Abstract
Nanomechanical motion of bacteria adhered to a chemically functionalized silicon surface is studied by means of a microcantilever. A non-specific binding agent is used to attach Escherichia coli (E. coli) to the surface of a silicon microcantilever. The microcantilever is kept in a liquid medium, and its nanomechanical fluctuations are monitored using an optical displacement transducer. The motion of the bacteria couples efficiently to the microcantilever well below its resonance frequency, causing a measurable increase in the microcantilever fluctuations. In the time domain, the fluctuations exhibit large-amplitude low-frequency oscillations. In corresponding frequency-domain measurements, it is observed that the mechanical energy is focused at low frequencies with a 1/fα-type power law. A basic physical model is used for explaining the observed spectral distribution of the mechanical energy. These results lay the groundwork for understanding the motion of microorganisms adhered to surfaces and for developing micromechanical sensors for bacteria.
Biological function in motile microorganisms is intimately coupled to mechanical motion,1–4 which is generated and sustained by a vast array of forces. Conversely, motile microorganisms exert forces on their surroundings as a result of their incessant motion and metabolism.5 A deeper physical understanding of biological processes in terms of these nanomechanical forces and motions is of value for both fundamental science and clinical medicine.6 For instance, bacterial communication through motion is believed to be an important factor for infections and for bacterial resistance to antibiotics.7,8,37 Consequently, the development of sensitive force and motion probes,9 which can match the time and length scales of microorganisms, has been an important element in the study of motile microorganisms.
The microcantilever has been one of the mainstays for probing the forces and motions of biological entities.10,11 Early microcantilever-based experiments were extensions of Atomic Force Microscopy (AFM). For instance, the AFM tip scanned over biomolecules adsorbed on a surface provided information on the conformational changes of the biomolecules.12,13 Similarly, nanomechanical motion and forces of microorganisms, such as Saccharomyces cerevisiae1 and bacteria, were investigated using contact-mode or dynamic-mode AFM under a number of different biological conditions. More recently, biological entities have been attached to bio-chemically functionalized microcantilevers in an effort to develop novel sensors. These micromechanical biosensors, which are typically integrated with microfluidics, utilize both the static deflection and the resonance frequency shift of the cantilever.14,15 In static and resonant sensing modalities, microcantilevers have allowed for the sensing of diverse biological entities, ranging from DNA to proteins to microorganisms16–18 (e.g., viruses and bacteria). In more recent work, Longo et al.19,20 adhered bacteria to a microcantilever, and measured the nanomechanical fluctuations of the cantilever before and after bacteria adhesion. With the bacteria present on the cantilever, the fluctuations increased significantly. Based on these results, a rapid detection scheme for bacterial antibiotic resistance was proposed.
The above-mentioned studies clearly demonstrate that small mechanical devices are capable of measuring signals from biological entities, given the attainable force sensitivities and response times. In this letter, we apply the microcantilever-based technique developed by Longo et al.19,20 to measurements of the nature (e.g., time scales and amplitudes) of the forces that bacteria exert on the microcantilever. We start with time domain measurements, in which we observe an increase in the variance of the microcantilever fluctuations due to bacterial motion. In complementary frequency domain measurements, we elucidate the spectral properties of the microcantilever fluctuations. The power of the fluctuations scale with frequency as 1/fα as well as with the surface density of the bacteria. We provide a basic physical model for the observed spectral distribution of the mechanical energy, and discuss implications of these results on biology and biosensor development.
We perform all the experiments in a custom liquid chamber (Fig. 1(a)); the chamber allows for optical access to the microcantilever. The optical beam deflection technique21,22 is used to measure the fluctuations of the microcantilever, which has linear dimensions l × w × t = 350 × 32.5 × 1 μm3. The voltage signal from the photodetector is converted to displacement units by using the thermal calibration technique23 (see thermal spectrum inset of Fig. 3(a)). The microcantilever fluctuations are measured in the time domain with a digital storage oscilloscope and in the frequency domain with a FFT spectrum analyzer.
FIG. 1.
