry (n = 3, *P < 0.05 compared with WT).
Interaction of collagen with HSCs regulates genes related with extracellular matrix dynamics.
Finally, we studied the functional response of HSCs to extracellular collagen. As a first step, we performed a targeted PCR array to determine changes in mRNA transcripts in HSCs in response to collagen. Multiple targets related with collagen degradation or synthesis were changed upon HSC interaction with collagen, including a prominent increase in MMP-9 (Table 2). We next performed quantitative real-time PCR for a number of individual matrix regulatory proteins to confirm the array data. This analysis confirmed that MMP-9 was upregulated by 3.7-fold (Fig. 5A) after HSCs interact with collagen. Several other changes were observed as well (Fig. 5, B–D), but we focused on MMP-9 for further confirmation and analysis owing to the known role of this protein in matrix degradation. Gelatin zymography confirmed increased MMP-9 activity in response to collagen (Fig. 5E). In addition to effects on MMP activity, interaction with collagen significantly decreased collagen Iα mRNA levels by 22% based on RT-PCR (Fig. 5D). Moreover, mRNA levels of collagen XVIII α1 were also diminished in the microarray analysis (Table 2). These data indicate that extracellular collagen initiates a response in HSCs that regulates matrix dynamics through mechanisms that include MMP-9 as well as collagen production. Additionally, gene ontology analysis suggested that genes regulating wound-healing response, cell survival/apoptosis, and those encoding proteins residing in the extracellular space were among the highly regulated genes (Tables 3, 4, 5, and 6). This analysis supports previous observations suggesting important survival signal provided by cellular collagen interaction and also supports our data that interaction with extracellular matrix (collagen) by cells is able to provide feedback to its surrounding extracellular components. However, it should be noted that the present analysis could not distinguish whether the observed signaling effects are secondary to collagen ligand interaction with a collagen membrane receptor, such as an integrin family member vs. the internalization process itself.
Table 2.
Changes in HSC mRNA levels upon collagen interaction
| Unigene | GeneBank | Symbol | Description | Fold Changes |
|---|---|---|---|---|
| Hs.227817 | NM_004049 | BCL2A1 | B-cell CLL/lymphoma 2 (BCL2)-related protein A1 | 11.79415 |
| Hs.591873 | NM_000880 | IL7 | Interleukin 7 | 7.889862 |
| Hs.1349 | NM_000758 | CSF2 | Colony stimulating factor 2 (granulocyte-macrophage) | 7.061624 |
| Hs.412707 | NM_000194 | HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | 4.756828 |
| Hs.126256 | NM_000576 | IL1B | Interleukin 1β | 4.169863 |
| Hs.303649 | NM_002982 | CCL2 | Chemokine (C-C motif) ligand 2 | 3.863745 |
| Hs.150749 | NM_000633 | BCL2 | B-cell CLL/lymphoma 2 | 3.837056 |
| Hs.141125 | NM_004346 | CASP3 | Caspase 3, apoptosis-related cysteine peptidase | 3.5801 |
| Hs.83169 | NM_002421 | MMP1 | Matrix metalloproteinase 1 (interstitial collagenase) | 3.555371 |
| Hs.591016 | NM_003805 | CRADD | Caspase 2 and RIPK1 domain containing adaptor with death domain | 3.24901 |
| Hs.297413 | NM_004994 | MMP9 | Matrix metalloproteinase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) | 3.197479 |
| Hs.522632 | NM_003254 | TIMP1 | Tissue inhibitor of metalloproteinase 1 | 3.182146 |
| Hs.534255 | NM_004048 | B2M | β2-microglobulin | 2.969047 |
| Hs.744837 | NM_031850 | AGTR1 | Angiotensin II receptor, type 1 | 2.928171 |
| Hs.480653 | NM_001154 | ANXA5 | Annexin A5 | 2.928171 |
| Hs.436873 | NM_002210 | ITGAV | Integrin-αV (vitronectin receptor, α-polypeptide, antigen CDS1) | 2.907945 |
| Hs.443914 | NM_000454 | SOD1 | Superoxide dismutase 1, soluble | 2.732081 |
| Hs.244139 | NM_000043 | FAS | Fas (TNF receptor superfamily, member 6) | 2.713209 |
| Hs.523185 | NM_012423 | RPL13A | Ribosomal protein L13a | 2.713209 |
