Abstract
SUMMARY
Human astroviruses (HAtVs) are positive-sense single-stranded RNA viruses that were discovered in 1975. Astroviruses infecting other species, particularly mammalian and avian, were identified and classified into the genera Mamastrovirus and Avastrovirus. Through next-generation sequencing, many new astroviruses infecting different species, including humans, have been described, and the Astroviridae family shows a high diversity and zoonotic potential. Three divergent groups of HAstVs are recognized: the classic (MAstV 1), HAstV-MLB (MAstV 6), and HAstV-VA/HMO (MAstV 8 and MAstV 9) groups. Classic HAstVs contain 8 serotypes and account for 2 to 9% of all acute nonbacterial gastroenteritis in children worldwide. Infections are usually self-limiting but can also spread systemically and cause severe infections in immunocompromised patients. The other groups have also been identified in children with gastroenteritis, but extraintestinal pathologies have been suggested for them as well. Classic HAstVs may be grown in cells, allowing the study of their cell cycle, which is similar to that of caliciviruses. The continuous emergence of new astroviruses with a potential zoonotic transmission highlights the need to gain insights on their biology in order to prevent future health threats. This review focuses on the basic virology, pathogenesis, host response, epidemiology, diagnostic assays, and prevention strategies for HAstVs.
INTRODUCTION
Using electron microscopy (EM), Appleton and Higgins reported in 1975 the occurrence of 28- to 30-nm particles in stools of children suffering from vomiting and mild diarrhea (1). The same year, Madeley and Cosgrove used the term astrovirus (AstV) to describe the small round viruses with a characteristic star-like appearance (“astron” means star in Greek) found in the feces of hospitalized infants with gastroenteritis (2). These particles were placed together with other viruses with a smooth entire edge within the category of “small round viruses” (SRVs), which differed from the “small round structured viruses” (SRSVs) that grouped particles with a rough, hairy, or irregular edge, such as caliciviruses (3).
The genomic and subgenomic organization of AstV and its polyprotein processing led to the proposal of the new family Astroviridae, separated from the families Picornaviridae and Caliciviridae, within the positive-sense single-stranded RNA (ssRNA) viruses (4). In 1995, the International Committee for the Taxonomy of Viruses (ICTV) definitively established the Astroviridae family in their sixth report (5). Table 1 depicts the main characteristics of the Astroviridae family.
TABLE 1.
Main characteristics of the Astroviridae family
| Characteristic |
|---|
| Virion structure |
| Icosahedral particles, 28–41 nm in diam |
| Nonenveloped |
| Distinctive five- or six-pointed star-like shape under the electron microscope in about 10% of the virions |
| Genome |
| A single molecule of infectious, positive-sense ssRNA, 6.4–7.7 kb, with a poly(A) tail |
| Three ORFs: ORF1a and ORF1b at the 5′ end encoding the nonstructural proteins and ORF2 at the 3′ end encoding the structural proteins |
| VPg protein linked to the 5′ end |
| A ribosomal frameshifting signal present between ORF1a and ORF1b |
| Lack of a helicase domain |
| Replicative cycle |
| Structural proteins are expressed from an ∼2.8-kb subgenomic RNA |
Initially, the Astroviridae family consisted of a single genus, Astrovirus, based on virion morphology (5). However, later on, two genera were established based on their hosts of origin: Mamastrovirus (MAstV) and Avastrovirus (AAstV), infecting mammalian and avian species, respectively. Although initially detected in children's stool, AstVs have been found in the feces of a wide variety of mammalian species, i.e., cats (6), cattle (7), deer (8), dogs (9), mice (10), rats (11), pigs (12), sheep (13), mink (14), bats (15), cheetahs (16), rabbits (17), and even sea lions and dolphins (18), as well as in avian species, i.e., turkeys (19), chickens (20), ducks (21), pigeons (22), and guinea fowl (23) and other wild aquatic birds (24).
