Abstract
The nonsense-mediated mRNA decay (NMD) pathway is a specialized mRNA degradation pathway that degrades select mRNAs. This pathway is conserved in all eukaryotes examined so far, and it triggers the degradation of mRNAs that prematurely terminate translation. Originally identified as a pathway that degrades mRNAs with premature termination codons as a result of errors during transcription, splicing, or damage to the mRNA, NMD is now also recognized as a pathway that degrades some natural mRNAs. The degradation of natural mRNAs by NMD has been identified in multiple eukaryotes, including Saccharomyces cerevisiae, Drosophila melanogaster, Arabidopsis thaliana, and humans. S. cerevisiae is used extensively as a model to study natural mRNA regulation by NMD. Inactivation of the NMD pathway in S. cerevisiae affects approximately 10% of the transcriptome. Similar percentages of natural mRNAs in the D. melanogaster and human transcriptomes are also sensitive to the pathway, indicating that NMD is important for the regulation of gene expression in multiple organisms. NMD can either directly or indirectly regulate the decay rate of natural mRNAs. Direct NMD targets possess NMD-inducing features. This minireview focuses on the regulation of natural mRNAs by the NMD pathway, as well as the features demonstrated to target these mRNAs for decay by the pathway in S. cerevisiae. We also compare NMD-targeting features identified in S. cerevisiae with known NMD-targeting features in other eukaryotic organisms.
INTRODUCTION
The amount of a particular mRNA present at a specific time in a cell is dependent on the rates of synthesis and decay. The nonsense-mediated mRNA decay (NMD) pathway is a translation-dependent mRNA degradation pathway that recognizes and elicits the rapid degradation of select mRNAs. This pathway is highly conserved in all eukaryotes examined, from yeast to humans. NMD targets mRNAs that prematurely terminate translation. Such mRNAs can be produced due to genomic mutations or errors in gene expression. In cases where the mRNAs have a premature termination codon (PTC), degradation of these mRNAs by NMD prevents the accumulation of potentially harmful truncated proteins.
The NMD pathway also regulates the expression of specific genes by degrading natural mRNAs (1–7). The natural mRNAs regulated by NMD are normal, cellular mRNAs that largely code for functional proteins. Importantly, this subset of mRNAs has features that can induce the translating ribosome to prematurely terminate translation, leading to degradation of the transcript by NMD. The regulation of natural mRNAs by NMD is fairly widespread. Genome-wide studies have shown that 5 to 10% of the Saccharomyces cerevisiae transcriptome is affected when NMD is inactivated (1). Similar percentages of the transcriptomes of the nematode Caenorhabditis elegans (8), the fruit fly Drosophila melanogaster (9), and humans are sensitive to NMD (10, 11). Natural mRNAs regulated by the pathway have also been identified in other eukaryotic organisms, including the fungi Yarrowia lipolytica, Aspergillus nidulans, and Schizosaccharomyces pombe and several plant species (12–15).
Three core trans-acting factors are required for a functional NMD pathway in all eukaryotes. These core NMD factors are the up-frameshift proteins Upf1, Upf2, and Upf3. The Upf proteins were originally identified in S. cerevisiae (16, 17). Mutation or silencing of any one of these three factors selectively stabilizes mRNAs that are regulated by NMD (18). Upf1p is the central regulator of the degradation pathway and is the most conserved of the Upf proteins (19–23). Upf2p and Upf3p are responsible for regulating Upf1p function. Upf1p is a group 1 RNA helicase with ATPase activity (24–26). S. cerevisiae Upf1p has been shown to interact with Upf2p, which in turn interacts with Upf3p (18, 27). Upf1p also associates with additional factors, including the eukaryotic translational release factors eRF1 and eRF3 (28). It is important to note that Upf1p plays additional roles distinct from NMD. These cellular processes include staufen 1-mediated mRNA decay (SMD), telomere maintenance, histone mRNA decay, genome stability, and advancement of the cell cycle (29–32).
THE NMD MECHANISM IN S. CEREVISIAE
NMD in S. cerevisiae is known to be triggered by the messenger ribonucleoprotein (mRNP) context surrounding the translation termination event. Models exist that explain the mechanism that causes a termination codon to be recognized as premature. In the most widely accepted model, premature translation termination is perceived to be an inefficient event because it is in an improper context compared to a normal translation termination event. Specifically, there is evidence in S. cerevisiae and other organisms that NMD targets can be recognized as targets due to the lack of factors bound downstream from the termination codon (23). In this model, known as the faux untranslated region (UTR) model, a ribosome terminating translation at a termination codon substantially upstream from the poly(A) tail terminates translation inefficiently. The faux-UTR model posits that NMD occurs because the Pab1p or other factor bound to the poly(A) tail is not in close proximity to the terminating ribosome to enable interaction of the Pab1p with release factor 3 (eRF3p), which is bound to the terminating ribosome and thus establishes the correct mRNP context for a normal translation termination event (33). In the absence of correct translation termination, the Upf factors interact with the release factors, eRF1p and eRF3p (28), resulting in an aberrant translation termination event and NMD activation (23, 34–36). NMD activation leads to the decapping of the mRNA by the Dcp1-Dcp2p complex, followed by 5′ to 3′ degradation of the mRNA by the exoribonuclease Xrn1p (37). The faux-UTR model explains how some natural mRNAs with known NMD-inducing features are targeted for degradation by the pathway; however, it does not explain the observation that the presence of Pab1p is not required to distinguish a normal termination event from a premature translation termination event in S. cerevisiae (38).
THE NMD MECHANISM IN MAMMALS
In metazoans, additional factors are required for NMD to operate normally. These factors include the suppressors with morphological effect on genitalia, or SMGs, in C. elegans. SMG proteins are also found in other multicellular organisms and perform a variety of functions. SMG-1, SMG-5, and SMG-7 regulate the phosphorylation and dephosphorylation of SMG-2, the C. elegans homolog of Upf1p, while SMG-6 is an endonuclease. Additionally, the exon-junction complex (EJC) is a multiprotein complex that enhances NMD of mRNAs that undergo splicing. The EJC is deposited 20 to 24 nucleotides upstream from exon-exon junctions. The core components of the EJC are eIF4AIII, Y14, MAGOH, barentsz, and additional effector proteins, such as Upf3p (39). Upf3p is reported to associate with the EJC during splicing, and subsequently, Upf2p associates with the EJC-bound Upf3p within the cytoplasm (40).