| Hs.74615 | NM_006206 | PDGFRA | Platelet-derived growth factor receptor α polypeptide | 2.639016 |
| Hs.516578 | NM_006287 | TFPI | Tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 2.566852 |
| Hs.77274 | NM_002658 | PLAU | Plasminogen activator, urokinase | 2.531513 |
| Hs.81564 | NM_002619 | PF4 | Platelet factor 4 | 2.42839 |
| Hs.713079 | NM_000602 | SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 2.411616 |
| Hs.544577 | NM_002046 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 2.394957 |
| Hs.2490 | NM_033292 | CASP1 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, β convertase) | 2.158456 |
| Hs.440848 | NM_000552 | VWF | Von Willebrand factor | 2.099433 |
| Hs.647092 | NM_000603 | NOS3 | Nitric oxide synthase 3 (endothelial cell) | 1.972465 |
| Hs.731912 | NM_003879 | CFLAR | Caspase 8 and Fas-associated death domain-like apoptosis regulator | 1.958841 |
| Hs.483635 | NM_000800 | FGF1 | Fibroblast growth factor 1 (acidic) | 1.931873 |
| Hs.491582 | NM_000930 | PLAT | Plasminogen activator, tissue | 1.765406 |
| Hs.502876 | NM_004040 | RHOB | Ras homolog gene family, member B | 1.765406 |
| Hs.654458 | NM_000600 | IL6 | Interleukin 6 (interferon β2) | 1.693491 |
| Hs.643447 | NM_000201 | ICAM1 | Intercellular adhesion molecule 1 | 1.624505 |
| Hs.631562 | NM_003706 | PLA2G4C | Phospholipase A2, group IVC (cytosolic, calcium-independent) | 1.591073 |
| Hs.203717 | NM_002026 | FN1 | Fibronectin 1 | 1.547565 |
| Hs.109225 | NM_001078 | VCAM1 | Vascular cell adhesion molecule 1 | 1.536875 |
| Hs.2030 | NM_000361 | THBD | Thrombomodulin | 1.526259 |
| Hs.591014 | NM_003006 | SELPLG | Selectin P ligand | 1.515717 |
| Hs.89499 | NM_000698 | ALOX5 | Arachidonate 5-lipoxygenase | 1.494849 |
| Hs.694 | NM_000588 | IL3 | Interleukin 3 (colony-stimulating factor, multiple) | 1.394744 |
| Hs.709191 | NM_000625 | NOS2 | Nitric oxide synthase 2, inducible | 1.394744 |
| Hs.82848 | NM_000450 | SELE | Selectin E | 1.394744 |
| Hs.728756 | NM_000655 | SELL | Selectin L | 1.328686 |
| Hs.512937 | NM_001872 | CPB2 | Carboxypeptidase B2 (plasma) | 1.319508 |
| Hs.713645 | NM_001955 | EDN1 | Endothelin 1 | 1.319508 |
| Hs.2007 | NM_000639 | FASLG | Fas ligand (TNF superfamily, member 6) | 1.319508 |
| Hs.404914 | NM_003183 | ADAM17 | ADAM metallopeptidase domain 17 | 1.245862 |
| Hs.592605 | NM_002538 | OCLN | Occludin | 1.197479 |
| Hs.219140 | NM_002521 | NPPB | Natriuretic peptide B | 1.189207 |
| Hs.730607 | NM_001953 | TYMP | Thymidine phosphorylase | 1.156688 |
| Hs.183713 | NM_001957 | EDNRA | Endothelin receptor type A | 1.140764 |
| Hs.298469 | NM_000789 | ACE | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | 1.123531 |
| Hs.113916 | NM_001716 | CXCR5 | Chemokine (C-X-C motif) receptor 5 | 1.117287 |
| Hs.479756 | NM_002253 | KDR | Kinase insert domain receptor (a type III receptor tyrosine kinase) | 1.117287 |
| Hs.164226 | NM_003246 | THBS1 | Thrombospondin 1 | 1.109569 |
| Hs.467304 | NM_000641 | IL11 | Interleukin 11 | 1.101905 |
| Hs.594454 | NM_002019 | FLT1 | Fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 1.071773 |
| Hs.654616 | NM_032992 | CASP6 | Caspase 6, apoptosis-related cysteine peptidase | 1.057018 |
| Hs.1407 | NM_001956 | EDN2 | Endothelin 2 | 1.021012 |
| Hs.143436 | NM_000301 | PLG | Plasminogen | 1.006956 |
| Hs.19383 | NM_000029 | AGT | Angiotensinogen (serpin peptidase inhibitor, clade A, member B) | −0.987509 |
| Hs.478275 | NM_003810 | TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | −1 |
| Hs.514821 | NM_002985 | CCL5 | Chemokine (C-C motif) ligand 5 | −1.05702 |
| Hs.302085 | NM_000961 | PTGIS | Prostaglandin I2 (prostacyclin) synthase | −1.10191 |
| Hs.655801 | NM_003841 | TNFRSF10C | Tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain | −1.10957 |
| Hs.68061 | NM_021972 | SPHK1 | Sphingosine kinase 1 | −1.12506 |
| Hs.76206 | NM_001795 | CDH5 | Cadherin 5, type 2 (vascular endothelium) | −1.1487 |
| Hs.479754 | NM_000222 | KIT | V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | −1.16473 |