Very recently, the complete picture of the AstV field has dramatically changed with the discovery after metagenomic surveillance studies of a variety of highly divergent novel AstVs able to infect different animal species, including humans, which are unrelated to the previously described 8 serotypes of HAstVs, now termed classic HAstV (25–29). The first novel HAstVs were identified in 2008 in pediatric stool specimens in Melbourne, Australia (26). They were termed HAstV-MLB, and so far several MLB-related strains (named MLB1, MLB2, and MLB3) have been detected in different parts of the world (27, 30, 31). In 2009, a second group of novel HAstVs was described in samples from children with diarrhea in Virginia (VA) (28) and in Nigeria, Pakistan, and Nepal (HMO, referring to human, mink, and ovine-like astroviruses) (29). In this review, the term HAstV-VA/HMO will be used to refer to this group. So far, up to four VA/HMO strains have been described (27, 31). Thus, a single host species may be susceptible to be infected by different AstVs (15, 32). Altogether, the number of AstVs associated with human infections has almost doubled in the last few years.
The present review focuses on HAstVs within the genus Mamastrovirus, which according to molecular surveillance data are one of the most important causes of pediatric acute gastroenteritis, after rotaviruses and arguably caliciviruses (33–36).
BASIC VIROLOGY
Virion Structure and Assembly
The AstV virion is an icosahedral nonenveloped particle with a smooth margin and a distinctive five- or six-pointed star identified on the surface of some (around 10%) virions (Fig. 1). Particles are assembled from the VP90 precursor protein (approximately 90 kDa), which is further processed by cellular caspases to generate the VP70 protein, losing an acidic C-terminal domain (37). Particles composed of the 70-kDa protein are immature and require trypsin cleavages, which occur extracellularly through a rather complicated pathway producing highly infectious particles with capsid proteins of 32 to 34, 27 to 29, and 25 to 26 kDa (VP34, VP27/29, and VP25/26) (38). VP34 is derived from the highly conserved N-terminal region of the polyprotein and builds up the capsid shell, while VP27/29 and VP25/26 are both derived from the variable C-terminal domain with a different N terminus and form the dimeric spikes (39, 40) (see below).
FIG 1.
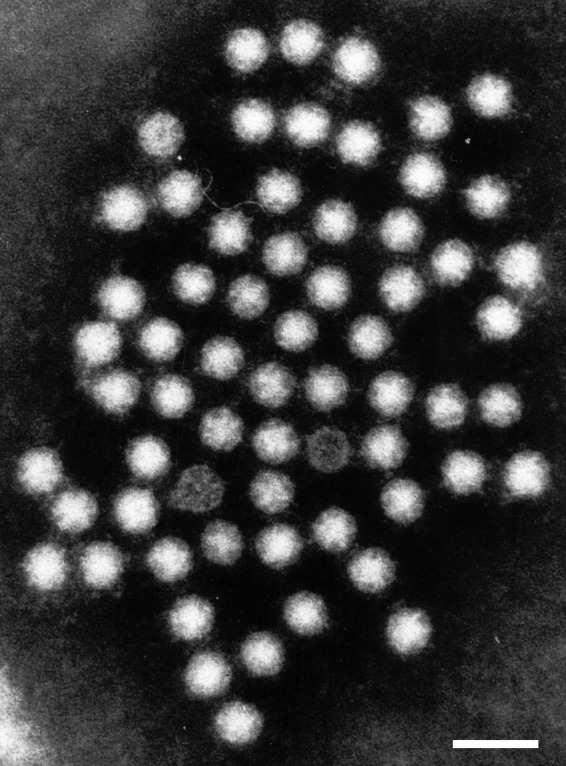
Human astrovirus particles observed by transmission immunoelectron microscopy in feces negatively stained with phosphotungstic acid. Bar, 50 nm.
Transmission EM observation reveals that viruses shed in stool are 28 to 30 nm in diameter, while viruses produced in cell monolayers are larger, with an external diameter of 41 nm, and distinct protruding spikes but without the characteristic star-like appearance (41). This surface star-like shape could, however, be observed after alkaline treatment of the particles. There is evidence that particle diameter could vary according to the source of the virus and sample processing for the preparation of EM suspensions.
