Abstract
OBJECTIVE:
Oral vaccine efficacy is low in less-developed countries, perhaps due to intestinal dysbiosis. This study determined if stool microbiota composition predicted infant oral and parenteral vaccine responses.
METHODS:
The stool microbiota of 48 Bangladeshi infants was characterized at 6, 11, and 15 weeks of age by amplification and sequencing of the 16S ribosomal RNA gene V4 region and by Bifidobacterium-specific, quantitative polymerase chain reaction. Responses to oral polio virus (OPV), bacille Calmette-Guérin (BCG), tetanus toxoid (TT), and hepatitis B virus vaccines were measured at 15 weeks by using vaccine-specific T-cell proliferation for all vaccines, the delayed-type hypersensitivity skin-test response for BCG, and immunoglobulin G responses using the antibody in lymphocyte supernatant method for OPV, TT, and hepatitis B virus. Thymic index (TI) was measured by ultrasound.
RESULTS:
Actinobacteria (predominantly Bifidobacterium longum subspecies infantis) dominated the stool microbiota, with Proteobacteria and Bacteroidetes increasing by 15 weeks. Actinobacteria abundance was positively associated with T-cell responses to BCG, OPV, and TT; with the delayed-type hypersensitivity response; with immunoglobulin G responses; and with TI. B longum subspecies infantis correlated positively with TI and several vaccine responses. Bacterial diversity and abundance of Enterobacteriales, Pseudomonadales, and Clostridiales were associated with neutrophilia and lower vaccine responses.
CONCLUSIONS:
Bifidobacterium predominance may enhance thymic development and responses to both oral and parenteral vaccines early in infancy, whereas deviation from this pattern, resulting in greater bacterial diversity, may cause systemic inflammation (neutrophilia) and lower vaccine responses. Vaccine responsiveness may be improved by promoting intestinal bifidobacteria and minimizing dysbiosis early in infancy.
Keywords: vaccine, intestinal, microbiota, Bifidobacterium, Actinobacteria, Proteobacteria, Bangladesh, T lymphocyte, antibody, polio, tetanus, tuberculosis, hepatitis
What’s Known on This Subject:
Oral vaccine responses are low in children from less-developed countries perhaps as a result of intestinal dysbiosis. New high-throughput DNA-based methods allow characterization of intestinal microbiota as a predictor of vaccine responses.
What This Study Adds:
High abundance of stool Actinobacteria, including Bifidobacterium, was associated with higher responses to oral and parenteral vaccines and a larger thymus in Bangladeshi infants. Conversely, high abundance of Clostridiales, Enterobacteriales, and Pseudomonadales was associated with neutrophilia and lower vaccine responses.
The composition of the community of microbes that inhabits the gastrointestinal tract (the gut microbiota) has a profound influence on the developing infant.1 Dysbiosis, defined as deviation from an optimal, health-promoting microbial community,2 may cause necrotizing enterocolitis and sepsis in premature infants3 and allergic disease in term infants.4 A gut microbiota dominated by appropriate commensal bacteria promotes infant health by a variety of mechanisms,5,6 including appropriate development of the immune system.7,8
Infant immunization is an important measure for decreasing morbidity and mortality from infectious diseases.9 However, oral vaccines are often less effective than expected when used in developing countries, as described in a recent review,10 perhaps as a result of malnutrition, intestinal dysbiosis, or the presence of other inhibitory factors related to the local environment. Evidence that dysbiosis may influence vaccine responses include direct effects of intestinal bacteria on the ability of polio virus to infect target cells in the intestine11 and direct impairment by dysbiosis of the host immune response.12 Studies showing that probiotic interventions increase immune responses to oral vaccines in adults13 and animals14 support this hypothesis, although results in infants and children are equivocal.15,16
New DNA-based methods that use direct sequencing of short regions of the bacterial 16S ribosomal RNA (rRNA) gene produce a detailed picture of the gut microbiota, providing the relative abundance of taxa at multiple phylogenetic levels (ie, phylum, class, order, family, and genus).17 The intestinal microbiota of term infants is dominated by 4 major phyla: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. The relative abundance of taxa within these phyla is influenced early in infancy by mode of delivery,18 gestational age,19 type of feeding,20 and locality.21,22 Early colonizers typically include commensal facultative anaerobes such as Escherichia coli and other Enterobacteriaceae (phylum Proteobacteria), followed by an increased relative abundance of strict anaerobes including Bifidobacterium (phylum Actinobacteria), Bacteroides (phylum Bacteroidetes), and Clostridium (phylum Firmicutes).1 Firmicutes includes several genera of importance to infant health including Streptococcus, Staphylococcus, and Lactobacillus, whereas the phylum Proteobacteria includes pathogens from the genera Escherichia, Shigella, and Campylobacter. The relative abundance of these taxa is used to define the gut microbiota.
In the current study we hypothesized that the composition of the gut microbiota would affect responses to oral and perhaps parenteral vaccines. To test this hypothesis, we characterized the microbiota from the stool of 48 Bangladeshi infants and measured responses to 4 vaccines: oral polio virus (OPV), bacille Calmette-Guérin (BCG; given to protect against tuberculosis), tetanus toxoid (TT), and hepatitis B virus (HBV) by using 2 methods for each vaccine. Vaccine-specific T-cell proliferation was measured for all vaccines; the delayed-type hypersensitivity (DTH) skin test to purified protein derivative (PPD) of Mycobacterium tuberculosis was measured as a second, functional indicator of response to BCG vaccination; and vaccine-specific immunoglobulin (Ig) G levels were measured for OPV, TT, and HBV using the antibody in lymphocyte supernatant assay23 as an index of the memory B-cell response.
Methods
Subjects
The 48 infants in this study were the first to be recruited in a larger trial (clinicaltrials.gov identifier: NCT01583972) and were included here because a relatively complete set of immunologic data were available when funding became available to analyze stool microbiota. Procedures are summarized in Supplemental Table 3. Parents of infants born at the Maternal and Child Health Training Institute in Dhaka, Bangladesh, were approached during the third trimester and informed consent was obtained within 48 hours of birth. The study was approved by the Ethical Review Committee of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B) and by the Human Studies Committee of the World Health Organization.