| Hs.93177 | NM_002176 | IFNB1 | Interferon β1, fibroblast | −1.20581 |
| Hs.218040 | NM_000212 | ITGB3 | Integrin β3 (platelet glycoprotein IIIa, antigen CD51) | −1.23114 |
| Hs.241570 | NM_000594 | TNF | Tumor necrosis factor | −1.33793 |
| Hs.376675 | NM_000442 | PECAM1 | Platelet/endothelial cell adhesion molecule | −1.34723 |
| Hs.252820 | NM_002632 | PGF | Placental growth factor | −1.36604 |
| Hs.211600 | NM_006290 | TNFAIP3 | Tumor necrosis factor, α-induced protein 3 | −1.43396 |
| Hs.505654 | NM_002205 | ITGA5 | Integrin α5 (fibronectin receptor, α polypeptide) | −1.45397 |
| Hs.643813 | NM_002211 | ITGB1 | Integrin β1 (fibronectin receptor, β polypeptide, antigen CD29 includes MDF2, MSK12) | −1.47427 |
| Hs.517356 | NM_030582 | COL1BA1 | Collagen, type XVIII, α1 | −1.61328 |
| Hs.490330 | NM_000906 | NPR1 | Natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) | −1.67018 |
| Hs.171995 | NM_001648 | KLK3 | Kallikrein-related peptidase 3 | −2.04202 |
| Hs.531668 | NM_002996 | CX3CL1 | Chemokine (C-X3-C motif) ligand 1 | −2.62079 |
| Hs.519842 | NM_003804 | RIPK1 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 | −3.16017 |
| Hs.516966 | NM_138578 | BCL2L1 | BCL2-like 1 | −3.36359 |
| Hs.369675 | NM_001146 | ANGPT1 | Angiopoietin 1 | −3.70635 |
| Hs.89640 | NM_000459 | TEK | TEK tyrosine kinase, endothelial | −3.83706 |
| Hs.73793 | NM_003376 | VEGFA | Vascular endothelial growth factor A | −3.86375 |
| Hs.624291 | NM_004324 | BAX | BCL2-associated X protein | −5.65685 |
| Hs.513617 | NM_004530 | MMP2 | Matrix metalloproteinase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase) | −11.7942 |
Hepatic stellate cells (HSCs) were treated with vehicle or collagen for 2 h before mRNA was extracted for PCR array. The data were arranged according to mRNA fold change upon collagen interaction.
Fig. 5.
Collagen and HSC interaction stimulates increase in matrix metalloproteinase (MMP)-9 mRNA levels and zymographic activity. A: human HSCs were incubated with BSA or DQ-collagen I (0.5 μg/ml) for 2 h, and RNA was extracted for real-time PCR for MMP-9, MMP-2 (B), smooth muscle actin (SMA) (C), and collagen Iα (D) (n = 3, *P < 0.05 compared with control). E: human HSCs were incubated with BSA or DQ-collagen I (0.5 μg/ml) for 48 h, and cell medium was collected for gelatin zymography. Representative data and quantification are shown from 3 independent zymographies (*P < 0.05).
Table 3.
Functional gene ontology of upregulated genes
| Term | Count | % | Adjusted P Value Using Benjamini-Hochberg Method |
|---|---|---|---|
| Response to wounding | 23 | 39 | 1.70e-13 |
| Regulation of cell proliferation | 25 | 42.4 | 2.30e-12 |
| Cell migration | 16 | 27.1 | 2.10e-10 |
| Regulation of body fluid levels | 13 | 22 | 2.30e-10 |
| Wound healing | 14 | 23.7 | 3.20e-10 |
| Localization of cell | 16 | 27.1 | 4.80e-10 |
| Cell motility | 16 | 27.1 | 4.80e-10 |
| Regulation of response to external stimulus | 13 | 22 | 5.50e-10 |
| Regulation of apoptosis | 22 | 37.3 | 1.10e-09 |
| Regulation of programed cell death | 22 | 37.3 | 1.20e-09 |
| Regulation of cell death | 22 | 37.3 | 1.10e-09 |
| Cell activation | 15 | 25.4 | 1.60e-09 |
| Blood coagulation | 11 | 18.6 | 1.80e-09 |
| Coagulation | 11 | 18.6 | 1.80e-09 |
| Negative regulation of apoptosis | 16 | 27.1 | 1.70e-09 |
| Response to oxygen levels | 12 | 20.3 | 1.70e-09 |
| Negative regulation of programmed cell death | 16 | 27.1 | 1.80e-09 |
| Negative regulation of cell death | 16 | 27.1 | 1.80e-09 |
| Blood vessel development | 14 | 23.7 | 2.20e-09 |
| Hemostasis | 11 | 18.6 | 2.20e-09 |
The GeneBank IDs of the genes from the PCR array were analyzed using DAVID v6.7 for gene ontology analysis of biological process and cellular components. The results were ranked based on adjusted P values using Benjamini-Hochberg statistical method for multiple-comparison corrections. The count represents the number of genes involved in that function group. % represents the percentage of genes in the input genes that are involved in that function.