Recent cryo-electron microscopy studies of immature (VP70) and mature (processed protein) particles of cell-adapted HAstV have revealed a number of distinctive features that provide insight on AstV structure and polyprotein processing (42). The cryo-EM reconstructions show a capsid shell of 35 nm organized as T = 3 icosahedral symmetry assembled from 180 protein subunits and decorated with either 30 (mature particles) or 90 (immature particles) globular dimeric spikes, resulting in a total diameter of 44 nm.
The crystal structure of the AstV spike without its C-terminal end has been elucidated (40). Remarkably, the shape, size, and architecture of the AstV spike show a notable similarity with those of hepatitis E virus, the only member of the recently established Hepeviridae family (43).
Despite the advances in the determination of virion structure, the AstV protein composition, which is critical to determine virus specific infectivity (38, 44), remains to be clearly defined.
Genome Organization
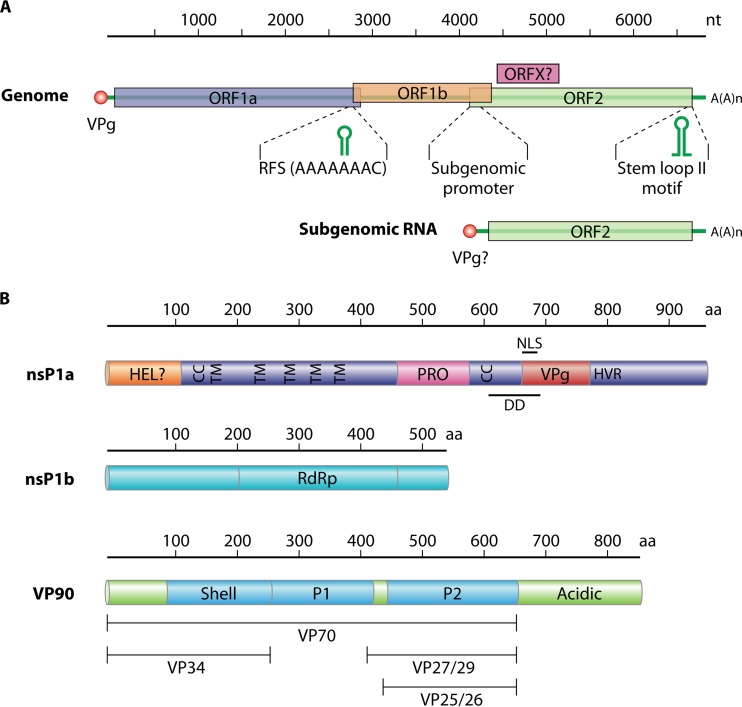
The AstV genome is a positive-sense ssRNA molecule of around 6.8 (6.2 to 7.8) kb, excluding the polyadenylated tail at the 3′ end (45). A VPg protein is covalently linked to the 5′ end of the genome (46). The genome contains three open reading frames (ORFs), named from the 5′ end to the 3′ end ORF1a, ORF1b, and ORF2. ORF1a and ORF1b encode the nonstructural proteins (nsPs) involved in RNA transcription and replication, while ORF2 encodes the structural proteins, which are expressed from a subgenomic RNA (Fig. 2) (4, 47). In the classic HAstVs and in other mammalian AstVs, a new ORF, termed ORFX, overlapping the 5′ end of ORF2 in the +1 reading frame, has been described (48). It has been suggested that ORFX could be translated through a leaking scanning mechanism. The translation product of this ORFX has not yet been experimentally confirmed.
FIG 2.
Genome organization and polyprotein products of human astrovirus. (A) Genomic and subgenomic RNA organization, with open reading frames (ORFs) ORF1a, ORF1b, ORF2, and putative ORFX represented as boxes. Nucleotide sequences represent highly conserved sequences located in the ribosomal frameshift (RFS) signal and upstream of the initiation site of subgenomic RNA transcription. Putative RNA secondary structures conserved in the RFS and in the 3′ end of the genome are depicted. (B) Characteristic motifs of the HAstV polyprotein products. HEL, putative helicase domain; TM, transmembrane domain; CC, coiled-coil domain; PRO, protease domain; VPg, coding region for a VPg protein; HVR, hypervariable region; NLS, putative nuclear localization signal; DD, putative death domain; RdRp, RNA dependent-RNA polymerase motif. The ORF2-encoded structural polyprotein (VP90) consists of conserved regions (shell and P1 domains), a variable region containing the P2 domain (capsid spikes), and an acidic C-terminal region which is cleaved by cellular caspases to result in the VP70 precursor. Particles containing VP70 are further cleaved by trypsin to yield the VP34, VP27/29, and VP25/26 proteins.
