Abstract
Package inserts of Food and Drug Administration (FDA) approved prescription drugs, including chemotherapy drugs, must follow a specific format imposed by the FDA. These inserts are created by unrelated pharmaceutical companies and as a result tend to be very different in the way the required information is reported. Chemical and pharmacokinetic properties including absorption, distribution, metabolism, excretion, and toxicity (ADME/Tox) are crucial elements to a prescribing information packet and are often missing from the reported data. This undergraduate research project analyzes the information packets of 85 randomly chosen chemically diverse chemotherapy drugs for four parameters important to patient care; viz, volume of distribution (VD), elimination half-life (t1/2), bioavailability, and water solubility. The prescribing information from the package inserts of each was analyzed in detail and pertinent information was consequently tabulated into a database using a commercial informatics platform. Then using a substructure search-tool, sixty-five chemotherapy drugs containing a carbonyl group in their chemical structure were selected and as hypothesized, it was found that many of these packets were significantly lacking in the reporting of the four parameters of interest. To further enhance this cataloged data, a freely available online database was consequently developed (http://annotation.dbi.udel.edu/CancerDB/) with the intention that the chemical, biological, and clinical community will now add some of the missing parameters.
Keywords: ADME/Tox, bioavailability, chemotherapy, database, half-life, elimination prescribing information packets, water solubility, volume of distribution, pharmaceutical drugs
I. INTRODUCTION
Developing new drugs is one of the most expensive, time demanding and laborious process in health care. It usually involves a standardized development, experimentation and implementation channel. This is instigated with an extensive chemically based, ADME/Tox dependent selection process followed by in-depth laboratory experimentation both in-vivo and in-vitro. The final clinical trial phase is the most criticized and along with the gathered experimental data, is the bases on which the FDA drug accreditation is conducted [1]. The FDA implements elaborate highly detailed standards to evaluate and report the information provided by the pharmaceutical company, and approves the drug if these standards are met [1]. On approval, the clinical information is subsequently compiled by the companies into prescribing information packets or package inserts in a standardized format approved by the FDA [2]. The material provided in these inserts includes, among other parameters, pharmacokinetic (ADME/Tox) properties, dosage and administration, adverse reactions, contraindications, and precautions. This drug information and warnings are required for patient education and are also essential for a physician to safely administer a drug to all patient populations [3].
High risk category medications such as chemotherapy drugs have highly variable pharmacokinetics and very narrow therapeutic windows i.e., very little room for error in dosage [4]. Hence, having available all of the pharmacokinetic properties of such drugs documented in the package inserts can help guide its selection in order to achieve efficacy while minimizing or avoiding toxicity. Also, this knowledge could improve patient-centered care practices by modifying dosage based on pharmacokinetics and pharmacodynamics [5].
Initial results from recent studies in our laboratory showed that some essential pharmacokinetic parameters necessary for directing patient care were significantly lacking in 30 FDA approved cancer drugs [6, 7] and 75 other common consumer drug packets [8, 9, 10].
Parameters of interest included: neurotoxicity, volume of distribution (VD), rate of absorption, water solubility, teratogenicity, human intestinal absorption, bioavailability, elimination half-life (t1/2), oncogenicity, plasma protein binding, pKa, and blood brain barrier. Comprehension of these properties is important for the determination of the route of administration, dosage, onset of action, time to peak concentration, duration of action, and frequency of administration.
Since a number of these parameters are also common to QSAR (quantitative structure activity relationship) modeling; in an earlier study, we extracted available information from 75 common consumer drug prescription inserts and created a drug database [8–10] using the commercial KnowItAll® informatics platform [11–14]. We showed that we could successfully predict and validate some of the missing properties on the basis of chemical structure using the commercial prediction tools [15] available in Bio-Rad’s KnowItAll® ADME/Tox edition [11].
A contemporary review [16] of web-based technologies has likewise shown that better communication and documentation of clinical information could enable rapid sharing across global boundaries. Frequently used examples of such ‘free-to-access’ structure based compound information systems are the Royal Society of Chemistry’s (RSC) ChemSpider database [17] and PubChem [18]. Notwithstanding, in such systems numerous experimentally determined clinical parameters are unknown.
A principal objective of this project is to further substantiate our earlier revelation of convincing shortcomings in the reporting of two crucial parameters (VD and t1/2) found in 30 randomly selected cancer drugs. In this project we have augmented our study to 85 indiscriminately chosen structurally diverse cancer drugs and in 65 of the drugs that contain at least one carbonyl group in their chemical structure have thoroughly assessed the reporting of 4 valuable parameters viz., bioavailability, water solubility, VD, and t1/2.