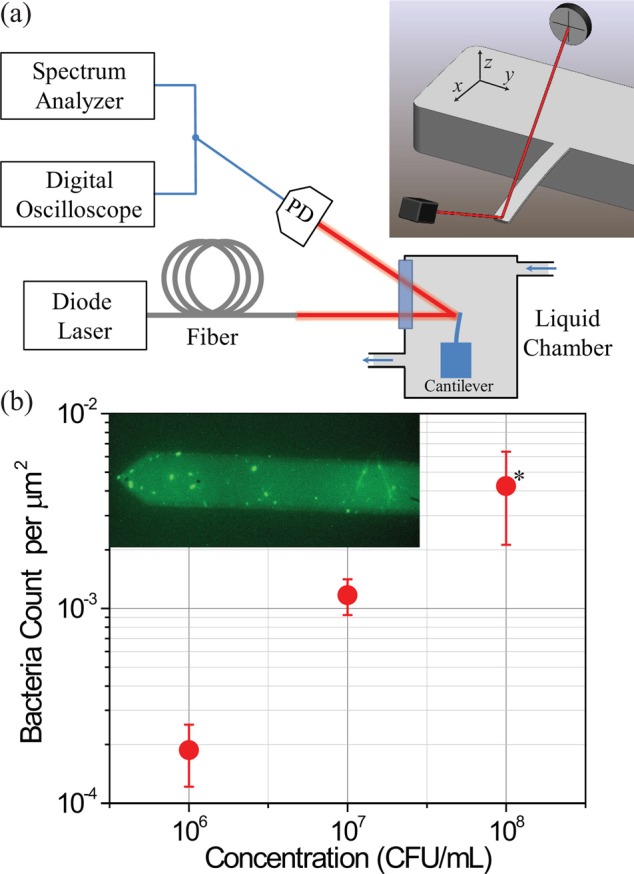
(a) Schematic of our experimental setup. The cantilever is housed in a liquid chamber; its motion is probed using an optical technique. PD: photodetector. The inset shows the cantilever, the optical beam, and the split photodetector. (b) Surface density of bacteria adhered to the surface of silicon for three different bacterial concentrations. Data are indicated as averages ± standard deviations. * represents statistical significance of this data point (see supplementary material,24 Sec. 2.1). The inset is a fluorescence image of the cantilever after bacterial adhesion. Both single bacterium and clumps of bacteria are visible.
FIG. 3.
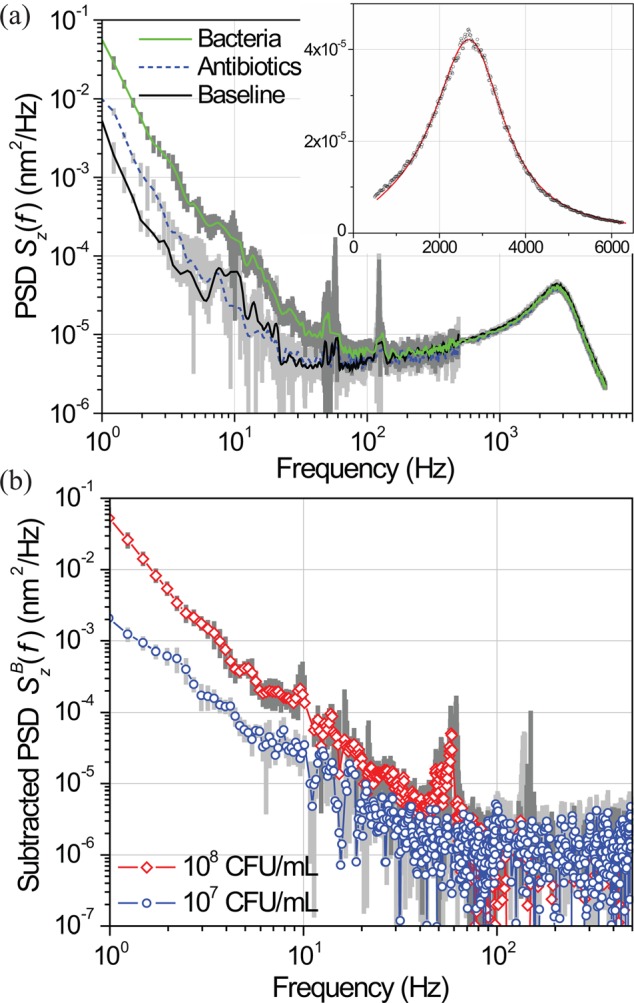
PSD of microcantilever fluctuations as a function of frequency in a double-logarithmic plot. Single standard deviations are shown in gray behind the data traces. The cantilever thermal resonance is at fR ≈ 2.5 kHz with Q ≈ 1.5. The inset shows this thermal peak in PBS in a linear plot. The data are fitted to a damped harmonic oscillator model driven by fluid fluctuations. (b) Subtracted PSD of the microcantilever fluctuations as a function of frequency. The PSD of the baseline is subtracted from that measured after bacterial adhesion.
