Abstract
Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics has been a subject of dramatic developments over the past decade. This review focuses on state of the art in LC-MS-based lipidomics, covering all the steps of global lipidomic profiling.
On the basis of review of 185 original papers and application notes, we can conclude that typical LC-MS-based lipidomics methods involve:
(1) extraction using chloroform/MeOH or MTBE/MeOH protocols, both with addition of internal standards covering each lipid class;
(2) separation of lipids using short microbore columns with sub-2-μm or 2.6–2.8-μm (fused-core) particle size with C18 or C8 sorbent with analysis time <30 min;
(3) electrospray ionization in positive- and negative-ion modes with full spectra acquisition using high-resolution MS with capability to MS/MS.
Phospholipids (phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols, phosphatidylserines, phosphatidylglycerols) followed by sphingomyelins, di- and tri-acylglycerols, and ceramides were the most frequently targeted lipid species.
Keywords: Acylglycerol, Biological system, Comprehensive analysis, Extraction method, Global lipidomic profiling, LC-MS, Lipidomics, Liquid chromatography-mass spectrometry, Metabolomics, Phospholipid
1. Introduction
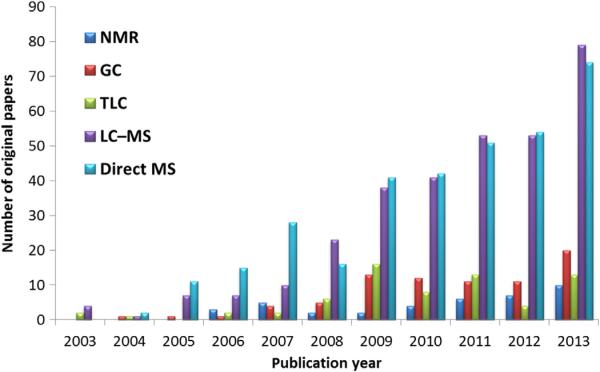
Since its introduction in 2003 [1], lipidomics has emerged as one of the most promising research fields as a result of advances in mass spectrometry (MS). Direct infusion (shotgun) techniques were prevalent in the beginning of lipidomics research due to their relative simplicity of operation, fast analysis, and possibility to detect various lipid classes within a single run. In most cases, these methods used tandem MS in a class-specific or targeted way, so detection and subsequent identification of unknowns were impossible. This was followed by rapid progress in liquid-chromatography (LC) separation and computational methods [2–4]. The popularity of LC-MS-based methods can be explained by several advantages over direct infusion techniques, such as more reliable identification of individual lipid species, even at trace level, separation of isomers and isobars, or reduced ion-suppression effects. In addition, current LC instruments permit more effective separation, reduce analysis time and solvent consumption [5,6]. Currently, direct infusion-MS(/MS) and LC-MS(/MS) methods are reported in the scientific literature in almost equal ratio, while complementary techniques and their combinations, such as gas chromatography (GC), thin-layer chromatography (TLC) and nuclear magnetic resonance (NMR), are less frequently used in lipidomics (Fig. 1).
Fig. 1.
Number of original papers published over 11 years dedicated to lipidomics and different instrumental platforms. Scopus (www.scopus.com) and Web of Knowledge (www.webofknowledge.com) databases used for citation analysis.
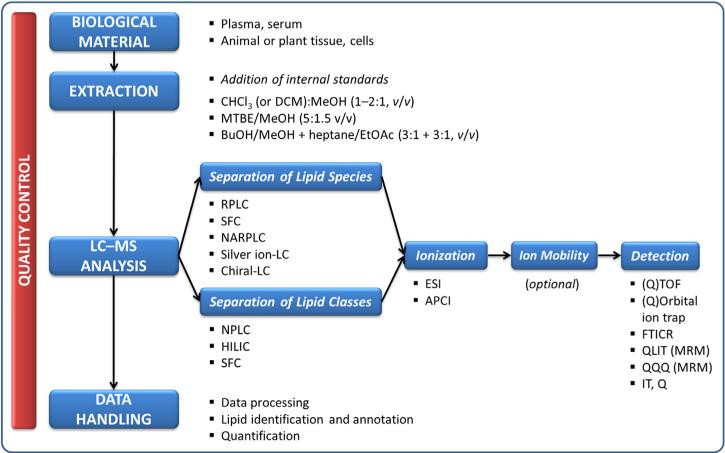
LC-MS-based lipidomic analyses (Fig. 2) typically start with extraction of the lipids from the biological sample followed by LC separation, which can be performed based on lipid species [e.g., reversed-phase LC (RPLC)] or classes [e.g., normal-phase LC (NPLC)]. Once chromatographically separated, the molecules enter the ion source where they undergo ionization followed by detection of particular ions using a mass analyzer. This can be conducted in an untargeted (full spectra acquisition), class-specific (product-ion scanning, precursor-ion scanning or neutral-loss scanning) or targeted (multiple-reaction monitoring) way [7]. The data handling represents a post-acquisition phase, which is focused on identification and (semi)-quantification of detected lipids, followed by statistical analysis if the primary focus of the study is to distinguish groups of samples.
Fig. 2.
LC-MS-based lipidomic analysis.
For this article, we reviewed 185 original LC-MS-based lipidomics papers and application notes published over the past decade (see references in Supplementary material, Tables S1 and S2). All the aspects, such as sample extraction, LC separation and MS detection are discussed in subsequent sections of this review. Since our primary focus was on the analysis of complex lipid mixtures in various biological systems, we omitted those papers dedicated to only a single lipid class (e.g., triacylglycerols or fatty acids).
2. Sample extraction
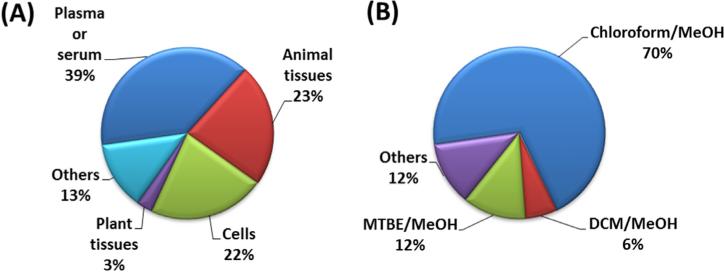
In general, lipidomics applications require sample-preparation methods that are fast, reproducible, and able to extract a wide range of analytes with different polarities, and that, at the same time, are compatible with the instrumental technique. Analytical strategies, which allow for increased coverage of metabolites determined in one sample, are therefore desirable [8,9]. In addition, samples may be available in only limited amounts, posing practical requirements to develop efficient, sample-saving experimental procedures. The reviewed studies were focused mainly on the analysis of lipids in plasma or serum, followed by animal tissues, cells, and plant tissues (Fig. 3A). Generally, 1–100 mg of tissue or 10–100 μL of biofluids (plasma/serum) per analysis were required in the reports reviewed here. Other matrices, which were less frequently studied, applied lipidomics to apicoplasts, microsomes, mitochondria, lipo-protein particles, milk, oxidized oils, Drosophila, microalgae, mesenteric lymph, cerebrospinal fluid, placental microvesicle, synovial fluid, tear samples, sebum, hetapocyte lipid droplets, urine, and solid fecal material (see references in Supplementary material, Table S1). Several sample-preparation methods were applied to biological samples with the goal of improving overall lipid coverage, including liquid-liquid extraction (LLE), organic solvent precipitation, and solid-phase extraction (SPE) [10].
Fig. 3.
Focus of LC-MS-based lipidomics studies: (A) analyzed matrices; and, (B) extraction protocols.
Introduced more than 70 years ago by Folch et al. [11] and Bligh– Dyer [12] (Fig. 3B), most lipidomics studies still rely on these general extraction procedures, often in modified versions. The Folch method employs roughly a 20-fold excess of a mixture of chloroform/ MeOH (2:1, v/v) for the extraction, while the Bligh–Dyer method is also based on a mixture of chloroform/MeOH (1:2, v/v), but uses a subsequent addition of 1 volume of chloroform and 1 volume of water. As a less toxic alternative, chloroform was replaced with dichloromethane (DCM) in some studies.
In 2008, Matyash et al. [13] introduced a novel sample-extraction procedure employing methyl tert-butyl ether (MTBE). The method involves addition of MeOH and MTBE (1.5:5, v/v) to the sample and phase separation is induced by adding water. The advantage of MTBE extraction over conventional two-phase chloroform-containing solvent systems came from the low density of the lipid-containing organic phase that forms the upper layer during phase separation. This greatly simplified its collection and minimized dripping losses. Furthermore, compared with chloroform, MTBE is non-toxic and non-carcinogenic, which reduces the environmental burden as well as the health risks for exposed personnel. Since its introduction, this method has a tendency to replace chloroform-containing solvent systems. Another chloroform-free extraction method has been developed by Lofgren et al. [14]. The protocol includes an initial one-phase extraction of sample (plasma) into a BuOH/MeOH mixture (3:1, v/v) followed by two-phase extraction into heptane/EtOAc (3:1, v/v) using 1% acetic acid as buffer.
As a simple procedure, organic solvent precipitation with MeOH or ACN (or mixtures with water) can be used, but, in this case, the extraction efficiency for certain lipids is lower than that of chloroform/MeOH or MTBE/MeOH extraction protocols. Last, but not least, SPE methods have also been used for lipidomic profiling, enabling fractionation of lipids. Use of SPE can be recommended if specific lipid fractions interfere with LC-MS measurements, or where in-depth characterizations of lipid classes are required in lieu of more comprehensive lipidomic profiling.
2.1. Combined extraction of amphiphilic and lipophilic metabolites
Comprehensive lipidomics targets amphiphilic lipids from palmitoyl lysophosphatidylcholine with a predicted octanol/ water partitioning coefficient (XlogP) of 5.6 over free fatty acids such as palmitic acid (XlogP 6.4) to very lipophilic compounds such as palmitoyl cholesterol (XlogP 16.4) or tripalmitoylglycerol (XlogP 21.9), covering more than 15 orders of magnitude on the octanol/ water coefficient scale. Lipidomic extractions using a single solvent mixture can therefore be expected to be less efficient than two separate extraction steps or use of separate extraction solvent mixtures; however, more elaborate extraction sequences may demand larger amounts of biological samples or require more time, impeding large-scale lipidomics analyses of clinical cohorts of biofluids.
Streamlining the sample-preparation step with subsequent analyses under different LC separation conditions was described recently. Giavalisco et al. [15] analyzed plant extracts obtained using a MeOH/ MTBE/H2O extraction protocol. Lipophilic metabolites included in the MTBE/MeOH fraction were separated on a C8 column, while amphiphilic metabolites in the MeOH/H2O fraction were separated on a C18 column.
The analysis of both phases obtained during MeOH/MTBE/H2O extraction was also reported for plasma and tissue. Specifically, Yang et al. [16] used C18 and hydrophilic interaction LC (HILIC) columns for separation of lipid and polar fractions, respectively, while Whiley et al. [9] and Godzien et al. [8] analyzed those two fractions on C8 and C18 columns. In these last two studies [8,9] the authors streamlined the procedure even further, performing the plasma extraction within an HPLC vial with a fixed 0.3-mL glass insert and using the same vial for analysis. After centrifugation and phase separation, the upper MTBE/MeOH phase and the lower MeOH/H2O phase were injected onto the LC-MS system directly from the vial/insert by adjusting the instrumental needle height in two separate runs.
