Abstract
Candida albicans vacuoles are central to many critical biological processes, including filamentation and in vivo virulence. The V-ATPase proton pump is a multisubunit complex responsible for organellar acidification and is essential for vacuolar biogenesis and function. To study the function of the V1B subunit of C. albicans V-ATPase, we constructed a tetracycline-regulatable VMA2 mutant, tetR-VMA2. Inhibition of VMA2 expression resulted in the inability to grow at alkaline pH and altered resistance to calcium, cold temperature, antifungal drugs, and growth on nonfermentable carbon sources. Furthermore, V-ATPase was unable to fully assemble at the vacuolar membrane and was impaired in proton transport and ATPase-specific activity. VMA2 repression led to vacuolar alkalinization in addition to abnormal vacuolar morphology and biogenesis. Key virulence-related traits, including filamentation and secretion of degradative enzymes, were markedly inhibited. These results are consistent with previous studies of C. albicans V-ATPase; however, differential contributions of the V-ATPase Vo and V1 subunits to filamentation and secretion are observed. We also make the novel observation that inhibition of C. albicans V-ATPase results in increased susceptibility to osmotic stress. Notably, V-ATPase inhibition under conditions of nitrogen starvation results in defects in autophagy. Lastly, we show the first evidence that V-ATPase contributes to virulence in an acidic in vivo system by demonstrating that the tetR-VMA2 mutant is avirulent in a Caenorhabditis elegans infection model. This study illustrates the fundamental requirement of V-ATPase for numerous key virulence-related traits in C. albicans and demonstrates that the contribution of V-ATPase to virulence is independent of host pH.
INTRODUCTION
The fungal pathogen Candida albicans is the fourth most common cause of hospital-acquired bloodstream infections and is a major cause of catheter-associated infections, sepsis, and device-related infections. It is also an extremely common cause of urinary and mucosal infections. Despite its clinical significance, the diagnosis and treatment of disseminated candidiasis remain limited by an incomplete understanding of its molecular pathogenesis. The fungal vacuole, a degradative organelle roughly equivalent to the mammalian lysosome, plays an important role in numerous biological processes in C. albicans, including key aspects of pathogenesis. Previous studies have established that C. albicans mutants compromised in vacuolar function are defective in yeast-to-hypha transitioning, a major virulence-related trait, and exhibit reduced virulence in vivo (1, 2).
An essential component of vacuolar biogenesis and function is the vacuolar H+-ATPase (V-ATPase) proton pump, which is a multisubunit complex responsible for the acidification of internal organelles. V-ATPase is located at the vacuolar membrane and throughout the endomembrane system, including prevacuolar compartments and the Golgi complex (1). The acidification of these organelles has several important functions: first, the protons pumped into the compartment by V-ATPase energize multiple secondary transporter systems, such as those involved in metal ion homeostasis, and second, the acidic pH created by V-ATPase is necessary for the activity of degradative enzymes. Accordingly, in both Saccharomyces cerevisiae and C. albicans, mutations in V-ATPase lead to the Vma− phenotype, which is characterized by poor growth at alkaline pH as well as defective metal sequestration, glycerol metabolism, and other responses to environmental stress conditions (1, 3–6). V-ATPase mutants are also impaired in vacuolar biogenesis due to defects in both vacuolar membrane fusion and fission (7). In addition to its requirement for intraorganellar pH homeostasis, V-ATPase function has been implicated in cytosolic and extracellular pH homeostasis (1, 8–11), underscoring the far-reaching importance of this pump to overall cellular homeostasis. V-ATPase inhibition also interferes with virulence-related traits in C. albicans, including the secretion of degradative enzymes and yeast-to-hypha transitioning (3, 4, 12, 13). Notably, V-ATPase previously has been shown to be important for in vivo virulence in C. albicans (3, 13); however, the requirement of acidic environmental pH for the growth of V-ATPase mutants complicates the interpretation of these studies due to the growth-limiting alkaline pH in the bloodstream of the murine host.
V-ATPase is composed of two multisubunit domains, Vo and V1. Vo is embedded in the organellar membrane and is the site of proton transport. V1 contains the catalytic subunits of the complex responsible for ATP hydrolysis at the cytosolic side of the membrane. The catalytic portion of V-ATPase is a hexamer composed of three copies of V1 subunit A (V1A) and three copies of V1 subunit B (V1B), which alternate in configuration. V1A is the primary site of ATP hydrolysis, whereas V1B (encoded by the VMA2 gene) plays a regulatory role in ATP hydrolysis and contributes to ATP-binding sites (6). Studies of S. cerevisiae have shown that disruption of VMA2 completely inhibits both ATPase activity and proton transport by the V-ATPase (14). We have previously investigated the contribution of several subunits of C. albicans V-ATPase to cell biology and virulence-related traits (4, 12). This is the first study analyzing a subunit of the catalytic hexamer in the V1 domain of V-ATPase in C. albicans. We generated a tetracycline-regulatable mutant of VMA2 in order to establish the contribution of the V1B subunit of the V-ATPase to pH and stress response, V-ATPase function, and vacuolar morphology. We also analyzed VMA2 contribution to C. albicans virulence-related traits, including filamentation and secretion of degradative enzymes. We next studied the effect of V-ATPase inactivation on autophagy, the recycling of cellular building blocks in response to starvation and stress, by monitoring long-term survival during nitrogen starvation and turnover of the autophagy-related protein Ape1p. Finally, we utilized a Caenorhabditis elegans model of infection in the first study of V-ATPase contribution to virulence in an acidic host environment.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table 1. Standard growth was completed at 30°C in yeast peptone dextrose (YPD; 1% yeast extract, 2% peptone, and 2% glucose) supplemented with 80 μg/ml uridine where required. When needed, doxycycline (DOX) was added to a final concentration of 20 μg/ml. Media were buffered to pH 4.0 to 5.0 using 50 mM succinic acid–50 mM Na2PO4 or to pH 7.5 to 8.5 using 50 mM morpholineethanesulfonic acid (MES) hydrate–50 mM morpholinepropanesulfonic acid (MOPS) where required. Unless otherwise specified, agar plates were prepared with 2% agar. For all experiments, cells were grown for 24 h in unbuffered YPD with or without DOX prior to the start of the experiment to ensure the complete turnover of extant Vma2p. Where the pH of the media is not explicitly stated, the media used were not buffered to a specific pH (i.e., unbuffered media). All plates were incubated at 30°C for 48 h unless otherwise stated.
TABLE 1.
List of strains used in this study
| Strain | Parent | Relevant genotype | Source |
|---|---|---|---|
| BWP17 | SC5314 | ura3Δ/ura3Δ arg4Δ/arg4Δ hisΔ/his1Δ VMA2/VMA2 | Wilson et al. (17) |
| BWP17-VMA2Δ/+ | BWP17 | ura3Δ/ura3Δ arg4Δ/arg4Δ hisΔ/his1Δ VMA2/VMA2Δ::dpl200-URA3-dpl200 | This study |
| THE1 | CAI8 | ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 VMA2/VMA2 | Nakayama et al. (18) |
| THE1-CIp10 | THE1 | ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 RP10/RP10::URA3 VMA2/VMA2 | Bernardo et al. (20) |
| VMA2Δ/+ | THE1 | ura3Δ::imm434/ura3Δ::imm434 VMA2/VMA2Δ::dpl200-URA3-dpl200 ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 | This study |
| VMA2Δ/+ FOA | VMA2Δ/+ | ura3Δ::imm434/ura3Δ::imm434 VMA2/VMA2Δ::dpl200 ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 | This study |
| tetR-VMA2 | VMA2Δ/+ FOA | ura3Δ::imm434/ura3Δ::imm434 VMA2Δ::dpl200::99t-VMA2-URA3 ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 | This study |
| tetR-VMA3 | VMA3Δ/+ FOA | ura3Δ::imm434/ura3Δ::imm434 VMA3Δ::dpl200::99t-VMA3-URA3 ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 | Rane et al. (4) |
| T-LAP41-GFP | THE1-CIp10 | ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 RP10/RP10::URA3 VMA2/VMA2 LAP41/LAP41-GFP-NAT1 | This study |
| tV2-LAP41-GFP | tetR-VMA2 | ura3Δ::imm434/ura3Δ::imm434 VMA2Δ::dpl200::99t-VMA2-URA3 ade2Δ::hisG/ade2Δ::hisG ura3Δ::imm434/ura3Δ::imm434 ENO1/eno1Δ::ENO1-tetR-ScHAP4AD-3× HA-ADE2 LAP41/LAP41-GFP-NAT1 | This study |
Statistical analyses.
Results were analyzed for statistical differences using one-way analysis of variance (ANOVA), followed by Tukey's multiple-comparison test (GraphPad Prism 6). A result was considered significant when P < 0.05 compared to all other treatments.
C. albicans sequence data.
Sequence data for C. albicans were obtained from the Candida Genome Database website at http://www.candidagenome.org.
Attempted disruption of C. albicans VMA2.
The primers used in this study are listed in Table 2. A BLASTp search of the Candida Genome Database (http://www.candidagenome.org/) revealed a single potential ortholog to S. cerevisiae Vma2p (http://www.yeastgenome.org/), C. albicans orf19.6634. We first attempted to generate a VMA2 null mutant in C. albicans using a PCR-based gene disruption strategy (15). Primers VMA2-5DR and VMA2-3DR were used to amplify the dpl200-URA3-dpl200-containing plasmid pDDB57 (from A. Mitchell, Carnegie Mellon University). C. albicans BWP17 was transformed with the VMA2Δ::dpl200-URA3-dpl200 PCR amplicon using the lithium acetate method. Because vma mutants exhibit poor growth at alkaline pH, selective media were buffered to pH 4.0 to 5.0 to facilitate VMA2 disruption. Genomic DNA was extracted from transformants as described previously (16). The transformants were screened for homologous reintegration via PCR with primers VMA2-5Det and VMA2-3Det. Disruption of the second allele of VMA2 was attempted using the VMA2-5DR and VMA2-3DR primers to amplify the plasmid pRS-ARG4ΔSpeI (from A. Mitchell, Carnegie Mellon University) (17). This VMA2Δ::ARG4 PCR amplicon was used to transform the BWP17-VMA2Δ/+ strain via the lithium acetate method using selective media, without arginine, buffered to pH 4.0 or 5.0. The genotype of the transformants was assessed by allele-specific PCR using primers VMA2-5Det and VMA2-3Det.
TABLE 2.
Primers used in this study
| Primer | Primer sequence (5′–3′) | Source |
|---|---|---|
| VMA2–5DR | ATGGAAAAAGAGAGAGTAATTGGCCGTTTCTTATCGTCCTTTTGTTGTACTCTTTTAAATTTTTCATTTAGTTTTCCCAGTCACGACGTT | This study |
| VMA2–3DR | ATAGATAATTTTTGTCAGAATTAAAAAAAATGAAAATAATAATGATAATAATAGTAAATAAAACTAATGATGTGGAATTGTGAGCGGATA | This study |
| VMA2–5Det | GCAACGAATGGGACATAGTG | This study |
| VMA2–3Det | GATATGATTGCCAAGGACGG | This study |
| tetVMA2–5DR | TGATCGAATCTATACCGAGTCTACTACGAATACGGAGGTTACTCAGATCCCAAAAAAATTTGACACTAAAGTAATACGACTCACTATAGGG | This study |
| tetVMA2–3DR | ATTTTAAACCCTTCTGTGACTGCTTTTTTGTTTAATTCAAACAATTCTTTATCAGAGAGTGACATTATTGCTAGTTTTCTGAGATAAAGCTG | This study |
| tetVMA2–5Det | GTCATCTGTCGAGCCTGCAG | This study |
| tetVMA2–3Det | CCATTGACACCACCAACAGT | This study |
| tetINS-3Det | CTAGTTTTCTGAGATAAAGCTG | Rane et al. (4) |
| RT-VMA2–5Det | CCACGTTTGGCCTTGACTAC | This study |
| RT-VMA2–3Det | TCTTCTGTAGACAATGCTTC | This study |
| LAP41-GFP-5DR | ATTAGGTATTAAATTCTTCTATGGTTTCTTCAAGAATTGGAGAGATGTCTATGATAATTTTGTTGATTTAGGTGGTGGTTCTAAAGGTGAAGAATTATT | This study |
| LAP41-GFP-3DR | TAAACTAACAAATAACTAATACTTGCAAATCAACTTTTAATGATTCCTTTTCCTGTTGTTCAATGTTCGACGTTAGTATCGAATCGACAGC | This study |
| LAP41-GFP-5Det | GCCTTGGCTGAAGAATTGGC | This study |
| GFP-UP | CACCTTCACCGGAGACAG | Milne et al. (33) |
Construction of a tetracycline-regulated VMA2 gene.