In the experiments, we used genetically modified E. coli which were transfected with a plasmid consisting of ampicillin resistance and green fluorescence protein (for more details, see supplementary material,24 Sec. 1.1). After incubation, we serially diluted the bacteria to obtain solutions of concentrations C = 106, 107, and 108 colony forming units per milliliter (CFU/ml) in phosphate-buffered saline (PBS). To prepare the microcantilever surface for adhesion of bacteria, we first cleaned it with acetone, methanol, and isopropanol and dried in nitrogen gas. The cantilever was then submerged in a reservoir containing 1% (3-aminopropyl)triethoxysilane (APTES) dissolved in molecular biology grade water for 15 min, and was rinsed with water afterward. The APTES solution provided functional amine groups for bacterial attachment. Next, the cantilever was submerged in the E. coli solutions of various concentrations and incubated for 15 min at room temperature. During incubation, the bacteria adhered to the cantilever surface.
We confirmed bacterial adhesion by immediately imaging the cantilever surface with a microscope in fluorescence and in bright-field modes (inset of Fig. 1(b)). To quantify the number of bacteria adhered onto the surface, we performed a separate study, in which we adhered bacteria to large pieces of silicon using the above-described steps and imaged in bright-field. We subsequently used these images to obtain the average surface density of bacteria for all bacterial concentrations tested. We plot the surface density of the bacteria as a function of the bacterial concentration in solution in Fig. 1(b). The error bars are due to the finite number of images analyzed (5 images of 300 × 300 μm2 for each concentration), clumping of bacteria, and the occasional non-uniformities in coverage. We performed statistical analysis on these data to confirm statistical significance (see supplementary material,24 Sec. 2.1).
A typical experiment began by measuring the fluctuations of the cantilever in PBS before bacterial adhesion. This determined the baseline for the fluctuations of the cantilever. After the collection of the baseline data, we proceeded with the bacterial adhesion process, allowed time for incubation, and flushed the liquid chamber with PBS. We then repeated our measurement. Finally, we introduced to the liquid chamber a 1 mg/ml solution of streptomycin dissolved in water, allowed time for incubation, flushed the chamber with PBS, and repeated the measurement once more.
Figure 2(a) depicts a typical time-domain measurement of the mechanical fluctuations of the tip of the microcantilever, z(t), under different conditions (for more details, see supplementary material,24 Sec. 1.2). The top (black) trace shows the fluctuations measured in PBS before bacterial adhesion. The middle (green) trace shows that the fluctuations after bacteria have been adhered to the cantilever as described above. Finally, the bottom (blue) trace shows the fluctuations after administration of the antibiotic streptomycin. The probability density function (PDF) of the three time signals is shown in Fig. 2(b) to demonstrate the Gaussian nature of the fluctuations. For these particular measurements, a concentration of C = 108 CFU/ml of bacteria was used, and we estimate, using the data in Fig. 1(b), that there were N ∼ 102 bacteria on the microcantilever. Qualitatively, there are differences between the middle trace with bacteria and the other two traces; the data taken after bacterial adhesion exhibit large-amplitude low-frequency fluctuations, which are not present in the no-bacteria case. The different characteristic frequencies in these data traces provided the motivation for measuring the spectrum of the fluctuations.
FIG. 2.