The AstV RNA is infectious and upon transfection produces infectious virus progeny (49). Infectious particles may also be obtained after in vitro transcription of a full-length cDNA clone of the HAstV genome and transfection into permissive cells (49, 50). During the replicative cycle, a positive-sense single-stranded subgenomic RNA corresponding to around 2.8 kb of the 3′ end of genomic RNA is produced in infected cells, coding for the virion structural proteins (51).
Regarding genome organization, two untranslated regions (UTR)s, the 5′ UTR and the 3′ UTR, of 11 to 85 and 80 to 85 bases, respectively, are located at the ends of the AstV genome. No internal ribosome entry site (IRES) has so far been described in AstV. This element is present at the 5′ ends of the genomes of other positive ssRNA viruses belonging to the families Picornaviridae (52) and Flaviviridae (53) but is absent in the family Caliciviridae (54). Besides the poly(A) tail consisting of around 30 adenines, a highly conserved secondary element is present at the 3′ end of the AstV genome. This secondary structure motif, termed the stem-loop II motif, is present in the genomes of classic HAstVs, HAstV-VA/HMO, and cat, ovine, porcine, and avian AstVs, but it is only partially conserved in the genome of HAstV-MLB (55–57). Interestingly this structure is also found at the 3′ ends of the genomes of some members of coronaviruses, noroviruses, and rhinoviruses (56, 58). This stem-loop II motif appears to be conserved at both the nucleotide sequence and RNA structure levels, indicating a strong evolutionary selection for its conservation. Although its function is yet to be determined, it has been suggested that it could provide stability to the secondary structure of RNA and interact with viral and cellular proteins essential for genome replication (56).
The lengths of ORF1a, ORF1b, and ORF2 vary depending on the AstV strain. This variation is largely dependent on the insertions and deletions present at the 3′ end of ORF1a (59, 60). Likewise, the overlapping regions of the different ORFs also vary in length depending on the AstV strain. In mammalian AstVs the overlapping region of ORF1a and ORF1b ranges from 10 to 148 nucleotides (nt), while the overlap is only 12 to 45 nt long in avian AstVs (61). A ribosomal frameshifting signal (RFS), essential for the translation of the RNA-dependent RNA polymerase (RdRp) encoded in ORF1b, is present precisely in this overlapping region (62, 63). The RFS has been found in all AstVs except in a newly described chicken astrovirus, in which ORF1b has its own start codon (64). The overlap found between ORF1b and ORF2 may also vary in length or be nonexistent, as occurs in duck AstV (65). A highly conserved sequence that could be part of the promoter for subgenomic RNA synthesis is located in this region (Fig. 2) (13).
ORF1a encodes a putative helicase domain (HEL), several transmembrane (TM) and coiled-coil (CC) domains, the protease domain (PRO), a VPg, a hypervariable region (HVR), a nuclear localization signal (NLS), and a putative death domain (DD). ORF1b encodes the RNA dependent-RNA polymerase (RdRp). ORF2 contains the region coding for the shell proteins, a P1 domain of unknown function, a variable region containing the P2 domain which includes the spike proteins, and an additional acidic domain at the C terminus (Fig. 2).