The initial model to explain mammalian NMD proposed that the round of translation an mRNA is undergoing determines whether an mRNA is an NMD target. In newly synthesized mRNAs, the 5′ cap structure is bound by the cap-binding complex, consisting of the proteins CBP80 and CBP20. While still bound by CBP80/20, mRNAs undergo the first or “pioneer” round of translation. mRNAs undergoing the pioneer round of translation are subject to NMD (41). These mRNAs are targeted to the pathway if the translating ribosomes terminate translation 50 to 55 nucleotides upstream from an EJC. In most mRNAs, the natural stop codon is found in the last exon, and all EJCs would be displaced from the mRNA before the translating ribosome encounters the natural stop codon. In contrast, a ribosome stalled at a PTC associates with the SURF complex, which is composed of the SMG-1 kinase, Upf1p, and release factors eRF1 and eRF3. The SURF complex-associated Upf1p interacts with the downstream EJC which is still bound to the mRNA. This association occurs via interactions between Upf2p and the EJC-bound Upf3p, consequently targeting the mRNA to NMD.
It is becoming apparent that NMD in mammalian cells is not always restricted to spliced transcripts undergoing the first round of translation. In fact, there are reports of intronless mammalian mRNAs undergoing NMD, demonstrating that mammalian mRNAs are subject to NMD in the absence of an EJC downstream from a stop codon (36, 42, 43). Furthermore, recent work showing that mRNAs bound by the cap-binding protein, eukaryotic initiation factor 4E (eIF4E), are subject to NMD suggests that mammalian mRNAs undergoing bulk translation can also be subject to NMD (44, 45).
The S. cerevisiae NMD model is applicable to mammalian NMD to some extent; however, apparent differences exist. S. cerevisiae may be a good model for EJC-independent NMD. The EJC mark deposited on mammalian mRNAs during splicing adds an additional layer of regulation and is absent in S. cerevisiae. In S. cerevisiae, NMD is splicing independent. Nevertheless, EJC-independent NMD has been demonstrated in mammalian cells that is akin to NMD in S. cerevisiae. Moreover, there have been reports of proteins such as the RNA helicase hrp1p, which marks S. cerevisiae mRNAs downstream from a premature stop codon and targets the mRNAs to the pathway, analogous to the EJC targeting mechanism in mammals (46, 47).
As we already noted, the core NMD factors are conserved in all eukaryotes examined so far. In addition, recent studies demonstrating that mammalian mRNAs are subject to NMD while undergoing bulk translation suggest that mammalian mRNAs targeted to the pathway may not be restricted to the first round of translation and that mRNAs can be targeted to the pathway at each round of translation, similar to NMD in S. cerevisiae (48). However, mRNAs that are subject to NMD in S. cerevisiae are primarily degraded by Xrn1p from the 5′ ends of the mRNAs after removal of the 5′ cap, while mammalian NMD is initiated by endonucleolytic cleavage of the target transcript, followed by exonucleolytic degradation of the 5′ and 3′ RNA fragments (49).
NATURAL mRNA REGULATION BY NMD
As previously stated, natural mRNAs regulated by the NMD pathway have been identified in multiple organisms. These natural NMD targets have been most extensively studied in S. cerevisiae and can be classified as either direct or indirect NMD targets. Direct NMD targets typically have significantly altered decay rates in cells with a functional NMD pathway relative to their decay rates in cells with a nonfunctional pathway. In most cases, indirect NMD targets have comparable decay rates in cells with a functional or nonfunctional NMD pathway. Indirect NMD targets may accumulate as a result of increased transcription, whereby a transcription factor that regulates the expression of a specific mRNA is the direct NMD target. An example of an indirect NMD target is the S. cerevisiae URA3 mRNA. The URA3 mRNA accumulates to higher levels in NMD mutants due to increased levels of its transcription activator Ppr1p (16). PPR1 mRNA is a direct NMD target. In S. cerevisiae, it is estimated that ∼48% of the natural NMD targets are direct targets with significantly altered decay rates between the wild type and NMD mutants (2). mRNAs that are direct NMD targets typically contain NMD-inducing features that can trigger their degradation by the pathway.
NMD-INDUCING FEATURES IN S. CEREVISIAE
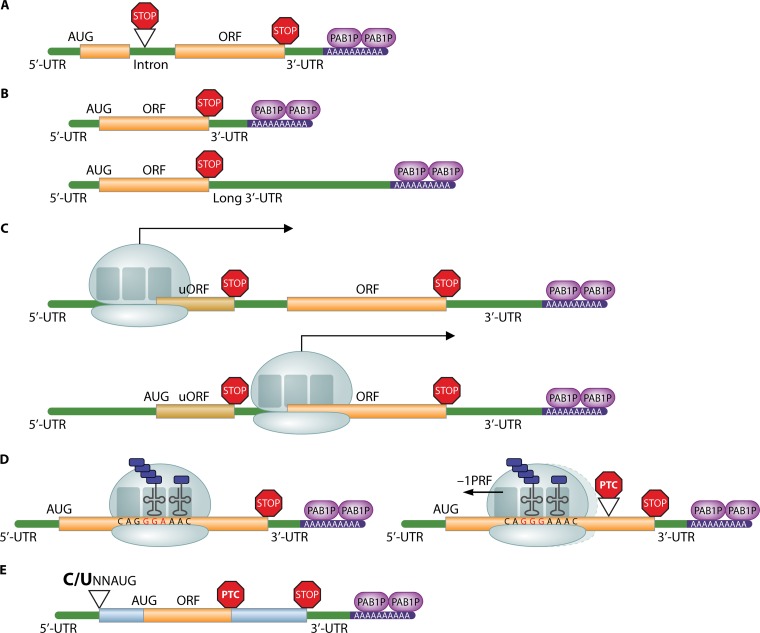
Initially, it was perceived that all targets of the NMD pathway contain a PTC. However, with the growing list of endogenous, error-free, natural mRNAs that are regulated by the pathway, it is becoming apparent that sequence features exist that can trigger the regulation of natural mRNAs by NMD. These NMD-inducing features have been identified in S. cerevisiae and in other organisms and activate NMD by directing translation to terminate prematurely. Here, we discuss each of the recognized NMD-inducing features in S. cerevisiae and their conservation in other eukaryotic species. These features are depicted schematically in Fig. 1 and include inefficiently or nonproductive alternatively spliced pre-mRNAs, mRNAs containing atypically long 3′ untranslated regions (UTRs), mRNAs containing upstream open reading frames (uORFs), mRNAs subject to programmed ribosomal frameshifting, and mRNAs subject to out-of-frame translation initiation caused by leaky ribosomal scanning (2, 4, 50–54). Although these features typically trigger degradation, the presence of these NMD-inducing features on an mRNA does not always activate the pathway. Some mRNAs with these features are immune to degradation by NMD (4, 34, 55). For example, the S. cerevisiae GCN4 and YAP1 mRNAs contain uORFs but evade degradation by NMD (55). Furthermore, these NMD-targeting features are not known to cause the degradation of all natural mRNAs that are regulated by NMD in S. cerevisiae, indicating that additional NMD-targeting features remain to be identified (2).