Immune Function
Thymic index (TI), blood leukocytes, vaccine-specific T-cell stimulation index (SI), vaccine-specific IgG responses using the antibody in lymphocyte supernatant assay, and the PPD-DTH skin test were measured as described in the Supplemental Information.
Bacterial DNA Methods
The relative abundance of stool bacteria at the phylum, class, order, family, and genus levels was characterized by sequence analysis of the V4 segment of the 16S rRNA gene, as described in the Supplemental Information. In addition, 3 methods were used to identify members of the genus Bifidobacterium: (1) a terminal restriction fragment length polymorphism (T-RFLP) assay24 was used to determine the relative abundance of Bifidobacterium species, (2) Bifidobacterium 16S rDNA copy numbers (per gram of stool) were measured by quantitative polymerase chain reaction (qPCR), and (3) the Bifidobacterium longum subspecies infantis and longum were identified by polymerase chain reaction (PCR). Methods are described in the Supplemental Information and Supplemental Table 4.
Statistical Analysis
Spearman rank-order correlation and Wilcoxon rank-sum analyses were used to identify associations between microbiota and immune variables. Correlation was used when ≥75% of samples had nonzero results, whereas the Wilcoxon test (using a normal adjustment) was used when ≥25% of samples had nonzero results to compare “high” and “low” responders defined as those above or below the median value for each of the immune variables. A P value of <.05 was considered statistically significant for all analyses. Data are presented as means ± SDs or as medians (interquartile range) unless otherwise indicated. Statistical analysis was performed by using SAS 9.2 (SAS Institute, Cary, NC).
Results
Subjects
Of the 48 infants studied, 28 (58%) were male, 16 (33%) had a birth weight <2500 g, and 40 (83%) were born by cesarian delivery. The prevalence of moderate wasting malnutrition (>2 z scores below the reference mean weight-for-length) was 4% to 8% depending on age, and prevalence of moderate stunting was 10% to 12% (Table 1). All infants were breastfed with some receiving supplemental foods (Table 2). The median (25th–75th percentiles) years of education for mothers and fathers were 9 (7–10) and 9 (5–10) years, respectively. Seventy-one percent of mothers (34 of 48) had completed at least some secondary education. All 48 households had electricity, 98% had paved flooring, 44% had glass in the windows, 92% cooked with gas (rather than wood), 75% had drinking water piped into the house, and 38% had flush toilets available.
TABLE 1.
Nutritional Status of Subjects by Age
| 6 Weeks | 10 Weeks | 15 Weeks | P | |
|---|---|---|---|---|
| Weight-for-length | ||||
| z Score, mean ± SD. | −0.314 ± 1.044 | −0.256 ± 1.051 | −0.317 ± 1.221 | .29a |
| < −2 z scores, n (%) | 2 (4.2) | 4 (8.3) | 4 (8.3) | .65b |
| Length-for-age | ||||
| z Score,mean ± SD. | −0.991 ± 1.091 | −0.914 ± 0.988 | −0.975 ± 0.869 | .62c |
| < −2 z scores, n (%) | 7 (14.6) | 7 (14.6) | 5 (10.4) | .78b |
| Weigh-for-age | ||||
| z Score,Mean ± SD. | −1.139 ± 0.970 | −0.981 ± 0.977 | −0.963 ± 1.004 | .16a |
| < −2 z scores, n (%) | 5 (10.4) | 5 (10.4) | 6 (12.5) | .93b |
N = 48.
Repeated-measures analysis of variance of ranks.
χ2 Test.
Repeated-measures analysis of variance.
TABLE 2.
Number (%) of infants by breastfeed status at 6, 11 and 15 wk of age.
| Breastfeeding Status | 6 Weeks | 11 Weeks | 15 Weeks |
|---|---|---|---|
| Exclusive breastfeedinga | 38 (79.2) | 29 (60.4) | 22 (45.8) |
| Breastfeeding + water or fruit juice | 3 (6.2) | 4 (8.3) | 11 (22.9) |
| Breastfeeding + non–human milk or other liquid food | 7 (14.6) | 15 (31.2) | 12 (25) |
| Breastfeeding + any solid food | 0 (0) | 0 (0) | 3 (6.2) |
| Total | 48 (100) | 48 (100) | 48 (100) |
Data are presented as n (%).
Allows use of oral medications, vitamin supplements, and oral rehydration therapy, as needed.
Actinobacteria, Particularly Bifidobacterium, Dominated the Stool Microbiota
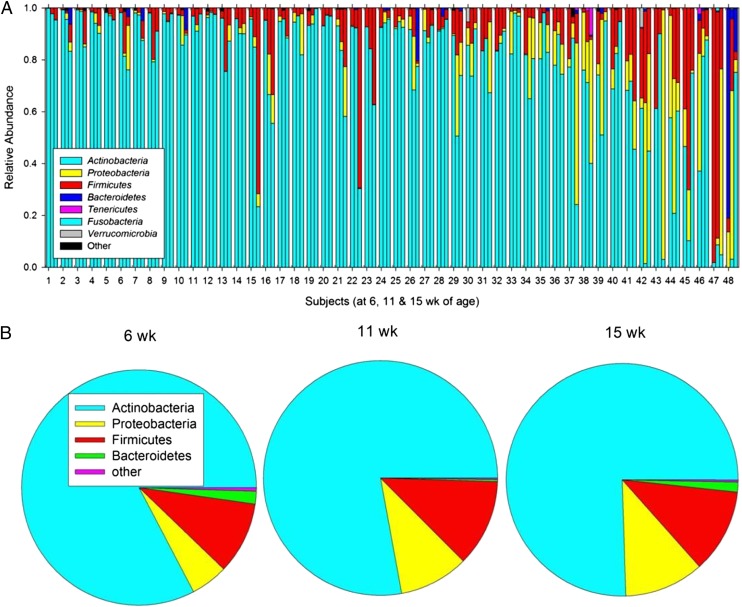
DNA sequence analysis of the V4 region of the 16S rRNA gene was used to characterize the relative abundance of bacteria from the phylum through the genus level in stool samples at 6, 11, and 15 weeks of age. Actinobacteria was the most abundant phylum at all ages, followed by Firmicutes, Proteobacteria, and Bacteroidetes (Fig 1; Supplemental Tables 5 and 6). The relative abundance of Proteobacteria and Bacteroidetes increased with age, resulting in greater bacterial diversity by 15 weeks (Supplemental Table 7). Within these 4 phyla, a total of 24 common genera (seen in at least 25% of infants) were identified at 15 weeks of age (Supplemental Table 8): 5 from Actinobacteria (Bifidobacterium being the most abundant), 3 from Bacteroidetes (Bacteroides, Parabacteroides, and Prevotella), 13 from Firmicutes (Streptococcus and Lactobacillus being the most abundant but also including Enterococcus, Staphylococcus, and Clostridium), and 3 from Proteobacteria (Campylobacter, Escherichia/Shigella, and Acinetobacter). Although diversity increased by 15 weeks, the median relative abundance of Actinobacteria remained quite high at 0.823 (0.329), with the median relative abundance of Bifidobacterium being 0.675 (0.282).