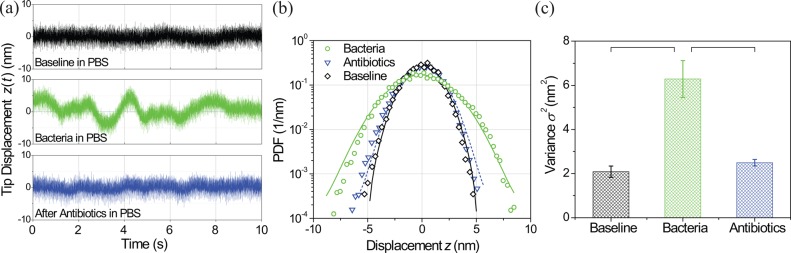
(a) Time domain measurement of the microcantilever fluctuations. The top (black) trace is the baseline cantilever fluctuations with no bacteria present in PBS, i.e., the equilibrium value. The middle (green) trace is the same measurement after bacteria have adhered to the cantilever surface in a 108 CFU/ml solution. The data in Fig. 1(b) and the cantilever area suggest that the signal comes from ∼102 bacteria on the microcantilever. The bottom (blue) trace is after the bacteria are killed in an antibiotic solution. (b) Probability density function of the three time signals shown in (a) with Gaussian fits. (c) Variance of the microcantilever fluctuations measured from the three data traces in (a). These values match those obtained from the Gaussian fits in (b). Data are indicated as averages ± standard deviations. Brackets represent groups which were compared in the statistical analysis (details in supplementary material,24 Sec. 2.2).
For each experiment, we repeated these measurements three times, waiting for ∼1/2 h between measurements, and computed the variance of the fluctuating time signal as , where is the mean (typically ). In Table I, we present these average variances from the cantilevers that were incubated in bacteria solutions with concentrations C = 107 CFU/ml and C = 108 CFU/ml, corresponding to N ∼ 20 and N ∼ 102 bacteria, respectively. The sample prepared using the lowest bacteria concentration (C = 106 CFU/ml) with N ∼ 4 was below the uncertainty of the experimental measurement. In Table I, and correspond to the variances for the baseline and bacteria experiments, respectively. We also subtracted from to obtain a quantitative measure of the additional cantilever fluctuations induced by the bacterial motion. The higher bacterial concentration case exhibits a larger increase in variance than the lower concentration case. The average variances for the C = 108 CFU/ml experiment are also depicted graphically in Fig. 2(c). These values match those found from the Gaussian fits of the PDFs in Fig. 2(b). More details regarding the statistical analysis of these data can be found in supplementary material24 Sec. 2.
TABLE I.
Variances of the displacement fluctuations. The number of bacteria N on the microcantilever was estimated from the concentration C using the data in Fig. 1(b). The variances for the baseline and bacteria experiments are and , respectively; quantifies the additional cantilever fluctuations induced by the bacterial motion.
| C (CFU/ml) | N | (nm2) | (nm2) | (nm2) |
|---|---|---|---|---|
| 107 | 27 | 2.01 | 3.39 | 1.38 |
| 108 | 96 | 2.08 | 6.29 | 4.21 |
Figure 3(a) depicts the power spectral density (PSD) Sz(f) of the fluctuations of the microcantilever tip in a double-logarithmic plot under three different conditions: The black trace (lower, solid) is obtained in PBS before bacterial adhesion; the green trace (upper, solid) is obtained after the adhesion of bacteria in the C = 108 CFU/ml bacteria solution; the blue line (lower, dashed) is obtained after administration of streptomycin. The PSD increases after bacterial adhesion, but only in the frequency range f ≲ 100 Hz. After incubation in antibiotics, we observe that the PSD returns to the baseline (no bacteria) level to within experimental error.
The presented data were averaged at two different time scales: To suppress the random noise, 1000 traces were averaged during collection; to assess longer-term drifts, the measurement was repeated three times with ∼1/2 h temporal separation. The standard deviation between the measurements obtained at different times is shown as the shaded regions of uncertainty in Fig. 3. (The error bars are shown only in the relevant frequency region.)
The calibration of the displacements was based on the thermal resonance peak visible around fR ≈ 2.5 kHz using standard AFM calibration practices. The inset shows the thermal peak in a linear plot. The (red) line is a fit to the damped harmonic oscillator model driven by the fluctuations in a liquid25 with the addition of white noise. The PSD has a resonance frequency fR ≈ 2.5 kHz and quality factor Q ≈ 1.5. The cantilever is assumed to be at room temperature, composed of silicon, and has a spring constant of K ≈ 0.03 N/m.