Furthermore, to create a framework that supports researchers in their data collection and submission activities, a freely available online database was developed with the intention that the chemical, biological, and clinical community will now add some of the missing parameters.
II. MATERIALS AND METHODS
A. Collect Information Packets
First, the Scott Hamilton CARES initiative website [19] was utilized as a reference for chemotherapy treatments currently being used on cancer patients. Then, eighty-five of the current available chemotherapy drugs were randomly selected. The PDF files of the drug information inserts were downloaded using the Drugs@FDA website [20] and the Physicians’ Desk Reference website [21]. Only packets containing published chemical 2-D drug structures were collected.
B. Isolate for Carbonyl-Containing Compounds
The 85 arbitrarily chosen packets were reviewed in detail to uncover the 14 chemical, pharmacokinetic, and toxicological parameters [6, 8–10] of general interest (viz., oncogenicity, teratogenicity, mutagenicity, human intestinal absorption, plasma protein binding, water solubility, volume of distribution, elimination half time, rate of absorption, blood brain barrier, neurotoxicity, pKa, bioavailability, and log P). As communicated earlier [6–10], there were significant differences in the methodology of reporting, hence to ensure data integrity; data normalizations were sometimes performed [6, 8–10]. Utilizing the KnowItAll® Informatics platform [11–14] each 2-D chemical structure was drawn with the available drawing tools. A database containing the extracted chemical, pharmacokinetic, and pharmacological data for all 85 chemotherapy drugs was created using the database building and management package in the KnowItAll® Informatics platform. Employing the substructure search tool SearchIT™ [11–14] within the platform, 65 compounds containing carbonyl groups were isolated in order to assess the magnitude of lacking data and for future SAR studies with this group of molecules. The brand names of the 65 carbonyl containing drugs shown in Table 1 are: Abraxane® Accutane® Afinitor® Agrylin® Alkeran® Aromasin® Camptosar® Carac® Casodex® CeeNu® Cosmegen® Dacogen® Depocyt® Depo-Medrol® Doxil® Eldisine® Eligard® Evista® Fareston® Fusilev® Gemzar® Gleevec® Hycamtin® Hydrea® Idamycin® Ixempra® Leukeran® Matulane® Megace® Mutamycin® Navelbine® Nexavar® Nilandron® Nolvadex® Novantrone® Orapred ODT® Oxsoralen® Panretin® Plenaxis® Revlimid® Rheumatrex® Sandostatin® Solu-cortef® Sprycel® Sutent® Targretin® Taxotere® Temodar® Thalomid® Tomudex® Toposar® Torisel® Treanda® Valstar® Velban® Velcade® Vesanoid® Vidaza® Vincasar® Vumon® Xeloda® Zanosar® Zinecard® Zoladex® Zolinza®
Table 1.
FDA chemotherapy drug targeted pharmacokinetics database. Includes alphabetically sorted sampled drugs with related brand names, and data for volume of distribution (L/Kg), t1/2 (hours), water solubility (mg/mL) and bioavailability (%). NR (not reported). N/A (not applicable).