FIG 1.
Schematic representation of NMD-targeting features in S. cerevisiae. (A) Intron containing pre-mRNA containing a stop codon. (B) mRNA with an atypically long 3′ UTR. (C) mRNA with an upstream open reading frame (uORF) in the 5′ UTR that can induce NMD. (D) mRNAs with sequence features that can induce a −1 ribosomal frameshift. (E) mRNA with a suboptimal start codon context that may be subject to out-of-frame translation initiation in an alternate AUG codon within the ORF, which can lead to a PTC and NMD activation. ORFs are illustrated by thick orange lines, and untranslated regions (UTRs) and introns are shown by thin green lines. The poly(A) tail is shown in blue, and ribosomes are illustrated in gray. AUG, start codon; PTC, premature termination codon; PRF, programmed (−1) ribosomal frameshift; PAB1P, poly(A)-binding protein.
SOME INTRON-CONTAINING PRE-mRNAS ARE NMD TARGETS
The generation of a functional mRNA depends on precise processing of the pre-mRNA. Pre-mRNAs containing introns undergo splicing prior to export to the cytoplasm. In some cases, pre-mRNA processing events result in the retention of an inefficiently spliced intron. The majority of intron-containing pre-mRNAs that are exported to the cytoplasm are degraded by the NMD pathway in S. cerevisiae, because these pre-mRNAs are likely to have an in-frame stop codon (Fig. 1A) (51, 56). This regulation prevents the accumulation of incompletely processed transcripts (57). Alternatively, if a retained intron does not contain a stop codon but is out of frame with the main open reading frame, it can cause a frameshift to a downstream exon. This can result in the introduction of a PTC and, consequently, degradation of the transcript by NMD. Several studies in S. cerevisiae have shown that inefficiently spliced pre-mRNAs accumulate in NMD mutants (51, 56), albeit only ∼5% of the genes in S. cerevisiae contain introns (58). For example, the S. cerevisiae CYH2 pre-mRNA is inefficiently spliced, transported to the cytoplasm, and degraded by NMD. Similar results have been observed in the hemiascomycete fungus Yarrowia lipolytica, which contains four times as many introns as S. cerevisiae (12). In some cases, the accumulation of unspliced pre-mRNAs in S. cerevisiae NMD mutants was found to occur under stress conditions (56). In addition, localization experiments demonstrated that the unspliced pre-mRNAs accumulate in cytoplasmic processing bodies (P bodies) in the absence of a functional NMD pathway (56).
SOME NONPRODUCTIVE ALTERNATIVELY SPLICED TRANSCRIPTS ARE NMD TARGETS
Pre-mRNA processing can also result in alternative splicing events, where exons are spliced together differentially to generate proteome diversity. Aberrant alternative splicing can generate NMD-sensitive transcripts by introducing PTCs. This mechanism of NMD targeting is often referred to as “alternative splicing coupled to NMD.” A recent study in S. cerevisiae demonstrated that nonproductive alternatively spliced transcripts that contain a PTC or produce nonfunctional proteins are regulated by NMD (59). In addition, genome-wide studies have shown that aberrantly spliced transcripts are regulated by the NMD pathway in C. elegans (8) and in D. melanogaster (60). Interestingly, in C. elegans, alternatively spliced transcripts of ribosomal protein genes are also regulated by the NMD pathway, linking the pathway to splicing regulation (61).
The NMD pathway is also linked to splicing regulation in mammalian systems. Studies using mouse cell lines found that downregulating the levels of the NMD factor UPF2 affected splicing, resulting in aberrant alternatively spliced mRNA isoforms that were sensitive to NMD (62). Consistent with these findings, Saltzman et al. found that in mammalian cells, pre-mRNAs that undergo alternative splicing coupled to NMD were enriched for core spliceosomal proteins, further linking the NMD pathway to regulation of splicing (63). Collectively, these studies demonstrate that NMD controls splicing regulators and aberrant alternatively spliced transcripts in mammalian systems.
In other systems, such as plants, NMD has also been found to regulate the expression of pre-mRNAs that retain introns due to alternative splicing. It has been reported that the retention of introns accounts for ∼41% of alternative splicing events in plants (64). In addition, the presence of an intron in the 3′ UTR has been shown to activate NMD, although only ∼3.6% of Arabidopsis thaliana genes contain introns in the 3′ UTR (65–67). The regulation of these intron-containing pre-mRNAs by NMD is reported to occur in a position-dependent manner (67). Specifically, if the intron is located at least 50 nt downstream from the stop codon, NMD is triggered. Furthermore, the efficiency of NMD was found to increase with increasing distance from the stop codon (67).
Consistent with the findings in S. cerevisiae, plants, and mammals, nonproductive alternatively spliced transcripts are regulated by the NMD pathway in other systems, such as zebrafish and Paramecium tetraurelia (68). Collectively, these studies show that pre-mRNA processing events that result in the generation of intron-containing pre-mRNAs, as well as aberrantly spliced transcripts, generate NMD targets in a wide range of organisms. Moreover, in some species, NMD has been found to have an additional level of regulation by directly affecting splicing factors.
ATYPICALLY LONG 3′ UTRS CAN TARGET mRNAS TO THE NMD PATHWAY
The presence of a long 3′ UTR has been shown to be sufficient to target mRNAs for NMD. The mechanism by which atypically long 3′ UTRs induce NMD has been extensively studied in S. cerevisiae. In S. cerevisiae, mRNA 3′ UTRs range in size from 50 to 200 nt in length, with a median length of 121 nt (69). The majority of the examined transcripts with 3′ UTRs of 350 nt or longer are degraded by the NMD pathway (4, 70). Interestingly, some mRNAs produce different isoforms of the same transcript that vary in their 3′ UTR lengths. Transcripts with varying 3′ UTR lengths can be produced as a result of alternative 3′-end processing, which can be sensitive to growth conditions (71).
Pre-mRNAs that undergo alternative 3′-end processing and thereby produce multiple mRNA isoforms with different 3′ UTR lengths can be differentially regulated. This differential regulation can be observed through mRNA stabilization or destabilization. Surprisingly, in some mRNAs, the half-lives can vary significantly between mRNA isoforms separated by <3 nt at the 3′ end (72). Furthermore, some mRNA isoforms produced by alternative 3′-end processing may be regulated by NMD, while others may not. For example, the S. cerevisiae MAK31 mRNA has two mRNA isoforms with differing 3′ UTR lengths, a short (200-nt) and a long (920-nt) 3′ UTR. The short, 200-nt 3′ UTR mRNA isoform is immune to degradation by NMD; however, the long, 920-nt 3′ UTR mRNA isoform is degraded by the pathway (4). On the other hand, MPA43 mRNA also produces two mRNA isoforms with differing 3′ UTR lengths of 300 nt and 600 nt. In this case, both 3′ UTRs lead to NMD-induced degradation (4). In S. cerevisiae, it appears that natural mRNAs targeted to the pathway by a long 3′ UTR are regulated by NMD due to the length of the 3′ UTR and not due to the specific sequence of the 3′ UTR. This is consistent with the faux UTR model, which states that increasing the 3′ UTR length increases the distance between the terminating ribosome and the Pab1p (Fig. 1B) (23).