Table 4.
Cellular component gene ontology of upregulated genes
| Term | Count | % | Adjusted P Value Using Benjamini-Hochberg Method |
|---|---|---|---|
| Extracellular space | 26 | 44.1 | 4.90e-16 |
| Extracellular region part | 29 | 49.2 | 3.70e-16 |
| Extracellular region | 34 | 57.6 | 1.70e-12 |
| External side of plasma membrane | 8 | 13.6 | 2.10e-04 |
| Platelet α granule | 5 | 8.5 | 2.20e-03 |
| Secretory granule | 7 | 11.9 | 2.20e-03 |
| Plasma membrane part | 22 | 37.3 | 2.30e-03 |
| Plasma membrane | 29 | 49.2 | 7.50e-03 |
| Extracellular matrix | 8 | 13.6 | 8.00e-03 |
| Cell surface | 8 | 13.6 | 7.50e-03 |
| Platelet α granule lumen | 4 | 6.8 | 7.60e-03 |
| Cytoplasmic membrane-bound vesicle lumen | 4 | 6.8 | 8.50e-03 |
| Vesicle lumen | 4 | 6.8 | 9.00e-03 |
| Integral to plasma membrane | 13 | 22 | 2.90e-02 |
| Intrinsic to plasma membrane | 13 | 22 | 3.30e-02 |
| Cytoplasmic vesicle | 9 | 15.3 | 3.90e-02 |
| Vesicle | 9 | 15.3 | 4.80e-02 |
| Cytoplasmic membrane-bound vesicle | 8 | 13.6 | 5.40e-02 |
| Cytosol | 13 | 22 | 5.30e-02 |
| Membrane-bound vesicle | 8 | 13.6 | 5.70e-02 |
Data are as described in Table 3.
Table 5.
Functional gene ontology of downregulated genes
| Term | Count | % | Adjusted P Value Using Benjamini-Hochberg Method |
|---|---|---|---|
| Regulation of cell proliferation | 12 | 46.2 | 4.80e-05 |
| Regulation of programed cell death | 12 | 46.2 | 3.30e-05 |
| Regulation of cell death | 12 | 46.2 | 2.30e-05 |
| Regulation of apoptosis | 11 | 42.3 | 1.80e-04 |
| Regulation of locomotion | 7 | 26.9 | 2.00e-04 |
| Cell adhesion | 10 | 38.5 | 3.90e-04 |
| Biological adhesion | 10 | 38.5 | 3.40e-04 |
| Blood vessel development | 7 | 26.9 | 5.30e-04 |
| Vasculature development | 7 | 26.9 | 5.40e-04 |
| Positive regulation of cell proliferation | 8 | 30.8 | 6.80e-04 |
| Cell migration | 7 | 26.9 | 7.60e-04 |
| Programmed cell death | 9 | 34.6 | 7.20e-04 |
| Positive regulation of apoptosis | 8 | 30.8 | 6.70e-04 |
| Positive regulation of programmed cell death | 8 | 30.8 | 6.50e-04 |
| Positive regulation of cell death | 8 | 30.8 | 6.30e-04 |
| Regulation of cell migration | 6 | 23.1 | 7.40e-04 |
| Localization of cell | 7 | 26.9 | 9.00e-04 |
| Cell motility | 7 | 26.9 | 9.00e-04 |
| Regulation of cell motion | 6 | 23.1 | 1.30e-03 |
| Cell death | 9 | 34.6 | 1.50e-03 |
Data are as described in Table 3.
Table 6.
Cellular component gene ontology of downregulated genes
| Term | Count | % | Adjusted P Value Using Benjamini-Hochberg Method |
|---|---|---|---|
| Extracellular space | 12 | 46.2 | 2.30e-06 |
| Extracellular region part | 12 | 46.2 | 3.60e-05 |
| Cell surface | 7 | 26.9 | 1.60e-03 |
| Extracellular region | 13 | 50 | 4.00e-03 |
| Integrin complex | 3 | 11.5 | 3.00e-02 |
| Receptor complex | 4 | 15.4 | 3.00e-02 |
| Plasma membrane part | 12 | 46.2 | 2.40e-02 |
| External side of plasma membrane | 4 | 15.4 | 5.50e-02 |
| Secretory granule | 4 | 15.4 | 5.70e-02 |
| Plasma membrane | 15 | 57.7 | 5.20e-02 |
| Platelet α granule | 3 | 11.5 | 4.90e-02 |
| Integral to plasma membrane | 8 | 30.8 | 5.40e-02 |
| Intrinsic to plasma membrane | 8 | 30.8 | 5.60e-02 |
| Cell fraction | 7 | 26.9 | 1.10e-01 |
| Platelet α granule membrane | 2 | 7.7 | 1.30e-01 |
| Cytoplasmic membrane-bound vesicle | 5 | 19.2 | 1.30e-01 |
| Soluble fraction | 4 | 15.4 | 1.30e-01 |
| Membrane-bound vesicle | 5 | 19.2 | 1.30e-01 |
| Ruffle membrane | 2 | 7.7 | 1.20e-01 |
| Cytoplasmic vesicle | 5 | 19.2 | 1.70e-01 |
Data are as described in Table 3.