| Volume of Distribution (L/Kg) | tl/2 (hours) | Water Solubility (mg/mL) | Bioavailability (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Drug Common Name | Brand Name | FDA data | Estimation (L/kg) | FDA data | Estimation | FDA data | Assumption | Admin. Route | ||
| 1 | Abarelix | Plenaxis | 2433L–5647 L | 34.7–80.7 | 240–393.6 | NR | N/A | NR | N/A | IM Injection |
| 2 | alitretinoin | Panretin | NR | N/A | NR | 0 | N/A | NR | N/A | Gel |
| 3 | Anagrelide HCl | Agrylin | NR | N/A | 1.3 | very slightly | 0.001 | NR | N/A | Capsules |
| 4 | azacitidine | Vidaza | 50–102 L | 0.714–1.46 | 4 | sparingly | 0.001 | 89* (SC) | 100 (IV) | SC or IV |
| 5 | bendamustine hydrochloride | Treanda | 25 L | 0.357 | 0.67 | NR | N/A | N/A | 100 | IV Injection |
| 6 | Bexarotene | Targretin | NR | N/A | 7 | 0 | N/A | NR | N/A | Capsules |
| 7 | Bicalutamide | Casodex | NR | N/A | 139.2 | 0 | N/A | NR | N/A | Tablets |
| 8 | bortezomib | Velcade | 498–1884 L/m2 | 12.3–46.52 | 40–193* | 3.3–3.8 | N/A | N/A | 100 | IV Injection |
| 9 | capecitabine | Xeloda | NR | N/A | 0.75 | 26 | N/A | NR | N/A | Tablets |
| 10 | chlorambucil | Leukeran | NR | N/A | 1.5 | NR | N/A | NR | N/A | Tablets |
| 11 | Cytarabine | Depocyt | NR | N/A | 100–263 | NR | N/A | NR | N/A | Intrathecal Inj. |
| 12 | Dactinomycin | Cosmegen | NR | N/A | 36 | NR | N/A | N/A | 100 | IV Injection |
| 13 | dasatinib | Sprycel | 2505 L | 35.79 | 3–5 | 0 | N/A | NR | N/A | Tablets |
| 14 | Decitabine | Dacogen | NR | N/A | 0.25–0.42 | sparingly | 0.001 | N/A | 100 | IV Injection |
| 15 | dexrazoxane | Zinecard | 25 L/m2 | 0.617 | 2.1–2.5* | sparingly | 0.001 | N/A | 100 | IV Injection |
| 16 | docetaxel | Taxotere | 113 L | 1.61 | 11.1 | practically insoluble | 0.001 | N/A | 100 | IV Injection |
| 17 | doxorubicin hydrochloride | Doxil | 2.6–2.975 L/m2* | 0.064–0.073 | 55 | NR | N/A | N/A | 100 | IV Injection |
| 18 | etoposide | Toposar | 18–29 L | 0.26–0.41 | 4 to 11 | sparingly | 0.001 | N/A | 100 | IV Injection |
| 19 | Everolimus | Afinitor | NR | N/A | 30 | NR | N/A | NR | N/A | Tablets |
| 20 | Exemestane | Aromasin | NR | N/A | 24 | 0 | N/A | NR | N/A | Tablets |
| 21 | fluorouracil | Carac | NR | N/A | N/A | NR | N/A | NR | N/A | Cream |
| 22 | Gemcitabine | Gemzar | 50–370 L/m2* | 1.23–9.14 | 1.7–19.4 | soluble | 1000 | N/A | 100 | IV Injection |
| 23 | goserelin | Zoladex | 16.2–58.7 L | 0.436–0.824 | 1.7–5.3 | soluble | 1000 | NR | N/A | Implant |
| 24 | hydro cortisone sodium succinate | Solu-cortef | NR | N/A | NR | very soluble | 1000 | NR | N/A | IV or IM |
| 25 | hydroxyurea | Hydrea | NR | N/A | NR | NR | N/A | NR | N/A | Capsules |
| 26 | Idarubicin Hcl | Idamycin | NR | N/A | 22 | NR | N/A | N/A | 100 | IV Injection |
| 27 | Imatinib Mesylate | Gleevec | NA | N/A | 18 | practically insoluble | 0.001 | 98 | N/A | Tablets |
| 28 | irinotecan hydrochloride | Camptosar | 61.5–303.6 L/m2* | 1.52–7.5 L/kg | 6.0–12.0 | slightly soluble | 0.01 | N/A | 100 | IV Injection |
| 29 | isotretinoin | Accutane | NR | N/A | 21 | NR | N/A | NR | N/A | Capsules |
| 30 | Ixabepilone | Ixempra | 1000 L | 14.29 | 52 | NR | N/A | N/A | 100 | IV Injection |
| 31 | lenalidomide | Revlimid | NR | N/A | 3 | NR | N/A | NR | N/A | Capsules |
| 32 | leucovorin calcium | Fusilev | NR | N/A | 5.1–6.8 | NR | N/A | N/A | 100 | IV Injection |
| 33 | leuprolide acetate | Eligard | 27 L | .386 1/KG | 3 | NR | N/A | NR | N/A | SC Injection |
| 34 | lomustine | Ceenu | NR | N/A | 16–48 | 0–0.0499999 | N/A | NR | N/A | Capsules |
| 35 | megestrol acetate | Megace | NR | N/A | 34.2 | 0.002 | N/A | NR | N/A | Tablets |
| 36 | melphalan hydrochloride | Alkeran | 0.5 L/kg | N/A | 1.25 | practically insoluble | 0.001 | N/A | 100 | IV Injection |