An interesting application was developed by Chen et al. [17], who used the MeOH/MTBE/H2O extraction protocol. After phase separation, the upper, non-polar fraction was evaporated, resuspended in a chloroform/MeOH mixture (2:1, v/v), diluted in ACN/IPA/H2O (65:30:5), and used for lipidomics analysis (C8 column). Besides this procedure, the equal relative volumes of the upper and lower phases were mixed, evaporated, resuspended in 20% MeOH, and subsequently separated on a C18 column. This mixture was comparable or superior in yield and reproducibility to a standard 80% MeOH extraction for profiling polar and lipophilic metabolites and led to an increase in lipidomics coverage by 30% relative to using the polar fraction alone.
While the previous studies relied on LC-MS analysis of two different extracts, studies focusing on the use of a single extract separated on different LC systems have also been reported. Garcia-Canaveras et al. [18] used an MeOH/chloroform/H2O extraction protocol for human liver followed by separation of lipids on a C8 column and polar metabolites on HILIC and C18 columns. A similar protocol was developed by Ivanisevic et al. [19], who used an MeOH/ACN/H2O extraction scheme for isolating cell metabolites with subsequent analysis on C18 and HILIC columns.
These studies have clearly demonstrated that lipidomics coverage can be significantly increased by analyzing both phases of bi-phase extractions or utilizing LC columns with different separation mechanisms from a single-phase extract. Overall, when using bi-phase extraction of blood, animal or plant tissues, the lipophilic phase (MTBE/MeOH) preferably comprised lipids of different classes such as phospholipids, acylglycerols, free cholesterol and cholesteryl esters, while the amphiphilic phase (MeOH/H2O) provided complementary information on hydrophilic lipids, such as acylcarnitines but also on hydrophilic metabolites, such as amino acids, monosaccha-rides, or flavonoids.
3. Liquid-chromatography separation
Coupling LC to MS substantially reduces some of the limitations linked to direct infusion MS, such as detection of isobars and isomers or ion-suppression effects caused by molecules competing for ionization [6,20]. Moreover, use of LC gives the possibility to separate or to concentrate different classes of compounds according to their physicochemical properties.
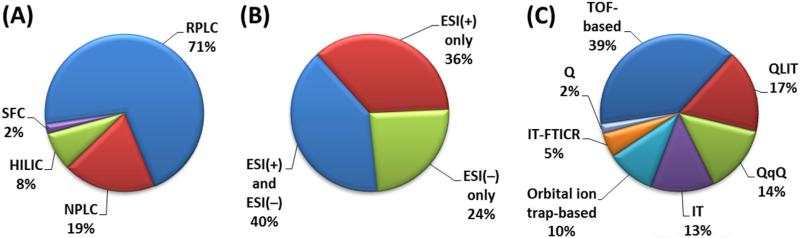
Several LC configurations were described for the analysis of complex lipid mixtures (Fig. 2). The three most important ones include RPLC, NPLC and HILIC (Fig. 4A). The mechanism of action in RPLC of lipids is based on lipophilicity, which is governed by the carbon-chain length and the number of double bonds. Thus, lipid species containing longer acyl chains are eluted from the LC column later than shorter chain lipids, and saturated acyl structures are eluted later than polyunsaturated analogs. However, NPLC and HILIC typically distinguish lipid species according to their hydrophilic functionalities, so lipids are separated according to their representative polar head-group classes [6,21]. Other LC techniques used for lipid separation include non-aqueous RPLC, silver-ion RPLC, chiral LC, and supercritical fluid chromatography (SFC). In addition, off-line and on-line two-dimensional LC systems can be considered for separation of complex lipids.
Fig. 4.
Use of (A) chromatographic systems, (B) ionization modes, and (C) mass analyzers in LC-MS-based lipidomics papers.
3.1. Reversed-phase LC
RPLC has been most widely used for the analysis of complex lipids according to our literature search (Fig. 4A) (see also references in Supplementary material, Table S2). Previous studies (before 2004) used relatively long (100–250 mm), narrow and normal-bore (2– 4.6 mm I.D.) columns with 3.5–5 μm particle size. Using this setup, HPLC systems operating typically at pressure <400 bar were sufficient. Starting in 2004, improvements in chromatographic performance were achieved by the introduction of ultrahigh-performance LC (UHPLC). Today's UHPLC systems are capable of operations at high back pressures of up to 1200 bar, so columns with sub-2 μm particles can be operated at high mobile-phase flow-rates without loss of resolution under optimized conditions [17,22–24]. Recently, “fused-core” technology was introduced, and produces 2.6–2.8-μm particles with a 0.35–0.5-μm porous shell fused to a solid core. The porous shell decreases the diffusional mass transfer path, compared to totally porous particles, so chromatographic resolution might be achieved similar to that with sub-2 μm totally porous particles at comparable or even higher speeds without high backpressures [25].
Typical methods in RPLC-based lipidomics use short (50– 150 mm; typically 100 mm) microbore columns (1–2.1 mm I.D.) with sub-2-μm or 2.6–2.8-μm (fused-core) particle size and C18 or C8-modified sorbents. Examples are the Acquity UPLC BEH C18, Zorbax Eclipse XDB-C18, Acquity UPLC HSS T3, Acquity UPLC BEH C8, Kinetex C18, or Ascentis Express C8 columns. These columns are operated at a flow-rate of 0.1–0.5 mL/min and maintained at temperatures of 40–55°C.
A few applications focused on miniaturization of lipidomics analyses, employing capillary (50–650 × 0.15–0.32 mm I.D.; 1.7–5 μm) and nanobore (50–170 × 0.075 mm I.D.; 3–5 μm) columns. Compared with conventional LC columns, LC separations with capillary or nanobore columns offer higher sensitivity and smaller sample requirements, and mobile-phase consumption is significantly lower since flow-rates of 0.3–10 μL/min are used for lipid elution.
As a weak mobile phase, water or mixtures with organic solvent are used, such as H2O/MeOH (50:50), H2O/ACN (40:60–60:40), H2O/ MeOH/ACN, H2O/IPA (95:5) or H2O/MeOH/tetrahydrofuran (50:30:20), while a strong mobile phase consists primarily of isopropanol (IPA) or tetrahydrofuran mixed with other solvents (IPA/ ACN (95:5–70:30), IPA/MeOH/H2O (using different ratios but high % of IPA), tetrahydrofuran/MeOH/H2O (75:20:5)). Use of mobile-phase additives is highly recommended to improve both LC separation and detection of lipids. Different combinations of ammonium formate or acetate (5–10 mM) and formic or acetic acid (0.05–0.1%) are used. When using MeOH or ACN as the only strong elution solvents, typically shorter LC columns and/or longer separation times are required to elute the triacylglycerols and cholesteryl esters retained most.
Special attention has to be paid to the extract resuspension prior to LC analysis. Samples need to be injected into the LC system in a solvent mixture that is
compatible with the initial composition of mobile phases to avoid peak distortion of early eluted lipids;
sufficient for dissolving the lipids in dry extracts; and, (3) able to prevent possible degradation of lipids after resuspension.
Acceptable solvents or mixtures involve MeOH, chloroform/ MeOH (1:1; 2:1; 1:3; 1:4 ratio), chloroform/MeOH/H2O (10:10:3), 80% MeOH, ACN/IPA/H2O (65:30:5), ACN/H2O (1:1; 7:3), ACN/ MeOH (75:25), BuOH/MeOH (1:1), chloroform, chloroform/IPA (1:1), chloroform/MeOH/H2O (10:10:3), MeOH/IPA/H2O (65:30:5), IPA/ hexane/H2O (10:5:2), IPA:ACN (1:1; 1:9). In some studies, lipid extracts were diluted with only IPA, ACN/IPA/H2O (65:30:5) or hexane/IPA (3:2), or injection of chloroform phase was even direct.
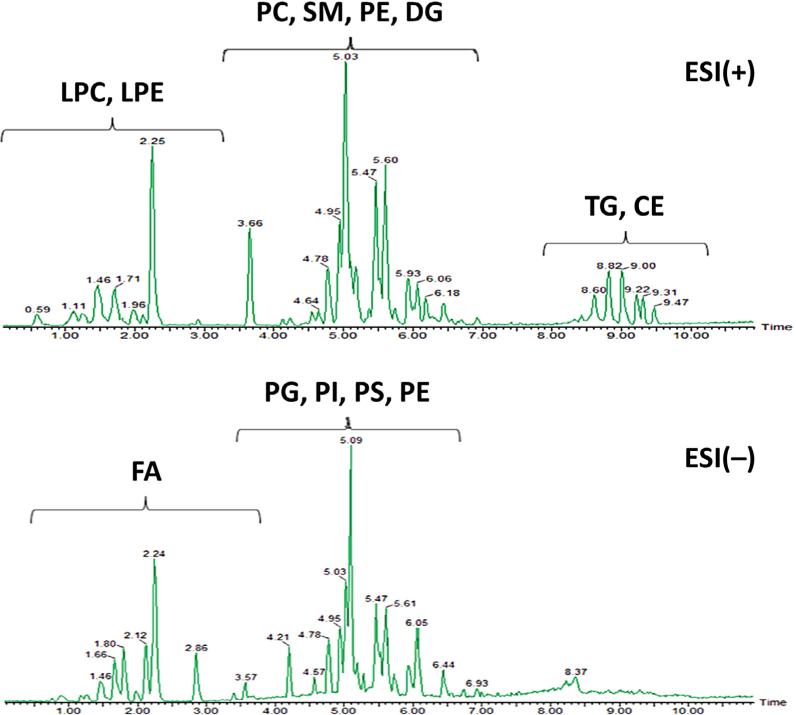
For large-scale lipidomics experiments, such as in clinical cohorts, the total cycle time for the analytical runs is important. Short analytical runs decrease peak capacity, and less retention time-mass spectral features are detected compared to longer LC runs. Hence, longer LC runs promise more comprehensive lipidomic profiles. However, for large-scale human cohort studies or if laboratories face a large number of smaller studies, a compromise has to be reached, balancing the required sample throughput and maximizing lipidome coverage. In 65% of the lipidomics studies reviewed here, reported analysis run times were less than 30 min, including column reequilibration time. In 40% of all reviewed lipidomics applications, further time reductions led to overall cycle times of less than 20 min. As an example, Fig. 5 shows the separation of human-plasma lipids using RP-UHPLC-ESI(+/–)-QTOF-MS with an injection-to-injection cycle of 15 min. Such turnaround times were about three times faster than regular HPLC methods under the same gradient conditions [26].
Fig. 5.
RP-UHPLC-ESI(+/–)-QTOFMS chromatograms of human plasma extract separated on an Acquity UPLC HSS T3 (100 × 2.1 mm I.D.; 1.8 μm) column with a gradient elution using mobile phase ACN/H2O (40:60), 10 mM ammonium acetate and IPA/ACN (90:10), 10 mM ammonium acetate at a flow-rate of 0.4 mL/min at 55°C. {Adapted and reproduced with permission from [26]}.