General Overview of the HAstV Replicative Cycle
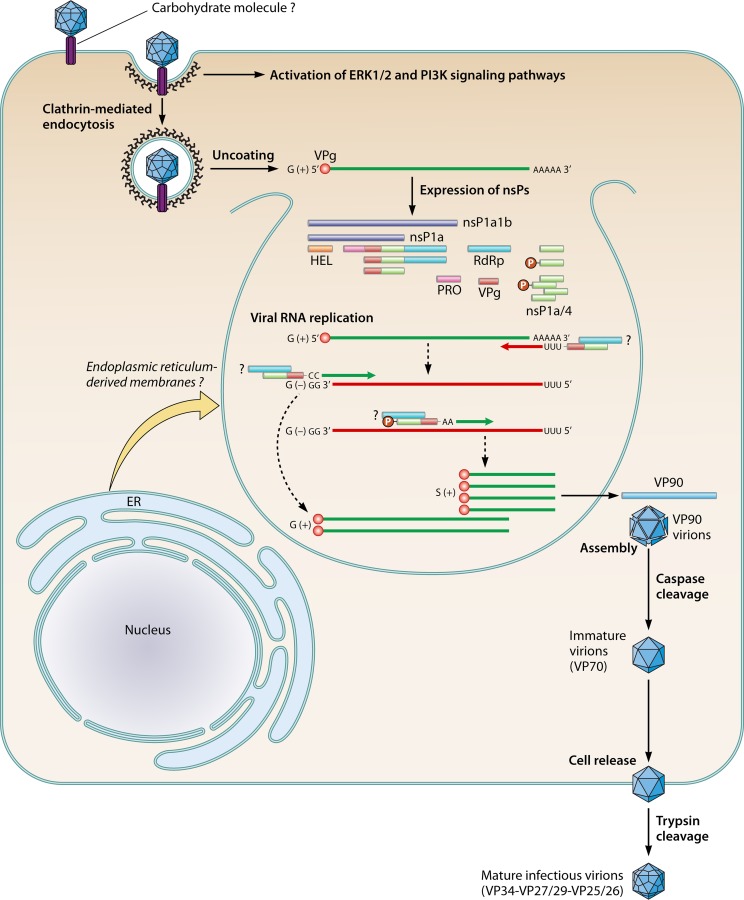
The following sections dealing with the replicative cycle of HAstVs essentially refers to the so-called classic HAstVs. The HAstV replication cycle shares many common features with the replication cycle of members of the Caliciviridae family (Fig. 3). After cell entry and uncoating, the two main nonstructural polyproteins, nsP1a (∼102 kDa) and nsP1a1b (∼160 kDa), are translated from the VPg-linked genomic RNA. Nonstructural polyprotein nsP1a1b is expressed thanks to the RFS that exists between ORF1a and ORF1b (62, 63). Cleavage of these polyproteins results in the individual nonstructural proteins required for genome replication, which takes place in replication complexes assembled in close association with intracellular membranes. This process results in the formation of both genomic and subgenomic RNAs, which are produced in large quantities to ensure the production of high yields of structural proteins. After encapsidation and maturation, virions are released from the cell. While it is known that both genomic and subgenomic RNAs are encapsidated into virions of animal caliciviruses (66), it is yet to be elucidated whether a similar process occurs in AstVs.
FIG 3.
Replication of human astroviruses. After binding to one or more cellular receptors, virus uptake occurs via clathrin-dependent endocytosis. A drop in the pH leads to viral uncoating. Two main nonstructural polyproteins, nsP1a and nsP1a1b, are translated from the VPg-linked genomic RNA and further cleaved by viral and cellular proteases, resulting in mature nonstructural proteins (nsPs), which are required for genome replication. Replication complexes assemble in close association with intracellular membranes. The nsP1a/4 protein or one of its precursors, which would include the VPg domain, may interact with the RdRp protein and contribute to the regulation of the synthesis of negative- and positive-sense RNA strands as well as of subgenomic RNA. The phosphorylation status of the nsP1a/4 protein may contribute to this regulation step. Subgenomic RNAs are produced in large quantities and are used for the expression of capsid proteins. The structural VP90 polyprotein initially assembles into immature virions in association with intracellular membranes. Several cellular caspases further cleave these VP90 polyproteins once they have dissociated from membranes, resulting in VP70 immature viral capsids. Release of VP70 particles into the medium seems to occur without cell lysis, and virions mature extracellularly by the action of trypsin.