DISCUSSION
Tissue collagen homeostasis is tightly regulated by collagen production and degradation. Imbalance of this homeostasis leads to tissue destruction or fibrosis. In the current study, we investigated whether HSCs internalize collagen as a means to sense their microenvironment and respond accordingly. We make the novel observation in this study that human HSCs internalize collagen through a macropinocytosis mechanism that is regulated by dynamin-2 and c-abl and that interaction of collagen with HSCs provides feedback on MMP-9 production.
In this study, we identify macropinocytosis, a nonselective endocytosis pathway, as a mechanism for collagen endocytosis by HSCs. This was based on colocalization studies with Dextran 10K as well as inhibition of matrix endocytosis by compounds that perturb macropinocytosis, including EIPA. Perturbation of actin cytoskeleton, PI3K/AKT signaling, and the large GTPase dynamin-2 also diminished HSC collagen internalization. Prior studies have implicated these pathways in macropinocytosis (27, 35). Innate immune cells such as macrophages continuously survey their environment in search of foreign particles and soluble antigens through macropinocytosis (24, 27). Our study suggests that HSCs also use this mechanism to examine and respond to environmental changes and highlight increasing evidence for innate immune surveillance function of HSCs, especially considering that some matrix proteins can stimulate Toll-like receptor 4 and other innate immune markers on HSCs (6, 45, 52).
The interaction between HSC and extracellular matrix is complex. Extracellular collagens can be recognized by cells through putative collagen receptors including integrins, the discoidin domain receptors (DDRs), and mannose receptor family, which upon ligand binding elicit different signaling pathways that respond to extracellular matrix (14, 25, 48, 56). Interaction of integrin with collagens provides multiple cues, including cell-extracellular matrix adhesion and cellular polarization that eventually regulate many aspects of cell behavior, including positive or negative feedback on synthesis of the extracellular proteins themselves. For example, prior observations suggest that negative feedback regulation of collagen synthesis is a major role of integrin α1β1 receptor stimulation in fibroblasts (23, 40). DDR receptors regulate cell adhesion, migration, proliferation, and remodeling of the extracellular matrix by controlling the expression and activity of MMPs (42, 55). Similarly, uPARAP/Endo180 from the mannose receptor family was found to be important for collagen internalization in activated rat HSCs (37) and other cultured cells (12, 29, 33, 37, 51). Our study provides evidence that collagen macropinocytosis is an additional communication mechanism between HSCs and extracellular matrix. Further studies will be required to elucidate the distinct or related roles of matrix-receptor binding and macropinocytosis when HSCs are exposed to collagen in the microenvironment.
Surprisingly, nonreceptor tyrosine kinase family members, c-abl and Src, differentially regulated HSC collagen uptake. MEF cells with c-abl and Arg double knockdown showed decreased collagen uptake compared with control MEF cells. C-abl tyrosine kinase likely regulates macropinocytosis by regulating Abi1 interactions with several actin polymerization regulatory complexes, including PI3K, whose involvement in actin dynamics is well established (53). In contradistinction, MEF cells with Src isoform deletion showed enhanced collagen uptake compared with control MEF. Indeed, CRK phosphorylation was increased in YSF MEF, indicating that lack of Src may lead to a compensatory upregulation of c-abl with ensuing increase in matrix macropinocytosis in these cells. Although most of our studies were performed in the absence of TGF-β1 stimulation, the signaling networks we studied are classically downstream from TGF-β1 stimulation, and this growth factor did stimulate collagen internalization in our studies and in prior studies (18).
Although recognized predominantly for their fibrogenic activity, HSCs also degrade matrix by producing proteolytic MMPs. Collagen degradation includes the cleavage of full-length extracellular collagen by collagenases (MMP-1, -2, -8, -13, and the membrane-bound MMP-14 and -15) followed by further byproduct cleavage by the gelatinases, MMP-2, or MMP-9 (28, 34, 47). Degraded collagen is then internalized by cells and targeted to lysosomes, a step that may account for the rheostatic function of HSCs, as proposed in the present study. Thus the proteolytic degradation of matrix may achieve the goal of allowing HSCs to “sample and respond” to extracellular matrix dynamics by modifying collagen production or collagen-degrading enzymes. Indeed, we did observe an increase MMP-9 in production in response to collagen endocytosis, supporting such a feedback loop. A prior study supporting this concept showed that HSCs generate MMP-2 in response to type I collagen fibril exposure (50). In addition to MMP, interaction with collagen significantly decreased HSC collagen Iα and collagen XVIII α1 mRNA transcripts, indicating a negative feedback on collagen production. In summary, the present studies identify a role for HSCs as a sensor and responder to the extracellular matrix microenvironment through their ability to endocytose extracellular matrix proteins and adjust their production of proteases in response to what they sample.
