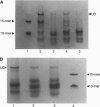
Abstract
Inordinate expansion and hypermethylation of the fragile X DNA triplet repeat, (GGC)n.(GCC)n, are correlated with the ability of the individual G- and C-rich single strands to form hairpin structures. Two-dimensional NMR and gel electrophoresis studies show that both the G- and C-rich single strands form hairpins under physiological conditions. This propensity of hairpin formation is more pronounced for the C-rich strand than for the G-rich strand. This observation suggests that the C-rich strand is more likely to form hairpin or "slippage" structure and show asymmetric strand expansion during replication. NMR data also show that the hairpins formed by the C-rich strands fold in such a way that the cytosine at the CpG step of the stem is C.C paired. The presence of a C.C mismatch at the CpG site generates local flexibility, thereby providing analogs of the transition to the methyltransferase. In other words, the hairpins of the C-rich strand act as better substrates for the human methyltransferase than the Watson-Crick duplex or the G-rich strand. Therefore, hairpin formation could account for the specific methylation of the CpG island in the fragile X repeat that occurs during inactivation of the FMR1 gene during the onset of the disease.
Full text
PDF


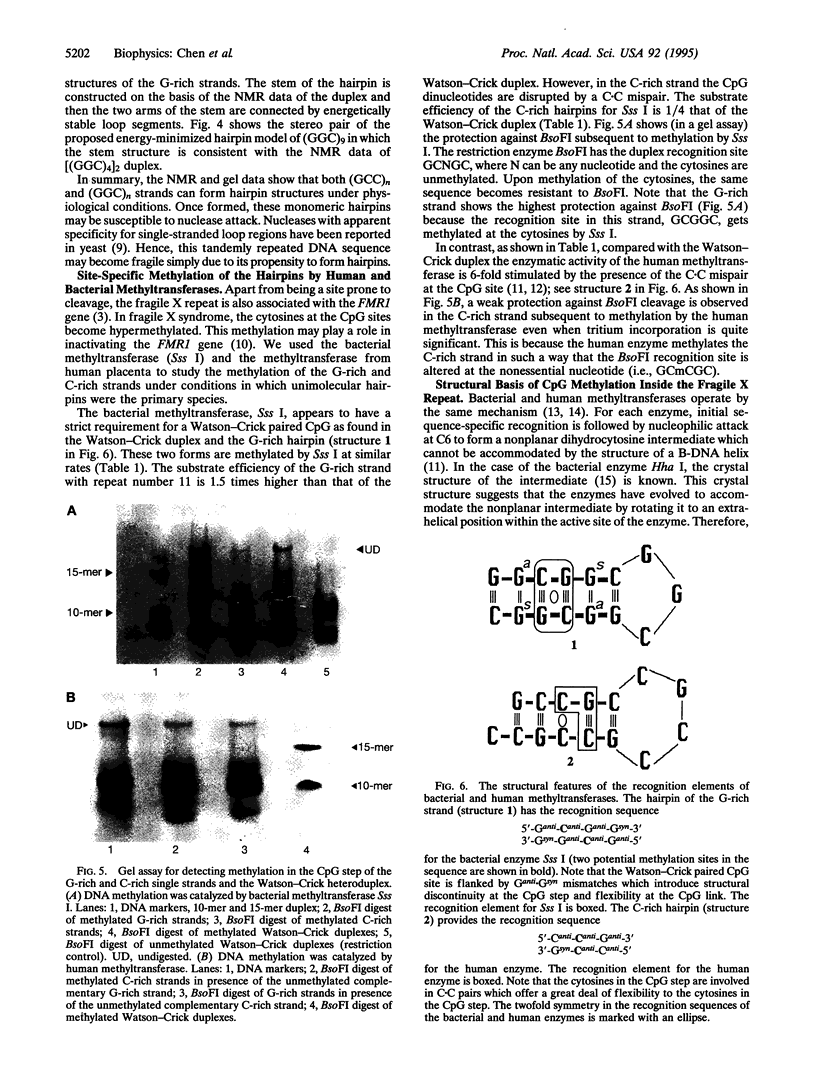
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Catasti P., Gupta G., Garcia A. E., Ratliff R., Hong L., Yau P., Moyzis R. K., Bradbury E. M. Unusual structures of the tandem repetitive DNA sequences located at human centromeres. Biochemistry. 1994 Apr 5;33(13):3819–3830. doi: 10.1021/bi00179a005. [DOI] [PubMed] [Google Scholar]
- Grady D. L., Ratliff R. L., Robinson D. L., McCanlies E. C., Meyne J., Moyzis R. K. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G., García A. E., Hiriyanna K. T. Sampling of the conformations of the d(CGCTGCGGC) hairpin in solution by two-dimensional nuclear magnetic resonance and theoretical methods. Biochemistry. 1993 Jan 26;32(3):948–960. doi: 10.1021/bi00054a028. [DOI] [PubMed] [Google Scholar]
- Guéron M., Kochoyan M., Leroy J. L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987 Jul 2;328(6125):89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Moyzis R. K., Torney D. C., Meyne J., Buckingham J. M., Wu J. R., Burks C., Sirotkin K. M., Goad W. B. The distribution of interspersed repetitive DNA sequences in the human genome. Genomics. 1989 Apr;4(3):273–289. doi: 10.1016/0888-7543(89)90331-5. [DOI] [PubMed] [Google Scholar]
- Nancarrow J. K., Kremer E., Holman K., Eyre H., Doggett N. A., Le Paslier D., Callen D. F., Sutherland G. R., Richards R. I. Implications of FRA16A structure for the mechanism of chromosomal fragile site genesis. Science. 1994 Jun 24;264(5167):1938–1941. doi: 10.1126/science.8009225. [DOI] [PubMed] [Google Scholar]
- Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988 Jul 12;27(14):5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. Simple repeat DNA is not replicated simply. Nat Genet. 1994 Feb;6(2):114–116. doi: 10.1038/ng0294-114. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Wells R. D. DNA structure, mutations, and human genetic disease. Curr Opin Biotechnol. 1992 Dec;3(6):612–622. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Hardy T. A., Baker D. J. Human DNA (cytosine-5)methyltransferase selectively methylates duplex DNA containing mispairs. Nucleic Acids Res. 1987 Sep 11;15(17):6899–6916. doi: 10.1093/nar/15.17.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Kan J. L., Baker D. J., Kaplan B. E., Dembek P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J Mol Biol. 1991 Jan 5;217(1):39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Kaplan B. E., Sowers L. C., Newman E. M. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Laayoun A., Lingeman R. G., Baker D. J., Riley J. Hypermethylation of telomere-like foldbacks at codon 12 of the human c-Ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J Mol Biol. 1994 Oct 21;243(2):143–151. doi: 10.1006/jmbi.1994.1640. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]