| 37 | methotrexate | Rheumatrex | 4.8 L/kg | N/A | 3.0–15.0* | NR | N/A | 60 | N/A | Tablets |
| 38 | methoxsalen | Oxsoralen | NR | N/A | 2 | NR | N/A | NR | N/A | Capsules |
| 39 | methylprednisolone acetate | Depo-Medrol | NR | N/A | NR | practically insoluble | 0.001 | NR | N/A | Injection* |
| 40 | mitomycin | Mutamycin | NR | N/A | NR | NR | N/A | N/A | 100 | IV Injection |
| 41 | mitoxantrone | Novantrone | 1000 L/m2 | 24.69 | 75 | NR | N/A | N/A | 100 | IV Injection |
| 42 | nilutamide | Nilandron | NR | N/A | 38–59.1 | slightly soluble | 0.01 | NR | N/A | tablets |
| 43 | octreotide acetate | Sandostatin | 13.6 L | 0.194 | 1.7–1.9 | NR | N/A | NR | N/A | SC injection |
| 44 | paclitaxel | Abraxane | 632 L | 15.6 | 27 | 0 | N/A | N/A | 100 | IV injection |
| 45 | prednisolone sodium phosphate | Orapred ODT | NR | N/A | 2.0–4.0 | freely soluble | 1000 | NR | N/A | tablets |
| 46 | procarbazine hydrochloride | Matulane | NR | N/A | NR | soluble | 1000 | NR | N/A | capsules |
| 47 | Raloxifene | Evista | 2348 L/Kg | N/A | 27.7 | very slightly | 0.001 | 2 | N/A | tablets |
| 48 | raltitrexed | Tomudex | 548 L | 7.83 | 198 | NR | N/A | N/A | 100 | IV injection |
| 49 | sorafenib tosylate | Nexavar | NR | N/A | 25–48 | practically insoluble | 0.001 | 38–49 | N/A | tablets |
| 50 | streptozocin | Zanosar | NR | N/A | NR | very soluble | 1000 | N/A | 100 | IV injection |
| 51 | sunitinib malate | Sutent | 2230 L | 31.86 | 40–60 | 25 | N/A | NR | N/A | capsules |
| 52 | tamoxifen citrate | Nolvadex | NR | N/A | 120–168 | 0.5 | N/A | NR | N/A | tablets |
| 53 | temozolomide | Temodar | 0.4L/Kg | 0.4 | 1.8 | NR | N/A | NR | N/A | capsules |
| 54 | temsirolimus | Torisel | 172 L | 2.46 | NR | practically insoluble | 0.001 | N/A | 100 | IV injection |
| 55 | teniposide | Vumon | 8–44L/m2 | 0.198–1.086 | 5 | 0 | N/A | N/A | 100 | IV injection |
| 56 | thalidomide | Thalomid | NR | N/A | 5.0–7.0 | sparingly | 0.001 | NR | N/A | capsules |
| 57 | topotecan hydrochloride | Hycamtin | NR | N/A | 2.0–3.0 | soluble | 1000 | N/A | 100 | IV Injection |
| 58 | toremifene | Fareston | 580 L | 8.29 | 120 | 0.63 | N/A | NR | N/A | tablets |
| 59 | tretinoin | Vesanoid | NR | N/A | 0.5–2 | NR | N/A | NR | N/A | capsules |
| 60 | valrubicin | Valstar | NR | N/A | NR | relatively insoluble | 0.001 | NR | N/A | Intravesical Inj. |
| 61 | vinblastine sulfate | Velban | NR | N/A | 24.8 | freely soluble | 1000 | N/A | 100 | IV Injection |
| 62 | vincristine sulfate | Vincasar | NR | N/A | 85 | freely soluble | 1000 | N/A | 100 | IV Injection |
| 63 | vindesine | Eldisine | 8.11 L/kg | N/A | 20–24 | NR | N/A | N/A | 100 | IV Injection |
| 64 | vinorelbine tartrate | Navelbine | 25.4–40.1 L/kg | N/A | 27.7–43.6 | 1000 | N/A | N/A | 100 | IV Injection |
| 65 | vorinostat | Zolinza | NR | N/A | NR | very slightly | 0.001 | NR | N/A | Capsules |
C. Collect Water Solubility Parameters
Water solubility information is important for orally administered drugs that will be absorbed across the intestinal walls. In general, the more water-soluble a drug, the quicker it will be absorbed and utilized by the body. Ideally water solubility parameters are reported as intrinsic solubility in mg/ml of unionized species (free form) and ionized species (salt forms), and documented at around 25 °C. However, many times non-numerical values were reported. In such cases, approximations were made: sparingly soluble, practically insoluble, relatively insoluble were documented as 0.001 mg/ml; slightly soluble were documented as 0.01 mg/ml; and freely soluble, highly soluble, very soluble, and soluble were recorded as 1000 mg/ml.