FIGURE 1.
A, Relative abundance of microbiota by 16S rRNA gene sequence analysis in each of the 48 subjects (numbered from highest to lowest abundance of Actinobacteria at 6 weeks) at all time points. For each subject, bars are ordered by age (6, 11, and 15 weeks) from left to right. B, Mean relative abundance at 6, 11, and 15 weeks of age.
Bifidobacterium-specific PCR amplification and T-RFLP analysis were used as additional methods to independently quantify Bifidobacterium levels and to identify and quantify Bifidobacterium species and subspecies. By using these methods, the median Bifidobacterium concentration in stool at 15 weeks was found to be 3.08 × 1010/g (2.74 × 1010). Several Bifidobacterium species were identified by T-RFLP (Supplemental Fig 8), including Bifidobacterium adolescentis, Bifidobacterium pseudocatenulatum, Bifidobacterium breve, and Bifidobacterium bifidum, as expected,1 but in these infants B longum was found to be by far the most abundant species, with a median relative abundance of 96.4% (8.9%) within the genus. Within B longum, the infantis subspecies predominated, at 68.1% (31.3%), with the longum subspecies contributing 31.9% (31.3%).
Environmental Factors and Stool Microbiota Composition at 15 Weeks of Age
The relative abundance of stool microbiota often differs by mode of delivery (vaginal versus cesarean),18 but in the current study no such differences were seen at 15 weeks (data not shown). Breastfeeding status also strongly affects stool microbiota20; and in these infants, who were all predominantly breastfed, more extensive breastfeeding was associated with a higher abundance of B longum, as might be expected, but few other differences were seen (Supplemental Table 9). Nutritional status (as measured by length-for-age) was also found to be positively associated with B longum and negatively associated with Escherichia/Shigella (Supplemental Table 10).
Stool Actinobacteria and Vaccine Responses
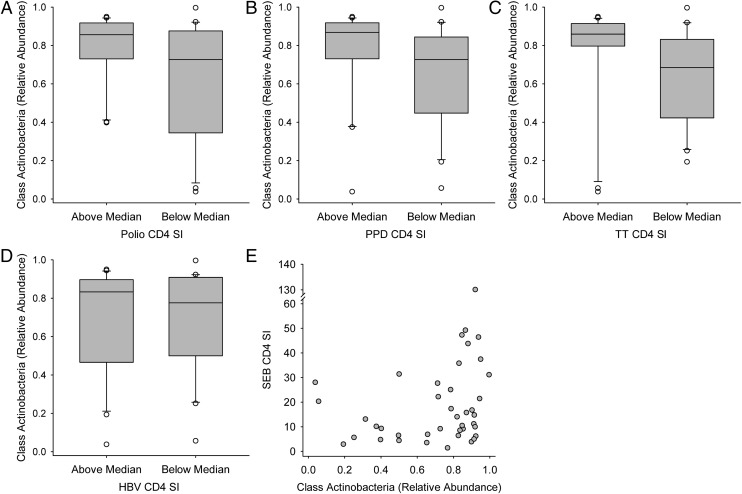
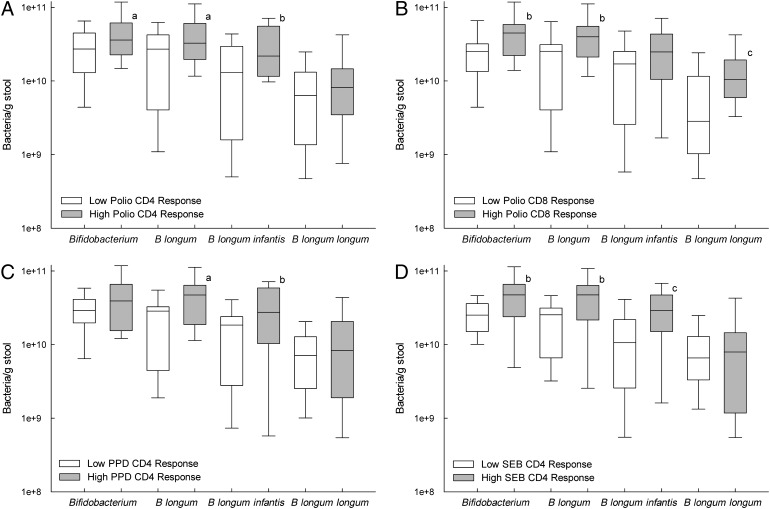
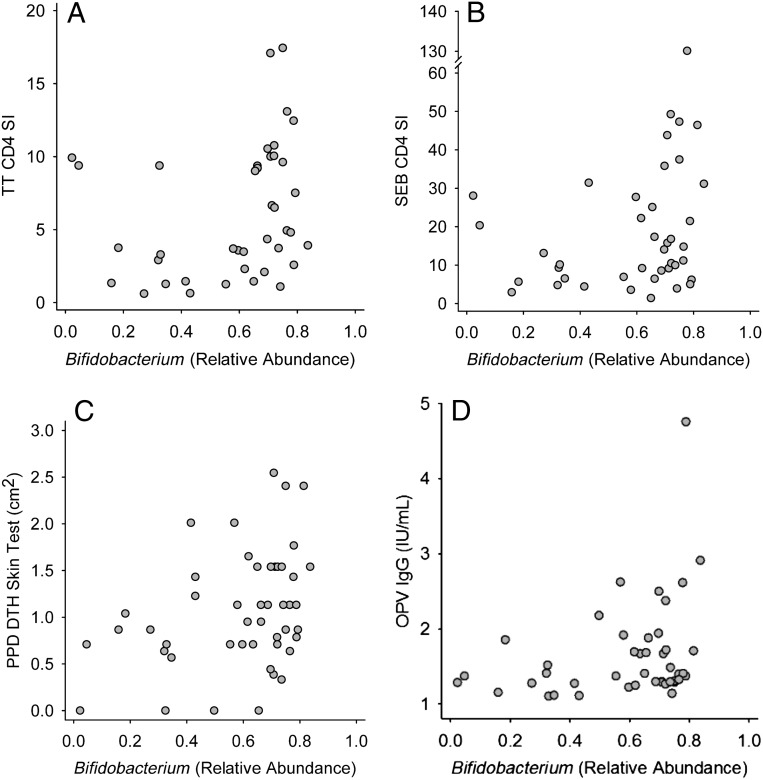
Vaccine responses are characterized in the Supplemental Information (including Supplemental Fig 9). Both OPV-specific T-cell proliferation (Supplemental Figs 10–12) and OPV-specific IgG responses (Supplemental Figs 13 and 14) were positively associated with members of the phylum Actinobacteria. For example, infants with a “high” CD4 T-cell response to OPV vaccination (ie, those above the median response) had a significantly greater relative abundance of Actinobacteria in 15-week stool samples than did infants with a “low” (ie, below the median) OPV-CD4 response (Fig 2A). At the family level, high OPV-CD4 responders also had a higher (P = .024) median relative abundance of