One C. albicans VMA2 allele was deleted from the THE1 strain background (18) using PCR-based gene disruption (15) and lithium acetate transformation. Primers VMA2-5DR and VMA2-3DR (Table 2) were used to amplify plasmid pDDB57. Strain THE1 then was transformed with the resulting PCR amplicon to generate VMA2Δ/+ (Table 1). Correct genomic integration of the gene disruption cassette was confirmed via PCR using primers VMA2-5Det and VMA2-3Det. The VMA2Δ/+ strain then was plated to 5-fluoroorotic acid (5-FOA) agar media, and the resultant 5-FOA-resistant colonies were screened via PCR for the VMA2/VMA2Δ::dpl200 genotype using primers VMA2-5Det and VMA2-3Det. The tetracycline-regulatable system described by Nakayama et al. (18), with modifications described by Bates et al. (19), was used to place the remaining VMA2 allele under a tetracycline-regulatable promoter. Plasmid p99CAU1 (from H. Nakayama, Suzuka University) was amplified using primers tetVMA2-5DR and tetVMA2-3DR. The resulting PCR amplicon was inserted upstream of the remaining VMA2 allele of the VMA2Δ/+ 5-FOA strain via lithium acetate transformation. Transformants were screened for the correct insertion of the tetR-VMA2 allele using primers tetVMA2-5Det and tetVMA2-3Det (Table 2). Reverse transcription-PCR (RT-PCR) was performed to assay expression of VMA2 in both the THE1-CIp10 control strain and the tetR-VMA2 strain after 24 h of growth in YPD with or without DOX. After 24 h of growth in YPD with or without DOX, RNA was extracted from cells using the RiboPure yeast kit (Ambion) according to the manufacturer's protocol, with 1 μl of the RNase inhibitor RNasin (Promega) added after the binding, elution, and DNase I steps to prevent RNA degradation. mRNA then was extracted from total RNA using the PolyATtract mRNA isolation system (Promega) according to the manufacturer's instructions. One μl RNasin was added to each treatment after the final elution step. Finally, RT-PCR was completed with the Access RT-PCR system (Promega) according to the manufacturer's instructions using primers RT-VMA2-5Det and RT-VMA2-3Det and 5 μg total mRNA as the template. The absence of contaminating DNA was tested in parallel PCR-based reactions.
pH and stress response.
The ability of tetR-VMA2 to respond to various pHs was tested on complete synthetic media (CSM; 0.67% yeast nitrogen base without amino acids [YNB], 0.079% complete synthetic mixture, 2% glucose) agar plates buffered to pH 4.0 to 8.5 with or without DOX. The THE1-CIp10 control strain, a strain from the THE1 background with URA3 reintegrated into the genome (20), was used as an additional control in this and all subsequent experiments. THE1-CIp10 also was studied with and without DOX to control for the effect of DOX on this and all subsequent experiments. The ability of tetR-VMA2 to grow on unbuffered media was tested on CSM with or without DOX; unbuffered CSM has a pH of ∼6. Cells from overnight cultures were washed and counted as previously described (21); next, 108 cells per ml were inoculated in phosphate-buffered saline (PBS). Five 5-fold dilutions then were completed in 96-well plates, and cells were stamped onto agar plates using a multiblot replicator per the manufacturer's instructions (VP 408H; VP Scientific). Plates were incubated at 30°C for 48 h. Growth at pH 4.0 to 8.5 also was tested in liquid media: cells from overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in CSM, pH 4.0 to 8.5, with or without DOX. Cells were grown at 30°C using a Synergy H1 microplate reader (Biotek) with double orbital shaking at fast speed and 2-mm frequency; OD600 readings were taken at 15-min intervals.
The ability of tetR-VMA2 to respond to various stress conditions was tested on agar plates with or without DOX. The calcium stress response was tested on unbuffered YPD plus 200 mM CaCl2. Plates used to test the response of strains to media containing glycerol as the sole carbon source contained unbuffered yeast extract plus peptone (YEP)–2% ethanol–3% glycerol. Plates used to test the response of strains to oxidative stress contained CSM, pH 4.0, with 5 mM H2O2; for the oxidative stress assay, plates were incubated at 30°C for 72 h rather than 48 h. The ability of strains to respond to challenge with antifungals was tested on YPD, pH 4.0, with or without DOX with 0.00625 to 0.5 μg/ml amphotericin B (AMB); 0.05 μg/ml caspofungin (CAS); 10 to 20 μg/ml flucytosine (FCT); or 0.6125, 1.25, 2.5, 5, or 10 μg/ml fluconazole (FLU). For fluconazole susceptibility studies, the tetR-VMA3 strain also was included (4). Temperature sensitivity was assessed by stamping cells onto unbuffered YPD with or without DOX agar plates and incubating at 25°C, 30°C, 37°C, or 42°C for 48 h. CSM, pH 4.0, with or without DOX plates containing either 1 M NaCl, 200 μg/ml Congo red, or 50 μg/ml calcofluor white also were tested. For all plates, cells were stamped onto media using a multiblot replicator as described above.
V-ATPase assembly and activity assays.
Starter cultures were grown for 6 to 8 h in YPD, pH 4.0, with or without DOX. Vacuoles were prepared by resetting cells in YPD, pH 4.0, with or without DOX and growing to them to an OD600 of 1.0 to 1.5 (18 h). Total growth time in YPD, pH 4, with or without DOX before vacuolar membrane purification was 24 h to 26 h. Ficoll density gradient centrifugation (22) was used to purify vacuolar membranes. For Western blotting, 80 μg of vacuolar protein was separated by SDS-PAGE and transferred to nitrocellulose at 150 mA overnight. The V1A subunit was visualized with a 1:1,000 dilution of anti-human V1A rabbit polyclonal antibody, which cross-reacts with C. albicans Vma1p (aka V1A) (23).
In order to quantify ATP hydrolysis, 15 μg purified vacuolar vesicles was used in an enzymatic assay in which the rate of ATP hydrolysis is coupled to the oxidation of NADH, measured as a loss of A340 over time (24) in the presence and absence of the V-ATPase inhibitor concanamycin A (12, 25). Proton transport of 30 μg purified vacuolar vesicles was measured via quenching of 1 μM 9-amino-6-chloro-2-methoxyacridine (ACMA) upon the addition of 0.5 mM ATP–1 mM MgSO4 (MgATP) as described previously (25, 26). Fluorescence at excitation and emission wavelengths of 410 nm and 490 nm, respectively, was monitored for 60 s prior to MgATP addition and for 40 s after. Proton transport was calculated as the change in fluorescence for the first 15 s following MgATP addition. For both assays, 100 nM concanamycin A, a specific V-ATPase inhibitor, was used to assess V-ATPase-specific activity.
Vacuolar acidification assays.
Quinacrine staining was performed to visualize acidified vacuoles as described previously (27), with modifications as indicated previously (4). Briefly, cells were grown for 24 h in unbuffered YPD with or without DOX and then reset in fresh unbuffered YPD with or without DOX and grown to early log phase. Cells were stained with 200 μM quinacrine in YPD buffered with 50 mM Na2PO4, pH 7.6, for 10 to 15 min. Cells were washed and resuspended in 100 mM HEPES–50 mM Na2PO4, pH 7.6–2% glucose before visualization via differential interference contrast (DIC) and fluorescence microscopy. Vacuolar pH was quantified as described previously (12) using 2′-7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein-acetoxymethyl ester (BCECF-AM; from Invitrogen).
Vacuolar morphology.
For all vacuolar staining, DOX was added to the appropriate treatments upon each medium change. To simultaneously stain vacuoles with FM4-64 [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide] and CMAC (7-amino-4-chloromethyl coumarin), cells were grown for 24 h in unbuffered YPD with or without DOX. Cells then were reset in fresh unbuffered YPD with or without DOX and grown to early log phase. Cells were resuspended to an OD600 of 2 to 4 in unbuffered YPD containing 40 μM FM4-64 and incubated for 15 min at 30°C, reset in fresh unbuffered YPD, and incubated for 45 min at 30°C. Cells were resuspended to an OD600 of 0.1 in 10 mM HEPES, 5% glucose, pH 7.4. CMAC was added to a concentration of 100 μM, and cells were incubated at room temperature for 15 min and examined via DIC microscopy and fluorescence microscopy with a Zeiss Axio Imager (Carl Zeiss AG, Germany) using Texas red (FM4-64) and 4′,6-diamidino-2-phenylindole (DAPI) (CMAC) filters.
Filamentation and secretion assays.
Filamentation was assessed on solid and in liquid media with or without DOX. Media tested were YPD plus 10% fetal calf serum (FCS), medium 199 supplemented with l-glutamine (M199), Spider medium as previously described (28), and RPMI–l-glutamine. All media were tested unbuffered as well as buffered to pH 4.0. Spider medium was prepared with 1.35% (wt/vol) agar. Three μl cells from overnight cultures were spotted to agar plates and incubated at 37°C for 120 h. Filamentation in liquid media was tested in RPMI–l-glutamine, pH 5.0, with or without DOX. Media were inoculated with cells from overnight cultures to a starting density of 5 × 106 cells per ml, and cells were grown at 37°C, 200 rpm for 2 to 24 h. Cells were visualized via DIC microscopy using a Zeiss EC Plan-NeoFluar 63×/1.25 oil objective (Carl Zeiss AG, Germany) at selected time points.
Aspartyl protease and lipase secretion was assessed on solid media. Extracellular protease secretion was assayed on unbuffered bovine serum albumin (BSA) plates (29), and lipase secretion was assayed on plates containing unbuffered YNB plus 2.5% Tween 80 (30). All plates were prepared with or without DOX. First, cells were grown in YPD with or without DOX for 24 h. Three μl cells then was spotted onto plates. BSA plates were incubated at 30°C for 48 h, and Tween 80 plates were incubated at 37°C for 120 h. For aspartyl protease secretion, the relative halo size was calculated using the software program TotalLab 100. First, THE1-CIp10 and tetR-VMA2 were grown in YPD with or without DOX for 24 h, and then 3 μl cells from each treatment was spotted onto six BSA plates and onto six BSA-plus-DOX plates. Plates were grown at 30°C for 48 h and scanned. For each colony spot, the areas, in pixels of the colony alone and of the halo and colony combined, were quantified using TotalLab 100. The relative area of the halo was calculated as the area of the halo and colony divided by the area of the colony alone; a relative halo size of 1 would indicate a lack of detectable halo. Relative halo size is given as the averages ± standard deviations from 6 replicates. One-way ANOVA was used to compare the relative halo area for each treatment.
Long-term survival during nitrogen starvation.
In order to assess the ability of tetR-VMA2 to survive long-time nitrogen starvation, we analyzed tetR-VMA2 resistance to nitrogen starvation using a previously described assay (31). Strains THE1-CIp10 and tetR-VMA2 first were grown for 24 h in unbuffered YPD ± DOX, and then 108 cells per treatment were transferred to a microcentrifuge tube and washed twice in 1 ml nitrogen starvation media (i.e., SD − N; 0.17% YNB without ammonium sulfate or amino acids plus 2% glucose) before resuspending in 1 ml SD − N. One hundred μl of this cell suspension was added to 4.9 ml SD − N with or without DOX for a final cell concentration of 2 × 106 cells per ml. At 24-h intervals, fresh DOX to a concentration of 20 μg/ml was added to appropriate treatments. Dilutions (1:1,000) were completed at selected time points. One hundred μl of the resulting dilution was plated to YPD in triplicate for CFU counts. On days 0, 10, 20, and 30 of the experiment, the pH of the media was tested by transferring 100 μl SD − N from each treatment to pH test strips (Sigma-Aldrich).