Returning to Fig. 3(a), we notice that, at the low frequency region, the noise data can be approximated by 1/fα with α ≈ 2. This is the noise signature of our measurement set up, possibly the laser.26 To find the power spectral density of bacterial motion, we naively subtract the PSDs, , i.e., we subtract the black curve from the green curve. The subtracted PSDs are shown in Fig. 3(b). The extra noise power due to the bacteria is not appreciable for f ≳ 80 Hz. It appears that the majority of the noise power stays at low frequencies; for 1 Hz ≲ f ≲ 80 Hz.
Although it is typically difficult to untangle 1/f noise sources,27 several features in the data suggest that subtraction of the PSDs as described above effectively removes the experimental 1/f noise. First, in Fig. 3(a), the PSD of cantilever fluctuations with adsorbed bacteria is almost an order of magnitude above the baseline. Second, in Fig. 3(b), the additional noise power coming from the bacteria, i.e., , grows with the number of bacteria on the microcantilever. Convinced that the observed 1/f-like behavior is indeed due to the bacteria, we turn to a discussion of possible explanations. Many diverse biological processes fluctuate with PSDs S(f) ∼ 1/f α. While the ubiquity of this 1/f behavior is intriguing, it may only be a reflection of the simple fact that the measured signal combines many processes that act on different time scales.28 Here, each bacterium exhibits motions with different characteristic time scales τi and amplitudes Ai. For instance, τi can pertain to the motion of the cilia; the motion of the flagella; and even the slow diffusive motion of the entire bacterium on the surface due to the breaking and re-forming of chemical bonds. The spectral density of the motion of a single bacterium can thus be considered to be a sum of these different spectral densities, . This argument suggests that the noise power spectral density of a single bacterium should be ∼ 1/f α with α determined by the interplay between the numerous time scales τi present in the problem. While this simple explanation may be satisfactory for a first-pass analysis, a comprehensive model should take into account further complexities due to the fact that the cantilever responds to input from many bacteria. For instance, bacteria will have a distribution of sizes and thus time scales; bacteria are positioned randomly on the cantilever; the coupling strength of the motion of each bacterium to the cantilever will be different; and so on.
Regarding sensor applications, we emphasize that conventional methods for studying bacterial behavior and detecting bacteria are time consuming and require expensive infrastructure.38 Recent biosensing approaches developed on microfluidic platforms based on electrical and optical sensing are not able to monitor bacterial motion and antibiotic resistance in real-time;29–34 similarly, micromechanical detection based on frequency shifts of a microcantilever resonator does not provide enough sensitivity in viscous liquids.35 For instance, there was no noticeable frequency shift in our experiments due to the mass of the bacteria, while fluctuations due to the bacteria were detectable. The theoretical limit for the minimum detectable mass based on frequency shift is of order Mc/Q ∼ 2 × 10−11 kg,36 where Mc is the cantilever mass. Given that the mass of a single E. coli is ∼ 1 × 10−15 kg, ∼104 bacteria are needed for a detectable frequency-shift based signal. This is well above the limit attained by monitoring the cantilever fluctuations. All these suggest that, by monitoring the fluctuations of a cantilever, one can develop functional and versatile sensors. It may be possible to enhance the fluctuation signal by lowering the spring constant of the microcantilever; this may reduce the resonance frequency and make the resonator overdamped in liquid. Careful modeling is required for finding the optimal design parameters for next generation sensors.
In this letter, we measured the nanomechanical spectrum of the forces that bacteria exert on a microcantilever from the fluctuations of the microcantilever. We observed that the amplitude of the fluctuations scales with frequency as 1/f α (2 ≤ α ≤ 3) as well as the surface density of the bacteria. Our physical model suggests that each bacterium provides fluctuations with multiple time scales and amplitudes, resulting in a collective 1/f-like spectrum.