Coriobacteriaceae (1.76 × 10−4 [1.77 × 10−2]) than did low OPV-CD4 responders (0.00 [6.18×10−5]). By using Bifidobacterium-specific PCR, high OPV-CD4 responders were found to have higher B longum subspecies infantis levels than low responders (Fig 3A) and high OPV-CD8 responders had higher Bifidobacterium, B longum, and B longum subspecies longum levels than did low OPV-CD8 responders (Fig 3B). A significant, positive correlation was also seen between Bifidobacterium relative abundance (assessed by 16S rRNA gene sequencing) and the OPV-IgG response (Fig 4D).
FIGURE 2.
Positive association of vaccine responses with stool Actinobacteria at 15 weeks of age. The relative abundance of stool microbiota was determined by sequence analysis of the 16S rRNA gene. Vaccine responses were characterized by the CD4 T-cell SI in response to vaccine antigens for OPV vaccine (A), BCG vaccine using PPD antigen (B), TT vaccine (C), HBV vaccine (D), and SEB (E), the positive control for T-cell stimulation. Box plots (A–D) show the median; 10th, 25th, 75th, and 90th percentiles; and individual outliers. P values (Wilcoxon rank-sum test) for comparisons between groups were as follows: A, .046; B, .036; C, .019; and D, 0.78. E, The scatterplot association was determined by Spearman correlation (R): R = 0.331, P = .034.
FIGURE 3.
Positive association of vaccine responses with stool Bifidobacterium, B longum, B longum subspecies infantis, and B longum subspecies longum at 15 weeks of age. Infant vaccine responses were characterized as being above (“high”) or below (“low”) the median vaccine responses for the CD4 SI for OPV (A), the CD8 SI for OPV (B), the CD4 SI for PPD as an index of response to the BCG vaccine (C), and for the positive control for polyclonal T-cell proliferation, the CD4 SI for SEB (D). Differences between those above and below the median were identified by the Wilcoxon rank-sum test as indicated by the superscript letters: aP < .10, bP < .05, cP < .01. The Bifidobacterium genus was identified by PCR, B longum species by T-RFLP, and B longum subspecies infantis and B longum subspecies longum as described in Methods.
FIGURE 4.
Positive association of vaccine responses with relative abundance of stool Bifidobacterium determined by sequence analysis of the 16S rRNA gene. Vaccine responses were characterized by CD4 stimulation index response to TT (A), CD4 stimulation index in response to the polyclonal T-cell mitogen SEB (B), the DTH skin-test response to PPD antigen as an index of the response to BCG immunization (C), and the IgG response to the OPV vaccine (D). Spearman correlation coefficients (R) and P values were as follows: A, R = 0.389, P = .013; B, R = 0.315, P = .045; C, R = 0.337, P = .020; and D, R = 0.301, P = .047.