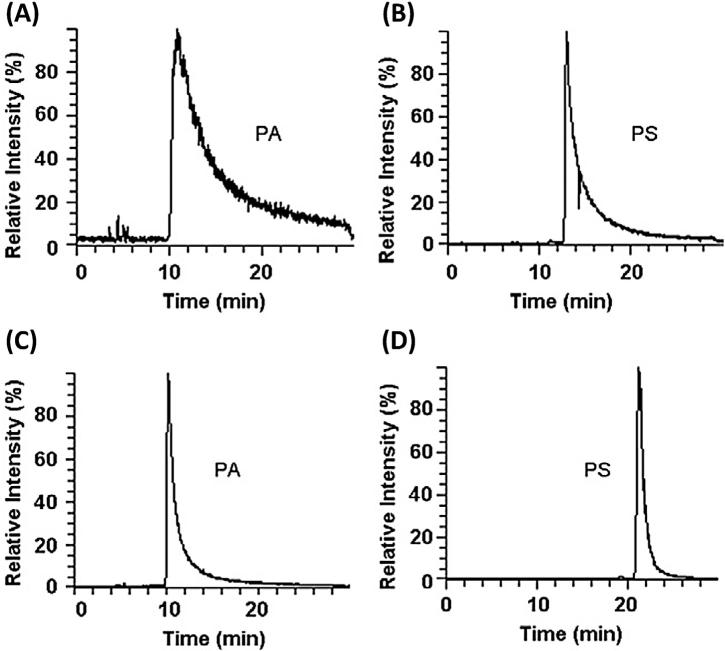
Among all phospholipid classes, phosphatidic acid (PA) and phosphatidylserine (PS) tend to elute as extensively broad peaks under the RPLC conditions generally used (Fig. 6A and B), so class-specific analyses of PA and PS have often been performed with NPLC or by omitting any separation column (flow-injection analysis). Ogiso et al. [27] developed RPLC conditions that reduced peak tailing of PA and PS by adding a small quantity of phosphoric acid into the mobile phase (5 μM) and the injector-rinsing solvent (0.1%) and using a relatively high percentage of water in the starting mobile phase. While phosphoric acid suppressed the unidentified interaction between PA and the flow passage materials within the separation column, a higher percentage of water at the beginning of the LC run enabled PS to be eluted as a narrower peak on RPLC-C18 column used (Fig. 6C and D). Since phosphoric acid may harm the ESI source, concentrations higher than 5 μM were not tested.
Fig. 6.
RP-HPLC-ESI(–)-orbital ion trap MS extracted ion chromatograms of the standard PA(34:1), m/z 673.5 (A + C) and PS(36:1), m/z 788.5 (B + D) separated on a Waters X-Bridge C18 (150 × 1.0 mm I.D.; 3.5 μm) column. (A + B) Mobile phase A: ACN/MeOH/H2O (19:19:2) with 0.2% formic acid and 0.028% ammonia; mobile phase B: IPA with 0.2% formic acid and 0.028% ammonia at a flow-rate of 20 μL/min and temperature 50°C. (C) Phosphoric acid added into the mobile phase A and the injector-rinsing solvent. (D) Mobile phase A: IPA/MeOH/H2O (5:1:4) with 0.2% formic acid, 0.028% ammonia, and 5 μM phosphoric acid; mobile phase B: IPA with 0.2% formic acid and 0.028% ammonia at a flow-rate of 20 μL/min and temperature 25°C. {Reproduced with permission from [27]}.
3.2. Normal-phase LC
NPLC represents a separation mechanism complementary to RPLC. The current approach in NPLC lipidomics involves using long (100– 250 mm; typically 150 mm), microbore columns (2 mm I.D.) with 3–5-μm particle size, such as Luna 3 μm Silica, LiChrospher Si60, Betasil Silica-100, and Nucleosil 100-5 OH (see references in Supplementary material, Table S2). These columns are operated at a flow-rate of 0.1–0.5 mL/min and maintained at temperatures of 20–35°C. If a higher flow-rate during the LC separation is required (1 mL/min), the column effluent is split (1:1–1:5) and only part of the total effluent enters the ion source.
Highly non-polar solvents with low ionization capacity have to be used during NPLC. The separation starts with a weak mobile phase, such as heptane, chloroform, chloroform/MeOH (9:1), hexane/ propan-1-ol (79:20), IPA/MTBE (100:50–100:83), hexane/IPA (3– 7:7–3), or MTBE/MeOH/IPA (80:10:7), while a strong mobile phase comprises heptane/IPA/EtOH (2:1:1), chloroform/MeOH (55:39), ACN/ MeOH (4:6), MeOH, IPA/MeOH (70:30) or MeOH/IPA (90:7). Additives used in NPLC involve 0.5% NH4OH; 5–15 mM ammonium acetate, ammonium formate, diethylamine, formic acid, or small amounts of water (0.5–3%). For the extract resuspension, various mixtures of solvents such as 4.5% MTBE in hexane, chloroform/MeOH (1:1; 1:2; 2:1), chloroform/MeOH (95:5), H2O/ACN/MeOH (10:55:35), heptane/chloroform/MeOH (95:2.5:2.5), hexane/IPA (70:30) are utilized. Using such lipophilic resuspension mixtures certainly help obtaining a complete transfer of the extracted lipidome onto the analytical column, whereas more hydrophilic resuspension mixtures (which are necessary in RPLC-based lipidomics) may introduce a bias against very lipophilic components, such as cholesteryl esters and triacylglycerols.
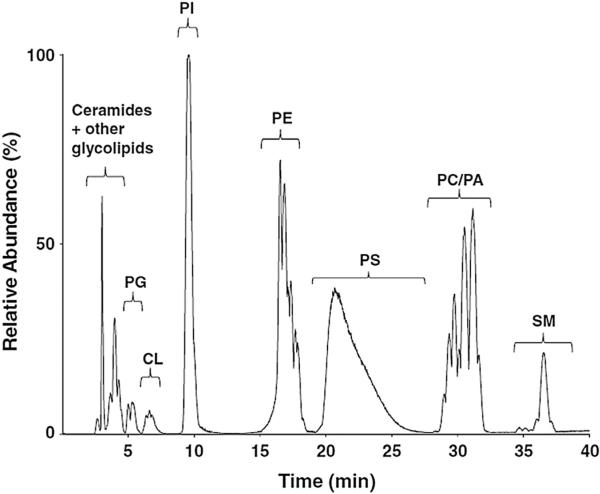
Compared to RPLC, the analysis time in NPLC is typically longer (30–60 min), which can be attributed to the separation on long columns with large particle-size sorbents and the focus on complete separation of lipid classes. NPLC is suitable for the separation of phospholipids, specifically if PAs are of a great interest, since, in RPLC, this lipid class gives poor peak shapes [27]. As an example, Fig. 7 shows a 40-min separation of rat-brain lipids analyzed by NPLC-ESI(–)-MS/MS [28].
Fig. 7.
NP-HPLC-ESI(–)-MS/MS (QqQ) chromatograms of rat-brain extract separated on a Phenomenex Luna 3 μm Silica (150 × 2 mm I.D.; 3 μm) column with a gradient elution using mobile phase chloroform/MeOH/triethylamine/acetic acid (80:19:0.5:0.5) and chloroform/MeOH/H2O/triethylamine/acetic acid (60:33.5:5.5:1:0.065) at a flow-rate of 0.2 mL/min at room temperature. {Reproduced with permission from [28]}.
3.3. Hydrophilic interaction chromatography (HILIC)
HILIC represents a separation alternative to NPLC. Unlike NPLC, which uses non-polar solvents and excludes water from the mobile phase, the percentage of water in the mobile phase during HILIC separation can vary (5–40%) and a minimum of 2% is essential to maintain a stagnant enriched water layer on the surface into which analytes may partition [29]. In addition, water-miscible organic solvents are used instead of the water-immiscible solvents used in NPLC. Compared to NPLC, HILIC provides better reproducibility and robustness, and is more compatible with MS [30].
Lipid separations under HILIC conditions are usually conducted on shorter (100–150 mm) micro-bore columns (2 mm I.D.) with 1.7–5-μm particle size, such as Atlantis HILIC Silica, Acquity UPLC BEH HILIC, Nucleosil 100-5 OH and Spherisorb Si (see references in Supplementary material, Table S2). These columns are operated at a flow-rate of 0.1–1 mL/min and maintained at temperatures of 25–40°C. The analysis time is typically in the range 15–60 min.
As a weak mobile phase, ACN is used or its mixtures (ACN/H2O (95–90:5–10), ACN/MeOH/H2O (55:35:10), ACN/IPA (80:20)) are used, while the strong mobile phase usually comprises water or mixtures of water with organic solvents [H2O/MeOH (1:1), H2O/ACN (95–50:5–50), H2O/IPA (80:20)]. Additives used in HILIC-based lipidomics involve 0.1–0.2% formic acid, 5–10 mM ammonium acetate, 20 mM ammonium formate, or 10 mM NH4OH. For dry-extract resuspension, ACN, ACN/H2O (1:1), ACN/H2O (7:3), ACN/ MeOH/H2O (55:35:10), chloroform, chloroform/IPA (1:1), chloroform/ MeOH (1:1; 4:1), MeOH, IPA/ACN (1:1), IPA/H2O (1:1) can be considered.
3.4. Supercritical fluid chromatography (SFC)
SFC using columns packed with sub-2-μm particles is an emerging technique in fast lipid profiling (<20 min analysis time). Supercritical fluids have higher diffusion coefficients and lower viscosities than regular liquids, thereby permitting higher throughput analysis than LC [31–33]. In addition, supercritical CO2 used as a mobile phase has almost the same polarity as hexane, and its polarity can be adjusted by adding a modifier (usually MeOH). These advantages make SFC suitable for the simultaneous analysis of diverse lipids with a wide range of polarities. It has been reported that the detection sensitivity in SFC was higher than that in LC, and several structural isomers were successfully separated using SFC but could not be separated by LC [32–35].
3.5. Two-dimensional liquid chromatography (2D-LC)
The fact that LC techniques can separate lipids according to two independent molecular properties [e.g., hydrophobic character of the molecule (RPLC) and electrostatic forces related to the compound polarity (HILIC)], offers great opportunities for 2D-LC to characterize further complex lipidomes [21]. 2D-LC of lipids can be performed in on-line or off-line modes.
The advantage of off-line 2D-LC is the full optimization of separation conditions in both dimensions. However, this approach is more laborious and more time consuming, involving the following steps: collection of each fraction, followed by solvent evaporation, resuspension of dry extracts in solvent(s) compatible with the second-dimension LC column, and, finally, using a second-dimension LC method. However, on-line 2D-LC can be automated, but the chromatographic resolution in the second dimension is compromised at the cost of sampling time (typically 1 min) for the first dimension [36]. In most cases, HILIC or NPLC is used in the first dimension to fractionate different lipid classes followed by analysis of collected fractions by RPLC in the second dimension. Since the analysis time can significantly increase when using off-line 2D-LC (1–2 h per single analysis), it is obvious that this instrumental technique is more suitable for comprehensive characterization of sample extracts rather than large-scale lipidomic screening studies.