Similar to the case for S. cerevisiae, C. elegans, Drosophila, and mammalian mRNAs with long 3′ UTRs are targeted for degradation by NMD. For example, in murine embryonic stem cells (mESCs), increased mRNA 3′ UTR length was found to correlate with increased sensitivity to the NMD pathway (73). Likewise, human transcripts with long 3′ UTRs are subject to degradation by the pathway. These transcripts were found to be upregulated in cells in which UPF1, SMG-6, and SMG-7 were downregulated (34). Interestingly, the NMD factors UPF1, SMG-5, and SMG-7 mRNA were also found to contain long 3′ UTRs that induce degradation by NMD. These studies point to a feedback loop where components of the NMD machinery are also regulated by the pathway (34). The regulation of NMD factors by the pathway has also been observed in zebrafish and C. elegans, further supporting the notion of a conserved NMD feedback loop (74).
A long 3′ UTR can also induce NMD in plants, providing additional support for the faux UTR model (65, 66). The average length of plant 3′ UTRs is reported to be 241 nt. A 3′ UTR of >350 nt has been shown to consistently elicit NMD in plants (75). Additionally, the efficiency of NMD in plants has been reported to increase with increasing length of the 3′ UTR (65).
Collectively, these studies demonstrate that a long 3′ UTR is a conserved NMD-inducing feature in multiple organisms. Furthermore, it appears that in several organisms, increasing the length of the 3′ UTR increases NMD efficiency, suggesting that the distance between the stop codon and the features downstream from the stop codon is the NMD-determining characteristic. If the distance between the stop codon and the poly(A) tail is the essential NMD determinant, then it follows that the length of the 3′ UTR itself, and not the 3′ UTR sequence, is the important NMD-inducing feature. In addition, transcripts that undergo alternative 3′-end processing may generate multiple mRNA isoforms that are differentially regulated by NMD. This would enable cells to selectively regulate mRNAs transcribed from the same gene. An important question for future studies is to determine whether physiological conditions can affect alternative 3′-end processing, resulting in the generation of transcripts with altered NMD sensitivity.
A SUBSET OF uORF-CONTAINING mRNAS IS SENSITIVE TO THE NMD PATHWAY
An upstream open reading frame, or uORF, is an open reading frame that is located in the 5′ UTR of a transcript (Fig. 1C). mRNAs can have one or more uORFs, some of which overlap the main protein coding region. These uORFs can play regulatory roles by affecting mRNA stability and protein synthesis (76). When actively translated, a uORF can interfere with the expression of the main ORF. Furthermore, if the uORF of an mRNA is translated, the stop codon of the uORF may be recognized as a PTC, resulting in degradation of the mRNA by NMD. The presence of a uORF has been shown to target mRNAs for degradation by NMD in S. cerevisiae and in the fission yeast, S. pombe. Transcriptome sequencing (RNA-Seq) analysis of the S. cerevisiae transcriptome predicted that 321 transcripts contain uORFs (77). A subset of these transcripts is expected to be regulated by NMD. An additional S. cerevisiae study identifying direct and indirect NMD targets estimated that ∼35% of direct NMD targets are sensitive to the pathway due to the presence of uORFs (2). An example of an S. cerevisiae mRNA targeted by NMD due to the presence of a uORF in the 5′ UTR is the CPA1 mRNA. CPA1 encodes the small subunit of arginine-specific carbamoyl phosphate synthetase (50). The sensitivity of the CPA1 mRNA to the NMD pathway is responsive to the levels of arginine in the medium. The addition of arginine causes ribosomal stalling at the uORF termination codon, resulting in destabilization and NMD-mediated degradation of the CPA1 mRNA (50). The S. cerevisiae ALR1 mRNA is also targeted to NMD due to the presence of a uORF (78). ALR1 encodes the major magnesium transporter in S. cerevisiae. Inactivation of the NMD pathway results in stabilization of the ALR1 mRNA and increased intracellular magnesium levels. This increase in intracellular magnesium levels promotes reduced translational termination fidelity and leads to an increase in read-through at PTCs. This study established a link between the NMD pathway, magnesium homeostasis, and translational termination fidelity (78). Consistent with studies in S. cerevisiae, a genome-wide study of S. pombe transcripts regulated by the NMD pathway also found that the majority of the transcripts that were upregulated upon NMD inactivation contained uORFs, suggesting that this is also a major NMD-targeting feature in S. pombe (14).
In addition, uORFs can trigger NMD in C. elegans (8) and mammals (10, 11, 73). In C. elegans, a genome-wide study of NMD-inducing features found that natural mRNAs regulated by the pathway are likely to have uORFs (8). In mammalian cells, it is estimated that ∼50% of transcripts contain uORFs (79). Many of these uORF-containing mRNAs are not regulated by NMD, demonstrating that some uORF-containing transcripts are capable of evading degradation by the pathway (80). This discrepancy is thought to be a result of varying translation efficiency. This view is supported by a study utilizing mESCs that found that translated uORFs were likely to target mRNAs to NMD, while nontranslated uORFs did not (73). The same study also found that translated NMD targets containing uORFs were enriched for transcriptional regulators, suggesting that genes regulated by these transcription factors would be indirect NMD targets (73).
Consistent with the studies described above, plant mRNAs containing uORFs are also regulated by NMD. An estimated 20 to 30% of plant genes have been reported to contain uORFs (65, 81, 82). In plants, it appears that the likelihood of a uORF inducing NMD depends on several factors. These factors include the length of the uORF and the distance between the stop codon of the uORF and the main start codon of the protein-coding region. Therefore, it has been proposed that longer plant uORFs induce NMD, while shorter uORFs do not (81).