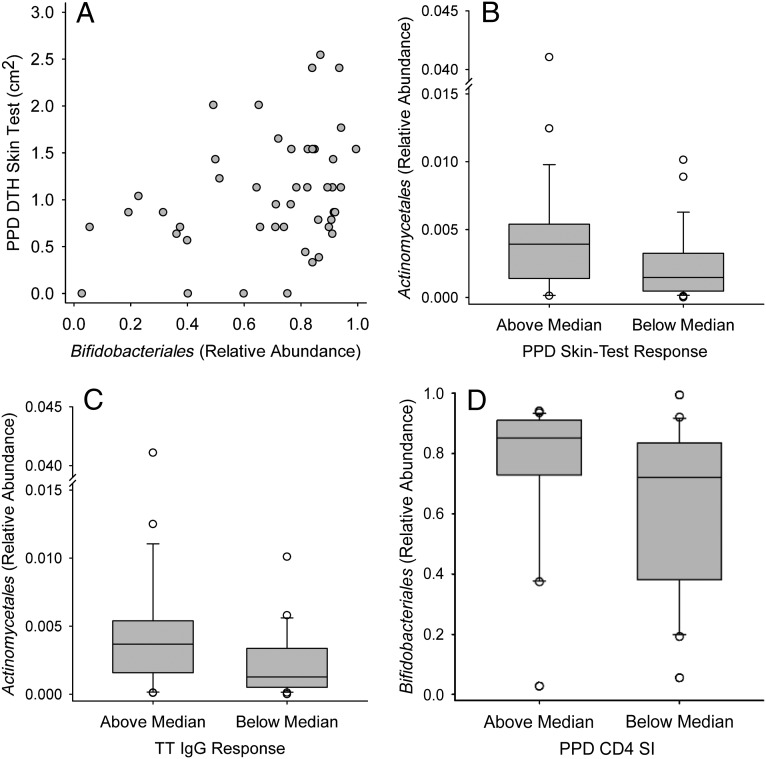
Positive associations were also seen between Actinobacteria, including Bifidobacterium, and BCG vaccine responses, including CD4 T-cell proliferation to PPD antigen and the skin-test response to PPD. High PPD-CD4 vaccine responders had significantly higher Actinobacteria (Fig 2B) and Bifidobacteriales (Fig 5D) relative abundance in stool than did low responders, and a similar association of marginal statistical significance (P = .055) was also seen for Bifidobacterium, with the relative abundance in high and low responders being 0.720 (0.128) and 0.619 (0.350), respectively. By using Bifidobacterium-specific PCR, B longum subspecies infantis levels were found to be higher in high versus low PPD-CD4 responders (Fig 3C). The PPD-DTH skin-test response also revealed a marginally significant positive correlation with Actinobacteria relative abundance (R = 0.284, P = .053) as well as significant, positive associations with Bifidobacteriales (Fig 5A) and Bifidobacterium (Fig 4C). Positive associations were also seen between the PPD-DTH response and another Actinobacteria order, Actinomycetales (Fig 5B), and genus, Rothia (high PPD-DTH responders: 2.48 × 10−3 [2.55 × 10−3]; low responders: 9.14 × 10−4 [1.19 × 10−3]; P = .034).
FIGURE 5.
Positive association of vaccine responses with stool Bifidobacteriales, Actinomycetales, and Coriobacteriales (from the class Actinobacteria) at 15 weeks of age. The relative abundance of stool microbiota at the order level in the class Actinobacteria was determined by sequence analysis of the 16S rRNA gene at 15 weeks of age. A and B, The response to BCG vaccination was determined by using the DTH skin-test response to PPD antigen. C, The response to TT vaccination was determined by using the antibody in lymphocyte supernatant assay to measure TT-specific IgG levels (TT IgG). D, The CD4 T-cell SI in response to PPD antigen. A, The scatterplot association was determined by Spearman correlation (R): R = 0.333, P = .025. Box plots (B, C, and D) show the median; 10th, 25th, 75th, and 90th percentiles; and individual outliers. P values (Wilcoxon rank-sum test) for comparisons between groups were as follows: B, .018; C, .041; and D, .034.
The TT-CD4 response was positively associated with Actinobacteria from the phylum (Fig 2C) through the genus level, including both Bifidobacterium (Fig 4A) and Corynebacterium (R = 0.325, P = .041; n = 40). Bifidobacterium-specific PCR also showed a positive correlation between the TT-CD4 response and B longum subspecies infantis (R = 0.319, P = .048; n = 39). Although the TT-IgG response was not associated with Bifidobacterium, it was positively associated with the Actinobacteria order Actinomycetales (Fig 5C). Thus, the response to TT immunization showed a consistent, positive association of Actinobacteria with both T-cell and IgG responses.
When the HBV-CD4 responses were evaluated, positive associations were not seen with Actinobacteria (Fig 2D, Supplemental Fig 10) or with other stool microbiota, although a positive association was seen between the HBV-IgG response and the Actinobacteria genus Rothia (R = 0.406, P = .029; n = 29).
Three of the four vaccines (OPV, BCG, and TT) showed positive associations between vaccine-specific T-cell proliferative and the relative abundance of taxa from the phylum Actinobacteria, particularly the genus Bifidobacterium. This positive association was also seen for the Staphylococcus enterotoxin B (SEB)–stimulated cultures, which were included as a positive control for polyclonal T-cell proliferation. Positive associations were seen from the phylum (Fig 2E) through the Bifidobacterium genus (Fig 4B) level and, using Bifidobacterium-specific PCR, again at the genus level but also at the species and subspecies level for B longum and B longum subspecies infantis (Fig 3D). Thus, Bifidobacterium relative abundance is also associated with polyclonal T-cell proliferation, in addition to vaccine-specific responses.
Gammaproteobacteria, Bacterial Diversity and Vaccine Responses
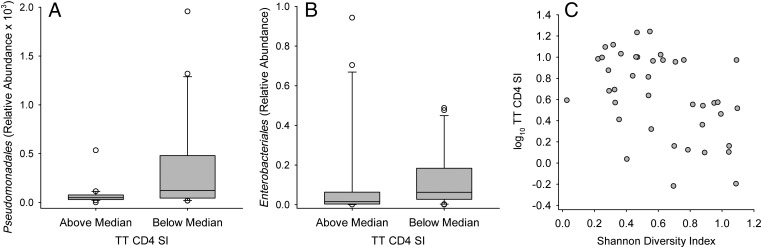
Several taxa of Gammaproteobacteria had negative associations with vaccine responses. Pseudomonadales and its members showed negative associations with T-cell SI responses for TT (Fig 6A), OPV, PPD, HBV, and SEB as well as a negative association with the OPV-IgG response (Supplemental Figs 10 and 13), whereas the order Enterobacteriales showed a negative association with the TT-CD4 SI response (Fig 6B) as well as with the PPD-DTH skin-test response (Supplemental Fig 13). Similar but less consistent associations (across phylogenetic levels) were seen with Firmicutes and Bacteroidetes (Supplemental Figs 10 and 13). High diversity was also negatively associated with the CD4 TT SI response (Fig 6C) and with the CD4-SEB response (R = −0.361, P = .021).
FIGURE 6.