4. Ionization and mass spectrometric detection
4.1. Ionization techniques
The choice of ionization mode used in LC-MS analyses plays a major role in the lipidome profile that will be obtained. One methodology cannot cover all types of molecules, since some lipids are better ionized in one ionization mode while other lipids are ionized more efficiently in another mode (Table 1) [23,37–40]. Ionization efficiency can be enhanced by the additives dissolved in the mobile phases leading to formation of different type of adducts. Electrospray ionization (ESI) in positive mode is the most common mode in LCMS because it can effectively ionize a wide range of lipids (Fig. 4B). However, negative ionization provides superior results for certain lipid classes, such as phosphatidylinositol, phosphatidylserine, and phosphatidic acid [41]. Atmospheric-pressure chemical ionization (APCI) is preferred for more non-polar lipids (e.g., triacylglycerols) and has been used in a smaller number of lipidomics investigations.
Table 1.
Molecular species formed during electrospray ionization of lipids
| Lipid class | Positive mode | Negative mode |
|---|---|---|
| LPC, PC | [M + H]+, [M + Na]+ | [M–H]–, [M + HCOO]–, [M + CH3COO]– |
| LPE, PE | [M + H]+, [M + Na]+ | [M–H]– |
| PG | [M + H]+, [M + NH4]+, [M + Na]+ | [M–H]– |
| PI | [M + H]+, [M + NH4]+ | [M–H]– |
| PS | [M + H]+ | [M–H]– |
| PA | [M–H]– | |
| CE | [M + NH4]+ | |
| SM | [M + H]+ | [M + HCOO]– |
| Cholesterol | [M–H2O+H]+ | |
| Cer, GluCer, LacCer | [M + H]+, [M + NH4]+, [M + Na]+ | [M–H]–, [M + HCOO]– |
| MG, DG, TG | [M + NH4]+, [M + Na]+ | |
| MGDG, DGDG, SQDG | [M + NH4]+, [M + Na]+ | [M–H]– |
| Fatty acids | [M–H]– | |
| CL | [M + H]+, [M + NH4]+, [M + Na]+ | [M–H]– |
| Cer | [M + H]+, [M + NH4]+, [M + Na]+ | [M–H]– |
In more detail, when using RPLC separation, 53% of studies acquired data in both ESI(+) and ESI(–) and 38% in ESI(+) only. However, in NPLC/HILIC, both ionization modes were used in 45% of applications, and 41% of studies acquired data in ESI(–) only. The explanation of these differences can be related to NPLC/HILIC methods being focused mainly on detection of phospholipids, where ESI(–) permitted effective ionization of these lipids, while RPLC analyses usually covered also lipids that effectively ionize in ESI(+), such as acylglycerols and cholesteryl esters.
4.2. Ion mobility mass spectrometry
Ion mobility-mass spectrometry (IM-MS) is an emerging technology that was only recently applied in lipid analysis [42,43]. In principle, IM-MS enables the differentiation of ions by size, shape, charge, and mass based on their different mobilities in low or high electric fields. This provides important supplementary information to the LC separation of molecules and MS separation of ions. IM-MS can be applied to the separation of isobaric compounds, the reduction of high background noise, and the separation of endogenous matrix interferences from target analytes to increase the selectivity.
Jackson et al. [44] observed several trends when using IM-MS in lipid analysis, including mobility differences based solely on phospholipid acyl-chain degree of unsaturation, type of linkage, and the type of polar head group. Distinguishable, and sometimes baseline resolvable, mobility drift-time differences between classes of phospholipids were observed.
The effect of the number of double bonds on the mobility of lipid ions was investigated by Kim et al. [45] for phosphatidylcholines. A single double bond was found to account for a 5% shift in the mobility-time deviation from the average mobility-mass correlation line of phosphatidylcholines, while each additional double bond appears to shift the mobility time further by ~1%.
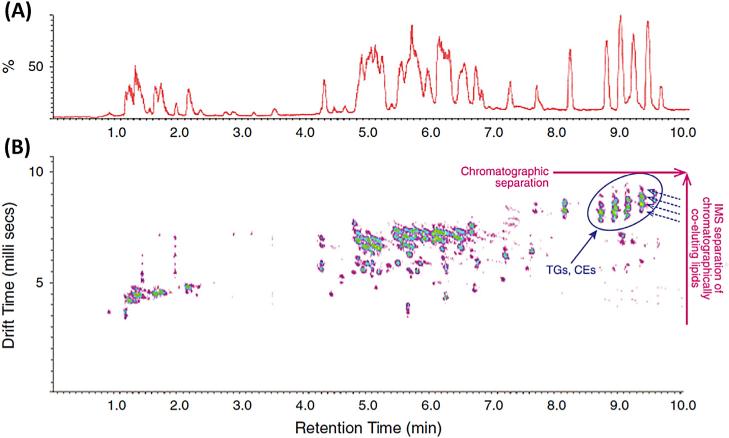
Shah et al. [46] took advantage of IM-MS in conjunction with UHPLC-ESI(+)-QTOFMS to gain quantitative and deeper qualitative structural insight from a single experiment (Fig. 8). The chromatographically co-eluting human-plasma lipids were further resolved by IM tube drift time and then subjected to low-energy and high-energy fragmentation without pre-selecting respective precursor species (MSE mode). The fragment ions produced in a high-energy mode were aligned with their precursor ions in a low-energy mode. By aligning intact molecular spectra and fragment spectra for these lipids at a given IM drift time and chromatographic retention time, much cleaner fragment-ion spectra were obtained for structural elucidation.
Fig. 8.
RP-UHPLC-ESI(+)-IM-QTOFMS analysis of a human plasma lipid extract separated on an Acquity UPLC HSS T3 (100 × 2.1 mm I.D.; 1.8 μm) column with a gradient elution using mobile phase ACN/H2O (60:40), 10 mM ammonium formate and IPA/ACN (90:10), 10 mM ammonium formate. (a) A chromatogram with ion mobility separation turned on. (b) Corresponding driftogram with orthogonal ion mobility separation. {Reproduced with permission from [46]}.
4.3. Mass spectrometric detection
Over the past decade, various mass spectrometers introduced were capable of selective, sensitive detection of lipids in a comprehensive way. Of all LC-MS lipidomics applications, 80% relied on a mass spectrometer capable of full mass spectra acquisition, usually in high-resolution (HR) mode, instead of target mass spectra acquisition using multiple reaction monitoring (MRM) or selected ion monitoring (SIM). The key limitation of MRM and SIM approaches is that monitoring only specific transitions or specific ions, respectively, excludes recording and post-acquisition data reprocessing of unidentified (untargeted) lipids.
The basic characteristics of the quality of a high-resolution mass analyzer are resolving power, mass accuracy, isotope-profile error, sensitivity, maximum spectral acquisition speed, linear dynamic range, and availability of tandem-MS function [43]. For MRM instruments, low dwell time is an important parameter because hundreds of transitions need to be collected within a short time. TOF-based mass spectrometers, such as time-of-flight (TOF) and quadrupole/time-of-flight (QTOF), dominate in lipidomics studies. Other instruments are less used, such as quadrupole/linear ion trap (QLIT), triple quadrupole (QqQ), quadrupole (3D) or linear (2D) ion trap (IT), orbital ion-trap-based MS (single orbital ion trap, ion trap/ orbital ion trap, or quadrupole/orbital ion trap), ion trap/Fourier transform ion cyclotron resonance (IT-FT-ICR), ion trap/time-offlight (IT-TOF) or even single quadrupole (Q) mass spectrometers (Fig. 4C).
Current HRMS instruments used in lipidomics are available with a broad range of resolving power (10,000–450,000 full width at half maximum, FWHM) with mass accuracy typically better than 1–5 ppm. At the same time, instruments have become more sensitive. This improvement in sensitivity permits us to obtain information-rich LC-MS data with minimal sample amounts to be injected. It is difficult to answer the frequently asked question: “How much mass resolving power is enough?”. In general, the higher the mass-resolving power the better, if it does not compromise sensitivity, linear dynamic range, and acquisition speed. The advantage of high mass-resolving power is that ions having the same nominal mass can be resolved partly or completely [47]. Although current MS systems are capable of sub-ppm mass accuracy, this is still insufficient to exclude enough candidates with complex elemental compositions. Kind and Fiehn demonstrated that use of isotopic abundance patterns as a single further constraint may remove >95% of false candidates in an untargeted search for the molecular formulas of unknowns in metabolomics [48].
For an MS/MS investigation, different collision energies are typically measured. The precursor ions can be selected manually based on previous MS1 data acquisition, using data-dependent acquisition (DDA) or data-independent acquisition (DIA) (also referred to as non-selective tandem MS). The last two approaches represent the most effective MS/MS data collection within a single LC-MS/MS analysis.
DDA consists of:
an initial survey scan (full MS scan), where precursor ion species exceeding a pre-defined threshold are isolated using the Q1 quadrupole;
their activation via collision-induced dissociation (CID); and,
detection of fragments (product ions).
To avoid repetitive acquisition of the same precursor ions, it is possible to create an exclusion list to exclude certain ions (e.g., background ions, and reference ions used for mass drift correction) or to exclude already acquired ions for a given time (dynamic exclusion).
The use of non-selective tandem MS methodology has been enabled on (Q)TOF (Waters, MSE; Agilent, All Ions MS/MS; and, Leco, MSc2) and orbital ion trap (Thermo Scientific; All Ion Fragmentation, AIF) instruments. This technique provides low-energy, intact, molecular spectra and corresponding high-energy fragment spectra simultaneously without preselecting any specific ions. However, when co-elution occurs, combining fragment ions with precursor molecules for structurally similar lipid species can be difficult, as they may share the same fragment ions [46]. This limitation can be overcome to some extent for partly overlapped chromatographic peaks by using software deconvolution algorithms that merge precursor ions from MS1 and corresponding fragment ions from MS/ MS analyses based on retention-time differences.
Recently, acquisition of all theoretical fragment-ion spectra (MS/ MSALL SWATH, AB SCIEX) using sequential windows for selection of precursor ions was presented [43]. The Q1 quadrupole is stepped in (typically) 25-amu increments across the mass range of interest, passing a 25-amu window through into the collision cell. The transmitted ions are fragmented and the resulting fragments are analyzed in TOF analyzer at high resolution. Setting narrower sequential mass windows for precursor-ion selections decreases the risk of spectral co-elutions during the analysis of complex lipid extracts in comparison to previous DIA approaches where all precursor ions over the given mass range (e.g., m/z 100–2000) are fragmented simultaneously.
Because certain lipid classes can be ionized in only one polarity mode, mass spectrometers capable of fast polarity switching are desirable because parallel measurements of both polarity modes are performed within a single run. The fast polarity switching in case of HRMS is demanding technically since the electronics need a certain time to stabilize high voltages, so only a few HRMS systems are capable of relatively fast polarity switching [43].
Gallart-Ayala et al. [24] explored the potential of a single-stage orbital ion-trap instrument (Thermo Scientific, Exactive) for lipidomic profiling. The instrument was operated in polarity-switching mode at a mass-resolving power of 50,000 FWHM. In addition, during the analysis, AIF experiments were carried out in two different regions: a high-energy collisional dissociation (HCD) cell and in-source collision-induced dissociation (CID). The best MS/MS conditions were obtained at 30 eV in the HCD cell and 50 eV in the in-source CID. Under these conditions the combination of the AIF MS mode with the polarity-switching mode allowed identification of several lipid classes (LPC, LPE, PC, PE, PI, SM, TG) within a single run.