D. Collect Bioavailability Parameters
Bioavailability refers to the amount of the administered drug that reaches the circulatory system unchanged. Bioavailability varies depending on the route of administration, incomplete absorption, first-pass gut and hepatic metabolism. This parameter is reported as a percentage. Thus, the bioavailability of a drug should be 100% if administered intravenously (IV). We have made this assumption for those chemotherapy drug inserts lacking bioavailability parameters and that were administered via the IV route. Bioavailability is a crucial piece of information necessary in oral medications because the bioavailability reported data must be considered for all non-IV routes when calculating doses.
E. Collect Volume of Distribution (VD) Parameters
VD is a useful pharmacokinetic parameter that quantifies the relationship between amounts of drug in the body to its distribution in the plasma after oral or parenteral dosing. Its numerical value is indicative of the extent of distribution of the drug since VD is used to estimate the amount of drug in the body, peak serum levels, and clearance. The magnitude of the apparent volume of distribution can be used as a guide for dosage regimens and hence the need to access this data from clinically controlled dose-optimization studies. Drugs that are highly lipid soluble have a very high VD value (in liters). In this project VD values are normalized to units of L/Kg. This was done by dividing the reported volume by 70 kg which is the average weight of the human subjects in a majority of the documented FDA file data [8]. When reported in units of L/m2, the VD value was divided by 40.5 to yield the necessary units in L/Kg [22].
F. Collect Elimination Half-life (t1/2) data
The half-life is the speed of elimination for a drug to lose half its pharmacologic or physiologic activity from plasma or serum. This process is determined on the results of chemical analyses that tend to follow first-order kinetics and can be rationalized using linear or non-linear pharmacokinetic models [23]. The smaller the half-life the larger the dose necessary, as the drug has a small residence time in the body. For its determination, researchers evaluate volume of distribution and clearance. In this project t1/2 values are reported in units of hours.
The data for each of the 4 parameters of interest was initially extracted from the prescription inserts of the 85 chemotherapy drugs and organized as an MS-Excel table. Previously described criteria [6, 8–10] were later employed to assess numerical estimations for the specific parameters studied in this project when only descriptive texts for each were reported. Each 2-D drug structure was redrawn using the drawing tools in the KnowItAll® informatics package, and with its database building capacity we built a database so that in future, we can predict and report the missing parameters using available SAR relationship models [24].
In order to accomplish our goal of developing a freely-available dynamic database to host these FDA cancer drugs, we aim to design a database, load all existing compounds, and implement a web application that will allow users to view the content of the database.
III. RESULTS
A. Data Analysis
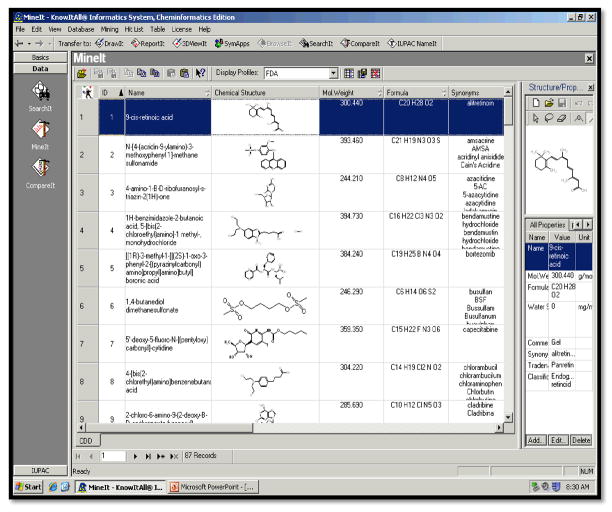
Figure 1 reveals the screen-shot of the created SAR relevant diverse chemical-structure searchable database for 85 FDA approved chemotherapy drugs using Bio-Rads KnowItAll® ADME/Tox Edition. Individual 2-D structures can be viewed in 3-D utilizing a 3-D ViewIt™ application present in the KnowItAll® Cheminformatics edition. All drugs are documented with their reported brand name, generic name, International Union of Pure and Applied Chemistry (IUPAC) name, molecular formula composition, chemical structure, and molecular weight. In Table 1 we report the numerical figures including appraisals for VD, t1/2, bioavailability, and water solubility. Reported values were obtained from the drug package inserts of the 65 cancer drugs that contain the common chemical structural motif, C=O (a carbonyl group). We did this because the physiochemical and biological activities of chemotherapy and other prescription drugs are partially due to the molecular structures of the compounds (SAR studies).