Negative association of stool Pseudomonadales (A), Enterobacteriales (B), and Shannon Diversity Index (measured at the phylum level) (C) with CD4 T-cell SI response to TT vaccine at 15 weeks of age. The relative abundance of stool microbiota was determined by sequence analysis of the 16S rRNA gene. Box plots show the median; 10th, 25th, 75th, and 90th percentiles; and individual outliers. P values (Wilcoxon rank-sum test) for comparisons between groups were as follows: A, 0.022; B, 0.047. C, The Spearman correlation coefficient was R = −0.490 with P = .0013.
Possible Mediators of Microbiota Associations With Vaccine Response
Gut microbiota may affect vaccine responses indirectly by affecting development of T cells. To examine this possibility, TI and peripheral blood T-cell concentrations were correlated with vaccine responses; positive associations were found between TI and OPV-CD4 (R = 0.377, P = .015), OPV-CD8 (R = 0.336, P = .032), and SEB-CD4 (R = 0.396, P = .010), but no associations were found with T-cell concentrations. In addition, systemic inflammation caused by enteric bacteria might disrupt a vaccine response. Peripheral blood neutrophil, eosinophil, and monocyte levels were used as indicators of such inflammation and correlated with vaccine responses: neutrophils correlated negatively with PPD-CD8 (R = −0.334, P = .023) and TT-CD4 (R = −0.358, P = .028). Given these results, we searched for associations between TI, neutrophil levels, and microbiota taxa associated with vaccine responses.
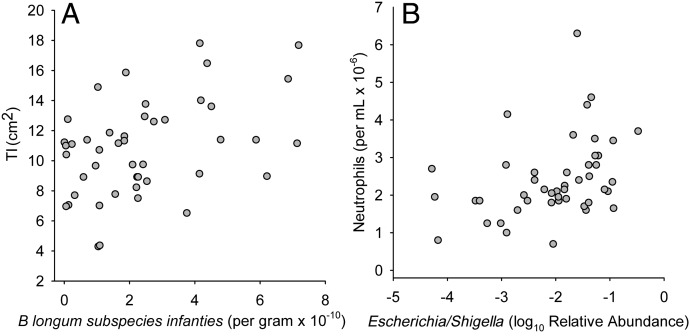
Among taxa that were positively associated with vaccine responses, positive associations were also found with TI but no associations were found with neutrophils. Specifically, the relative abundance of Bifidobacterium (from 16S rRNA gene sequencing) was positively correlated with TI (R = 0.293, P = .046). In addition, stool concentrations of Bifidobacterium (R = 0.388, P = .0070), B longum (R = 0.394, P = .0075), and B longum subspecies infantis (Fig 7A) were positively associated with TI, and these associations were shown to be independent of length-for-age (Supplemental Table 11), which also correlated with TI.
FIGURE 7.
Correlation of Bifidobacterium longum subspecies infantis in stool determined by qPCR with TI (A) and relative abundance of Escherichia/Shigella genera in stool determined by sequence analysis of the 16S rRNA gene with blood neutrophil levels (B). Spearman correlation coefficients (R) and P values were as follows: A, R = 0.389, P = .0083; B, R = 0.431, P = .0028.
Among taxa that were negatively associated with vaccine responses, only 1 association was seen with TI (a negative association of Lactococcus: R = −0.389, P = .006), whereas multiple positive associations were seen with neutrophils in phylum Firmicutes, including Clostridia and Clostridiales (R = 0.418 and P = .0038 for both), and phylum Proteobacteria, including Proteobacteria (R = 0.389, P = .008), Gammaproteobacteria (R = 0.346, P = .019), Enterobacteriales (R = 0.350, P = .017), Enterobacteriaceae (R = 0.350, P = .017), and Escherichia/Shigella (Fig 7B). Bacterial diversity (measured as the Shannon Diversity Index at the phylum level) was also positively correlated with neutrophils (R = 0.293, P = .048).
Discussion
A principal finding of our study was that a high relative abundance of Actinobacteria, particularly of Bifidobacterium, was positively associated with several measures of both oral and parenteral vaccine responses, including T-cell proliferative responses to OPV, PPD, and TT; the PPD-DTH skin-test response; and vaccine-specific IgG responses. At the genus level, Actinomyces, Corynebacterium, Rothia, and Bifidobacterium all showed positive associations with at least 1 of these responses. B longum was the most abundant species in infant stool and B longum subspecies infantis was its predominant subspecies and was positively associated with CD4 SI responses to OPV, PPD, and TT, whereas the subspecies longum was positively associated with the CD8 SI response to OPV. It is well established that gut microbiota can affect mucosal immune function, as discussed below, but the present observation of an equally strong association of stool microbiota with response to parenteral vaccination is, we believe, novel.
The importance of intestinal microbiota for the development of the mammalian immune system has been well documented in animal models.12,25 The absence of flora in germ-free mice affects both local12,25–27 and systemic12,25 lymphoid compartments, whereas colonization with specific gut bacteria such as segmented filamentous bacteria,28 Clostridium,29 and bacteria that produce short-chain fatty acids30 (which include Bifidobacterium) can affect development of CD4 T-cell subsets. In humans, the hygiene hypothesis suggests that quantitative or qualitative “deficiencies” in microbial exposure in “hygienic” environments increase subsequent risk of atopic diseases including asthma,31 whereas the administration of probiotic bacteria is associated with a decreased risk of atopic eczema.32 Perhaps most compelling, administration of Bifidobacterium as a probiotic has increased responses to oral vaccines in both animal and human studies,33 including OPV-specific IgA.34 Relatively few human studies have examined the relationship of naturally occurring gut bacteria to immune function. Bifidobacteria have been associated with development of IgA- and IgM-secreting plasma cells,35 memory B cells,36 and higher salivary IgA.37 Overall, these data are consistent with the present findings that high Bifidobacterium abundance is positively associated with response to immunization.