Collectively, these studies establish that uORFs are also conserved NMD-inducing features in all of the organisms that have been examined so far. These studies of uORFs also point to the facts that not all uORF-containing mRNAs are subject to NMD and that other factors, such as the level of translation the uORF is undergoing, the length of the uORF, and the distance between the uORF stop codon and the main ORF start codon, can influence NMD targeting of these mRNAs. It is also apparent that some physiological conditions can affect the NMD targeting of uORF-containing transcripts by influencing the efficiency by which the uORF is translated.
mRNAS SUBJECT TO −1 RIBOSOMAL FRAMESHIFT SIGNALS MAY BE REGULATED BY NMD
Programmed ribosomal frameshifting (PRF) has been observed in multiple organisms. Analyses of multiple genomes indicated that ∼8 to 12% of genes contain a probable −1 ribosomal frameshift signal (83). A study showed that −1 ribosomal frameshifting plays a role in mRNA stability in S. cerevisiae through utilization of the NMD pathway (83). Specifically, −1 PRF signals were reported to cause destabilization of mRNAs by driving translating ribosomes into an alternate reading frame. In mRNAs undergoing −1 PRF, the ribosome moves toward the 5′ end of the mRNA, shifting the reading frame by one nucleotide (Fig. 1D). As a result of the −1 frameshift, the translating ribosome is out of frame and may encounter a PTC, thus triggering NMD.
The majority of the PRF signals contain three features: a “slippery site,” a spacer sequence, and an mRNA pseudoknot (83). Currently, −1 ribosomal frameshift signals have not been reported as an NMD-inducing feature in eukaryotic organisms other than in S. cerevisiae. These signals appear to be common in S. cerevisiae, and as many as 11% of genes have a likely −1 PRF (83). Although PRF signals are present across a wide range of yeast species, specific signals are not entirely conserved. For example, strong candidate −1 ribosomal frameshift signals are present in many gene orthologs in multiple yeast species but not across all species examined (83). It remains to be determined whether −1 ribosomal frameshift signals target natural transcripts to the NMD pathway in other eukaryotic organisms.
mRNAS SUBJECT TO OUT-OF-FRAME TRANSLATION INITIATION CAUSED BY LEAKY RIBOSOMAL SCANNING MAY BE NMD TARGETS
A number of S. cerevisiae mRNAs are targeted to the NMD pathway due to out-of-frame initiation of translation within the open reading frame. This mechanism is referred to as leaky scanning. During leaky scanning, ribosomes scan past the main ORF start codon, which is in a suboptimal context, and initiate translation with an alternate start codon within the ORF (Fig. 1E). This alternate start codon can be out of frame and lead the translating ribosome to a stop codon, causing NMD-induced degradation of the transcript. Whether a translation initiation codon is considered to be in an optimal or a suboptimal context is based on the sequence context surrounding the initiator AUG (84). Nucleotides at the −6 to +6 positions in relation to the initiator AUG are important for translation initiation. Of particular importance is the −3 position, where an A or a G results in more efficient translation initiation than a C or a U (2, 52, 84).
Presently, leaky scanning has only been recognized as an NMD-inducing feature in S. cerevisiae, and an estimated 3% of S. cerevisiae mRNAs may be subject to leaky scanning (2, 52). One such mRNA is the S. cerevisiae SPT10 mRNA, which encodes a putative histone acetylase and has a role in transcriptional silencing. The SPT10 start codon is in a suboptimal context, resulting in degradation of the mRNA by the pathway (52). Additional S. cerevisiae mRNAs targeted by NMD due to leaky scanning were also identified by Guan et al. (2). However, how widely distributed this NMD-inducing feature is among other eukaryotic organisms remains to be determined.
CONSERVATION OF THE NMD-INDUCING FEATURES
Of the five identified NMD-inducing features discussed here, three appear to be broadly distributed across a wide range of eukaryotic organisms. Intron-containing and nonproductive alternatively spliced transcripts have been shown to be NMD targets in multiple yeast species, as well as in C. elegans, D. melanogaster, plants, and mammalian systems. Atypically long 3′ UTRs also induce NMD in multiple eukaryotic organisms, suggesting that this mechanism is conserved as well. Similarly, a subset of natural transcripts containing uORFs is regulated by NMD in multiple eukaryotes.
It is also apparent that the presence of known NMD-inducing features on transcripts does not always activate NMD. Some transcripts containing NMD-inducing features escape degradation by the pathway. For example, multiple transcripts containing uORFs and atypically long 3′ UTRs appear to be immune to the NMD pathway. This observation raises the question as to whether the functionality of NMD-inducing features is condition specific. Specifically, are these NMD-inducing features only functional under particular physiological conditions and not in others?
The conservation of specific NMD-inducing features across eukaryotic ancestors has been debated. These ancestors, often referred to as stem eukaryotes, are believed to have had a functional NMD pathway. However, some suggest that the NMD pathway in stem eukaryotes degraded mRNAs with a long-3′-UTR-centered approach (85). This notion is supported by the observation that long 3′ UTRs as NMD-inducing features are observed in most eukaryotes. Furthermore, this model supports the idea that an intron-centered approach developed in NMD in vertebrates (85). Alternatively, a second model advocates the idea that both a long-3′-UTR-centered approach and an intron-based approach were present in eukaryotic ancestors (65, 66). This model is supported by the study of plant NMD. In plants, both long-3′-UTR-based and intron-based NMD occur (65, 66). Specifically, long-3′-UTR-based NMD in plants shows similarities to that in yeast and Drosophila, while intron-based NMD shows similarities to mammalian NMD (66). Since plants are distantly related to both animals and fungi, these results support the presence of both types of NMD in eukaryotic ancestors.
PHYSIOLOGICAL CONSEQUENCES RESULTING FROM THE REGULATION OF SPECIFIC NATURAL mRNAS BY NMD
The regulation of natural mRNAs by NMD raises several questions. Why are specific transcripts regulated by NMD? Is this regulation physiologically significant? Furthermore, is the regulation of specific transcripts condition specific? There are reports showing that the regulation of specific natural mRNAs by the pathway can be of functional significance. In S. cerevisiae, NMD mutants have reduced growth rates on nonfermentable carbon sources (86). The growth defect on nonfermentable carbon sources, such as lactate, is partly due to overexpression of ADR1 mRNA (87). In addition, S. cerevisiae NMD mutants are sensitive to Calcofluor white, a cell wall disruptor. This sensitivity is partially attributed to the regulation of the PGA1 mRNA by NMD (4). PGA1 encodes an essential component involved in glycosylphosphatidylinositol (GPI) anchor synthesis. Pga1p functions in association with Gpi18p to add the second mannose residue in the GPI anchor synthesis. It appears that a functional NMD pathway maintains the PGA1 mRNA at appropriate levels for GPI anchor synthesis (2). Moreover, NMD mutants have recently been shown to be more tolerant of toxic levels of copper partly due to the regulation of CTR2 mRNA by the pathway. CTR2 encodes a copper transporter of the vacuolar membrane that controls the flux of copper into the vacuole (88, 89). As previously stated, NMD has also been found to regulate magnesium homeostasis. Thus, S. cerevisiae NMD is involved in both copper and magnesium homeostasis by regulation of specific mRNAs (2, 3, 50, 88).