In order to further study the contribution of VMA2 to autophagy, we utilized the commonly used autophagy marker aminopeptidase I (Ape1), encoded by the C. albicans LAP41 gene (31, 32). Ape1p was C-terminally tagged with green fluorescent protein (GFP) in the THE1-CIp10 and tetR-VMA2 backgrounds to generate strains T-LAP41-GFP and tV2-LAP41-GFP (Table 1). First, primers LAP41-GFP-5DR and LAP41-GFP-3DR (Table 2) were used to amplify the GFP-NAT1 cassette from plasmid pGFP-NAT1 (from S. Bates, University of Exeter) (33); these primers use 70 nucleotides of LAP41 homologous targeting sequences as previously described (31), combined with sequences for amplification of the GFP-NAT1 cassette (33). Strains THE1-CIp10 and tetR-VMA2 were transformed with the resulting PCR amplicon as previously described (33), and transformants were selected for on Sabouraud-dextrose agar (BD, Franklin Lakes, NJ) containing 200 μg/ml nourseothricin (Gold Biotechnology, St. Louis, MO). Primers LAP41-GFP-5Det and GFP-UP (Table 2) were used to screen for correct integration of the LAP41-GFP allele. For visualization of Ape1-GFP, strains T-LAP41-GFP and tV2-LAP41-GFP were grown for 24 h in unbuffered YPD with or without DOX prior to the start of the experiment. Cells then were washed twice in PBS, resuspended to an OD600 of 0.2 in fresh SD − N with or without DOX, and grown at 30°C for 1 h at 200 rpm. FM4-64 was added to a concentration of 40 μM. Cells were grown at 30°C for 15 min at 200 rpm, spun down, and resuspended in fresh SD − N with or without DOX media to remove excess FM4-64 stain. Cells were grown at 30°C for 20 days at 200 rpm, with fresh DOX added to appropriate treatments to a concentration of 20 μg/ml at 24-h intervals. After 2, 4, 6, 24, 48, 120, and 240 h in nitrogen starvation media, cells were visualized with a Zeiss Axio Imager and AxioVision 4.7 software (Carl Zeiss AG, Germany). Ape1-GFP fluorescence images were acquired using a GFP cube (400-ms exposure time), and FM4-64 fluorescence images were acquired using a Texas red cube (100-ms exposure time).
C. elegans infection assays.
C. elegans solid medium killing assays were performed as described previously (34), with some important modifications. First, all infection assays were completed at 30°C rather than 25°C due to the cold sensitivity of the tetR-VMA2 strain upon DOX repression (see Fig. 2A). Further, previous studies have utilized either C. elegans glp-4;sek-1 nematodes rather than wild-type nematodes (35) or treatment with 5-fluoro-2′-deoxyuridine (34) to avoid the complicating presence of eggs and larvae during the infection assay. However, because our experiments were conducted at 30°C, a temperature at which C. elegans reproduction, but not viability, is completely inhibited (36), we were able to use the C. elegans wild-type strain, N2. To ensure that exposure to chronic heat stress or doxycycline was not the primary cause of worm mortality, we utilized negative controls in which nematodes were exposed to heat-killed C. albicans cells, which have previously been shown to be avirulent (34). Negative controls were maintained at an elevated temperature (30°C) with or without DOX for the duration of the experiment.
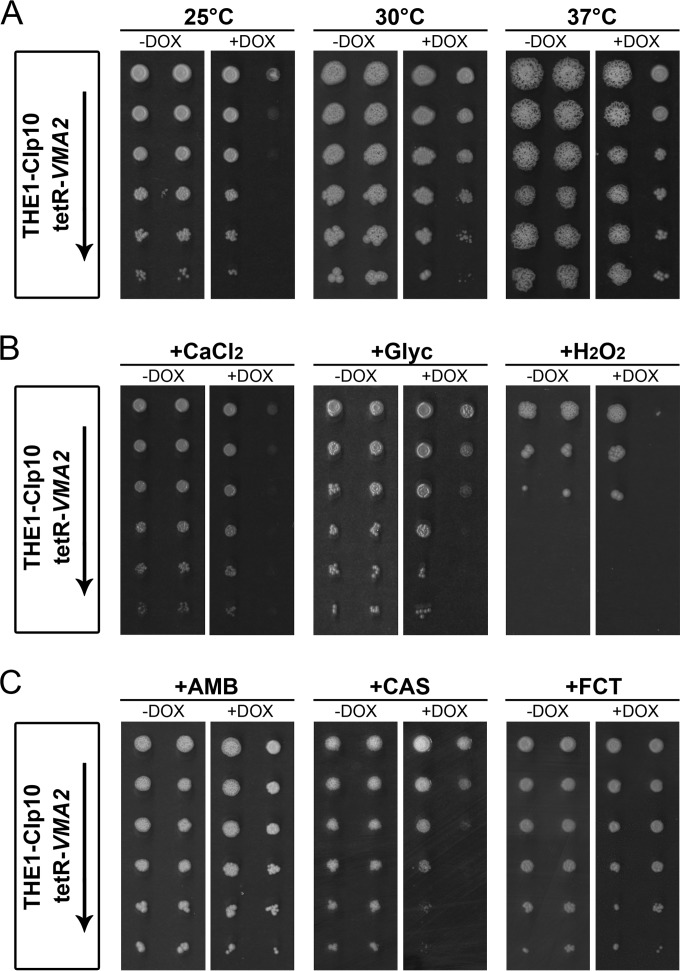
FIG 2.
tetR-VMA2 has decreased tolerance for various stress conditions. Serial dilutions of cells grown in YPD with or without DOX for 24 h were spotted onto different agar media with or without DOX and incubated for 48 h at 30°C. For the oxidative stress assay, plates of CSM, pH 4, plus 5 mM H2O2 were incubated at 30°C for 72 h rather than 48 h. Strains are indicated on the left, with an arrow signifying decreasing cell densities (1 × 108, 2 × 107, 4 × 106, 8 × 105, 1.6 × 105, and 3.2 × 104 cells per ml) of the strains spotted onto each plate. (A) tetR-VMA2 grows poorly on unbuffered YPD plus DOX at 25°C. From left to right, growth was tested on unbuffered YPD agar with or without DOX incubated at 25°C, 30°C, and 37°C. (B) Repression of VMA2 leads to reduced tolerance to calcium, nonfermentable carbon sources, and oxidative stress. From left to right, tetR-VMA2 growth was tested on unbuffered CSM with or without DOX with 200 mM CaCl2 added, on unbuffered yeast extract plus peptone (YEP) plus 2% ethanol plus 6% glycerol with or without DOX, and on CSM, pH 4.0, with or without DOX with 5 mM H2O2 added. (C) tetR-VMA2 has increased susceptibility to caspofungin. From left to right, growth was tested on CSM, pH 4.0, with or without DOX with 0.00625 μg/ml amphotericin B, 0.05 μg/ml caspofungin, and 10 μg/ml flucytosine.
Routine maintenance of C. elegans N2 nematodes was performed at 15°C as described previously (37). Prior to the start of the infection assay, nematodes were synchronized to the same life stage via egg axenization as described previously (37) and then incubated on plates of nematode growth media (NGM; 3 g NaCl, 2.5 g peptone, 17 g agar supplemented with 1 mM CaCl2, 5 μg/ml cholesterol, 1 mM MgSO4, and 25 mM KPO4 buffer, as described previously [37]) plus E. coli OP50 at 15°C until worms reached the L4 stage. For the infection assay, C. albicans THE1-CIp10 and tetR-VMA2 cells first were grown in YPD with or without DOX for 24 h to ensure complete repression of VMA2. Plates of YPD (pH 4) with or without DOX then were seeded with a lawn of 50 μl THE1-CIp10 or tetR-VMA2 cells that had been pretreated with DOX or left untreated. For negative controls, lawns of heat-killed cells were prepared on agar plates of YPD, pH 4, with or without DOX as described previously (34). The plates were incubated at 30°C for approximately 16 h. Synchronized L4 worms were washed from plates with sterile M9 buffer and collected via centrifugation for 1 min at 6,000 rpm. Approximately 500 worms were transferred to the center of the C. albicans lawns. For negative controls, approximately 500 worms were transferred to the center of lawns containing heat-killed yeast. All plates were incubated at 30°C for 4 h. After the 4-h infection, C. elegans nematodes were washed from the plate, taking care not to disturb the yeast lawn, and were washed in sterile M9 buffer and collected via centrifugation for 1 min at 6,000 rpm. Worms in M9 buffer were transferred to the center of unseeded NGM with or without DOX plates and incubated at 30°C to allow the M9 buffer to dry. Once dry, 80 to 100 nematodes per treatment were pick transferred using a platinum wire to a plate of fresh, unseeded NGM with or without DOX. Worms were monitored twice daily for survival, and surviving nematodes were transferred via platinum wire pick to plates of fresh, unseeded NGM with or without DOX once daily. Transfer to fresh NGM with or without DOX served two purposes: first, to avoid DOX oxidation due to light exposure, and second, to minimize the growth of C. albicans originating from either the original infection plate or from the worms' intestines. C. albicans lawns are opaque under a dissecting microscope, making monitoring worm survival difficult. Nematodes on heat-killed yeast lawns were not transferred daily. Worms were scored as dead if they did not respond to stimulation with a platinum wire pick. GraphPad Prism 6.0 was used to plot survival curves, calculate differences in survival (log-rank and Wilcoxon tests) via the Kaplan-Meier method, and calculate the LT50 (defined as the time for half of the worms to die). Dead worms were visualized via light microscopy at selected time points.
RESULTS
VMA2 is repressed by DOX in the tetR-VMA2 strain.
Our initial attempt to disrupt C. albicans VMA2 by generating a vma2Δ homozygous null mutant on acidic media was unsuccessful; after screening over 150 transformants from three separate lineages of heterozygous null mutants, a homozygous null mutant was not recovered. These results are consistent with previous attempts to disrupt V-ATPase genes in C. albicans (4). Therefore, we constructed the strain tetR-VMA2, a tetracycline-regulatable VMA2 mutant in which the VMA2 gene is repressed upon the addition of DOX (18). A strain from the THE1 background with URA3 reintegrated into the genome, THE1-CIp10, was used as an additional wild-type control (20). In order to confirm repression of VMA2 with DOX, we utilized RT-PCR to analyze VMA2 expression in the tetR-VMA2 strain (see Fig. S1 in the supplemental material). We have previously shown that, in a tetracycline-regulatable VMA3 mutant, the VMA3 transcript remained present for up to 18 h after treatment with DOX but disappeared completely after 24 h (4). Therefore, we pretreated THE1-CIp10 cells and tetR-VMA2 cells for 24 h in YPD with or without DOX before extracting mRNA for use in our transcriptional analysis. Treatment with DOX did not affect the VMA2 transcript level in the THE1-CIp10 control (see Fig. S1 in the supplemental material). Elevated levels of VMA2 transcript were detected in tetR-VMA2 without DOX compared to the levels of VMA2 transcript in the controls of THE1-CIp10 with or without DOX; this is consistent with higher levels of expression observed in other genes when placed under the tetracycline-regulatable (tetR) promoter (18). After 24 h of treatment with DOX, no VMA2 transcript was detected in the tetR-VMA2 strain (see Fig. S1). Thus, for all subsequent experiments, cells from the THE1-CIp10 and tetR-VMA2 strains were grown for 24 h in YPD with or without DOX to ensure complete repression of the VMA2 gene.
tetR-VMA2 exhibits the Vma− phenotype upon repression of VMA2.