Figure 1.
A screen-shot of the database containing 85 chemotherapy drugs that was built using the KnowItAll® Informatics System.
Zoladex® was the only drug whose package insert chronicled the VD and t1/2 values based on gender differences. Velcade®, Zinecord®, and Rheumatrex® reported t1/2 values that were contingent on dose-dependent pharmacokinetics. Doxil®, Gemzar®, and Camptosar® reported VD values that included specific drug dose adjustment guidelines.
As shown in Table 2, only six drugs reported their VD data in the necessary units of L/Kg. For 22 drugs we estimated the VD value from the expressed units of L/m2 or L. 27 of the 65 carbonyl containing drugs had explanatory phrases for their water solubility information. This verbiage was standardized using techniques described earlier [6, 8–10]. The 31 given bioavailability values shown in Table 2 include the intravenously administered drugs, where it was assumed that the bioavailability value would be 100%.
Table 2.
Results obtained after analyzing the drug package inserts of the 65 chemotherapy drugs for the four parameters of interest.
| Parameter | Given | Not Given | Estimation | Given Unit |
|---|---|---|---|---|
| Volume of Distribution | 6 | L/Kg | ||
| 7 | L/m2 | |||
| 15 | L | |||
| 37 | ||||
| Elimination Half-Life | 54 | h | ||
| 11 | ||||
| Water Solubility | 14 | mg/mL | ||
| 27 | N/A | |||
| 24 | ||||
| Bioavailability | 31 | % | ||
| 34 |
B. Database Design
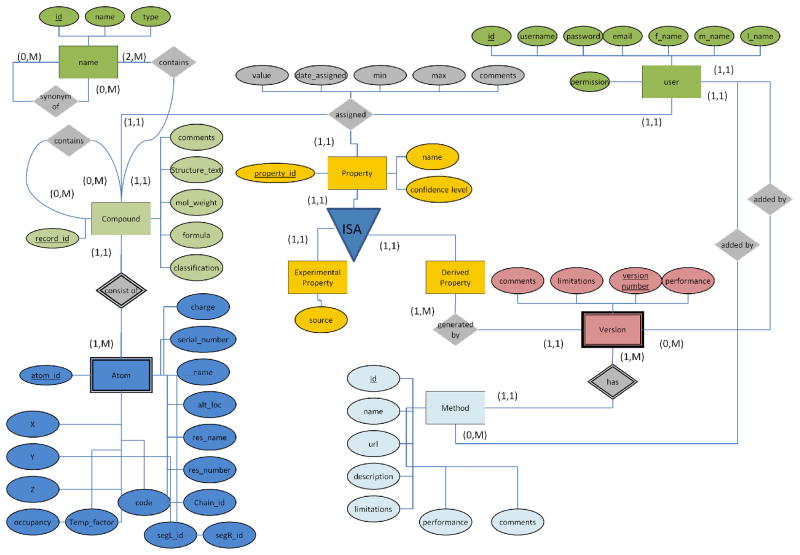
We have designed a schema of the normalized relational database (Figure 2). This design allows storing the various properties of the compounds. The database not only provides a framework for the existing 85 compounds but involves a community annotation services thus enriching existing information. The enrichment is possible through adding new compounds to the database as well as adding new properties to all cancer drugs stored in the database. While the properties or compounds are added into the database, the evidence attribution is kept allowing to track the information quality and facilitate analysis or prediction of pharmacokinetic properties.
Figure 2.
Database E/R diagram
The information for the existing compounds was loaded into the database. Even though, the physicochemical parameters of the initial compounds did not have evidence attribution, the database allowed the properties to be stored. After the initial batch loading of the data, the users will be required to provide evidence attribution in order to add new compounds or properties.
IV. DISCUSSION AND CONCLUSIONS
From Table 2, we demonstrate that 57% of the VD values, 17% of the t1/2 data, 37% of the water solubility information and 52% of the bioavailability values are not reported. These results clearly show that the FDA approved chemotherapy drug prescribing information inserts lack important pharmacokinetic information. In 2006 the FDA established a new set of rules for pharmaceuticals companies to follow when putting together a package insert [2]. This change in the insert requirements was influenced by the approximate 300,000 mishaps occurring each year relating to misleading information collected from the package inserts by health care providers [25].