Cell Binding and Entry
The main cellular receptor for HAstVs still remains unknown, but the susceptibilities of different cell lines to HAstV infection differ depending on the serotype (67), suggesting that these viruses could take advantage of having more than one attachment or receptor molecules, some of which may be of a polysaccharide nature and common to all HAstVs (40). Besides revealing a close relationship with hepatitis E virus, determination of the structure of the HAstV dimeric surface spikes has identified a putative receptor binding site that is conserved in all HAstV serotypes and which would recognize a polysaccharide molecule (40). In the same study, it was noted that dextran sulfate, heparin sulfate, and particularly heparin could partially block the infectivity of HAstVs, while sialic acid did not seem to take part in the virus-cell recognition event. Thus, as has been hypothesized for other related viruses such as noroviruses, a carbohydrate molecule may act as cellular receptor or at least as an initial recognition factor on the cell surface. Interestingly, binding of HAstVs to in vitro-differentiated intestinal epithelial cell monolayers results in an increase in cell barrier permeability, and this phenomenon occurs independently of viral replication (68). Whether these changes are required for the viruses to reach putative receptors present in the basolateral membrane or whether they are a consequence of the activation of cellular signaling pathways after initial binding is still uncertain.
Subsequent steps of the life cycle have been recently studied on the highly permissive CaCo-2 cells (69), confirming previous observations on HEK293 cells (70) which indicated that HAstVs use a clathrin-mediated endocytosis pathway as the viral entry mechanism. Drugs that inhibit clathrin-mediated endocytosis and actin filament polymerization, as well as those that reduce the presence of cholesterol in the cell membrane, decrease the infectivity of the virus (69). Furthermore, entry also depends on the acidification and maturation of endosomes where membrane permeabilization and RNA uncoating would occur. Overall, it has been estimated that while the half time of virus binding to the cell surface is about 10 min, virus uncoating takes around 130 min (69).
Cellular signaling pathways activated after HAstV initial infection have been analyzed using specific drug inhibitors, and it is known that interaction of HAstVs with cells results in the activation of the extracellular signal-regulated kinase (ERK1/2) and the phosphoinositide 3-kinase (PI3K) pathways, both being required for effective entry and establishment of a productive infection (71, 72). It seems that the PI3K-mediated cascade would act independently or downstream of that mediated by ERK1/2 (72). Finally, it has also been observed that activation of the ERK1/2 pathway does not require binding of infectious viruses and can be achieved after binding of inactivated viruses and/or virus-like particles (71). Although it is clear that ERK1/2 pathway activation is required for a productive HAstV infection and it has been speculated that ERK could phosphorylate the HAstV RdRp or other nonstructural proteins, the function of activated ERK still remains unknown, and it may apply an indirect mechanism.
Translation/Replication
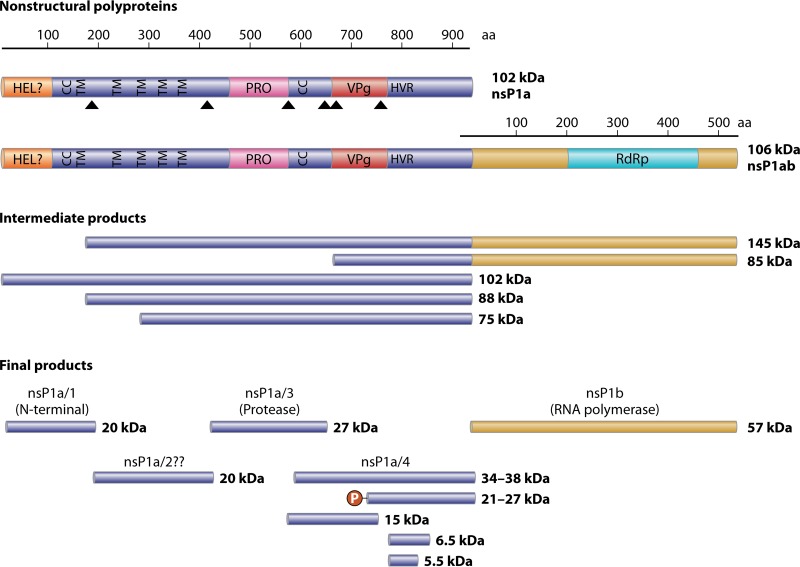
The VPg protein linked to the HAstV genome plays an essential role in infectivity following viral RNA uncoating. Removal of the VPg protein by proteolytic treatment of viral RNA completely abolishes infectivity (46), indicating that the VPg protein may play a role in the recruitment of cellular translation initiation factors, analogous to what has been described for caliciviruses (73). The genome is then translated, giving rise to nsP1a1b and nsP1a polyproteins, which are then cleaved by the viral serine protease (in nsP1a) as well as some cellular proteases, resulting in the individual nonstructural proteins (Fig. 4). The crystal structure of the viral protease has been resolved, showing properties of trypsin-like enzymes with a catalytic Asp-His-Ser triad typical of the serine proteases (74).