All of the vaccines used in this study require T-cell help, and the finding that B longum subspecies infantis was positively associated with both thymus size (as measured by TI) and with a generic measure of peripheral T-cell function (the SI response to SEB) suggests a direct effect of Bifidobacterium on the T-cell compartment at sites distant from the intestine. If this is true then 3 mechanisms seem plausible: soluble or cell-associated bacterial metabolites or other molecules could act at a distance on the thymus and other lymphoid tissue via the lymph and blood, they could act locally in the gut to trigger neuroendocrine stimuli that affect distant lymphoid tissue, or cells of the immune system that circulate through systemic and intestinal lymphoid tissue (as do naive T-cells) could be exposed to bacterial products in intestinal lymph nodes and carry this “experience” to systemic sites where they may then respond differently as a result. Mechanistic studies are needed to evaluate these possibilities.
Although bacterial diversity of the gut microbiota is thought to be a benefit in promoting health of the adult host,38 diversity in the current study in breastfed infants at 15 weeks was associated with higher neutrophilia and a lower response to the TT vaccine and may be a manifestation of dysbiosis. Breastfeeding provides optimal nutrition and immunologic protection through the first 6 months of life39 and human milk oligosaccharides selectively stimulate growth of Bifidobacterium,40 perhaps acting to limit diversity. Thus, Bifidobacterium predominance with low diversity may be optimal for infant health early in infancy, whereas high diversity may be beneficial later in life when the diet is also more diverse.
Dysbiosis of the gut microbiome causes local inflammation.2 The current study did not examine local inflammation but found that high levels of Clostridiales, Enterobacteriales, and Pseudomonadales, bacterial orders that contain enteric pathogens, were associated with neutrophilia and lower vaccine responses, suggesting that the presence of such organisms in healthy infants may cause systemic inflammation and immune suppression. Consistent with this hypothesis, neutrophil development in the bone marrow is driven by interleukin-17A derived from T-helper 17 cells,41 which could develop in response to such bacteria. In addition, Bifidobacterium can directly protect against enteropathogenic E coli infection in mice,42 data that are consistent with the idea presented here that Bifidobacterium predominance early in infancy promotes health by limiting “diversity” resulting from the presence of enteric pathogens.
Extrapolation of these results to the Bangladeshi population is limited by several factors, as seen by reference to countrywide data.43 First, the study was in an urban population, whereas 79% of Bangladeshi women live in rural areas. Second, 71% of mothers had completed some secondary education, higher than the 35% rate for Bangladesh. Third, study infants were delivered in the hospital; whereas only 15% of births in 2007 were in medical facilities, 31% of births in urban areas were in facilities. The rate of cesarean deliveries was high in this study, but the 2007 national rate was 7.5%, accounting for half of all facility-based deliveries. Families in this study were not affluent because most did not have flush toilets and a quarter did not have drinking water piped into their residences. Data from infants delivered at home and in rural areas would help make this study more representative.
Conclusions
We report the novel observations that a high abundance of Actinobacteria, particularly Bifidobacterium, in early infancy is associated with a higher TI, greater vaccine-specific and polyclonal T-cell proliferation, and a greater PPD-DTH skin-test response. Although this study is observational and not designed to assess causality, the data do suggest that Bifidobacterium colonization enhances both oral and systemic vaccine responses by supporting T-cell immunity. Other Actinobacteria were positively associated with the TT-specific IgG response. This study raises the possibility that supporting an “optimal” microbial community early in infancy (eg, by supporting breastfeeding) might enhance vaccine responses. Alternatively, a strategy has recently been suggested that could involve using probiotic microbes to enhance vaccine responses.44
Supplementary Material
Glossary
- BCG
bacille Calmette-Guérin
- DTH
delayed-type hypersensitivity
- HBV
hepatitis B virus
- Ig
immunoglobulin
- OPV
oral polio virus
- PCR
polymerase chain reaction
- PPD
purified protein derivative of Mycobacterium bovis
- qPCR
quantitative polymerase chain reaction
- rRNA
ribosomal RNA
- SEB
Staphylococcus enterotoxin B
- SI
stimulation index
- T-RFLP
terminal restriction fragment length polymorphism
- TI
thymus index
- TT
tetanus toxoid
Footnotes
Dr Stephensen, working with coauthors, conceptualized and implemented the major hypothesis of the study (evaluating the association of microbiota with immune function), conceptualized and implemented the work to evaluate immune function, conducted statistical analysis, and wrote the first draft of the manuscript; Dr Mills worked to conceptualize, design, and implement the major hypothesis of the study (evaluating the association of microbiota with immune function); supervised acquisition of microbiota data; and participated in analysis and interpretation of the overall data set; Dr Underwood worked to conceptualize, design, and implement the major hypothesis of the study (evaluating the association of microbiota with immune function) and participated in analysis and interpretation of the overall data set; Drs Ahmad, Raqib, and Qadri and Mr Rashid contributed substantially to acquisition of immunologic data and participated in the analysis and interpretation of the overall data set; Mr Huda carried out laboratory studies to analyze microbiota and to characterize immune function, conducted bioinformatic analysis of microbiota data, conducted statistical analysis, and collaborated in writing the first draft of the manuscript; Mr Lewis and Dr Kalanetra carried out laboratory and bioinformatic analysis of stool microbiota data and contributed to the first draft of the manuscript; all authors participated in drafting the manuscript and/or revising it critically for important intellectual content, had final approval authority for the version to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
This trial has been registered at www.clinicaltrials.gov (identifier NCT01583972).