While these new FDA rules may have improved the ability of people to understand the prescribing information inserts, the requirements did not include the necessary implementation of the important pharmacokinetic properties discussed in this article. According to the FDA website [1] in 2009 alone, 76 existing drug and therapeutic biological properties were issued new safety alerts. This suggests that although prescribing information packets created since 2006 may be easier to understand and provide more detailed safety information [2], they still lack important information needed for physicians to safely provide patients with care.
In a broad sense, most chemotherapy patients are weak and more vulnerable to severe side-effects due to multiple organ problems. Such derangement in organ function common in elderly cancer patients [26] and often coupled with decreases in skeletal muscle mass, can alter the pharmacokinetic processes of absorption, distribution, metabolism, and excretion of drugs.
For three capsules, Hydrea®, Matulane®, and Zolinza®, listed in Table 1, data for VD, t1/2, and bioavailability are unreported in their package inserts. Hence, duration of action and peak plasma concentration of a drug would be difficult to estimate. We also cannot assess the ability of these drugs to reach their target organs in an effective concentration. This makes toxicity less predictable.
In June 2010, pharmaceutical giant Pfizer voluntarily withdrew the acute myeloid leukemia (AML) drug Mylotarg® off the market over concerns of safety and the drug’s failure to extend survival time in patients [27]. A decade ago Mylotarg® was the first drug that was approved under the “accelerated approval” rules of the FDA. Such speedy FDA drug approvals under “special protocol assessments” are appraised on preliminary drug data and a promise by the company to follow up with more extensive clinical trials.
To overcome such inadequate clinical data, one option would be to utilize in-silico tools to predict the missing pharmacokinetic parameters thus giving us a better ability to estimate how these cancer drugs will act in the human body. Such tools available in commercial platforms such as KnowItAll® are able to predict the ADME/tox properties of drugs based on the compounds chemical structure [6–10]. We will report the results of such a task elsewhere.
A second option is to develop a freely-accessible online database where clinicians, researchers, and healthcare providers can add real-world data for future healthcare research and analytics. This will address the reporting challenges seen with the prescription inserts and will provide better therapeutic dosing strategies resulting in a higher quality care to beneficiaries. The implementation of the web application is currently under development. We are in the process to allow the browsing of the database, the loading of the new compounds, and the modification of the evidence attributions available to each user. The initial browsing of the database is currently available online -http://annotation.dbi.udel.edu/CancerDB/.
Acknowledgments
This research was supported by grant number 2 P2O RR016472-10 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). This IDeA Network of Biomedical Research Excellence (INBRE) grant to the state of Delaware was obtained under the leadership of the University of Delaware, and the authors sincerely appreciate their efforts. Additionally, J.M.W. acknowledges the receipt of an Undergraduate Tuition Scholarship from the NASA Grant NNG05GO92H Delaware Space Grant College and Fellowship Program.
Biographies
Ghada J. Alabed and Jordan M. Wheatley were Wesley College Biology majors who completed this undergraduate project as part of their Senior Research Capstone Experience. This project was mentored by Dr. Malcolm J. D’Souza, Professor of Chemistry, Wesley College. Both undergraduates received INBRE supported Undergraduate Research Assistantships in the Directed Research Program in Chemistry at Wesley. Additionally, Jordan received an Undergraduate Tuition Scholarship through the NASA funded Delaware Space Grant Consortium program at the University of Delaware. In April 2010, this project was one of just 60 from across the USA that was chosen to be showcased as a poster at the 2010 Council of Undergraduate Research (CUR) Posters on the Hill event.
Christopher Hart Continisio is an undergraduate and Yogasudha Veturi and Xia Bi are graduate students at the University of Delaware, who completed the development of the freely available online database as part of a project for a class taught by Dr. Natalia Roberts, Research Assistant Professor, University of Delaware.
Footnotes
Dedicated to Professor Dennis N Kevill on the occasion of his 76th birthday and in recognition of his many contributions to correlation analysis in chemistry and related areas.
Contributor Information
Malcolm J. D’Souza, Email: dsouzama@wesley.edu.
Natalia Roberts, Email: petrova@udel.edu.
REFERENCES AND NOTES
- 1.http://www.fda.gov/drugs/resourcesforyou/consumers/ucm143534.htm retrieved September 2008 – November 2010.