FIG 4.
Putative processing of the astrovirus nonstructural proteins expressed from ORF1a and ORF1b. HEL, putative helicase domain; CC, coiled-coil domain; TM, transmembrane domain; PRO, protease domain; VPg, viral protein genome-linked coding region; HVR, hypervariable region; RdRp, RNA-dependent RNA polymerase motif; P, posttranslational phosphorylation. Triangles depict proteolytic cleavage sites described in the literature (75, 77, 78, 80).
The cleavage process of the virus nonstructural polyproteins is rather unknown. None of the processing products has been confirmed by N-terminal sequencing, and studies performed in different experimental systems with a variety of classic HAstVs reported different intermediate and final products (75–79). It appears, however, that proteolytic processing of polyproteins nsP1a1b (around 160 kDa) produces the nsP1b protein (RdRp) (around 57 kDa) and the nsP1a protein (around 102 kDa), which is subsequently cleaved to yield several mature products (Fig. 4). Several putative transmembrane domains that could help in driving and anchoring the nonstructural replication complexes on cellular membranes have been identified in the nsP1a polyproteins (13, 62). Four potential cleavage sites have been proposed in the nsP1a protein, specifically between residues Ala174 and His175, between Val409 and Ala410, between Glu567 and Thr568, and between Glu654 and Ile655. Although discrepancies exist in regard to the exact cleavage sites, it seems clear that a viral protease would be responsible for all proposed proteolytic cleavages except for the one between Ala174 and His175, yielding at least four proteins, nsP1a/1 to -4 (75, 77, 78, 80). It has been shown that some of the resulting nonstructural proteins are posttranslationally modified by phosphorylation mechanisms (81, 82), and these modifications may modulate the interaction among them. Whether the proteolytic processing of these nonstructural polyproteins takes place before or after anchoring to intracellular membranes is still unknown, but large membrane rearrangements are observed in HAstV-infected cells both in vitro (82, 83) and in vivo (84). Since both viral nonstructural proteins and viral RNA have been found colocalizing with the endoplasmic reticulum and an endoplasmic reticulum retention signal has been predicted at the end of nsP1a (82), it is believed that endoplasmic reticulum-derived intracellular membranes would be the cellular site for HAstV genome replication.
The HAstV genome replication process has not been characterized in detail and is inferred from those of other positive-sense RNA viruses. Thus, a full-length negative-sense genomic RNA would be synthesized and used as a template for the production of both positive-sense genomic RNA and positive-sense subgenomic RNA, but the existence of a negative-sense subgenomic RNA template cannot be completely ruled out. Regarding the kinetics of this process, Jang et al. (85) observed that de novo synthesis of positive-sense RNAs occurs almost instantly following negative-sense RNA synthesis. They also estimated the total amount of HAstV negative-sense RNA synthesized in cells as nearly 0.7 to 4.0% of that of the HAstV positive-sense RNA. Many other aspects of the HAstV genome replication process, such as the role of the VPg protein as a primer for initiation of RNA strands, have not been addressed yet. Involvement of cellular host proteins in the process and the mechanisms used by the virus to switch between replication and transcription are other issues that remain to be elucidated. The expression of one of the nonstructural proteins (the nsP1a/4 protein) and the RdRp in the baculovirus expression system has suggested that oligomerization and phosphorylation states of nsP1a/4 protein may regulate its interaction with the RdRp protein (81), and although not confirmed, it is likely that these changes may contribute to the regulation of the synthesis of negative- and positive-sense RNA strands as well as of subgenomic RNA. In addition, variability in the nsP1a/4 gene, which contains a hypervariable region, also affects the virus replication phenotype (60). Percentages of genomic, subgenomic, and antigenomic RNAs in total cell-associated RNAs produced during infection vary depending on the nsP1a/4 gene. While the amount of subgenomic RNA produced by viruses containing certain nsP1a/4 variants represents only between 0.8 and 17% of the total RNA, the proportion of subgenomic RNA produced by other nsP1a/4 mutants can reach up to 85 to 95%, and this also results in significantly higher numbers of infectious progeny (60). Accordingly, the viral load in feces from children with gastroenteritis correlates with nsP1a/4 variability, and this phenotype can be typed based on the hypervariable region contained in the nsP1a/4 gene (86) (see below).