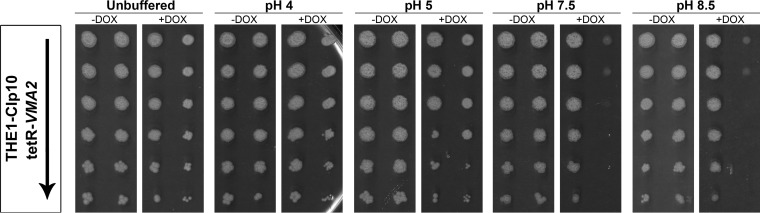
V-ATPase inhibition is lethal under certain conditions, including alkaline pH, the presence of heavy metals, and nonfermentable carbon sources; these traits are referred to collectively as the Vma− phenotype (6). To test whether inhibition of C. albicans VMA2 results in the Vma− phenotype, we tested the growth of tetR-VMA2 cells that had been pretreated with DOX on CSM with or without DOX plates that were either unbuffered or buffered to pH 4 to 8.5. Wild-type C. albicans cells grew as well on media buffered to pH 4 to 8.5 as on unbuffered media (Fig. 1). tetR-VMA2 cells grew comparably to wild-type cells without DOX under all conditions tested but grew poorly on alkaline media with DOX (Fig. 1, pH 7.5 and pH 8.5). It is worth noting that although tetR-VMA2 cells with DOX grow as well as the wild type on unbuffered media and media buffered to pH 4 or 5, wild-type cells but not tetR-VMA2 cells with DOX treatment form hyphae under these conditions, as evidenced by a wrinkly rather than smooth appearance caused by filamentous structures radiating from the colony. Therefore, inhibition of C. albicans VMA2 expression leads to the Vma− growth phenotype, indicating that V-ATPase function is significantly impaired. The Vma− phenotype also was observed in liquid cultures of CSM, pH 4.0 to 8.5 (data not shown).
FIG 1.
tetR-VMA2 exhibits the Vma− phenotype. Serial dilutions of cells grown in YPD with or without DOX for 24 h were spotted onto CSM agar media with or without DOX and incubated for 48 h at 30°C. Strains are indicated on the left, with an arrow signifying decreasing cell densities (1 × 108, 2 × 107, 4 × 106, 8 × 105, 1.6 × 105, and 3.2 × 104 cells per ml) of the strains spotted onto each plate. From left to right, growth was tested on unbuffered CSM, CSM buffered to pH 4 and 5 with 50 mM succinic acid–50 mM Na2PO4, and CSM buffered to pH 7.5 and 8.5 with 50 mM MES hydrate–50 mM MOPS.
Repression of VMA2 inhibits various stress responses in C. albicans.
V-ATPase is involved in multiple stress responses, partially due to the formation of a membrane potential across the vacuolar membrane that drives secondary transporters, resulting in the sequestration of harmful compounds inside the vacuole. To test VMA2 contribution to stress response in C. albicans, we grew tetR-VMA2 cells under various stress conditions. First, we tested the temperature response by plating DOX-pretreated tetR-VMA2 cells on YPD with or without DOX plates and incubating them at 25°C, 30°C, and 37°C. The THE1-CIp10 control strain with or without DOX and the tetR-VMA2 strain without DOX grew robustly at all three temperatures (Fig. 2A). These strains began to filament at 30°C and formed filamentous structures at 37°C; high temperature is one of many filamentation-induction signals in C. albicans (38). Upon repression of VMA2 with DOX, the tetR-VMA2 strain grew poorly at 25°C compared to controls, indicating poor tolerance of low temperature (Fig. 2A). We also observed the absence of filamentous structures in tetR-VMA2 plus DOX at both 30°C and 37°C (Fig. 2A).
V-ATPase function also is necessary for stress response under conditions such as growth on media containing high concentrations of calcium chloride and media containing glycerol as the sole carbon source (6). Therefore, we tested the response of the tetR-VMA2 strain to these stress conditions. Compared to the controls (THE1-CIp10 with or without DOX), tetR-VMA2 grew poorly upon repression of VMA2 on CSM plates plus 200 mM CaCl2 and on YEP plates plus 2% ethanol plus 6% glycerol (Fig. 2B). The tetR-VMA2 strain grew at wild-type levels on plates containing high concentrations of sodium chloride and on plates containing high concentrations of the cell wall stressors Congo red and calcofluor white (data not shown); these growth conditions have not been associated with the Vma− phenotype.
V-ATPase also has been shown to be important for the response to hypoxia in S. cerevisiae; S. cerevisiae V-ATPase mutants are hypersensitive to oxidative stress, and a S. cerevisiae vma2Δ mutant exhibits high levels of reactive oxygen species and oxidative protein damage even in the absence of an oxidant (39). Oxidative stress response is of particular interest in fungal pathogenesis, as it has been shown to be important for host colonization in C. albicans (40–42). Therefore, we tested tetR-VMA2 response to oxidative stress by growing strains on plates of CSM, pH 4, containing 5 mM H2O2. In the presence of 5 mM H2O2, repression of VMA2 led to a dramatic decrease in cell viability (Fig. 2B).
V-ATPase has been hypothesized to contribute to the activity of two classes of antifungal drugs: azoles, such as fluconazole, and polyenes, such as amphotericin B (43, 44). We asked whether the tetR-VMA2 strain was susceptible to four classes of antifungal drugs: echinocandins, polyenes, the antimetabolite flucytosine, and azoles. We tested tetR-VMA2 growth in the presence of sublethal concentrations of a polyene-class drug, amphotericin B; an echinocandin, caspofungin; and an antimetabolite, flucytosine. THE1-CIp10 and tetR-VMA2 cells were plated on CSM, pH 4, with or without DOX with 0.00625 to 0.5 μg/ml amphotericin B, 0.05 μg/ml caspofungin, or 10 to 20 μg/ml flucytosine. VMA2 repression decreased the resistance of the tetR-VMA2 strain to caspofungin (Fig. 2C) in accordance with previous results (4). In the presence of either 0.00625 μg/ml amphotericin B or 10 μg/ml flucytosine, tetR-VMA2 cells treated with DOX grew at levels comparable to those of the wild type (Fig. 2C). No difference in growth in the presence of higher levels of either amphotericin B or flucytosine was observed (data not shown).
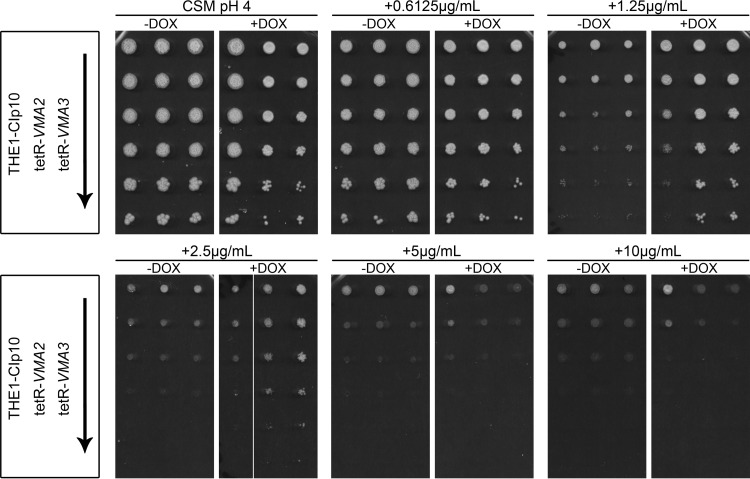
Because fluconazole activity has been linked previously to V-ATPase function (43), we studied tetR-VMA2 response to fluconazole in further detail. We tested various concentrations of fluconazole (0.6125 to 10 μg/ml) in tetR-VMA2 and the previously studied V-ATPase Voc-deficient strain tetR-VMA3 (4). Interestingly, repression of VMA2 and VMA3 in tetR-VMA2 and tetR-VMA3 cells, respectively, increased C. albicans growth on plates containing fluconazole at concentrations ranging from 1.25 to 2.5 μg/ml (Fig. 3). At higher concentrations, the growth of the tetR-VMA2 and tetR-VMA3 strains was suppressed to levels comparable to those of wild-type cells regardless of the absence or presence of DOX (Fig. 3). Therefore, repression of either V-ATPase Vo or V1 components results in increased resistance to fluconazole.
FIG 3.
tetR-VMA2 and tetR-VMA3 have decreased susceptibility to fluconazole. Serial dilutions of THE1-CIp10, tetR-VMA2, and tetR-VMA3 cells grown in YPD with or without DOX for 24 h were spotted onto CSM agar media with or without DOX and incubated for 48 h at 30°C. Strains are indicated on the left, with an arrow signifying decreasing cell densities (1 × 108, 2 × 107, 4 × 106, 8 × 105, 1.6 × 105, and 3.2 × 104 cells per ml) of the strains spotted onto each plate. Conditions tested in the top row are, from left to right, CSM, pH 4.0, as a control, CSM, pH 4, plus 0.6125 μg/ml fluconazole (FLU), and CSM, pH 4, plus 1.25 μg/ml FLU. The bottom row shows, from left to right, CSM, pH 4, plus 2.5 μg/ml FLU; CSM, pH 4, plus 5 μg/ml FLU; and CSM, pH 4, plus 10 μg/ml FLU.
Repression of VMA2 inhibits V-ATPase assembly and activity and leads to vacuolar alkalinization.
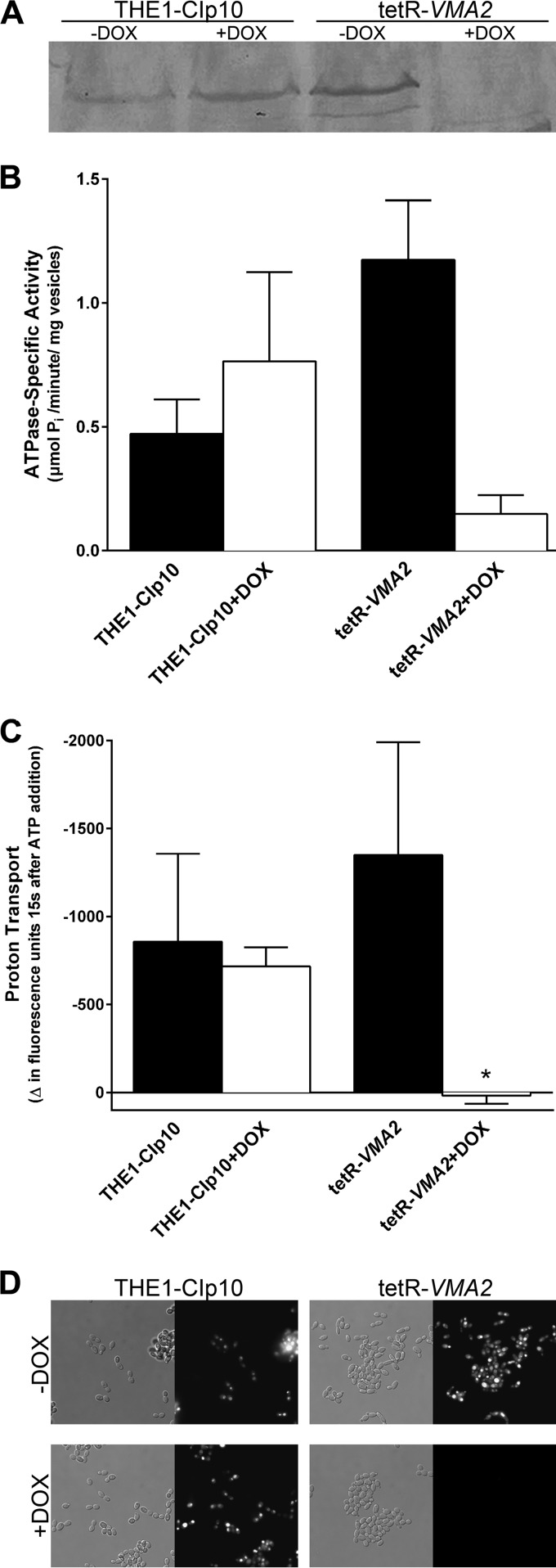
To establish the effect of VMA2 suppression on V-ATPase assembly and catalytic activity, we purified vacuolar membrane vesicles from cells grown in YPD, pH 4.0, with or without DOX for 24 h by density gradient centrifugation. Western blots using an antibody against the catalytic subunit A of V1 (V1A or Vma1p) did not detect the V1A subunit in vacuolar membrane fractions under repressing conditions (Fig. 4A), indicating that V-ATPase complexes are not fully assembled. As expected, deletion of the V1B subunit in C. albicans prevented assembly of the V1 domain at the membrane.