Acknowledgments
We thank Dr. N. Gozde Durmuş for discussions. We acknowledge funding from the following sources: US NSF Grant Nos. CMMI-0970071 and DGE-1247312 and NIH Grant Nos. RO1 AI093282, RO1 A1081534, U54EB15408, and R21 AI087107. Dr. U. Demirci is a founder of, and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic and imaging technologies for point-of-care diagnostic solutions, and (ii) Koek Biotech, a company that is developing microfluidic IVF technologies for clinical solutions. U.D.'s interests were reviewed and are managed by the Brigham and Women's Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- 1.Pelling A. E., Sehati S., Gralla E. B., Valentine J. S., and Gimzewski J. K., Science 305, 1147 (2004). 10.1126/science.1097640 [DOI] [PubMed] [Google Scholar]
- 2.Arnoldi M., Fritz M., Bäuerlein E., Radmacher M., Sackmann E., and Boulbitch A., Phys. Rev. E 62, 1034 (2000). 10.1103/PhysRevE.62.1034 [DOI] [PubMed] [Google Scholar]
- 3.Zhang P. C., Keleshian A. M., and Sachs F., Nature 413, 428 (2001). 10.1038/35096578 [DOI] [PubMed] [Google Scholar]
- 4.Jiang G., Giannone G., Critchley D. R., Fukumoto E., and Sheetz M. P., Nature 424, 334 (2003). 10.1038/nature01805 [DOI] [PubMed] [Google Scholar]
- 5.Bao G. and Suresh S., Nat. Mater. 2, 715 (2003). 10.1038/nmat1001 [DOI] [PubMed] [Google Scholar]
- 6.Frauenfelder H., Wolynes P. G., and Austin R. H., Rev. Mod. Phys. 71, S419 (1999). 10.1103/RevModPhys.71.S419 [DOI] [Google Scholar]
- 7.Visick K. L. and Fuqua C., J. Bacteriol. 187, 5507 (2005). 10.1128/JB.187.16.5507-5519.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callegan M. C., Kane S. T., Cochran D. C., Novosad B., Gilmore M. S., Gominet M., and Lereclus D., Invest. Ophthalmol. Visual Sci. 46, 3233 (2005). 10.1167/iovs.05-0410 [DOI] [PubMed] [Google Scholar]
- 9.Moffitt J. R., Chemla Y. R., Smith S. B., and Bustamante C., Annu. Rev. Biochem. 77, 205 (2008). 10.1146/annurev.biochem.77.043007.090225 [DOI] [PubMed] [Google Scholar]
- 10.Lavrik N. V., Sepaniak M. J., and Datskos P. G., Rev. Sci. Instrum. 75, 2229 (2004). 10.1063/1.1763252 [DOI] [Google Scholar]
- 11.Ekinci K. L. and Roukes M. L., Rev. Sci. Instrum. 76, 061101 (2005). 10.1063/1.1927327 [DOI] [Google Scholar]
- 12.Radmacher M., Fritz M., Hansma H. G., and Hansma P. K., Science 265, 1577 (1994). 10.1126/science.8079171 [DOI] [PubMed] [Google Scholar]
- 13.Thomson N. H., Fritz M., Radmacher M., Cleveland J. P., Schmidt C. F., and Hansma P. K., Biophys. J. 70, 2421 (1996). 10.1016/S0006-3495(96)79812-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos D., Tamayo J., Mertens J., Calleja M., and Zaballos A., J. Appl. Phys. 100, 106105 (2006). 10.1063/1.2370507 [DOI] [Google Scholar]
- 15.Burg T. P., Godin M., Knudsen S. M., Shen W., Carlson G., Foster J. S., Babcock K., and Manalis S. R., Nature 446, 1066 (2007). 10.1038/nature05741 [DOI] [PubMed] [Google Scholar]
- 16.Arlett J. L., Myers E. B., and Roukes M. L., Nat. Nanotechnol. 6, 203 (2011). 10.1038/nnano.2011.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G., Datar R. H., Hansen K. M., Thundat T., Cote R. J., and Majumdar A., Nat. Biotechnol. 19, 856 (2001). 10.1038/nbt0901-856 [DOI] [PubMed] [Google Scholar]
- 18.Gfeller K. Y., Nugaeva N., and Hegner M., Biosens. Bioelectron. 21, 528 (2005). 10.1016/j.bios.2004.11.018 [DOI] [PubMed] [Google Scholar]
- 19.Longo G. and Kasas S., Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 6, 230 (2014) 10.1002/wnan.1258 [DOI] [PubMed] [Google Scholar]
- 20.Longo G., Alonso-Sarduy L., Rio L. M., Bizzini A., Trampuz A., Notz J., Dietler G., and Kasas S., Nat. Nanotechnol. 8, 522 (2013). 10.1038/nnano.2013.120 [DOI] [PubMed] [Google Scholar]
- 21.Meyer G. and Amer N. M., Appl. Phys. Lett. 53, 1045 (1988). 10.1063/1.100061 [DOI] [Google Scholar]
- 22.Azak N. O., Shagam M. Y., Karabacak D. M., Ekinci K. L., Kim D. H., and Jang D. Y., Appl. Phys. Lett. 91, 093112 (2007). 10.1063/1.2776981 [DOI] [Google Scholar]
- 23.Butt H. J. and Jaschke M., Nanotechnology 6, 1 (1995). 10.1088/0957-4484/6/1/001 [DOI] [Google Scholar]
- 24.See supplementary material at http://dx.doi.org/10.1063/1.4895132E-APPLAB-105-014437 for more details regarding the experimental methods and statistical analysis of our data.