Polarity switching in combination with data-dependent MS/ MS analysis on a quadrupole/orbital ion trap MS (Thermo Scientific, Q-Exactive) was also employed for lipidomic profiling by Yamada et al. [49]. Using LipidSearch software, 495 lipids belonging to different lipid classes (LPC, LPE, LPI, LPG, PC, PE, PS, PI, PG, PA, SM, TG) in mouse plasma were identified. However, any instrument has a limited dynamic range for quantification, beyond which lipids may still be identified but saturation of the detectors occurs (e.g., multi-channel plates), limiting the capability to perform valid quantifications. It may therefore be advisable to run positive and negative ESI lipidomic profiles on two instruments with different sensitivities, because, often, lipids that are amenable to negative ESI conditions are at far lower concentrations than lipids that perform best at positive ESI techniques.
5. Data processing
Raw data processing is the first important step following data acquisition. In LC-MS analysis, three dimensions represent retention time, m/z value, and signal intensity. The data-processing pipeline usually proceeds through multiple stages, including:
filtering;
feature detection;
alignment; and,
normalization.
Filtering methods process the raw signal with the aim of removing the noise or baseline. Feature detection is conducted to identify all signals caused by true ions and avoid detection of false positives. This step may also involve a deconvolution method and isotope-pattern detection. Alignment methods are needed for correcting retention-time differences between runs and combining data from different samples. An important part of data processing is the “gap filling” algorithm. If this is not the part of data-processing software, many zero values in the final extracted feature set are present, obscure later statistical and chemometric data analyses and may result in incorrect biological interpretations. Normalization steps remove factors causing unwanted systematic bias in ion intensities between measurements or during sample-preparation steps.
Final reports are generated by merging the three-dimensional data into a two-dimensional data matrix in the form of a peak-table containing m/z value, retention time, and corresponding intensity for every detected peak. Detailed description of these approaches can be found in specialized review papers [50,51].
The application of different peak-processing software can have a significant effect on the end result of a study. There are four possible approaches:
using vendor software (e.g., MarkerLynx, MarkerView, Mass Profiler Professional, MassHunter/Genespring, MetQuest, SIEVE);
using special software from independent developers that can handle most of MS data (e.g., GeneData);
using open access software (e.g., XCMS, MZmine, MetAlign, IDEOM); and,
developing their own script (e.g., Matlab, R) [52].
The high impact of the choice and the parameters in various data-processing platforms has raised questions concerning the ability to compare results from different laboratories. To study this problem, Gurdeniz et al. [53] compared three different software applications (MarkerLynx, MZmine, and XCMS) in finding plasma biomarkers in fed and fasted rats. Overall, the degree of false positive and false negative peak detections, and the capability and the accuracy of gap-filling approaches (for false negative peak finding) showed large differences between the data-processing software tools and parameters that were evaluated in this study.
Good laboratory guidelines, community reference samples, and thorough software comparisons are needed to move untargeted LCMS based lipidomics to the realm of accepted protocols in clinical and preclinical research.
6. Lipid identification and automated annotation
Compound identification is still the bottleneck in LC-MS-based metabolomics. By utilizing MS/MS, the lipid class, carbon-chain length, and degree of unsaturation of fatty-acid components of lipid can be annotated. Although library matches for some of those spectra may be found in MS/MS databases of pure chemical standards, the identification rates are usually low because libraries, such as Metlin, MassBank, and the US National Institutes of Standards and Technology (NIST) database, cover fewer than 20,000 compounds in total. On-line databases and computational tools have been developed for mass spectral lipid analysis (e.g., LipidQA, LIMSA, FAAT, lipID, LipidSearch, LipidView, LipidInspector, LipidXplorer, and ALEX), but they do not provide stand-alone MS/MS spectral libraries [54,55]. Several commercial MS/MS databases are currently available (e.g., LipidView, AB SCIEX; and, Lipid Search, Thermo Scientific) but there is urgent need for an independent platform.
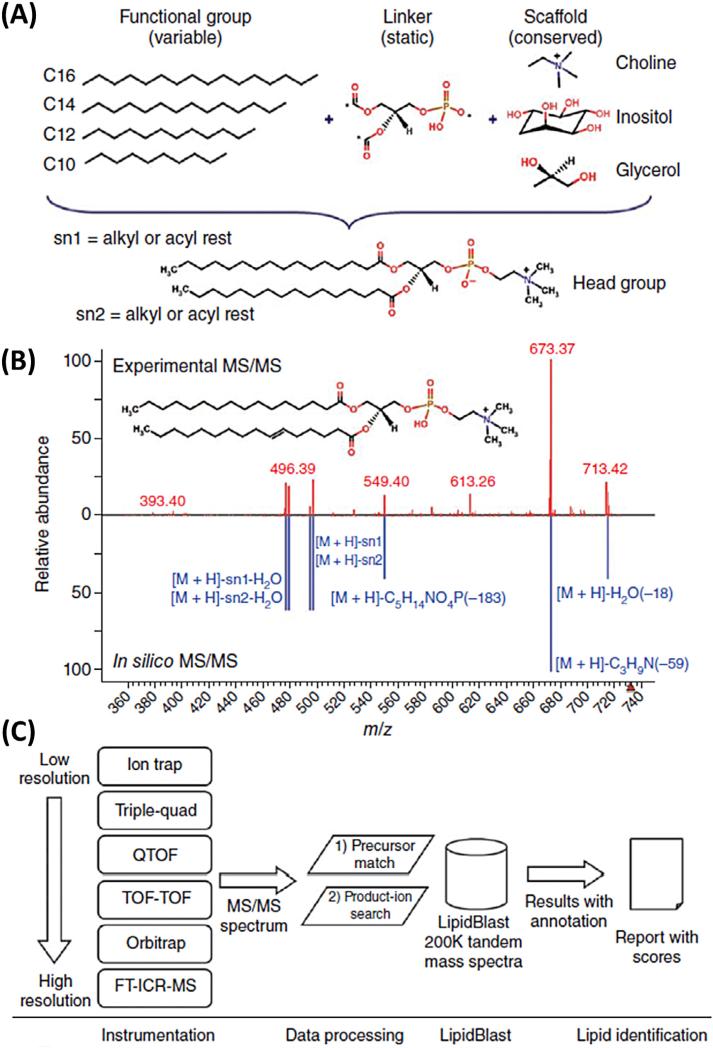
Recently, LipidBlast was presented as a large, platform-independent MS/MS database, which is freely available for commercial and non-commercial use [56]. This in-silico MS/MS database contains 212,516 spectra covering 119,200 compounds from 26 lipid-compound classes, including phospholipids, glycerolipids, bacterial lipoglycans, and plant glycolipids. The authors have shown that LipidBlast can be successfully applied to analyze MS/MS data from more than 40 different types of mass spectrometer. Fig. 9 illustrates the creation, the validation, and the application of insilico-generated MS/MS spectra in LipidBlast.
Fig. 9.
Creation, validation, and application of in-silico generated MS/MS spectra in LipidBlast. (a) New lipid compound structures were generated using in-silico methods. Lipid core structure scaffolds were connected via a linker to fatty acyls with different chain lengths and different degrees of unsaturation. Asterisks denote connection points. (b) Reference tandem spectra (top) were used to simulate mass spectral fragmentations and ion abundances of the in-silico spectra (bottom). The compound shown is PC(16:0/ 16:1) at precursor m/z 732.55 [M + H]+. (c) For lipid identification, MS/MS spectra obtained from LC-MS/MS or direct-infusion experiments were submitted to LipidBlast. An m/z precursor-ion filter first filtered the data, and a subsequent product-ion match generated a library hit score that reflects the level of confidence for compound annotation. {Reproduced with permission from [56]}.
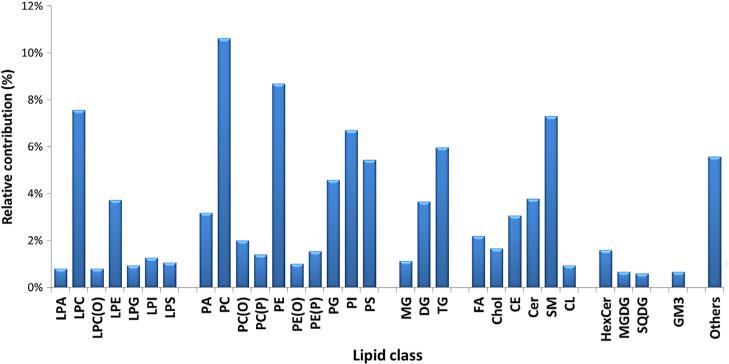
In the 185 reviewed papers, the number of annotated/identified lipids was in the range 32–722. This significant difference can be explained by some studies being focused on in-depth qualitative characterization of lipid extracts instead of quantitative aspects of the method. Considering that, typically, current ESI-MS systems offer only 3–4 orders of magnitude linear dynamic range, it is obvious that, for quantitative methods covering a broad range of lipid classes/ species and concentrations in biological samples, a compromise has to be made with respect to number of detected lipids and the linear dynamic range of the method, unavoidably leading to a lower number of detected and identified lipids compared to qualitative methods where, usually, more concentrated extracts are injected. Specifically, those few studies where the number of annotated/ identified lipids exceeded 350 species were focused mainly on in-depth characterization of lipid extracts [32,38,49,57–60], while only 50–300 annotated lipid species were reported when quantification was mentioned as major aim in studies. A deeper examination of particular lipid classes showed that LC-MS-based lipidomics methods most frequently covered phosphatidylcho-lines, followed by other phospholipids (phosphatidylethanolamines, phosphatidylinositols, phosphatidylserines, phosphatidylglycerols), sphingomyelins, di- and tri-acylglycerols, and ceramides (Fig. 10).
Fig. 10.
Focus on particular lipid classes in LC-MS-based lipidomics.
Double-bond positions and sn-positional isomers are difficult to distinguish by MS, so structures are often reported as a single isomer [61]. Although demanding, several approaches exist to differentiate and to identify double-bond isomers in lipids:
high-energy CID;
multi-stage fragmentation approaches using highly metallated lipids; and,
ozone-induced dissociation [62].
Double-bound positional isomers, cis-/trans-isomers, or regioisomers of TG can be separated using silver-ion LC based on formation of weak complexes of silver ions impregnated on the silica or bonded to the ion-exchange stationary phase with π electrons of double-bounds of unsaturated lipids. Chiral LC and non-aqueous RPLC methods have also been used for separation of sn-positional isomers of TG [63,64]. The drawback of these separation methods was that 1.5–2.5 h were needed for complete separation of lipid mixtures.
7. Lipid quantification
Subtle variations during extraction and data acquisition cause analytical errors. To compensate for these errors, non-naturally occurring standards can be spiked into all the samples at different stages of the sample processing [6]. In most cases, internal standards are added prior to the extraction by adding a small volume of concentrated stock solution of these standards, or they can be present already as part of the extraction solvents. Alternatively, internal standards can be added just prior to LC-MS analysis, but, in this case, these standards do not correct for possible lower recoveries of specific lipids during the extraction and sample-preparation steps, including the resolubilization prior to LC-MS injection.