FIG 4.
V-ATPase assembly and activity assays. (A) The V-ATPase is not fully assembled at the vacuolar membrane upon VMA2 repression. Western blots of vacuolar membrane fractions isolated from THE1-CIp10 and tetR-VMA2 with or without DOX were probed with anti-human-V1A rabbit polyclonal antibody. (B) DOX repression of VMA2 leads to decreased ATPase activity. V-ATPase-specific activity (concanamycin A-sensitive ATP hydrolysis) was measured in isolated vacuolar membrane vesicles from THE1-CIp10 and tetR-VMA2 cells with or without DOX in an enzymatic assay. (C) Proton transport is absent upon repression of VMA2. Proton transport in isolated vacuolar membrane vesicles isolated from THE1-CIp10 and tetR-VMA2 with or without DOX cells was measured using ACMA. Fluorescence was monitored for 60 s prior to MgATP addition and for 40 s afterwards. Proton transport was calculated as the change in fluorescence for the first 15 s following MgATP addition. The asterisk denotes statistical significance compared to all other treatments (P < 0.05). (D) DOX-treated tetR-VMA2 vacuoles do not stain with quinacrine in vivo. Cells were grown for 24 h in unbuffered YPD with or without DOX before staining with 200 mM quinacrine and observation via DIC and fluorescence microscopy.
The V1 domain contains the three catalytic sites where ATP binds and is hydrolyzed, whereas the Vo domain contains the sites of proton binding and transport. Thus, structural coupling of V1 and Vo is necessary for V-ATPase function (1). The assembly defect of the V-ATPase complex upon depletion of Vma2p is predicted to significantly reduce ATP hydrolysis and proton transport. Therefore, we measured both ATP hydrolysis and proton transport in purified vacuolar membrane vesicles. ATP hydrolysis was measured spectrophotometrically using a coupled enzymatic assay (24). Upon repression of VMA2, concanamycin A-sensitive ATP hydrolysis decreased by 91.5% (Fig. 4B). Proton transport was measured fluorometrically using ACMA (25, 26). Repression of VMA2 with DOX in tetR-VMA2 completely abrogated proton transport (101.2%) (Fig. 4C). These results are in agreement with the alkalinization of the vacuole in tetR-VMA2 plus DOX and show that C. albicans VMA2 is required for both ATPase hydrolysis and proton transport by the V-ATPase.
To confirm that VMA2 repression inhibits vacuolar acidification by V-ATPase, THE1-CIp10 and tetR-VMA2 cells with or without DOX were stained with quinacrine, a basic dye that accumulates in the vacuole and fluoresces in acidic environments. The THE1-CIp10 control with or without DOX and the tetR-VMA2 strain grown without DOX exhibited wild-type quinacrine staining of an acidic vacuole (Fig. 4D). In contrast, the tetR-VMA2 strain grown in the presence of DOX lacked quinacrine staining of the vacuoles, indicating an alkalinized vacuole (Fig. 4D). In order to quantitatively confirm this phenotype, cells were stained with BCECF, a pH-sensitive fluorophore that accumulates in the vacuole. By comparing the fluorescence profile of BCECF-treated cells to that of a calibration curve, vacuolar pH can be quantitatively determined (10, 25, 45). We previously studied vacuolar pH in the THE1-CIp10 strain both with and without DOX; the vacuolar pH was approximately 6.25 under both conditions (4). The vacuolar pH in the tetR-VMA2 strain without DOX was 6.099 ± 0.1475. Repression of VMA2 with DOX resulted in an increase in pH to 6.924 ± 0.05126, indicating vacuolar alkalinization.
Repression of VMA2 leads to abnormal vacuolar morphology.
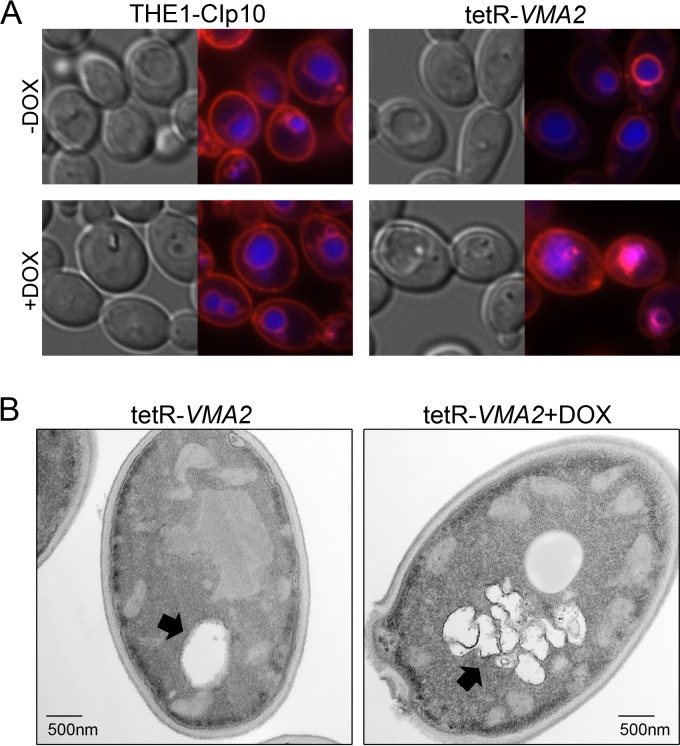
V-ATPase has been implicated in vacuolar biogenesis, a phenotype attributable to the role of V-ATPase in vacuolar membrane fission and fusion (7, 46, 47). Therefore, we assessed the effect of VMA2 suppression on vacuolar morphology by simultaneously utilizing two vacuolar stains, FM4-64 and CMAC. FM4-64 is a vital dye that is endocytosed to the vacuole, where it stains the vacuolar membrane (48). CMAC passively accumulates in the vacuolar lumen and stains the vacuolar lumen under both acidic and alkaline conditions. THE1-CIp10 cells both with and without DOX showed well-defined vacuoles, with the FM4-64 membrane stain and the CMAC lumenal stain remaining distinct from one another (Fig. 5A). Without DOX, the tetR-VMA2 cells showed vacuolar morphology indistinguishable from that of the wild type (Fig. 5A). However, upon DOX repression of VMA2, the FM4-64 membrane and the CMAC lumenal stain showed considerable overlap, with membranous structures on the interior of the vacuole revealed by FM4-64 staining (Fig. 5A). We additionally analyzed vacuolar morphology in tetR-VMA2 cells with or without DOX using thin-section electron microscopy. Vacuolar morphology in the THE1-CIp10 strain, both with and without DOX, has been analyzed previously (20). The tetR-VMA2 cells without DOX addition displayed wild-type vacuolar morphology, as indicated by the presence of a single, large vacuole (Fig. 5B). However, when treated with DOX for 24 h, the tetR-VMA2 cells had enlarged vacuoles with excessive internal membranous structures (Fig. 5B). This phenotype is consistent with a defect in vacuolar membrane fission (7).
FIG 5.
Vacuolar morphology. (A) FM4-64 and CMAC dual staining of vacuoles. To simultaneously stain vacuoles with FM4-64 and CMAC, cells were grown for 24 h in unbuffered YPD with or without DOX. Cells then were reset in fresh unbuffered YPD with or without DOX and grown to early log phase before costaining with FM4-64 and CMAC. Cells were examined via DIC and fluorescence microscopy using Texas red (FM4-64) and DAPI (CMAC) filters. (B) Thin-section electron microscopy showing the tetR-VMA2 strain after growth in unbuffered YPD with or without DOX for 24 h. In each panel, the black arrow is pointing to the vacuole.
VMA2 contributes to C. albicans virulence-related traits.
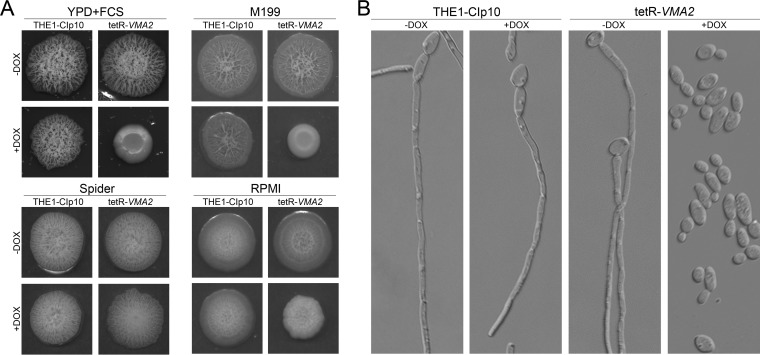
Vacuolar function has been implicated in several key traits of C. albicans pathogenesis, including the ability to switch between hyphal and yeast forms of growth. Therefore, we assessed the ability of the tetR-VMA2 strain to form hyphae in vitro. On solid media (YPD plus FCS, M199, and Spider media), the tetR-VMA2 strain exhibited dramatically reduced filamentation (data not shown). However, because the tetR-VMA2 strain has reduced growth at alkaline pH, these results were difficult to interpret. Therefore, we assessed in vitro filamentation by tetR-VMA2 on solid and in liquid media buffered to pH 4.0 or pH 5.0. The THE1-CIp10 control strain readily formed hyphae even under acidic conditions (4). On all types of media tested, the tetR-VMA2 strain grown under nonrepressing conditions produced hyphae at levels comparable to that of the THE1-CIp10 control strain. In contrast, under repressing conditions, the tetR-VMA2 strain did not produce hyphae on serum or M199 plates and produced less robust hyphal structures on RPMI plates (Fig. 6A). Interestingly, on Spider media, which causes induction of filamentation in response to nutrient-poor conditions (49), the tetR-VMA2 strain produced hyphae to the same degree as the THE1-CIp10 control even upon repression of VMA2. Lastly, we tested in vitro filamentation in liquid RPMI, pH 5. Under repressing conditions, tetR-VMA2 cells were present predominantly in the yeast form even after 24 h of incubation in liquid RPMI, pH 5, at 37°C (Fig. 6B).
FIG 6.
Filamentation on solid and in liquid media. (A) Filamentation on hypha-inducing agar plates. Cells were grown in unbuffered YPD with or without DOX for 24 h. Three μl cells was spotted onto agar plates containing YPD plus FCS, M199, Spider, or RPMI with or without DOX. All plates were buffered to pH 4 with 50 mM succinic acid–50 mM Na2HPO4. Plates were incubated at 37°C for 5 days. (B) Filamentation in liquid RPMI. Cells were grown in unbuffered YPD with or without DOX for 24 h, and then 5 × 106 cells/ml were added to RPMI, pH 5.0, with or without DOX and grown for 24 h at 37°C. Filaments were visualized via DIC microscopy. A representative image from each treatment is shown.