In addition, regulation of natural mRNAs by NMD or perturbation of NMD factors has physiological consequences in other organisms. For example, in C. elegans, mutations in the genes required for NMD result in morphogenetic effects on the genitalia and reduced numbers of offspring (90). In Arabidopsis, upf1 and upf3 mutants have multiple developmental phenotypes linking the NMD pathway to the regulation of genes during normal plant development and responses to pathogens (91, 92). A functional NMD pathway is also required for cell growth and differentiation in Drosophila (9, 93). In fact, NMD is essential for normal development in mice. Mice with defects in UPF1 and UPF2 die during embryogenesis (94, 95). Similarly, it has been shown that NMD effectors are essential for zebrafish embryonic development and survival (13). As previously stated, UPF1 plays additional roles in cells distinct from NMD, and downregulation of UPF1 could affect other cellular processes. Nevertheless, these studies show that, although a functional NMD pathway is not required for viability of S. cerevisiae, a functional pathway is required for the development and viability of multiple organisms.
Studies have shown that NMD has physiological consequences in humans as well. It is estimated that ∼30% of disease-associated mutations lead to the production of transcripts containing PTCs (96–98). The majority of these PTC-containing transcripts would not be categorized as natural mRNAs. Nevertheless, these transcripts are subject to regulation by the NMD pathway. Thus, NMD can play a critical role in whether or not these diseases manifest themselves in patients. In instances where the location of the PTC results in the generation of a nonfunctional truncated protein, NMD is beneficial to the patients. In other cases, where the location of the PTC results in the generation of a protein with partial function, NMD worsens the diseases by degrading the transcripts. Hence, NMD can be beneficial or detrimental depending on the specific mutations and the genes mutated (99). Additionally, mutations in the pathway or in any of the factors involved can have serious consequences. For example, in humans, mutated UPF3B has been linked to mental retardation, autism, and schizophrenia (100–102).
CONCLUSION
The NMD pathway exists as both a quality control mechanism and a mechanism to fine tune gene expression. As noted in this review, a subset of natural mRNAs with specific NMD-inducing features are recognized and rapidly degraded by the NMD pathway, demonstrating that the pathway plays an important role in the regulation of gene expression. A significant study discussed here that demonstrates the consequences of NMD regulation of specific natural transcripts is the ALR1 study. In S. cerevisiae, NMD regulates intracellular magnesium levels and, consequently, translational fidelity (78). NMD mutants have elevated ALR1 mRNA levels and, as a result, increased intracellular magnesium levels, which leads to reduced translational fidelity and read-through at PTCs.
Much is known about the factors required for NMD in S. cerevisiae and other eukaryotes. The repertoire of NMD-targeted transcripts is also beginning to be recognized. As discussed here, natural mRNAs regulated by NMD have been identified in multiple eukaryotes, and the NMD-inducing features for a subset of these natural mRNAs have been identified. These NMD-inducing features appear to be recognized in the same way in different organisms. Several studies suggest that the recognition of NMD-inducing features is conserved to some extent. In most cases, the presence of the NMD-targeting feature induces a premature translation termination event and activates NMD. However, there are apparent differences in the recognition of mRNA targets and the identity of the NMD targets themselves across species.
Two of the NMD-inducing features discussed have only been recognized in S. cerevisiae. These features are the −1 ribosomal frameshift signals and out-of-frame initiation of translation. Future studies are needed to determine the functionality of these features in other eukaryotic organisms. In addition, not all natural mRNAs regulated by the pathway possess recognizable NMD-inducing features, indicating that other NMD-targeting features exist. Furthermore, some natural mRNAs regulated by the pathway possess multiple NMD-inducing features. It is possible that these transcripts are subject to differential levels of NMD. It remains to be determined whether the presence of multiple NMD-targeting features on an mRNA could have an additive effect. If so, then these transcripts would be more efficiently degraded by the NMD pathway.
In summary, further investigation into the regulation of natural mRNAs by NMD will identify novel NMD-inducing features and enhance our understanding of how the pathway regulates transcripts with multiple NMD-inducing features. In addition, further investigation of the consequences that result from NMD regulation of specific transcripts will enhance our understanding of the extent to which NMD regulates particular cellular processes.
ACKNOWLEDGMENTS
We apologize for any related research not cited in this minireview due to space limitations. We acknowledge the helpful suggestions from three anonymous reviewers.
Research in the author's laboratory is supported by the Texas Higher Education Coordinating Board's Norman Hackerman Advanced Research Program and start-up funds from Baylor University.
Footnotes
Published ahead of print 18 July 2014
REFERENCES
- 1.Lelivelt MJ, Culbertson MR. 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19:6710–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, Tsui KW, Yandell BS, Culbertson MR. 2006. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2:e203. 10.1371/journal.pgen.0020203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson MJ, He F, Spatrick P, Li C, Jacobson A. 2007. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc. Natl. Acad. Sci. U. S. A. 104:20872–20877. 10.1073/pnas.0709257105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kebaara BW, Atkin AL. 2009. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 37:2771–2778. 10.1093/nar/gkp146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rebbapragada I, Lykke-Andersen J. 2009. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 21:394–402. 10.1016/j.ceb.2009.02.007 [DOI] [PubMed] [Google Scholar]
- 6.He F, Jacobson A. 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 21:1515–1530. 10.1128/MCB.21.5.1515-1530.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12:1439–1452. 10.1016/S1097-2765(03)00446-5 [DOI] [PubMed] [Google Scholar]
- 8.Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG. 2009. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 10:R101. 10.1186/gb-2009-10-9-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. 2005. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11:1530–1544. 10.1261/rna.2160905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36:1073–1078. 10.1038/ng1429 [DOI] [PubMed] [Google Scholar]
- 11.Wittmann J, Hol EM, Jäck HM. 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 26:1272–1287. 10.1128/MCB.26.4.1272-1287.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mekouar M, Blanc-Lenfle I, Ozanne C, Da Silva C, Cruaud C, Wincker P, Gaillardin C, Neuvéglise C. 2010. Detection and analysis of alternative splicing in Yarrowia lipolytica reveal structural constraints facilitating nonsense-mediated decay of intron-retaining transcripts. Genome Biol. 11:R65. 10.1186/gb-2010-11-6-r65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M, Izaurralde E. 2009. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 29:3517–3528. 10.1128/MCB.00177-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez-Gabriel MA, Watt S, Bähler J, Russell P. 2006. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol. Cell. Biol. 26:6347–6356. 10.1128/MCB.00286-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morozov IY, Negrete-Urtasun S, Tilburn J, Jansen CA, Caddick MX, Arst HN. 2006. Nonsense-mediated mRNA decay mutation in Aspergillus nidulans. Eukaryot. Cell 5:1838–1846. 10.1128/EC.00220-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leeds P, Peltz SW, Jacobson A, Culbertson MR. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303–2314. 10.1101/gad.5.12a.2303 [DOI] [PubMed] [Google Scholar]