GRANTS
This work was supported by National Institutes of Health (NIH) Grants DK59615 and AA021171 (V. Shah), the Cell Biology Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567), and NIH K12 CA90628 (B. Ji).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: Y.B. conception and design of research; Y.B., D.M., M.D., and S.C. performed experiments; Y.B., D.M., M.D., B.J., X.L., S.C., and V.H.S. analyzed data; Y.B., D.M., M.D., S.C., and V.H.S. interpreted results of experiments; Y.B. prepared figures; Y.B. and V.H.S. drafted manuscript; Y.B. and V.H.S. edited and revised manuscript; Y.B. and V.H.S. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors acknowledge Mrs. Terri Johnson for excellent editing.
REFERENCES
- 1.Arora PD, Wang Y, Bresnick A, Dawson J, Janmey PA, McCulloch CA. Collagen remodeling by phagocytosis is determined by collagen substrate topology and calcium-dependent interactions of gelsolin with nonmuscle myosin IIA in cell adhesions. Mol Biol Cell 24: 734–747, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atorrasagasti C, Aquino JB, Hofman L, Alaniz L, Malvicini M, Garcia M, Benedetti L, Friedman SL, Podhajcer O, Mazzolini G. SPARC downregulation attenuates the profibrogenic response of hepatic stellate cells induced by TGF-β1 and PDGF. Am J Physiol Gastrointest Liver Physiol 300: G739–G748, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest 115: 209–218, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bissell DM, Friedman SL, Maher JJ, Roll FJ. Connective tissue biology and hepatic fibrosis: report of a conference. Hepatology 11: 488–498, 1990 [DOI] [PubMed] [Google Scholar]
- 5.Blomhoff R, Wake K. Perisinusoidal stellate cells of the liver: important roles in retinol metabolism and fibrosis. FASEB J 5: 271–277, 1991 [DOI] [PubMed] [Google Scholar]
- 6.Brunn GJ, Bungum MK, Johnson GB, Platt JL. Conditional signaling by Toll-like receptor 4. FASEB J 19: 872–874, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee S, Cao S, Petersen T, Simari R, Shah V. Inhibition of GTP-dependent vesicle trafficking impairs internalization of plasmalemmal eNOS and cellular nitric oxide production. J Cell Sci 116: 3645–3655, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497: 633–637, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels C, Wilkes M, Edens M, Kottom T, Murphy S, Limper A, Leof E. Imatinib mesylate inhibits the profibrogenic activity of TGF-beta and prevents bleomycin-mediated lung fibrosis. J Clin Invest 114: 1308–1316, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das A, Shergill U, Thakur L, Sinha S, Urrutia R, Mukhopadhyay D, Shah VH. Ephrin B2/EphB4 pathway in hepatic stellate cells stimulates Erk-dependent VEGF production and sinusoidal endothelial cell recruitment. Am J Physiol Gastrointest Liver Physiol 298: G908–G915, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A, Yaqoob U, Mehta D, Shah VH. FXR Promotes Endothelial Cell Motility Through Coordinated Regulation of FAK and MMP-9. Arterioscler Thromb Vasc Biol 29: 562–570, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelholm LH, List K, Netzel-Arnett S, Cukierman E, Mitola DJ, Aaronson H, Kjoller L, Larsen JK, Yamada KM, Strickland DK, Holmbeck K, Dano K, Birkedal-Hansen H, Behrendt N, Bugge TH. uPARAP/Endo180 is essential for cellular uptake of collagen and promotes fibroblast collagen adhesion. J Cell Biol 160: 1009–1015, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SJ, Parola M. Liver fibrogenic cells. Best Pract Res Clin Gastroenterol 25: 207–217, 2011 [DOI] [PubMed] [Google Scholar]
- 14.Fu HL, Valiathan RR, Arkwright R, Sohail A, Mihai C, Kumarasiri M, Mahasenan KV, Mobashery S, Huang P, Agarwal G, Fridman R. Discoidin domain receptors: unique receptor tyrosine kinases in collagen-mediated signaling. J Biol Chem 288: 7430–7437, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaca MD, Zhou X, Issa R, Kiriella K, Iredale JP, Benyon RC. Basement membrane-like matrix inhibits proliferation and collagen synthesis by activated rat hepatic stellate cells: evidence for matrix-dependent deactivation of stellate cells. Matrix Biol 22: 229–239, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Heuser JE, Anderson RG. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J Cell Biol 108: 389–400, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Ikenaga N, Ohuchida K, Mizumoto K, Akagawa S, Fujiwara K, Eguchi D, Kozono S, Ohtsuka T, Takahata S, Tanaka M. Pancreatic cancer cells enhance the ability of collagen internalization during epithelial-mesenchymal transition. PLoS One 7: e40434, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang JX, Torok NJ. Liver injury and the activation of the hepatic myofibroblasts. Curr Pathobiol Rep 1: 215–223, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawelke N, Vasel M, Sens C, Au A, Dooley S, Nakchbandi IA. Fibronectin protects from excessive liver fibrosis by modulating the availability of and responsiveness of stellate cells to active TGF-beta. PLoS One 6: e28181, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerr MC, Teasdale RD. Defining macropinocytosis. Traffic 10: 364–371, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Kjoller L, Engelholm LH, Hoyer-Hansen M, Dano K, Bugge TH, Behrendt N. uPARAP/endo180 directs lysosomal delivery and degradation of collagen IV. Exp Cell Res 293: 106–116, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Langholz O, Rockel D, Mauch C, Kozlowska E, Bank I, Krieg T, Eckes B. Collagen and collagenase gene expression in three-dimensional collagen lattices are differentially regulated by alpha 1 beta 1 and alpha 2 beta 1 integrins. J Cell Biol 131: 1903–1915, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanzavecchia A. Mechanisms of antigen uptake for presentation. Curr Opin Immunol 8: 348–354, 1996 [DOI] [PubMed] [Google Scholar]
- 25.Leitinger B, Hohenester E. Mammalian collagen receptors. Matrix Biol 26: 146–155, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Lemoinne S, Cadoret A, El Mourabit H, Thabut D, Housset C. Origins and functions of liver myofibroblasts. Biochim Biophys Acta 1832: 948–954, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Lim JP, Gleeson PA. Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol Cell Biol 89: 836–843, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Madsen DH, Engelholm LH, Ingvarsen S, Hillig T, Wagenaar-Miller RA, Kjoller L, Gardsvoll H, Hoyer-Hansen G, Holmbeck K, Bugge TH, Behrendt N. Extracellular collagenases and the endocytic receptor, urokinase plasminogen activator receptor-associated protein/Endo180, cooperate in fibroblast-mediated collagen degradation. J Biol Chem 282: 27037–27045, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Madsen DH, Ingvarsen S, Jurgensen HJ, Melander MC, Kjoller L, Moyer A, Honore C, Madsen CA, Garred P, Burgdorf S, Bugge TH, Behrendt N, Engelholm LH. The non-phagocytic route of collagen uptake: a distinct degradation pathway. J Biol Chem 286: 26996–27010, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen DH, Leonard D, Masedunskas A, Moyer A, Jurgensen HJ, Peters DE, Amornphimoltham P, Selvaraj A, Yamada SS, Brenner DA, Burgdorf S, Engelholm LH, Behrendt N, Holmbeck K, Weigert R, Bugge TH. M2-like macrophages are responsible for collagen degradation through a mannose receptor-mediated pathway. J Cell Biol 202: 951–966, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallat A, Lotersztajn S. Cellular mechanisms of tissue fibrosis. Five novel insights into liver fibrosis. Am J Physiol Cell Physiol 305: C789–C799, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Martin-Vilchez S, Sanz-Cameno P, Rodriguez-Munoz Y, Majano PL, Molina-Jimenez F, Lopez-Cabrera M, Moreno-Otero R, Lara-Pezzi E. The hepatitis B virus X protein induces paracrine activation of human hepatic stellate cells. Hepatology 47: 1872–1883, 2008 [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Pomares L, Wienke D, Stillion R, McKenzie EJ, Arnold JN, Harris J, McGreal E, Sim RB, Isacke CM, Gordon S. Carbohydrate-independent recognition of collagens by the macrophage mannose receptor. Eur J Immunol 36: 1074–1082, 2006 [DOI] [PubMed] [Google Scholar]
- 34.McKleroy W, Lee TH, Atabai K. Always cleave up your mess: targeting collagen degradation to treat tissue fibrosis. Am J Physiol Lung Cell Mol Physiol 304: L709–L721, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol 11: 510–520, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Mousavi SA, Fonhus MS, Berg T. Up-regulation of uPARAP/Endo180 during culture activation of rat hepatic stellate cells and its presence in hepatic stellate cell lines from different species. BMC Cell Biol 10: 39, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mousavi SA, Sato M, Sporstol M, Smedsrod B, Berg T, Kojima N, Senoo H. Uptake of denatured collagen into hepatic stellate cells: evidence for the involvement of urokinase plasminogen activator receptor-associated protein/Endo180. Biochem J 387: 39–46, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murata K, Kudo M, Onuma F, Motoyama T. Changes of collagen types at various stages of human liver cirrhosis. Hepatogastroenterology 31: 158–161, 1984 [PubMed] [Google Scholar]