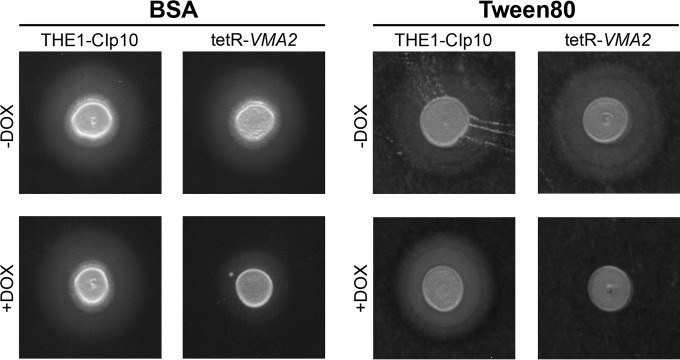
The secretion of degradative enzymes also is involved in C. albicans pathogenesis by assisting in the invasion of host tissues (50, 51). We assayed in vitro secretion of lipases and aspartyl proteases on YNB-Tween 80 and BSA media, respectively (Fig. 7). Wild-type C. albicans cells spotted onto media containing lipids, such as YNB-Tween 80 media, secrete lipases resulting in a halo of precipitation around the colony. Under repressing conditions, tetR-VMA2 exhibited decreased lipolytic activity on YNB-Tween 80 agar (Fig. 7). C. albicans cells spotted on media containing BSA as the sole nitrogen source secrete secreted aspartyl proteases (SAPs) that digest the BSA, resulting in a halo of proteolysis around the colony (29). The THE1-CIp10 control strain and the tetR-VMA2 strain exhibited normal proteolytic activity under derepressing conditions. DOX repression partially inhibited extracellular proteolytic activity in the tetR-VMA2 strain (Fig. 7). Because the defect in protease secretion was partial, we quantified the size of the proteolytic halo relative to the size of the colony. For reference, a relative halo size of 1 indicates a lack of detectable halo. For the THE1-CIp10 strain with and without DOX, the relative sizes of the proteolytic halos were 3.605 ± 0.4848 and 3.655 ± 0.8016, respectively. For the tetR-VMA2 strain without DOX, the relative size of the proteolytic halo was 2.915 ± 0.5017. Upon repression of VMA2, the relative size of the halo decreased to 1.639 ± 0.1371; this decrease was statistically significant compared to all three controls (P < 0.01). Because BSA media cannot be buffered to acidic pH without denaturing the BSA, we were unable to utilize growth-permissive media for these experiments. However, because the BSA medium utilized was not buffered to a specific pH (i.e., unbuffered) and tetR-VMA2 grows similarly to the wild type on unbuffered media (Fig. 1), this defect likely is not due solely to slow growth by the tetR-VMA2 strain under repressing conditions.
FIG 7.
SAP and lipase secretion on Tween 80 and BSA agar. Cells were grown in unbuffered YPD with or without DOX for 24 h, and then 3 μl of cells was spotted onto unbuffered Tween 80 agar plates and unbuffered bovine serum albumin (BSA) agar plates with or without DOX. Unbuffered plates of Tween 80 with or without DOX were incubated at 37°C for 5 days, and plates of unbuffered BSA with or without DOX were incubated at 30°C for 48 h. The relative amount of lipolytic and proteolytic degradation is indicated by the halo surrounding the fungal colony.
VMA2 is involved in autophagy.
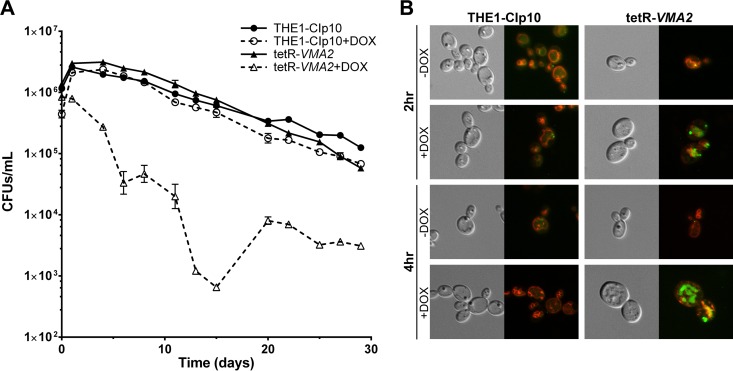
V-ATPase activity has previously been implicated in survival under conditions of nitrogen starvation (52); however, its exact role remains unclear (53). Therefore, we assessed the contribution of VMA2 to long-term survival under conditions of nitrogen starvation using a 30-day assay to monitor resistance to nitrogen starvation (31). Because the growth of V-ATPase-deficient cells is pH sensitive, we measured the initial pH of the nitrogen starvation media (pH 4.5) and that over the course of the experiment. Periodic measurements of the medium pH in all treatments revealed no significant changes (pH 4.0 to 5.0; data not shown). The cells were grown in nitrogen starvation media with or without DOX for a period of 30 days, with samples taken at select time points for CFU counts. The THE1-CIp10 control strain with or without DOX and the tetR-VMA2 strain without DOX exhibited a gradual decrease in CFU over the 30-day period (Fig. 8). However, repression of VMA2 with DOX caused a dramatic decrease in CFU that persisted over the 30-day period (Fig. 8). Interestingly, while repression of VMA2 impaired resistance to nitrogen starvation, it did not lead to complete cell death.
FIG 8.
tetR-VMA2 contribution to autophagy. (A) Resistance to nitrogen starvation in tetR-VMA2. Long-term survival of autophagy was determined by CFU counts. Cells were grown for 24 h in unbuffered YPD with or without DOX. Cells then were reset in nitrogen starvation media (SD − N) and grown for 30 days, with samples taken at select time points for CFU counts. (B) Ape1-GFP turnover in tetR-VMA2. T-LAP41-GFP and tV2-LAP41-GFP cells were transferred to liquid nitrogen starvation media (SD − N with or without DOX) after 24 h of growth in rich media (YPD with or without DOX). After 115 min of growth at 30°C, cells were coincubated with FM4-64 for 15 min, resuspended in fresh nitrogen starvation media, and grown at 30°C. Cells were visualized via DIC and fluorescence microscopy to detect Ape1-GFP and FM4-64 fluorescence 2 h and 4 h after transfer to nitrogen starvation media.
Survival of C. albicans under conditions of nitrogen starvation is accomplished via autophagic recycling of cellular material; therefore, resistance to nitrogen starvation is an indirect measure of autophagy (31). To confirm the role of VMA2 in autophagy, we sought to monitor the fate of an autophagic marker. The hydrolase aminopeptidase I (Ape1) is encoded by the C. albicans LAP41 gene (orf19.1628). Ape1p makes up the cytosol-to-vacuole targeting complex, which is engulfed by autophagosomes under starvation conditions (54, 55). Consequently, Ape1-GFP has been used commonly as an autophagic marker in C. albicans (31) and S. cerevisiae (55). Upon translation, the preform of Ape1 aggregates in the cytosol to form a homododecameric complex, also known as the cytosol-to-vacuole targeting (Cvt) complex. Under autophagic conditions, the Cvt complex is selectively shuttled to the vacuole by autophagosomes, where maturation of Ape1 occurs (54–56). Visualization of Ape1-GFP localization and processing upon nitrogen starvation can be used to identify defects in the autophagic pathway; Ape1-GFP originates in the cytosol and can be observed as intense fluorescent dots. Ape1-GFP then is trafficked to the vacuole, where it is visible first as intense spots and over time disperses throughout the vacuolar lumen as autophagosomes are degraded. We used a nourseothricin selection method to construct C. albicans strains, in both the THE1-CIp10 and tetR-VMA2 backgrounds, in which the C terminus of Ape1p was tagged with GFP. The two anticipated localizations of Ape1-GFP (cytosol and vacuolar lumen) and its subsequent vacuolar dispersion were observed in THE1-CIp10 cells with or without DOX as well as tetR-VMA2 cells without DOX 2 h after the transfer to nitrogen starvation media (Fig. 8B). We noted the presence of dispersed Ape1-GFP in all vacuoles, indicating active autophagic processes. In contrast, upon repression of VMA2 with DOX in tetR-VMA2 cells, intense spots of Ape1-GFP accumulated in the vacuole after 2 h (Fig. 8B). In THE1-CIp10 cells with or without DOX, the Ape1-GFP signal decreased from 2 h to 4 h (Fig. 8C). Without DOX, Ape1-GFP levels in tetR-VMA2 cells were indistinguishable from those of the wild type (Fig. 8C). Unlike the control strains, Ape1-GFP remained present in vacuoles of the tetR-VMA2 cells plus DOX after 4 h in nitrogen starvation media (Fig. 8C). At all time points, tetR-VMA2 cells treated with DOX exhibited markedly higher levels of Ape1-GFP (Fig. 8B and C and data not shown). These data indicate a delay in autophagy upon repression of VMA2. Of note, a defined vacuolar compartment was no longer visible in tetR-VMA2 cells treated with DOX past 24 h in nitrogen starvation media (data not shown). In contrast, an enlarged vacuolar compartment was clearly visible in control cells (both THE1-CIp10 with or without DOX and tetR-VMA2 without DOX).
VMA2 is required for virulence in an acidic infection model.
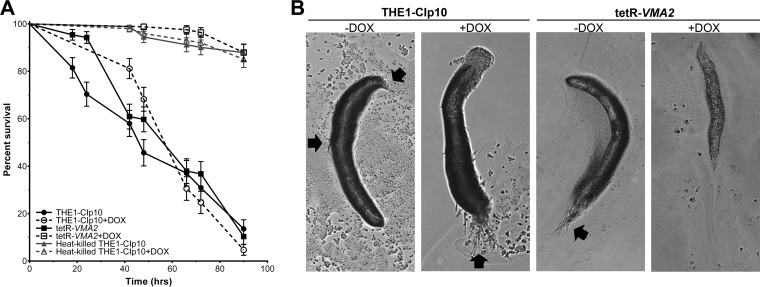
C. albicans vma7Δ, which is expected to lack all V-ATPase activity and exhibited the Vma− growth phenotype, is avirulent in a mouse model of systemic candidiasis (3). However, the pH-dependent phenotype of vma mutants makes interpretation of virulence models difficult, as in most model systems the physiological pH is restrictive for the growth of vma mutants. Therefore, we tested tetR-VMA2 virulence in a C. elegans model of infection (34). C. elegans is an ideal organism for the study of virulence in C. albicans vma mutants, as the pH of the larynx and intestine of C. elegans nematodes, where C. albicans infection occurs (34, 35), ranges from 3.5 to 5.5 (57). For these studies, we analyzed infection at the permissive pH and temperature, under acidic pH conditions (pH 4.0) and at 30°C, because tetR-VMA2 is unable to grow at neutral pH and low temperatures (i.e., 25°C). The wild-type worms were infected at 30°C and maintained at 30°C postinfection, with daily monitoring for survival. To ensure that neither high temperature (30°C) nor treatment with DOX were responsible for worm mortality, we included negative controls where worms were not infected with live C. albicans but rather were fed heat-killed C. albicans cells, which have been shown previously to be avirulent (34). Upon repression with DOX, tetR-VMA2 exhibited attenuated virulence in the C. elegans model (Fig. 9A). Notably, survival of the worms infected with doxycycline-repressed tetR-VMA2 was not significantly different from survival of untreated worms after 90 h (88.0% ± 3.6% survival for tetR-VMA2 plus DOX compared to 87.9% ± 3.4% and 85.1% ± 3.5% survival for the negative controls with and without DOX, respectively). C. elegans nematodes were killed by infection with the THE1-CIp10 controls, with an LT50 of 66 h. Without DOX, nematode killing by the tetR-VMA2 strain was not statistically significant (P < 0.001) compared to that of the THE1-CIp10 controls, with an LT50 of 66 h. In contrast, the LT50 of worms infected with the tetR-VMA2 strain with DOX was 144 h, comparable to the negative-control nematodes fed heat-killed yeast. Further, worms scored as dead in the assay were subjected to analysis via light microscopy (Fig. 9B). Worms infected with THE1-CIp10 with or without DOX or tetR-VMA2 without DOX cells had visible hyphae piercing the worm cuticle, indicative of hypha-mediated killing. In contrast, worms infected with DOX-repressed tetR-VMA2 cells had no visible hyphal protrusions. We conclude that tetR-VMA2 is avirulent in the acidic C. elegans infection model.
FIG 9.
tetR-VMA2 contribution to virulence in the in vivo C. elegans model. (A) Survival curve of nematodes infected with C. albicans strains THE1-CIp10 and tetR-VMA2, with and without DOX, or fed with heat-killed C. albicans THE1-CIp10 with and without DOX. Worm survival was monitored at select time points; worms that did not respond to stimulation with a platinum wire pick were scored as dead. Inhibition of VMA2 leads to avirulence; survival of nematodes infected with DOX-repressed tetR-VMA2 was not significantly different from survival of nematodes fed heat-killed C. albicans cells. (B) Light microscopy of nematodes infected with C. albicans strains. Black arrows indicate C. albicans hyphae piercing the C. elegans cuticle.