- 17.Leeds P, Wood JM, Lee BS, Culbertson MR. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2165–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He F, Brown AH, Jacobson A. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17:1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page MF, Carr B, Anders KR, Grimson A, Anderson P. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19:5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culbertson MR, Leeds PF. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13:207–214. 10.1016/S0959-437X(03)00014-5 [DOI] [PubMed] [Google Scholar]
- 21.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O. 2010. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67:677–700. 10.1007/s00018-009-0177-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kervestin S, Jacobson A. 2012. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13:700–712. 10.1038/nrm3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112–118. 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6:1226–1235. 10.1017/S1355838200000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czaplinski K, Weng Y, Hagan KW, Peltz SW. 1995. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1:610–623 [PMC free article] [PubMed] [Google Scholar]
- 26.He F, Ganesan R, Jacobson A. 2013. Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast. Mol. Cell. Biol. 33:4672–4684. 10.1128/MCB.01136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng Y, Czaplinski K, Peltz SW. 1996. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 16:5491–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665–1677. 10.1101/gad.12.11.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YK, Furic L, Desgroseillers L, Maquat LE. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120:195–208. 10.1016/j.cell.2004.11.050 [DOI] [PubMed] [Google Scholar]
- 30.Kaygun H, Marzluff WF. 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12:794–800. 10.1038/nsmb972 [DOI] [PubMed] [Google Scholar]
- 31.Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM. 2011. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J. 30:4047–4058. 10.1038/emboj.2011.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azzalin CM, Lingner J. 2006. The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle 5:1496–1498. 10.4161/cc.5.14.3093 [DOI] [PubMed] [Google Scholar]
- 33.Amrani N, Dong S, He F, Ganesan R, Ghosh S, Kervestin S, Li C, Mangus DA, Spatrick P, Jacobson A. 2006. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 34:39–42. 10.1042/BST0340039 [DOI] [PubMed] [Google Scholar]
- 34.Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17:2108–2118. 10.1261/rna.030247.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 6:e92. 10.1371/journal.pbio.0060092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh G, Rebbapragada I, Lykke-Andersen J. 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 6:e111. 10.1371/journal.pbio.0060111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao D, Parker R. 2003. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113:533–545. 10.1016/S0092-8674(03)00353-2 [DOI] [PubMed] [Google Scholar]
- 38.Meaux S, van Hoof A, Baker KE. 2008. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol. Cell 29:134–140. 10.1016/j.molcel.2007.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popp MW, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 47:139–165. 10.1146/annurev-genet-111212-133424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lykke-Andersen J. 2001. mRNA quality control: marking the message for life or death. Curr. Biol. 11:R88–R91. 10.1016/S0960-9822(01)00036-7 [DOI] [PubMed] [Google Scholar]
- 41.Ishigaki Y, Li X, Serin G, Maquat LE. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106:607–617. 10.1016/S0092-8674(01)00475-5 [DOI] [PubMed] [Google Scholar]
- 42.Metze S, Herzog VA, Ruepp MD, Mühlemann O. 2013. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 19:1432–1448. 10.1261/rna.038893.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bühler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. 2006. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol. 13:462–464. 10.1038/nsmb1081 [DOI] [PubMed] [Google Scholar]
- 44.Rufener SC, Mühlemann O. 2013. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 20:710–717. 10.1038/nsmb.2576 [DOI] [PubMed] [Google Scholar]
- 45.Durand S, Lykke-Andersen J. 2013. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol. 20:702–709. 10.1038/nsmb.2575 [DOI] [PubMed] [Google Scholar]
- 46.González CI, Ruiz-Echevarría MJ, Vasudevan S, Henry MF, Peltz SW. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5:489–499. 10.1016/S1097-2765(00)80443-8 [DOI] [PubMed] [Google Scholar]
- 47.González CI, Bhattacharya A, Wang W, Peltz SW. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274:15–25. 10.1016/S0378-1119(01)00552-2 [DOI] [PubMed] [Google Scholar]
- 48.Maderazo AB, Belk JP, He F, Jacobson A. 2003. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol. Cell. Biol. 23:842–851. 10.1128/MCB.23.3.842-851.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mühlemann O, Lykke-Andersen J. 2010. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol. 7:28–32. 10.4161/rna.7.1.10578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaba A, Jacobson A, Sachs MS. 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20:449–460. 10.1016/j.molcel.2005.09.019 [DOI] [PubMed] [Google Scholar]
- 51.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc. Natl. Acad. Sci. U. S. A. 90:7034–7038. 10.1073/pnas.90.15.7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welch EM, Jacobson A. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18:6134–6145. 10.1093/emboj/18.21.6134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker R. 2012. RNA degradation in Saccharomyces cerevisae. Genetics 191:671–702. 10.1534/genetics.111.137265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. 2013. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 1829:612–623. 10.1016/j.bbagrm.2013.02.005 [DOI] [PubMed] [Google Scholar]
- 55.Ruiz-Echevarría MJ, Peltz SW. 2000. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101:741–751. 10.1016/S0092-8674(00)80886-7 [DOI] [PubMed] [Google Scholar]
- 56.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell 31:360–370. 10.1016/j.molcel.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sayani S, Chanfreau GF. 2012. Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae. RNA 18:1563–1572. 10.1261/rna.033779.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Juneau K, Palm C, Miranda M, Davis RW. 2007. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc. Natl. Acad. Sci. U. S. A. 104:1522–1527. 10.1073/pnas.0610354104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawashima T, Douglass S, Gabunilas J, Pellegrini M, Chanfreau GF. 2014. Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae. PLoS Genet. 10:e1004249. 10.1371/journal.pgen.1004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, Brenner SE. 2009. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 5:e1000525. 10.1371/journal.pgen.1000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitrovich QM, Anderson P. 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 14:2173–2184. 10.1101/gad.819900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. 2012. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 13:R35. 10.1186/gb-2012-13-5-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 28:4320–4330. 10.1128/MCB.00361-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbazuk WB, Fu Y, McGinnis KM. 2008. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 18:1381–1392. 10.1101/gr.053678.106 [DOI] [PubMed] [Google Scholar]