- 25.Paul M. R., Clark M. T., and Cross M. C., Nanotechnology 17, 4502 (2006). 10.1088/0957-4484/17/17/037 [DOI] [Google Scholar]
- 26.Labuda A., Bates J. R., and Grütter P. H., Nanotechnology 23, 025503 (2012). 10.1088/0957-4484/23/2/025503 [DOI] [PubMed] [Google Scholar]
- 27.Dutta P. and Horn P. M., Rev. Mod. Phys. 53, 497 (1981). 10.1103/RevModPhys.53.497 [DOI] [Google Scholar]
- 28.Hausdorff J. M. and Peng C. K., Phys. Rev. E 54, 2154 (1996). 10.1103/PhysRevE.54.2154 [DOI] [PubMed] [Google Scholar]
- 29.Wang S., Esfahani M., Gurkan U. A., Inci F., Kuritzkes D. R., and Demirci U., Lab Chip 12, 1508 (2012). 10.1039/c2lc20706k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., Inci F., Chaunzwa T. L., Ramanujam A., Vasudevan A., Subramanian S., Ip A. C. F., Sridharan B., Gurkan U. A., and Demirci U., Int. J. Nanomed. 7, 2591 (2012) 10.2147/IJN.S29629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inci F., Tokel O., Wang S., Gurkan U. A., Tasoglu S., Kuritzkes D. R., and Demirci U., ACS Nano 7, 4733 (2013). 10.1021/nn3036232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokel O., Inci F., and Demirci U., Chem. Rev. 114, 5728 (2014). 10.1021/cr4000623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mani V., Wang S., Inci F., Libero G. D., Singhal A., and Demirci U., “ Emerging technologies for monitoring drug-resistant tuberculosis at the point-of-care,” Adv. Drug Deliv. Rev. (to be published) 10.1016/j.addr.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafiee H., Jahangir M., Inci F., Wang S., Willenbrecht R. B. M., Giguel F. F., Tsibris A. M. N., Kuritzkes D. R., and Demirci U., Small 9, 2553 (2013). 10.1002/smll.201202195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekinci K. L., Yakhot V., Rajauria S., Colosqui C., and Karabacak D. M., Lab Chip 10, 3013 (2010). 10.1039/c003770m [DOI] [PubMed] [Google Scholar]
- 36.Ekinci K. L., Yang Y. T., and Roukes M. L., J. Appl. Phys. 95, 2682 (2004). 10.1063/1.1642738 [DOI] [Google Scholar]
- 37.Wang S. Q., Inci F., De Libero G., Singhal A., and Demirci U., Biotechnol. Adv. 31(4), 438–449 (2013). 10.1016/j.biotechadv.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durmus N. G., Taylor E. N., Inci F., Kummer K. M., Tarquinio K. M., and Webster T. J., Int. J. Nanomed. 7, 537 (2012). 10.2147/IJN.S27957 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4895132E-APPLAB-105-014437 for more details regarding the experimental methods and statistical analysis of our data.