The use of reference standards for each target lipid species is not feasible due to their unavailability or prohibitive price, so a compromise has to be made with respect to quantification of lipids in biological samples. Fortunately, a variety of unlabeled (native or non-naturally occurring) and isotopically-labeled (13C, 2H) lipid standards are now available commercially for this purpose [10]. It should be kept in mind that different lipid species usually have quantitatively different MS responses. For example, instrument responses for both saturated and unsaturated phospholipid species decrease with increasing acyl-chain length. This effect becomes increasingly prominent with increasing overall lipid concentration. Moreover, the degree of acyl-chain unsaturation has a significant effect on instrument response at higher concentrations with polyunsaturated species giving higher intensities than their fully saturated counterparts. Last, but not least, quantitative calibration curves for the different head group classes vary markedly, depending on the solvent mixtures at the point of electrospray ionization [65].
For analysis of phospholipids, it is acceptable to use synthetic di-saturated phospholipids (di-C14, di-C15 or di-C17), or preferably mixed odd carbon number phospholipids (C13-C15, C15-C17) for each phospholipid class [10]. For MG, DG and TG, mono-, di- and tri-saturated species with odd carbon numbers, respectively, are commonly used. The use of odd carbon-chain lipids is also preferred for free fatty acids. For all lipid species, the alternative is use of isotopically-labeled standards. In this case, we are not limited to odd carbon number recommendation.
To achieve lipid quantifications, it is a common practice to normalize the individual molecular ion-peak intensities using an internal standard for each lipid class. The calculated ratio of analyte and internal standard is then multiplied by the concentration of the internal standard to obtain the concentration of a particular analyte. In spite of issues mentioned above, including the problems associated with single-point calibrations, the results can be sufficiently accurate to answer biochemical questions, such as differences between diseased or treated and normal samples [6]. Table 2 summarizes typical internal standards used in lipidomics studies to demonstrate not only the availability of these compounds for different lipid classes, but also the variability within the lipid classes. The use of internal standards has become more and more popular in lipidomics studies. Reviewing the lipidomics papers cited here, we found that at least one internal standard was used in 63% of all studies referenced here. More often, however, authors extended this concept by including at least one internal standard per lipid class.
Table 2.
Internal standards used in LC-MS-based lipidomics studies
| Lipid class | Available and/or used internal standards |
|---|---|
| LPA | LPA(14:0), LPA(17:0) |
| LPC | LPC(12:0), LPC(l3:0), LPC(14:0), LPC(15:0), LPC(16:0), LPC(l7:0), LPC(19:0) |
| LPE | LPE(14:0), LPE(17:1) |
| LPG | LPG(17:1) |
| LPS | LPS(14:0), LPS(17:0) |
| PA | PA(10:0/10:0), PA(12:0/13:0), PA(14:0/14:0), PA(17:0/17:0), d31-PA(34:1) |
| PC | d3-PC(16:1/0:0), PC(9:0/9:0), PC(17:0/0:0), PC(19:0/0:0), PC(10:0/10:0), PC(11:0/11:0), PC(13:0/13:0), PC(14:0/14:0), d54-PC(14:0/14:0), PC(14:1/14:1), PC(15:0/15:0), d9-PC(16:0/16:0), d6-PC(16:1/16:1), PC(17:0/14:1), PC(17:0/17:0), d31-PC(16:0/18:1), PC(19:0/19:0) |
| PE | PE(10:0/10:0), PE(12:0/12:0), PE(12:0/13:0), PE(14:0/14:0), PE(15:0/15:0), PE(16:0/16:0), PE(17:0/14:1), PE(17:0/17:0), d31-PE(34:1) |
| PG | PG(10:0/10:0), PG(12:0/12:0), PG(12:0/13:0), PG(14:0/14:0), PG(17:0/17:0) |
| PI | PI(8:0/8:0), PI(12:0/13:0), PI(17:0/14:1), PI(16:0/16:0), d31-PI(18:1/16:0), PI(21:0/22:6) |
| PS | PS(10:0/10:0), PS(12:0/13:0), PS(14:0/14:0), PS(17:0/17:0), d31-PS(16:0/18:1) |
| MG | MG(17:0/0:0/0:0) |
| DG | DG(18:1/2:0/0:0), DG(12:0/12:0/0:0), DG(14:0/14:0/0:0), DG(15:0/15:0/0:0), 4ME DG(16:0/16:0/0:0), d5-DG(15:0/18:1/0:0), DG(17:0/17:0/0:0), d8–DG(20:0/20:0/0:0) |
| TG | TG(15:0/15:0/15:0), 13C3-TG(16:0/16:0/16:0), TG(17:0/17:0/17:0), d5–TG(17:0/17:1/17:0), TG(19:0/19:0/19:0) |
| Cholesterol | d6-Cholesterol, d7-cholesterol |
| CE | CE(15:0), CE(17:0), d6-CE(18:0), 13C18-CE(18:1), CE(22:1) |
| Fatty acids | d3-Palmitic acid (16:0), heptadecanoic acid (17:0), d35-oleic acid (18:0), d8–eicosanoic acid (20:0), heneicosanoic acid (21:0), d47-lignoceric acid (24:0) |
| SM | SM(d18:1/12:0), SM(17:0) |
| Sphingosines | Sphingosine(17:1) |
| Cer | C17 Cer(d18:1/17:0), d31-Cer[EOS], d47-Cer[NS] |
| GluCer | C8 GluCer(d18:1/8:0), C12 GluCer(d18:1/12:0) |
| LacCer | C8 LacCer(d18:1/8:0), C12 LacCer(d18:1/12:0) |
| CL | CL(14:0/14:0/14:0/14:0), CL(18:1/18:1/18:1/18:1), CL(18:2/18:2/18:2/18:2) |
8. Quality control in large-scale lipidomics studies
Keeping up high quality of acquired data within a large-scale lipidomics study of more than 1000 samples represents a real challenge for laboratory practice. When performing a lipidomic profiling analysis, changes in instrument sensitivity caused by degradation of the extracts, contamination of ion source by non-volatiles or retention-time shifts may be observed over time. In order to avoid false expectations raised by stating a comprehensive metabolomics approach, it is important that samples are analyzed in a random order, with sample classes randomly spread out across the whole run. By adding quality control (QC) samples and adding internal standards into samples, method-accuracy drifts can be followed adequately over time and be corrected for using both the internal standards and the QC samples [66].
QC samples should be theoretically identical biological samples, with a metabolic and sample matrix composition similar to those of the biological samples under study. Two types of QC sample can be considered:
pooled QC in which small aliquots of each (or a few) biological sample to be studied are pooled and thoroughly mixed; and,
commercially available biofluids composed of multiple biological samples not present in the study [67].
With respect to the frequency of QC-sample injections, 3–25 injections of samples between each QC injection were reported [67–75]. As a compromise, the injection of QC samples after every 10 samples seems to be reasonable.
QC samples provide a measure of within-series and between-series repeatability. Using such QC-drift analysis, inconsistent metabolic features with excessive drifts in signal, retention times or accurate masses can be removed in the data-processing step prior to statistical analyses. The QC sample is applied for four reasons:
to test for method suitability within control limits before samples are analyzed;
to provide data to calculate technical precision within each analytical series;
to provide data to use for signal correction within and between analytical series; and,
for standardization if community QC samples, such as NIST plasma, are used.
In addition, each series of samples should contain procedural blanks as well as “no injection” analyses. Procedural or method blanks enable monitoring of potential contamination originating from extraction or resuspension solvents or from extraction materials (e.g., tubes or pipette tips). However, “no injection” analyses can be used to check the quality of solvents used for mobile phases as well as during the real acquisition for carry-over monitoring. During the method-development phase, maximum injections per LC column should be evaluated by monitoring the peak shape of the internal standard and selected native lipids. For such native lipids, it is convenient to evaluate column performance based on partially separated isomers. If chromatographic resolution of some “critical pairs” decreases, the column should be replaced. To protect the LC column, the use of guard columns is highly recommended. Nevertheless, information on the maximum number of injections per column should be determined prior to starting large-scale lipidomics cohort studies, so that columns can be exchanged before problems with data acquisitions occur. Instrument maintenance, such as ion-source cleaning, must be conducted on regular basis to avoid decreases of signal intensities in large-scale lipidomics studies.
9. Highlights of recent lipidomics studies
Here, we briefly highlight some recent applications of lipidomics conducted with the technologies described above. The lipidomic profiles from body fluids, animal and plant tissues, or cells provide a global snapshot of lipid concentrations in a particular biological sample at specific physiological state, time or intervention response.
Orešič et al. [76] focused on UHPLC-MS analysis of lipids in blood samples of 679 well-characterized individuals in whom liver-fat content was measured using proton magnetic resonance spectroscopy or liver biopsy. Individuals with non-alcoholic fatty-liver disease (NAFLD) had increased TGs with low carbon number and double-bond content while LPCs and ether PLs were diminished in those with NAFLD. A serum-lipid signature comprising three molecular lipids [TG(48:0), PC(O-24:1/20:4) and PC(18:1/22:6)] was developed to estimate the percentage of liver fat. It had a sensitivity of 69.1% and specificity of 73.8% when applied for diagnosis of NAFLD in the validation series. The usefulness of the lipid triplet was demonstrated in a weight-loss intervention study, in which patients were treated with placebo or the cannabinoid receptor type 1 blocker rimonabant. Liver fat determined using the lipid triplet decreased significantly in the rimonabant group. The estimated liver fat correlated closely with the lipid triplet concentration in both intervention arms before, and after, the intervention.
In a second study, the effect of trans fatty-acid intake on LC-MS plasma profiles was studied by Gurdeniz et al. [77] in a double-blinded randomized controlled parallel-group study involving 52 overweight post-menopausal women. This study demonstrated that intake of trans fatty acids affects phospholipid metabolism. The trans fatty-acid markers, PC(trans18:1/20:3), PC(trans18:1/22:4) and PC(trans18:1/22:5), preferentially integrated trans18:1 into the sn-1 position while PUFA was in the sn-2 position. This observation could be explained by a general up-regulation in the formation of long-chain PUFAs after trans fatty-acid intake and/or by specific mobilization of these fatty acids into PCs.
The hypothesis whether lipid profiling can inform the onset and the progression of type 2 diabetes was investigated by Rhee et al. [78]. LC-MS-based plasma-lipid profiling was performed on samples from 189 individuals, who developed type 2 diabetes, and 189 matched disease-free individuals. Lipids with lower carbon number and lower double bond content were associated with an increased risk of diabetes, whereas lipids with higher carbon number and double-bond content were associated with decreased risk. This pattern appeared to be most prevalent with TGs and did not change after multivariable adjustment for age, sex, body mass index, fasting glucose, fasting insulin, total TGs, and high-density lipoprotein cholesterol. Integrating the positive and negative risk captured by a TG of relatively lower carbon number and double-bond content [TG(50:0)] and a TG of relatively higher carbon number and double-bond content [TG(58:10)] further improved diabetes prediction.
LC-MS plasma lipidomic profiling was used by Kulkarni et al. [79] to characterize lipid species in 1192 individuals from 42 large and extended Mexican American families. The DG(16:0/22:5) and DG(16:0/22:6) lipid species were significantly associated with systolic blood pressure, diastolic blood pressure, and mean arterial pressure. Significant genetic correlations with the liability of hypertension in bivariate trait analyses were observed for, in total 14, PC, PI, DG and monohexosylceramide lipid species.