- 65.Kertész S, Kerényi Z, Mérai Z, Bartos I, Pálfy T, Barta E, Silhavy D. 2006. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34:6147–6157. 10.1093/nar/gkl737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kerényi Z, Mérai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. 2008. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27:1585–1595. 10.1038/emboj.2008.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nyikó T, Kerényi F, Szabadkai L, Benkovics AH, Major P, Sonkoly B, Mérai Z, Barta E, Niemiec E, Kufel J, Silhavy D. 2013. Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 41:6715–6728. 10.1093/nar/gkt366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Ségurens B, Le Mouël A, Lepère G, Schächter V, Bétermier M, Cohen J, Wincker P, Sperling L, Duret L, Meyer E. 2008. Translational control of intron splicing in eukaryotes. Nature 451:359–362. 10.1038/nature06495 [DOI] [PubMed] [Google Scholar]
- 69.Graber JH, McAllister GD, Smith TF. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30:1851–1858. 10.1093/nar/30.8.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deliz-Aguirre R, Atkin AL, Kebaara BW. 2011. Copper tolerance of Saccharomyces cerevisiae nonsense-mediated mRNA decay mutants. Curr. Genet. 57:421–430. 10.1007/s00294-011-0356-0 [DOI] [PubMed] [Google Scholar]
- 71.Kim Guisbert KS, Li H, Guthrie C. 2007. Alternative 3′ pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo. PLoS Biol. 5:e6. 10.1371/journal.pbio.0050006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. 2014. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156:812–824. 10.1016/j.cell.2013.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 23:1636–1650. 10.1101/gr.157354.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Longman D, Hug N, Keith M, Anastasaki C, Patton EE, Grimes G, Cáceres JF. 2013. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res. 41:8319–8331. 10.1093/nar/gkt585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, Dinh HQ, Barta A, Brown JW. 2012. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40:2454–2469. 10.1093/nar/gkr932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arribere JA, Gilbert WV. 2013. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 23:977–987. 10.1101/gr.150342.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349. 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johansson MJ, Jacobson A. 2010. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 24:1491–1495. 10.1101/gad.1930710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U. S. A. 106:7507–7512. 10.1073/pnas.0810916106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stockklausner C, Breit S, Neu-Yilik G, Echner N, Hentze MW, Kulozik AE, Gehring NH. 2006. The uORF-containing thrombopoietin mRNA escapes nonsense-mediated decay (NMD). Nucleic Acids Res. 34:2355–2363. 10.1093/nar/gkl277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nyikó T, Sonkoly B, Mérai Z, Benkovics AH, Silhavy D. 2009. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 71:367–378. 10.1007/s11103-009-9528-4 [DOI] [PubMed] [Google Scholar]
- 82.Kochetov AV, Syrnik OA, Rogozin IB, Glazko GV, Komarova ML, Shumnyi VK. 2002. Context organization of mRNA 5′-untranslated regions of higher plants. Mol. Biol. (Mosk) 36:649–656 (In Russian.) [PubMed] [Google Scholar]
- 83.Belew AT, Advani VM, Dinman JD. 2011. Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 39:2799–2808. 10.1093/nar/gkq1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamilton R, Watanabe CK, de Boer HA. 1987. Compilation and comparison of the sequence context around the AUG start codons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 15:3581–3593. 10.1093/nar/15.8.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rehwinkel J, Raes J, Izaurralde E. 2006. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem. Sci. 31:639–646. 10.1016/j.tibs.2006.09.005 [DOI] [PubMed] [Google Scholar]
- 86.Altamura N, Groudinsky O, Dujardin G, Slonimski PP. 1992. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J. Mol. Biol. 224:575–587. 10.1016/0022-2836(92)90545-U [DOI] [PubMed] [Google Scholar]
- 87.Taylor R, Kebaara BW, Nazarenus T, Jones A, Yamanaka R, Uhrenholdt R, Wendler JP, Atkin AL. 2005. Gene set coregulated by the Saccharomyces cerevisiae nonsense-mediated mRNA decay pathway. Eukaryot. Cell 4:2066–2077. 10.1128/EC.4.12.2066-2077.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang X, Okonkwo O, Kebaara BW. 2013. Physiological basis of copper tolerance of Saccharomyces cerevisiae nonsense-mediated mRNA decay mutants. Yeast 30:179–190. 10.1002/yea.2950 [DOI] [PubMed] [Google Scholar]
- 89.Rees EM, Lee J, Thiele DJ. 2004. Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J. Biol. Chem. 279:54221–54229. 10.1074/jbc.M411669200 [DOI] [PubMed] [Google Scholar]
- 90.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123:301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. 2006. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47:480–489. 10.1111/j.1365-313X.2006.02802.x [DOI] [PubMed] [Google Scholar]
- 92.Rayson S, Arciga-Reyes L, Wootton L, De Torres Zabala M, Truman W, Graham N, Grant M, Davies B. 2012. A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS One 7:e31917. 10.1371/journal.pone.0031917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Metzstein MM, Krasnow MA. 2006. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2:e180. 10.1371/journal.pgen.0020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10:99–105. 10.1093/hmg/10.2.99 [DOI] [PubMed] [Google Scholar]
- 95.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. 2008. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 22:1381–1396. 10.1101/gad.468808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. 2010. NMD: RNA biology meets human genetic medicine. Biochem. J. 430:365–377. 10.1042/BJ20100699 [DOI] [PubMed] [Google Scholar]
- 97.Kuzmiak HA, Maquat LE. 2006. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 12:306–316. 10.1016/j.molmed.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 98.Linde L, Kerem B. 2008. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 24:552–563. 10.1016/j.tig.2008.08.010 [DOI] [PubMed] [Google Scholar]
- 99.Nguyen LS, Wilkinson MF, Gecz J. 2013. Nonsense-mediated mRNA decay: inter-individual variability and human disease. Neurosci. Biobehav. Rev. 2013:S0149-7634(13)00270-4. 10.1016/j.neubiorev.2013.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, Tossell J, Bakalar J, Inoff-Germain G, Germain G, Gochman P, Long R, Rapoport JL, Rouleau GA. 2011. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol. Psychiatry 16:238–239. 10.1038/mp.2010.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, Briault S, Fryns JP, Hamel B, Chelly J, Ropers HH, Ronce N, Blesson S, Moraine C, Gécz J, Raynaud M. 2010. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol. Psychiatry 15:767–776. 10.1038/mp.2009.14 [DOI] [PubMed] [Google Scholar]
- 102.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O'Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J, Varian J, West S, Widaa S, Teague J, Dicks E, Butler A, Menzies A, Richardson D, Jenkinson A, Shepherd R, Raine K, Moon J, Luo Y, Parnau J, Bhat SS, Gardner A, Corbett M, Brooks D, Thomas P, Parkinson-Lawrence E, Porteous ME, Warner JP, Sanderson T, Pearson P, Simensen RJ, Skinner C, Hoganson G, Superneau D, Wooster R, Bobrow M, Turner G, Stevenson RE, Schwartz CE, Futreal PA, Srivastava AK, Stratton MR, Gécz J. 2007. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat. Genet. 39:1127–1133. 10.1038/ng2100 [DOI] [PMC free article] [PubMed] [Google Scholar]