DISCUSSION
The contribution of several V-ATPase subunits to C. albicans cell biology and virulence has been studied previously (3, 4, 12, 13). Here, we present the study of C. albicans VMA2, which encodes the B subunit of the V-ATPase V1 domain; this is the first component of the catalytic hexamer of the V1 complex to be studied in depth in C. albicans. Repression of VMA2 in C. albicans results in the Vma− phenotype, characterized by poor growth at alkaline pH and on nonfermentable carbon sources as well as reduced ability to respond to environmental stressors. This is in accord with previous studies of V-ATPase subunits encoded by a single gene in C. albicans, such as the V-ATPase structural genes VMA7 and VMA3 (3, 4). Genetic repression of C. albicans VPH1 or STV1, which encode isoforms of the V-ATPase subunit Voa, does not result in the Vma− phenotype (12, 13). As expected, repression of VMA2 leads to complete inhibition of V-ATPase-specific (concanamycin A-sensitive) ATP hydrolysis and proton transport activities, similar to findings in studies of VMA3 and VPH1 (4, 12).
We previously studied susceptibility to fluconazole in a tetracycline-repressible VMA3 strain and reported no difference in fluconazole susceptibility upon repression of VMA3 (4). However, only one concentration of fluconazole was reported (5 μg/ml). Here, we extended those studies by testing both the tetR-VMA2 and tetR-VMA3 strains under a range of fluconazole concentrations (0.6125 to 10 μg/ml). Interestingly, repression of both VMA2 and VMA3 decreased C. albicans susceptibility to subinhibitory concentrations of fluconazole despite an overall impairment in the ability of these mutants to respond to stress. More than one possibility exists to explain this phenomenon. Upon repression of VMA2 or VMA3, cells may compensate for V-ATPase loss by unknown mechanisms that may also lead to a decrease in the susceptibility to fluconazole. Alternatively, the dose-dependent antifungal activity of fluconazole may be directly attributable to effects on V-ATPase activity, as previously suggested (43). It is worth noting that null mutants of genes involved in ergosterol biosynthesis exhibit a similar decrease in fluconazole susceptibility (58, 59). The similar phenotype regarding fluconazole response in ergosterol and V-ATPase mutants further supports the notion that there is a link between V-ATPase and ergosterol.
Our phenotypic analyses of VMA2 also revealed its contribution to filamentation on M199, YPD plus FCS, and RPMI media, but not to filamentation on Spider medium, even at acidic pH. Spider medium induces filamentation under conditions of carbon deprivation (28). These results are in contrast to those of our previous study of VMA3, which was unable to produce filaments on any medium tested, including Spider (4). It is conceivable that the Vo domain plays a role in filamentation under nutrient deprivation conditions; further work is required to address this possibility. This concept is supported by the fact that S. cerevisiae vma2Δ cells assemble Vo domains at the membrane but vma3Δ cells do not (60).
In addition to the differential contribution of VMA2 and VMA3 to filamentation under nutrient-poor conditions, the secretion of protease enzymes is also different following DOX-mediated suppression of VMA2 and VMA3. Although inhibition of VMA2 and VMA3 completely abolished lipase secretion (4), only inhibition of VMA3 fully impairs aspartyl protease secretion (4), whereas inhibition of VMA2 results in a moderate decrease in aspartyl protease secretion. Notably, the V-ATPase Vo domain, but not V1, has been shown previously to play a role in membrane fusion; SNARE complexes tether fusing membranes together, and the proteolipid ring of V-ATPase Vo complexes located on the vesicular membrane enlarges to create a fusion pore through which materials can flow (61). Our results could be explained if the Vo proteolipid ring and its involvement in membrane fusion are necessary for the secretion of aspartyl proteases but not lipases in C. albicans. Notably, we have shown that deletion of VPH1, a vacuolar isoform of Voa that does not contribute to the Vo proteolipid ring, results in alkalinized vacuoles, complete elimination of lipase activity, and partial reduction of aspartyl protease activity (12) in a manner strikingly similar to that of VMA2 repression. These overlapping results suggest that secretion of lipases depends on vacuolar acidification, whereas secretion of aspartyl proteases is maintained partially even in the presence of alkalinized vacuoles.
During both infection and colonization, C. albicans must survive robust oxidative stress conditions, and this ability to respond to oxidative stress previously has been correlated with virulence (62). Studies in S. cerevisiae V-ATPase mutants indicate that vma mutants cannot tolerate H2O2 and that the cells exhibit chronic oxidative stress (39). In the present study, we showed a role for C. albicans V-ATPase in response to oxidative stress, indicated by increased sensitivity to H2O2 upon repression of VMA2 at acidic pH. This is a novel mechanism by which V-ATPase may contribute to virulence in C. albicans. Further studies are needed to clarify the exact role of V-ATPase in the C. albicans oxidative stress response pathway. Nonetheless, this finding emphasizes the multifactorial nature of the roles played by V-ATPase in fungal pathogenesis.
Autophagy is the process by which cells degrade cellular material in order to recycle cellular building blocks and is essential for survival under starvation conditions. Autophagy also is important for stress response, development and aging, and programmed cell death. In yeast, nutrient depletion in the cell causes a signaling cascade that induces autophagosome formation. The newly forming autophagosomal membrane elongates to enclose cellular components, such as cytoplasmic material, mitochondria, and nuclei. Autophagosomes then are transported to the vacuole and subsequently broken down. In S. cerevisiae, V-ATPase previously has been implicated in autophagic processes; V-ATPase mutants accumulate autophagosomes in the vacuole (52). In C. albicans, autophagy-deficient mutants are not impaired in filamentation or virulence in a mouse model of systematic infection (63, 64). However, autophagy has been implicated in survival within neutrophils and also may be important for commensalism by allowing long-term survival within the host (65). We showed that V-ATPase inhibition by repression of VMA2 leads to defective autophagy, even under growth-permissive acidic conditions. Cells repressed for VMA2 failed to survive nitrogen starvation and accumulated ApeI-GFP in the vacuole. Since the last step in autophagy is altered, our results suggest that lack of vacuolar acidification upon repression of VMA2 is responsible for this effect.
V-ATPase is emerging as a potential antifungal target due to its far-reaching cellular importance (1, 5, 66). C. albicans V-ATPase mutants have been studied in mouse virulence models (3, 13). For example, C. albicans vma7Δ has reduced virulence, upholding the potential value of V-ATPase as a drug target (3). However, the pH-dependent growth of V-ATPase mutants can be a confounding factor in studies of V-ATPase and underlines the importance of studying phenotypes of interest under permissive pH conditions (pH 4.0 to 5.0) whenever possible. Whether reduced virulence is due merely to decreased growth at alkaline physiological pH rather than a role of V-ATPase in virulence-specific processes has remained unclear to date. Here, we present the first study of V-ATPase inhibition in an acidic model of infection. Caenorhabditis elegans is a soil-dwelling nematode that feeds on bacteria and fungi and is exposed in situ to a wide range of ingested pathogens (67). C. elegans is a useful system to study fungal pathogenesis due to its sophisticated, pathogen-specific immune system that includes immune response pathways conserved with mammals (34, 67–69). In fact, bacterial and fungal genes identified as important for pathogenesis in C. elegans infection assays have considerable overlap with genes previously identified as important for pathogenesis in mammalian infection models (69, 70). These characteristics underline the utility of C. elegans as a model host organism. In the C. albicans-C. elegans infection assay, C. elegans nematodes ingest C. albicans cells, which then persist within the C. elegans gastrointestinal tract and cause an infection. At this point, C. elegans immune defenses are induced, including fungus-specific transcriptional changes involving antimicrobial, secreted, or detoxification proteins (34). The infection culminates in C. albicans hyphae piercing the C. elegans cuticle and killing the worm (35). Because the C. elegans gastrointestinal tract and pharynx have very low pH, ranging from pH 5.5 in the pharynx to pH 3.5 in the lower intestine (57), C. elegans is an ideal model to study V-ATPase-mediated pathogenesis in vivo. We tested the contribution of VMA2 to C. albicans virulence in an acidic environment, i.e., under pH conditions permissible for growth of V-ATPase-deficient cells. To establish this growth-permissive, acidic environment, we utilized both acidic media and an infection model where the site of infection is highly acidic. Upon repression of VMA2, the tetR-VMA2 strain is avirulent. Therefore, we can conclude that the contributions of V-ATPase to stress responses, filamentation, and virulence are not attributable to its pH-dependent growth phenotype. Rather, intact V-ATPase function has a direct role in C. albicans pathogenesis. Ongoing studies in our laboratories to further elucidate the cell biology underpinning that role are in progress.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hironobu Nakayama (Suzuka University of Medical Science, Japan) for providing strain THE1 and plasmid p99CAU1, Aaron P. Mitchell (Carnegie Mellon University) for providing plasmid pDDB57, Steven Bates (University of Exeter) for providing plasmid pGFP-NAT1, and Barbara Hunter (University of Texas Health Science Center at San Antonio) for assistance with transmission electron microscopy.
The C. elegans N2 strain and the E. coli OP50 strain were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Sequence data for C. albicans were obtained from the Candida Genome Database. Sequencing of C. albicans was accomplished with the support of the NIDCR, NIH, and the Burroughs Wellcome Fund.
This work was supported by funding from the Department of Veterans' Affairs (Merit Award 5 I01 BX001130 to S.A.L.), Biomedical Research Institute of New Mexico (to S.A.L.), National Institutes of Health grant R01 GM086495 (to K.J.P.), UNM IDIP T32 institutional training grant NIH 5 T32 AI007538 (to S.M.B.), and National Institutes of Health grant K12 GM088021 (to S.R.H.).
Footnotes
Published ahead of print 18 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00135-14.