Degenkolbe et al. [80] presented a comprehensive LC-MS analysis of the natural variation in the Arabidopsis thaliana lipidome and of the effects of cold acclimation in relation to leaf freezing tolerance. After 14 days of cold acclimatization at 4°C, the plants from most accessions had accumulated massive amounts of storage lipids, with most of the changes in long-chain unsaturated TGs, while the total amount of membrane lipids was only slightly changed. Nevertheless, major changes in the relative amounts of different membrane lipids were also evident. The relative abundance of several lipid species was highly correlated with the freezing tolerance of the accessions, allowing the identification of possible marker lipids for plant freezing tolerance.
10. Conclusions
Recent advances in LC-MS have revolutionized lipidomics analysis by simplifying the analytical protocol and by increasing the chromatographic separation power and sensitivity of detection. RPLC with positive electrospray and HRMS can now be seen as the “gold standard” for many lipidomics studies. The regular use of internal standards for quantifications means that lipidomics has a real chance of achieving a level of stability enabling inter-laboratory comparison in ring trials and direct comparisons of biological outcomes from different studies.
Future improvements in analytical sensitivity and in analytical instrumentation, including ion mobility, may reveal the presence of lipids that currently remain undetected. Automated data processing, data mining, and the accuracy of lipid annotations represent current challenges in untargeted LC-MS-based lipidomics, which need to be resolved using commonly accepted guidelines and pre-competitive ring trials.
Supplementary Material
Acknowledgements
This study was supported by the US National Institutes of Health (NIH) (Grants P20 HL113452 and U24 DK097154).
Abbreviations
- 2D-LC
Two-dimensional liquid chromatography
- ACN
Acetonitrile
- APCI
Atmospheric-pressure chemical ionization
- BuOH
Butanol
- CE
Cholesteryl esters
- Cer
Ceramide
- CID
Collision-induced dissociation
- CL
Cardiolipin
- DG
Diacylglycerol
- DGDG
Digalactosyldiacylglycerol
- ESI
Electrospray ionization
- FA
Fatty acids
- FWHM
Full width at half maximum
- FT-ICR
Fourier transform ion cyclotron resonance
- GM3
Monosialodihexosylganglioside
- HCD
High-energy collisional dissociation
- HexCer
Hexosylceramides
- HILIC
Hydrophilic interaction chromatography
- HPLC
High-performance liquid chromatography
- HRMS
High-resolution mass spectrometry
- IM-MS
Ion mobility mass spectrometry
- IT
Ion trap
- LacCer
Lactosylceramide
- LLE
Liquid-liquid extraction
- (L)PA
(Lyso)phosphatidic acid
- (L)PC
(Lyso)phosphatidylcholine
- LPC(O)
Lysoalkylphosphatidylcholine
- (L)PE
(Lyso)phosphatidylethanolamine
- (L)PG
(Lyso)phosphatidylglycerol
- (L)PS
(Lyso)phosphatidylserine
- MeOH
Methanol
- MG
Monoacylglycerol
- MGDG
Monogalactosyldiacylglycerol
- MRM
Multiple reaction monitoring
- MTBE
Methyl-tert-butyl ether
- NARPLC
Non-aqueous reversed-phase LC
- NMR
Nuclear magnetic resonance
- NPLC
Normal-phase LC
- PC(O)
Alkylphosphatidylcholine
- PC(P)
Phosphatidylcholine plasmalogen
- PI
Phosphatidylinositol
- QLIT
Quadrupole/linear ion trap
- QqQ
Triple quadrupole
- QTOF
Quadrupole/time-of-flight
- RPLC
Reversed-phase LC
- SFC
Supercritical fluid chromatography
- SIM
Selected ion monitoring
- SM
Sphingomyelin
- SPE
Solid-phase extraction
- SQDG
Sulfoquinovosyl diacylglycerol
- TG
Triacylglycerol
- TOF
Time-of-flight
- UHPLC
Ultrahigh-performance liquid chromatography
Footnotes
Disclaimer
Mention of brand or firm names in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the University of California, Davis, USA.
Appendix: Supplementary Material
Supplementary data to this article can be found online at doi:10.1016/j.trac.2014.04.017.
References
- 1.Han XL, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Hou W, Zhou H, Elisma F, Bennett SA, Figeys D. Technological developments in lipidomics. Brief. Funct. Genomic Proteomic. 2008;7:395–409. doi: 10.1093/bfgp/eln042. [DOI] [PubMed] [Google Scholar]
- 3.Loizides-Mangold U. On the future of mass-spectrometry-based lipidomics. FEBS J. 2013;280:2817–2829. doi: 10.1111/febs.12202. [DOI] [PubMed] [Google Scholar]
- 4.Wenk MR. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Zhou ZG, Nie HG, Bai Y, Liu HW. Recent advances of chromatography and mass spectrometry in lipidomics. Anal. Bioanal. Chem. 2011;399:243–249. doi: 10.1007/s00216-010-4327-y. [DOI] [PubMed] [Google Scholar]
- 6.Astarita G. New frontiers for mass spectrometry in lipidomics, part II. LC GC North Am. 2012;30:482. [Google Scholar]
- 7.Navas-Iglesias N, Carrasco-Pancorbo A, Cuadros-Rodriguez L. From lipids analysis towards lipidomics, a new challenge for the analytical chemistry of the 21st century. Part II: analytical lipidomics. TrAC Trends Anal. Chem. 2009;28:393–403. [Google Scholar]
- 8.Godzien J, Ciborowski M, Whiley L, Legido-Quigley C, Ruperez FJ, Barbas C. In-vial dual extraction liquid chromatography coupled to mass spectrometry applied to streptozotocin-treated diabetic rats. Tips and pitfalls of the method. J. Chromatogr. A. 2013;1304:52–60. doi: 10.1016/j.chroma.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 9.Whiley L, Godzien J, Ruperez FJ, Legido-Quigley C, Barbas C. In-vial dual extraction for direct LC-MS analysis of plasma for comprehensive and highly reproducible metabolic fingerprinting. Anal. Chem. 2012;84:5992–5999. doi: 10.1021/ac300716u. [DOI] [PubMed] [Google Scholar]
- 10.Wolf C, Quinn PJ. Lipidomics: practical aspects and applications. Prog. Lipid Res. 2008;47:15–36. doi: 10.1016/j.plipres.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 12.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 13.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lofgren L, Stahlman M, Forsberg GB, Saarinen S, Nilsson R, Hansson GI. The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 2012;53:1690–1700. doi: 10.1194/jlr.D023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, et al. Elemental formula annotation of polar and lipophilic metabolites using C-13, N-15 and S-34 isotope labelling, in combination with high-resolution mass spectrometry. Plant J. 2011;68:364–376. doi: 10.1111/j.1365-313X.2011.04682.x. [DOI] [PubMed] [Google Scholar]
- 16.Yang YH, Cruickshank C, Armstrong M, Mahaffey S, Reisdorph R, Reisdorph N. New sample preparation approach for mass spectrometry-based profiling of plasma results in improved coverage of metabolome. J. Chromatogr. A. 2013;1300:217–226. doi: 10.1016/j.chroma.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SL, Hoene M, Li J, Li YJ, Zhao XJ, Haring HU, et al. Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. J. Chromatogr. A. 2013;1298:9–16. doi: 10.1016/j.chroma.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Canaveras JC, Donato MT, Castell JV, Lahoz A. A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J. Proteome Res. 2011;10:4825–4834. doi: 10.1021/pr200629p. [DOI] [PubMed] [Google Scholar]
- 19.Ivanisevic J, Zhu ZJ, Plate L, Tautenhahn R, Chen S, O'Brien PJ, et al. Toward ‘omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal. Chem. 2013;85:6876–6884. doi: 10.1021/ac401140h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanksby SJ, Mitchell TW. Advances in mass spectrometry for lipidomics. Annu. Rev. Anal. Chem. 2010;3:433–465. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- 21.Sandra K, Sandra P. Lipidomics from an analytical perspective. Curr. Opin. Chem. Biol. 2013;17:847–853. doi: 10.1016/j.cbpa.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Nygren H, Seppänen-Laakso T, Castillo S, Hyötyläinen T, Orešič M, Metz TO, editors. Methods Mol. Biol. Humana Press; New York: 2011. pp. 247–257. [DOI] [PubMed] [Google Scholar]
- 23.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal. Chem. 2011;83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallart-Ayala H, Courant F, Severe S, Antignac JP, Morio F, Abadie J, et al. Versatile lipid profiling by liquid chromatography-high resolution mass spectrometry using all ion fragmentation and polarity switching. Preliminary application for serum samples phenotyping related to canine mammary cancer. Anal. Chim. Acta. 2013;796:75–83. doi: 10.1016/j.aca.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Guillarme D, Ruta J, Rudaz S, Veuthey JL. New trends in fast and high-resolution liquid chromatography: a critical comparison of existing approaches. Anal. Bioanal. Chem. 2010;397:1069–1082. doi: 10.1007/s00216-009-3305-8. [DOI] [PubMed] [Google Scholar]
- 26.Castro-Perez JM, Kamphorst J, DeGroot J, Lafeber F, Goshawk J, Yu K, et al. MSE lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J. Proteome Res. 2010;9:2377–2389. doi: 10.1021/pr901094j. [DOI] [PubMed] [Google Scholar]
- 27.Ogiso H, Suzuki T, Taguchi R. Development of a reverse-phase liquid chromatography electrospray ionization mass spectrometry method for lipidomics, improving detection of phosphatidic acid and phosphatidylserine. Anal. Biochem. 2008;375:124–131. doi: 10.1016/j.ab.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 28.Samhan-Arias AK, Ji J, Demidova OM, Sparvero LJ, Feng W, Tyurin V, et al. Oxidized phospholipids as biomarkers of tissue and cell damage with a focus on cardiolipin. Biochim. Biophys. Acta Biomembr. 1818;2012:2413–2423. doi: 10.1016/j.bbamem.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gama MR, Silva RGD, Collins CH, Bottoli CBG. Hydrophilic interaction chromatography. TrAC Trends Anal. Chem. 2012;37:48–60. [Google Scholar]
- 30.Novakova L, Vlckova H. A review of current trends and advances in modern bio-analytical methods: chromatography and sample preparation. Anal. Chim. Acta. 2009;656:8–35. doi: 10.1016/j.aca.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Q, Gao B, Zhang X, Xu Y, Shi H, Yu L. Chemical profiling of triacylglycerols and diacylglycerols in cow milk fat by ultra-performance convergence chromatography combined with a quadrupole time-of-flight mass spectrometry. Food Chem. 2014;143:199–204. doi: 10.1016/j.foodchem.2013.07.114. [DOI] [PubMed] [Google Scholar]
- 32.Yamada T, Uchikata T, Sakamoto S, Yokoi Y, Nishiumi S, Yoshida M, et al. Supercritical fluid chromatography/Orbitrap mass spectrometry based lipidomics platform coupled with automated lipid identification software for accurate lipid profiling. J. Chromatogr. A. 2013;1301:237–242. doi: 10.1016/j.chroma.2013.05.057. [DOI] [PubMed] [Google Scholar]
- 33.Bamba T, Shimonishi N, Matsubara A, Hirata K, Nakazawa Y, Kobayashi A, et al. High throughput and exhaustive analysis of diverse lipids by using supercritical fluid chromatography-mass spectrometry for metabolomics. J. Biosci. Bioeng. 2008;105:460–469. doi: 10.1263/jbb.105.460. [DOI] [PubMed] [Google Scholar]