- 39.Priya S, Sudhakaran PR. Cell survival, activation and apoptosis of hepatic stellate cells: modulation by extracellular matrix proteins. Hepatol Res 38: 1221–1232, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Ravanti L, Heino J, Lopez-Otin C, Kahari VM. Induction of collagenase-3 (MMP-13) expression in human skin fibroblasts by three-dimensional collagen is mediated by p38 mitogen-activated protein kinase. J Biol Chem 274: 2446–2455, 1999 [DOI] [PubMed] [Google Scholar]
- 41.Rothberg K, Heuser J, Donzell W, Ying Y, Glenney J, Anderson R. Caveolin, a protein component of caveolae membrane coats. Cell 68: 673–682, 1992 [DOI] [PubMed] [Google Scholar]
- 42.Ruiz PA, Jarai G. Discoidin domain receptors regulate the migration of primary human lung fibroblasts through collagen matrices. Fibrogenesis Tissue Repair 5: 3, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupper AC, Rodriguez-Paris JM, Grove BD, Cardelli JA. p110-related PI 3-kinases regulate phagosome-phagosome fusion and phagosomal pH through a PKB/Akt dependent pathway in Dictyostelium. J Cell Sci 114: 1283–1295, 2001 [DOI] [PubMed] [Google Scholar]
- 44.Schnitzer JE, Oh P, Pinney E, Allard J. Filipin-sensitive caveolae-mediated transport in endothelium: reduced transcytosis, scavenger endocytosis, and capillary permeability of select macromolecules. J Cell Biol 127: 1217–1232, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 13: 1324–1332, 2007 [DOI] [PubMed] [Google Scholar]
- 46.Semela D, Das A, Langer DA, Kang N, Leof E, Shah VH. Platelet-derived growth factor signaling through ephrin-B2 regulates hepatic vascular structure and function. Gastroenterology 135: 671–679, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song F, Wisithphrom K, Zhou J, Windsor LJ. Matrix metalloproteinase dependent and independent collagen degradation. Front Biosci 11: 3100–3120, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Tenner AJ. Membrane receptors for soluble defense collagens. Curr Opin Immunol 11: 34–41, 1999 [DOI] [PubMed] [Google Scholar]
- 49.Thomsen P, Roepstorff K, Stahlhut M, van Deurs B. Caveolae are highly immobile plasma membrane microdomains, which are not involved in constitutive endocytic trafficking. Mol Biol Cell 13: 238–250, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang DR, Sato M, Li LN, Miura M, Kojima N, Senoo H. Stimulation of pro-MMP-2 production and activation by native form of extracellular type I collagen in cultured hepatic stellate cells. Cell Struct Funct 28: 505–513, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Wienke D, MacFadyen JR, Isacke CM. Identification and characterization of the endocytic transmembrane glycoprotein Endo180 as a novel collagen receptor. Mol Biol Cell 14: 3592–3604, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winau F, Quack C, Darmoise A, Kaufmann SH. Starring stellate cells in liver immunology. Curr Opin Immunol 20: 68–74, 2008 [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Ziemnicka D, Merz GS, Kotula L. Human spectrin Src homology 3 domain binding protein 1 regulates macropinocytosis in NIH 3T3 cells. J Cell Sci 113: 3805–3814, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu L, Hui A, Albanis E, Arther M, O'Byrne S, Blaner W, Mukherjee P, Friedman S, Eng F. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut 54: 142–151, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu L, Peng H, Wu D, Hu K, Goldring MB, Olsen BR, Li Y. Activation of the discoidin domain receptor 2 induces expression of matrix metalloproteinase 13 associated with osteoarthritis in mice. J Biol Chem 280: 548–555, 2005 [DOI] [PubMed] [Google Scholar]
- 56.Yeh YC, Lin HH, Tang MJ. A tale of two collagen receptors, integrin β1 and discoidin domain receptor 1, in epithelial cell differentiation. Am J Physiol Cell Physiol 303: C1207–C1217, 2012 [DOI] [PubMed] [Google Scholar]
- 57.Zhou X, Murphy FR, Gehdu N, Zhang J, Iredale JP, Benyon RC. Engagement of alphavbeta3 integrin regulates proliferation and apoptosis of hepatic stellate cells. J Biol Chem 279: 23996–24006, 2004 [DOI] [PubMed] [Google Scholar]
- 58.Zou L, Cao S, Kang N, Huebert RC, Shah VH. Fibronectin induces endothelial cell migration through beta1 integrin and Src-dependent phosphorylation of fibroblast growth factor receptor-1 at tyrosines 653/654 and 766. J Biol Chem 287: 7190–7202, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]