REFERENCES
- 1.Hayek SR, Lee SA, Parra KJ. 2014. Advances in targeting the vacuolar proton-translocating ATPase (V-ATPase) for anti-fungal therapy. Front. Pharmacol. 5:4. 10.3389/fphar.2014.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer GE, Kelly MN, Sturtevant JE. 2005. The Candida albicans vacuole is required for differentiation and efficient macrophage killing. Eukaryot. Cell 4:1677–1686. 10.1128/EC.4.10.1677-1686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poltermann S, Nyugen M, Gunther J, Wendland J, Hartl A, Kunkel W, Zipfel PF, Eck R. 2005. The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development and virulence. Microbiology 151:1645–1655. 10.1099/mic.0.27505-0 [DOI] [PubMed] [Google Scholar]
- 4.Rane HS, Bernardo SM, Raines SM, Binder JL, Parra KJ, Lee SA. 2013. Candida albicans VMA3 is necessary for V-ATPase assembly and function and contributes to secretion and filamentation. Eukaryot. Cell 12:1369–1382. 10.1128/EC.00118-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parra KJ. 2012. Vacuolar ATPase: a model proton pump for antifungal drug discovery, p 89–100 In Tegos G, Mylonakis E. (ed), Antimicrobial drug discovery: emerging strategies. CAB International, Wallingford, England [Google Scholar]
- 6.Kane PM. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70:177–191. 10.1128/MMBR.70.1.177-191.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baars TL, Petri S, Peters C, Mayer A. 2007. Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol. Biol. Cell 18:3873–3882. 10.1091/mbc.E07-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Chang A. 2011. pH-dependent cargo sorting from the Golgi. J. Biol. Chem. 286:10058–10065. 10.1074/jbc.M110.197889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarsio M, Zheng H, Smardon AM, Martínez-Muñoz GA, Kane PM. 2011. Consequences of loss of Vph1 protein-containing vacuolar ATPases (V-ATPases) for overall cellular pH homeostasis. J. Biol. Chem. 286:28089–28096. 10.1074/jbc.M111.251363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Muñoz GA, Kane P. 2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283:20309–20319. 10.1074/jbc.M710470200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perzov N, Nelson H, Nelson N. 2000. Altered distribution of the yeast plasma membrane H+-ATPase as a feature of vacuolar H+-ATPase null mutants. J. Biol. Chem. 275:40088–40095. 10.1074/jbc.M007011200 [DOI] [PubMed] [Google Scholar]
- 12.Raines SM, Rane H, Bernardo SM, Binder JL, Lee SA, Parra KJ. 2013. Deletion of V-ATPase Voa isoforms clarifies the role of vacuolar pH as a determinant of virulence-associated traits in C. albicans. J. Biol. Chem. 288:6190–6201. 10.1074/jbc.M112.426197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patenaude C, Zhang Y, Cormack B, Köhler J, Rao R. 2013. Essential role for vacuolar acidification in Candida albicans virulence. J. Biol. Chem. 288:26256–26264. 10.1074/jbc.M113.494815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, Kane PM, Newman PR, Forgac M. 1996. Site-directed mutagenesis of the yeast V-ATPase B subunit (Vma2p). J. Biol. Chem. 271:2018–2022. 10.1074/jbc.271.4.2018 [DOI] [PubMed] [Google Scholar]
- 15.Wilson RB, Davis D, Enloe BM, Mitchell AP. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70. [DOI] [PubMed] [Google Scholar]
- 16.Bernardo SM, Lee SA. 2010. Candida albicans SUR7 contributes to secretion, biofilm formation, and macrophage killing. BMC Microbiol. 10:133. 10.1186/1471-2180-10-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, Aoki Y. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68:6712–6719. 10.1128/IAI.68.12.6712-6719.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bates S, Hughes HB, Munro CA, Thomas WPH, MacCallum DM, Bertram G, Atrih A, Ferguson MAJ, Brown AJP, Odds FC, Gow NAR. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281:90–98. 10.1074/jbc.M510360200 [DOI] [PubMed] [Google Scholar]
- 20.Bernardo SM, Khalique Z, Kot J, Jones JK, Lee SA. 2008. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet. Biol. 45:861–877. 10.1016/j.fgb.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramage G, López-Ribot JL. 2005. Techniques for antifungal susceptibility testing of Candida albicans biofilms. Methods Mol. Med. 118:71–79. 10.1385/1-59259-943-5:071 [DOI] [PubMed] [Google Scholar]
- 22.Owegi MA, Pappas DL, Finch MW, Jr, Bilbo SA, Resendiz CA, Jacquemin LJ, Warrier A, Trombley JD, McCulloch KM, Margalef KLM, Mertz MJ, Storms JM, Damin CA, Parra KJ. 2006. Identification of a domain in the V0 subunit d that is critical for coupling of the yeast vacuolar proton-translocating ATPase. J. Biol. Chem. 281:30001–30014. 10.1074/jbc.M605006200 [DOI] [PubMed] [Google Scholar]
- 23.Michel V, Licon-Munoz Y, Trujillo K, Bisoffi M, Parra KJ. 2012. Inhibitors of vacuolar ATPase proton pumps inhibit human prostate cancer cell invasion and prostate-specific antigen expression and secretion. Int. J. Cancer 132:E1–E10. 10.1002/ijc.27811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owegi MA, Carenbauer AL, Wick NM, Brown JF, Terhune KL, Bilbo SA, Weaver RS, Shircliff R, Newcomb N, Parra-Belky KJ. 2005. Mutational analysis of the stator subunit E of the yeast V-ATPase. J. Biol. Chem. 280:18393–18402. 10.1074/jbc.M412567200 [DOI] [PubMed] [Google Scholar]
- 25.Chan C-Y, Prudom C, Raines SM, Charkhzarrin S, Melman SD, De Haro LP, Allen C, Lee SA, Sklar LA, Parra KJ. 2012. Inhibitors of V-ATPase proton transport reveal uncoupling functions of tether linking cytosolic and membrane domains of V0 subunit a (Vph1p). J. Biol. Chem. 287:10236–10250. 10.1074/jbc.M111.321133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forgac M, Cantley L, Wiedenmann B, Altstiel L, Branton D. 1983. Clathrin-coated vesicles contain an ATP-dependent proton pump. Proc. Natl. Acad. Sci. U. S. A. 80:1300–1303. 10.1073/pnas.80.5.1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perzov N, Padler-Karavani V, Nelson H, Nelson N. 2002. Characterization of yeast V-ATPase mutants lacking Vph1p or Stv1p and the effect on endocytosis. J. Exp. Biol. 205:1209–1219 [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Köhler J, Fink GR. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726. 10.1126/science.7992058 [DOI] [PubMed] [Google Scholar]
- 29.Crandall M, Edwards JE., Jr 1987. Segregation of proteinase-negative mutants from heterozygous Candida albicans. J. Gen. Microbiol. 133:2817–2824 [DOI] [PubMed] [Google Scholar]
- 30.Fu Y, Ibrahim AS, Fonzi W, Zhou X, Ramos CF, Ghannoum MA. 1997. Cloning and characterization of a gene (LIP1) which encodes a lipase from the pathogenic yeast Candida albicans. Microbiology 14:331–340 [DOI] [PubMed] [Google Scholar]
- 31.Palmer GE. 2008. Autophagy in Candida albicans. Methods Enzymol. 451:311–322. 10.1016/S0076-6879(08)03221-7 [DOI] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin L-S, Choi A, Chu CT, Chung J, Clarke PGH, Clark RSB, Clarke SG, Clavé C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Dröge W, Dron M, Dunn WA, Jr, Duszenko M, et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175. 10.4161/auto.5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milne SW, Cheetham J, Lloyd D, Aves S, Bates S. 2011. Cassettes for PCR-mediated gene tagging in Candida albicans utilizing nourseothricin resistance. Yeast 28:833–841. 10.1002/yea.1910 [DOI] [PubMed] [Google Scholar]
- 34.Pukkila-Worley R, Ausubel FM, Mylonakis E. 2011. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 7:e1002074. 10.1371/journal.ppat.1002074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3:e18. 10.1371/journal.ppat.0030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMullen PD, Aprison EZ, Winter PB, Amaral LAN, Morimoto RI, Ruvinsky I. 2012. Macro-level modeling of the response of C. elegans reproduction to chronic heat stress. PLoS Comput. Biol. 8:e1002338. 10.1371/journal.pcbi.1002338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stiernagle T. 11 February 2006. Maintenance of C. elegans, p 1–11 In WormBook: the online review of C. elegans biology. http://www.ncbi.nlm.nih.gov/books/NBK19649/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- 39.Milgrom E, Diab H, Middleton F, Kane PM. 2007. Loss of vacuolar proton-translocating ATPase activity in yeast results in chronic oxidative stress. J. Biol. Chem. 282:7125–7136. 10.1074/jbc.M608293200 [DOI] [PubMed] [Google Scholar]
- 40.De Dios CH, Román E, Monge RA, Pla J. 2010. The role of MAPK signal transduction pathways in the response to oxidative stress in the fungal pathogen Candida albicans: implications in virulence. Curr. Protein Pept. Sci. 11:693–703. 10.2174/138920310794557655 [DOI] [PubMed] [Google Scholar]
- 41.Enjalbert B, MacCallum DM, Odds FC, Brown AJP. 2007. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 75:2143–2151. 10.1128/IAI.01680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Missall TA, Lodge JK, McEwen JE. 2004. Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot. Cell 3:835–846. 10.1128/EC.3.4.835-846.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y-Q, Gamarra S, Garcia-Effron G, Park S, Perlin DS, Rao R. 2010. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6:e1000939. 10.1371/journal.ppat.1000939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, Burke MD. 2012. Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. U. S. A. 109:2234–2239. 10.1073/pnas.1117280109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plant PJ, Manolson MF, Grinstein S, Demaurex N. 1999. Alternative mechanisms of vacuolar acidification in H+-ATPase-deficient yeast. J. Biol. Chem. 274:37270–37279. 10.1074/jbc.274.52.37270 [DOI] [PubMed] [Google Scholar]
- 46.Bayer MJ, Reese C, Buhler S, Peters C, Mayer A. 2003. Vacuole membrane fusion: V0 functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel. J. Cell Biol. 162:211–222. 10.1083/jcb.200212004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strasser B, Iwaszkiewicz J, Michielin O, Mayer A. 2011. The V-ATPase proteolipid cylinder promotes the lipid-mixing stage of SNARE-dependent fusion of yeast vacuoles. EMBO J. 30:4126–4141. 10.1038/emboj.2011.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vida TA, Emr SD. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792. 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastidas RJ, Heitman J, Cardenas ME. 2009. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5:e1000294. 10.1371/journal.ppat.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400–428. 10.1128/MMBR.67.3.400-428.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calderone RA, Fonzi WA. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327–335. 10.1016/S0966-842X(01)02094-7 [DOI] [PubMed] [Google Scholar]
- 52.Nakamura N, Matsuura A, Wada Y, Ohsumi Y. 1997. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J. Biochem. 121:338–344. 10.1093/oxfordjournals.jbchem.a021592 [DOI] [PubMed] [Google Scholar]
- 53.Mijaljica D, Prescott M, Devenish RJ. 2011. V-ATPase engagement in autophagic processes. Autophagy 7:666–668. 10.4161/auto.7.6.15812 [DOI] [PubMed] [Google Scholar]
- 54.Lynch-Day MA, Klionsky DJ. 2010. The Cvt pathway as a model for selective autophagy. FEBS Lett. 584:1359–1366. 10.1016/j.febslet.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki K, Kamada Y, Ohsumi Y. 2002. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell 3:815–824. 10.1016/S1534-5807(02)00359-3 [DOI] [PubMed] [Google Scholar]
- 56.Morales Quinones M, Winston JT, Stromhaug PE. 2012. Propeptide of aminopeptidase 1 protein mediates aggregation and vesicle formation in cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 287:10121–10133. 10.1074/jbc.M111.311696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chauhan VM, Orsi G, Brown A, Pritchard DI, Aylott JW. 2013. Mapping the pharyngeal and intestinal pH of Caenorhabditis elegans and real-time luminal pH oscillations using extended dynamic range pH-sensitive nanosensors. ACS Nano 7:5577–5587. 10.1021/nn401856u [DOI] [PubMed] [Google Scholar]
- 58.Jia N, Arthington-Skaggs B, Lee W, Pierson CA, Lees ND, Eckstein J, Barbuch R, Bard M. 2002. Candida albicans sterol C-14 reductase, encoded by the ERG24 gene, as a potential antifungal target site. Antimicrob. Agents Chemother. 46:947–957. 10.1128/AAC.46.4.947-957.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanglard D, Ischer F, Parkinson T, Falconer D, Bille J. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404–2412. 10.1128/AAC.47.8.2404-2412.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kane PM, Kuehn MC, Howald-Stevenson I, Stevens TH. 1992. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267:447–454 [PubMed] [Google Scholar]
- 61.Peters C, Bayer MJ, Bühler S, Andersen JS, Mann M, Mayer A. 2001. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature 409:581–588. 10.1038/35054500 [DOI] [PubMed] [Google Scholar]
- 62.Chauhan N, Latge J-P, Calderone R. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4:435–444. 10.1038/nrmicro1426 [DOI] [PubMed] [Google Scholar]
- 63.Palmer GE. 2007. Autophagy in the invading pathogen. Autophagy 3:251–253. 10.4161/auto.3820 [DOI] [PubMed] [Google Scholar]
- 64.Palmer GE, Kelly MN, Sturtevant JE. 2007. Autophagy in the pathogen Candida albicans. Microbiology 153:51–58. 10.1099/mic.0.2006/001610-0 [DOI] [PubMed] [Google Scholar]
- 65.Palmer GE, Askew DS, Williamson PR. 2008. The diverse roles of autophagy in medically important fungi. Autophagy 4:982–988. 10.4161/auto.7075 [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Rao R. 2012. The V-ATPase as a target for antifungal drugs. Curr. Protein Pept. Sci. 13:134–140. 10.2174/138920312800493205 [DOI] [PubMed] [Google Scholar]
- 67.Schulenburg H, Kurz CL, Ewbank JJ. 2004. Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198:36–58. 10.1111/j.0105-2896.2004.0125.x [DOI] [PubMed] [Google Scholar]
- 68.Irazoqui JE, Urbach JM, Ausubel FM. 2010. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10:47–58. 10.1038/nri2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mylonakis E, Ausubel FM, Tang RJ, Calderwood SB. 2003. The art of serendipity: killing of Caenorhabditis elegans by human pathogens as a model of bacterial and fungal pathogenesis. Expert Rev. Anti Infect. Ther. 1:167–173. 10.1586/14787210.1.1.167 [DOI] [PubMed] [Google Scholar]
- 70.Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:15675–15680. 10.1073/pnas.232568599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.