- 34.Lee JW, Uchikata T, Matsubara A, Nakamura T, Fukusaki E, Bamba T. Application of supercritical fluid chromatography/mass spectrometry to lipid profiling of soybean. J. Biosci. Bioeng. 2012;113:262–268. doi: 10.1016/j.jbiosc.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Bamba T, Lee JW, Matsubara A, Fukusaki E. Metabolic profiling of lipids by supercritical fluid chromatography/mass spectrometry. J. Chromatogr. A. 2012;1250:212–219. doi: 10.1016/j.chroma.2012.05.068. [DOI] [PubMed] [Google Scholar]
- 36.Lisa M, Cifkova E, Holcapek M. Lipidomic profiling of biological tissues using off-line two-dimensional high-performance liquid chromatography mass spectrometry. J. Chromatogr. A. 2011;1218:5146–5156. doi: 10.1016/j.chroma.2011.05.081. [DOI] [PubMed] [Google Scholar]
- 37.Myers DS, Ivanova PT, Milne SB, Brown HA. Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim. Biophys. Acta. 2011;1811:748–757. doi: 10.1016/j.bbalip.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bird SS, Marur VR, Stavrovskaya IG, Kristal BS. Qualitative characterization of the rat liver mitochondrial lipidome using LC–MS profiling and high energy collisional dissociation (HCD) all ion fragmentation. Metabolomics. 2013;9:S67–S83. doi: 10.1007/s11306-012-0400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: focus on triglyceride detection and characterization. Anal. Chem. 2011;83:6648–6657. doi: 10.1021/ac201195d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hummel J, Segu S, Li Y, Irgang S, Jueppner J, Giavalisco P. Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front. Plant Sci. 2011;2:54. doi: 10.3389/fpls.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seppanen-Laakso T, Oresic M. How to study lipidomes. J. Mol. Endocrinol. 2009;42:185–190. doi: 10.1677/JME-08-0150. [DOI] [PubMed] [Google Scholar]
- 42.Kliman M, May JC, McLean JA. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim. Biophys. Acta. 2011;1811:935–945. doi: 10.1016/j.bbalip.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holcapek M, Jirasko R, Lisa M. Recent developments in liquid chromatography-mass spectrometry and related techniques. J. Chromatogr. A. 2012;1259:3–15. doi: 10.1016/j.chroma.2012.08.072. [DOI] [PubMed] [Google Scholar]
- 44.Jackson SN, Ugarov M, Post JD, Egan T, Langlais D, Schultz JA, et al. A study of phospholipids by ion mobility TOFMS. J. Am. Soc. Mass Spectrom. 2008;19:1655–1662. doi: 10.1016/j.jasms.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HI, Kim H, Pang ES, Ryu EK, Beegle LW, Loo JA, et al. Structural characterization of unsaturated phosphatidylcholines using traveling wave ion mobility spectrometry. Anal. Chem. 2009;81:8289–8297. doi: 10.1021/ac900672a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah V, Castro-Perez JM, McLaren DG, Herath KB, Previs SF, Roddy TP. Enhanced data-independent analysis of lipids using ion mobility-TOFMSE to unravel quantitative and qualitative information in human plasma. Rapid Commun. Mass Spectrom. 2013;27:2195–2200. doi: 10.1002/rcm.6675. [DOI] [PubMed] [Google Scholar]
- 47.Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole-time-of-flight mass spectrometry. J. Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 48.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada T, Uchikata T, Sakamoto S, Yokoi Y, Fukusaki E, Bamba T. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J. Chromatogr. A. 2013;1292:211–218. doi: 10.1016/j.chroma.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 50.Boccard J, Veuthey JL, Rudaz S. Knowledge discovery in metabolomics: an overview of MS data handling. J. Sep. Sci. 2010;33:290–304. doi: 10.1002/jssc.200900609. [DOI] [PubMed] [Google Scholar]
- 51.Katajamaa M, Oresic M. Data processing for mass spectrometry-based metabolomics. J. Chromatogr. A. 2007;1158:318–328. doi: 10.1016/j.chroma.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 52.Theodoridis GA, Gika HG, Want EJ, Wilson ID. Liquid chromatography-mass spectrometry based global metabolite profiling: a review. Anal. Chim. Acta. 2012;711:7–16. doi: 10.1016/j.aca.2011.09.042. [DOI] [PubMed] [Google Scholar]
- 53.Gurdeniz G, Kristensen M, Skov T, Dragsted LO. The effect of LC-MS data preprocessing methods on the selection of plasma biomarkers in fed vs. fasted rats. Metabolites. 2012;2:77–99. doi: 10.3390/metabo2010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song H, Hsu FF, Ladenson J, Turk J. Algorithm for processing raw mass spectrometric data to identify and quantitate complex lipid molecular species in mixtures by data-dependent scanning and fragment ion database searching. J. Am. Soc. Mass Spectrom. 2007;18:1848–1858. doi: 10.1016/j.jasms.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forrester JS, Milne SB, Ivanova PT, Brown HA. Computational lipidomics: a multiplexed analysis of dynamic changes in membrane lipid composition during signal transduction. Mol. Pharmacol. 2004;65:813–821. doi: 10.1124/mol.65.4.813. [DOI] [PubMed] [Google Scholar]
- 56.Kind T, Liu KH, Lee do Y, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sysi-Aho M, Katajamaa M, Yetukuri L, Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao XL, Zhang QB, Meng D, Isaac G, Zhao R, Fillmore TL, et al. A reversed-phase capillary ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) method for comprehensive top-down/bottom-up lipid profiling. Anal. Bioanal. Chem. 2012;402:2923–2933. doi: 10.1007/s00216-012-5773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nie HG, Liu RR, Yang YY, Bai Y, Guan YF, Qian DQ, et al. Lipid profiling of rat peritoneal surface layers by online normal- and reversed-phase 2D LC QToF-MS. J. Lipid Res. 2010;51:2833–2844. doi: 10.1194/jlr.D007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Li J, Shi X, Qiao L, Lu X, Xu G. A novel stop-flow two-dimensional liquid chromatography-mass spectrometry method for lipid analysis. J. Chromatogr. A. 2013;1321:65–72. doi: 10.1016/j.chroma.2013.10.069. [DOI] [PubMed] [Google Scholar]
- 61.Mitchell TW, Pham H, Thomas MC, Blanksby SJ. Identification of double bond position in lipids: from GC to OzID. J. Chromatogr. B. 2009;877:2722–2735. doi: 10.1016/j.jchromb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 62.Poad BLJ, Pham HT, Thomas MC, Nealon JR, Campbell JL, Mitchell TW, et al. Ozone-induced dissociation on a modified tandem linear ion-trap: observations of different reactivity for isomeric lipids. J. Am. Soc. Mass Spectrom. 2010;21:1989–1999. doi: 10.1016/j.jasms.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 63.Lisa M, Netusilova K, Franek L, Dvorakova H, Vrkoslav V, Holcapek M. Characterization of fatty acid and triacylglycerol composition in animal fats using silver-ion and non-aqueous reversed-phase high-performance liquid chromatography/mass spectrometry and gas chromatography/flame ionization detection. J. Chromatogr. A. 2011;1218:7499–7510. doi: 10.1016/j.chroma.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 64.Lisa M, Holcapek M. Characterization of triacylglycerol enantiomers using chiral HPLC/APCI-MS and synthesis of enantiomeric triacylglycerols. Anal. Chem. 2013;85:1852–1859. doi: 10.1021/ac303237a. [DOI] [PubMed] [Google Scholar]
- 65.Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- 66.Denoroy L, Zimmer L, Renaud B, Parrot S. Ultra high performance liquid chromatography as a tool for the discovery and the analysis of biomarkers of diseases: a review. J. Chromatogr. B. 2013;927:37–53. doi: 10.1016/j.jchromb.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 68.Weir JM, Wong G, Barlow CK, Greeve MA, Kowalczyk A, Almasy L, et al. Plasma lipid profiling in a large population-based cohort. J. Lipid Res. 2013;54:2898–2908. doi: 10.1194/jlr.P035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen S, Yin P, Zhao X, Xing W, Hu C, Zhou L, et al. Serum lipid profiling of patients with chronic hepatitis B, cirrhosis, and hepatocellular carcinoma by ultra fast LC/IT-TOF MS. Electrophoresis. 2013;34:2848–2856. [PubMed] [Google Scholar]
- 70.Zhu C, Dane A, Spijksma G, Wang M, van der Greef J, Luo GA, et al. An efficient hydrophilic interaction liquid chromatography separation of 7 phospholipid classes based on a diol column. J. Chromatogr. A. 2012;1220:26–34. doi: 10.1016/j.chroma.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 71.Meikle PJ, Wong G, Tsorotes D, Barlow CK, Weir JM, Christopher MJ, et al. Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2011;31:2723–2732. doi: 10.1161/ATVBAHA.111.234096. [DOI] [PubMed] [Google Scholar]
- 72.Hu CX, Wei H, van den Hoek AM, Wang M, van der Heijden R, Spijksma G, et al. Plasma and liver lipidomics response to an intervention of rimonabant in ApoE*3Leiden.CETP transgenic mice. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0019423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Michopoulos F, Lai L, Gika H, Theodoridis G, Wilson I. UPLC-MS-based analysis of human plasma for metabonomics using solvent precipitation or solid phase extraction. J. Proteome Res. 2009;8:2114–2121. doi: 10.1021/pr801045q. [DOI] [PubMed] [Google Scholar]
- 74.Draisma HHM, Reijmers TH, Bobeldijk-Pastorova I, Meulman JJ, Burk GFEV, Bartels M, et al. Similarities and differences in lipidomics profiles among healthy monozygotic twin pairs. OMICS. 2008;12:17–31. doi: 10.1089/omi.2007.0048. [DOI] [PubMed] [Google Scholar]
- 75.Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal. Chem. 2006;78:567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- 76.Oresic M, Hyotylainen T, Kotronen A, Gopalacharyulu P, Nygren H, Arola J, et al. Prediction of non-alcoholic fatty-liver disease and liver fat content by serum molecular lipids. Diabetologia. 2013;56:2266–2274. doi: 10.1007/s00125-013-2981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gurdeniz G, Rago D, Bendsen NT, Savorani F, Astrup A, Dragsted LO. Effect of trans fatty acid intake on LC-MS and NMR plasma profiles. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0069589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Barlow CK, Jowett JB, et al. Plasma lipidomic profile signature of hypertension in Mexican American families: specific role of diacylglycerols. Hypertension. 2013;62:621–626. doi: 10.1161/HYPERTENSIONAHA.113.01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Degenkolbe T, Giavalisco P, Zuther E, Seiwert B, Hincha DK, Willmitzer L. Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. Plant J. 2012;72:972–982. doi: 10.1111/tpj.12007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.