- 2.http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/LawsActsandRules/ucm188665.htm retrieved September 2008 – November 2010.
- 3.Shrank WH, Avron J. Educating Patients About Their Medications: The Potential and Limitations of Written Drug Information. Health Affairs. 2007;26(3):731–740. doi: 10.1377/hlthaff.26.3.731. [DOI] [PubMed] [Google Scholar]
- 4.Tu T. Review: Pharmacogenomic Frontiers in Medicine and Race. The Journal of Young Investigators. 2005;13(5) retrieved November 2010 Available: http://www.jyi.org/research/re.php?id=595. [Google Scholar]
- 5.Duconge J. Applying Organ Clearing Concepts in a Clinical Setting. American Journal of Pharmaceutical Education. 2008;72(5):Article 121. doi: 10.5688/aj7205121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Souza MJ, Ghada AJ. Deficiencies in the Reporting of VD and t1/2 in the FDA approved chemotherapy drug inserts. Pharmaceutical Reviews. 2010;8(1) [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza MJ, Gerges FJ. Raising a Red Flag: Deficiencies found in the Reporting of Important Parameters in FDA Approved Drug Profiles. Pharma IQ. 2010 Jul; Available: http://www.pharma-iq.com/article.cfm?externalID=2850.
- 8.D’Souza MJ, Koyoshi F. Extracting Relevant Information From DFA Drug Files to Create a Structurally Diverse Drug Database Using KnowItAll®. Pharmaceutical Reviews. 2008;7(3) [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza MJ, Koyoshi F, Everett LM. Bioinformatics, Computational Biology, Genomics, and Chemoinformatics. 2009. Structure Activity Relationship (SAR) Patterns Observed Within a Series of Unrelated Common Consumer Drugs; pp. 1–6. [Google Scholar]
- 10.D’Souza MJ, Koyoshi F, Everett LM. Structure Activity Relationships (SARs) Using a Structurally Diverse Drug Database: Validating Success of Predictor Tools. Pharmaceutical Reviews. 2009;7(5) [PMC free article] [PubMed] [Google Scholar]
- 11.KnowItAll®. Informatics System Desktop Solutions. Retrieved Available: http://www.knowitall.com/
- 12.KnowItAll®. U System. Retrieved Available: http://www.knowitallu.com/
- 13.D’Souza MJ. KnowItAll® - Software Reviews. Chemistry World. 2005;2(9):70–71. [Google Scholar]
- 14.D’Souza MJ. KnowItAll® U System - Software Reviews. Chemistry World. 2007;4(11):70–72. [Google Scholar]
- 15.Dearden J, Worth A. In Silico Prediction of Physicochemical Properties. JRC Scientific and Technical Reports, EUR 23051, EN-2007. :1–68. [Google Scholar]
- 16.Ekins S, Williams AJ. Precompetitive Preclinical ADME/Tox Sata: Set it Free on the Web to Facilitate Computational Model Building and Assist Drug Development. Lab Chip. 2010;10:13–22. doi: 10.1039/b917760b. [DOI] [PubMed] [Google Scholar]
- 17.http://www.chemspider.com/ retrieved November 2010
- 18.http://pubchem.ncbi.nlm.nih.gov/ retrieved November 2010
- 19.http://www.chemocare.com/bio/ retrieved September 2008 – November 2010.
- 20.http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm retrieved September 2008 – November 2010.
- 21.http://www.pdr.net/ retrieved September 2009 – November 2010.
- 22.Bauer LA. Applied Clinical Pharmacokinetics. 2. McGraw Hill; 2008. pp. 504–509. [Google Scholar]
- 23.Perrier D, Ashley JJ, Levy G. Effect of Product Inhibition on Kinetics of Drug Elimination. Journal of Pharmacokinetic and Pharmacodynamics. 1973;1(3):231–242. [Google Scholar]
- 24.Benfenati E. Predicting Toxicity Through Computers: A Changing World. Chemistry Central Journal. 2007;1:32. doi: 10.1186/1752-153X-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.FDA Prescribes New Package Insert Rules to Prevent Mishaps. Drug Store News. 2006 Feb 13; Available: http://findarticles.com/p/articles/mi_m3374/is_2_28/ai_n26692083/
- 26.Balducci L, Beghé C. Pharmacology of Chemotherapy in the Older Cancer Patient. Cancer Control Journal. 1999;6(5):Article 4. [PubMed] [Google Scholar]
- 27.http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm216448.htm retrieved November 2010