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: This work was funded by grant UL1 RR024146 from the National Center for Research Resources, US Department of Agriculture–Agricultural Research Service project 5306-51530-018-00, and World Health Organization project 2010168947, which was funded by the Bill and Melinda Gates Foundation. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013;21(4):167–173 [DOI] [PubMed] [Google Scholar]
- 2.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330(6012):1768–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torrazza RM, Ukhanova M, Wang X, et al. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PLoS ONE. 2013;8(12):e83304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisgaard H, Li N, Bonnelykke K, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128(3):646–652, e641–e645 [DOI] [PubMed]
- 5.Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012;129(5):950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isolauri E. Development of healthy gut microbiota early in life. J Paediatr Child Health. 2012;48(suppl 3):1–6 [DOI] [PubMed] [Google Scholar]
- 7.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okwo-Bele JM, Cherian T. The expanded programme on immunization: a lasting legacy of smallpox eradication. Vaccine. 2011;29(suppl 4):D74–D79 [DOI] [PubMed] [Google Scholar]
- 10.Qadri F, Bhuiyan TR, Sack DA, Svennerholm AM. Immune responses and protection in children in developing countries induced by oral vaccines. Vaccine. 2013;31(3):452–460 [DOI] [PubMed] [Google Scholar]
- 11.Kuss SK, Best GT, Etheredge CA, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334(6053):249–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vrese M, Rautenberg P, Laue C, Koopmans M, Herremans T, Schrezenmeir J. Probiotic bacteria stimulate virus-specific neutralizing antibodies following a booster polio vaccination. Eur J Nutr. 2005;44(7):406–413 [DOI] [PubMed] [Google Scholar]
- 14.Brisbin JT, Gong J, Orouji S, et al. Oral treatment of chickens with lactobacilli influences elicitation of immune responses. Clin Vaccine Immunol. 2011;18(9):1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maidens C, Childs C, Przemska A, Dayel IB, Yaqoob P. Modulation of vaccine response by concomitant probiotic administration. Br J Clin Pharmacol. 2013;75(3):663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda F, Chowdhury MI, Saha A, et al. Evaluation of a probiotics, Bifidobacterium breve BBG-01, for enhancement of immunogenicity of an oral inactivated cholera vaccine and safety: a randomized, double-blind, placebo-controlled trial in Bangladeshi children under 5 years of age. Vaccine. 2011;29(10):1855–1858 [DOI] [PubMed] [Google Scholar]
- 17.Fraher MH, O’Toole PW, Quigley EM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol. 2012;9(6):312–322 [DOI] [PubMed] [Google Scholar]
- 18.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(suppl 1):13–15 [DOI] [PubMed] [Google Scholar]
- 19.Karlsson CL, Molin G, Cilio CM, Ahrné S. The pioneer gut microbiota in human neonates vaginally born at term—a pilot study. Pediatr Res. 2011;70(3):282–286 [DOI] [PubMed] [Google Scholar]
- 20.Tsuji H, Oozeer R, Matsuda K, et al. Molecular monitoring of the development of intestinal microbiota in Japanese infants. Benef Microbes. 2012;3(2):113–125 [DOI] [PubMed] [Google Scholar]
- 21.Echarri PP, Graciá CM, Berruezo GR, et al. Assessment of intestinal microbiota of full-term breast-fed infants from two different geographical locations. Early Hum Dev. 2011;87(7):511–513 [DOI] [PubMed] [Google Scholar]
- 22.Fallani M, Young D, Scott J, et al. Members of the INFABIO Team . Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51(1):77–84 [DOI] [PubMed] [Google Scholar]
- 23.Savino W, Dardenne M. Nutritional imbalances and infections affect the thymus: consequences on T-cell-mediated immune responses. Proc Nutr Soc. 2010;69(4):636–643 [DOI] [PubMed] [Google Scholar]
- 24.Lewis ZT, Bokulich NA, Kalanetra KM, Ruiz-Moyano S, Underwood MA, Mills DA. Use of bifidobacterial specific terminal restriction fragment length polymorphisms to complement next generation sequence profiling of infant gut communities. Anaerobe. 2013;19:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335 [DOI] [PubMed] [Google Scholar]
- 26.Chung H, Pamp SJ, Hill JA, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69 [DOI] [PubMed] [Google Scholar]
- 28.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savino W, Dardenne M, Velloso LA, Dayse Silva-Barbosa S. The thymus is a common target in malnutrition and infection. Br J Nutr. 2007;98(suppl 1):S11–S16 [DOI] [PubMed] [Google Scholar]
- 31.Frei R, Lauener RP, Crameri R, O’Mahony L. Microbiota and dietary interactions: an update to the hygiene hypothesis? Allergy. 2012;67(4):451–461 [DOI] [PubMed] [Google Scholar]
- 32.Kuitunen M. Probiotics and prebiotics in preventing food allergy and eczema. Curr Opin Allergy Clin Immunol. 2013;13(3):280–286 [DOI] [PubMed] [Google Scholar]
- 33.Licciardi PV, Tang ML. Vaccine adjuvant properties of probiotic bacteria. Discov Med. 2011;12(67):525–533 [PubMed] [Google Scholar]
- 34.Holscher HD, Czerkies LA, Cekola P, et al. Bifidobacterium lactis Bb12 enhances intestinal antibody response in formula-fed infants: a randomized, double-blind, controlled trial. JPEN J Parenter Enteral Nutr. 2012;36(1 suppl):106S–117S [DOI] [PubMed] [Google Scholar]
- 35.Grönlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Arch Dis Child Fetal Neonatal Ed. 2000;83(3):F186–F192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundell AC, Björnsson V, Ljung A, et al. Infant B cell memory differentiation and early gut bacterial colonization. J Immunol. 2012;188(9):4315–4322 [DOI] [PubMed] [Google Scholar]
- 37.Sjögren YM, Tomicic S, Lundberg A, et al. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy. 2009;39(12):1842–1851 [DOI] [PubMed] [Google Scholar]
- 38.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mussap M, Noto A, Cibecchini F, Fanos V. The importance of biomarkers in neonatology. Semin Fetal Neonatal Med. 2013;18(1):56–64 [DOI] [PubMed] [Google Scholar]
- 40.German JB, Freeman SL, Lebrilla CB, Mills DA. Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:205–218; discussion 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krstic A, Mojsilovic S, Jovcic G, Bugarski D. The potential of interleukin-17 to mediate hematopoietic response. Immunol Res. 2012;52(1–2):34–41 [DOI] [PubMed] [Google Scholar]
- 42.Jiang W, Lederman MM, Hunt P, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199(8):1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamal SM. Preference for institutional delivery and caesarean sections in Bangladesh. J Health Popul Nutr. 2013;31(1):96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moingeon P. Adjuvants for allergy vaccines. Hum Vaccin Immunother. 2012;8(10):1492–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.