Expression of Structural Proteins, Virion Assembly, and Cell Release
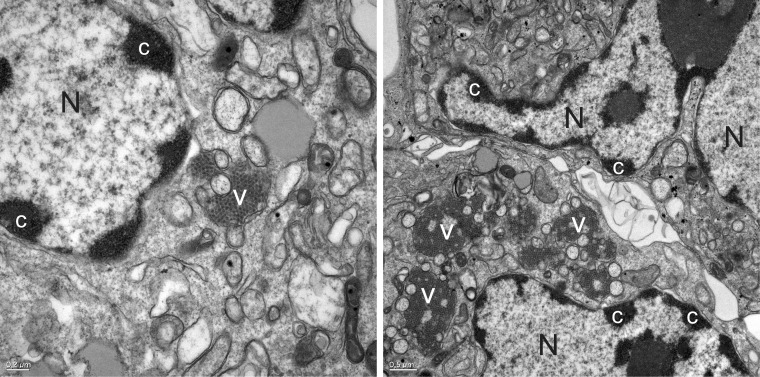
Structural proteins are expressed from the subgenomic RNAs as an ∼90 kDa polyprotein (VP90) through a mechanism that it is likely to involve the 5′-end VPg (46). Cell fractionation studies have shown that structural protein synthesis takes place in the same cellular compartments where the replication complexes form (83). The model of HAstV morphogenesis suggests that VP90 polyprotein initially assembles into immature virions in association with intracellular membranes through its C terminus and that several cellular caspases further cleave these VP90 polyproteins close to their C termini once they have dissociated from membranes, resulting in viral capsids composed of 70-kDa polyproteins (VP70) (83, 87). In infected cells, virus aggregates surrounded by a large quantity of double-membrane vacuoles may be observed in the vicinity of the nucleus (Fig. 5) (82, 83). Finally, VP70-containing viral particles would be released from cells and proteolytically processed by trypsin to enhance infectivity. Processing of VP90 to VP70 by caspases is required for viruses to exit the cell, and release of VP70 particles into the medium seems to occur without cell lysis (37). Virus release from cells is more dependent on caspase activity than on cell death itself (37).
FIG 5.
HAstV particles are associated with membranes in infected cells. Ultrastructural analysis of CaCo-2 cells infected with HAstV-4 at 48 h postinfection shows aggregates of virions accumulated in the cytoplasm in close association with double-membrane vesicles (V). Nuclei (N) of infected cells show masses of condensed chromatin dispersed at the periphery (c).
PATHOLOGY, PATHOGENESIS, AND HOST RESPONSE
Clinical Disease Correlates
HAstVs, especially classic HAstVs, are considered gastrointestinal pathogens affecting children worldwide, with very few reports of HAstV-mediated disease in normal healthy adults (88, 89). Serological studies indicate that most children are infected with HAstV and develop antibodies to the virus early in life (90, 91), which are thought to provide protection against future infections. Immunocompromised individuals and the elderly also represent high-risk groups. Typically, HAstV infection induces a mild, watery diarrhea that lasts 2 to 3 days, associated with vomiting, fever, anorexia, and abdominal pain. Vomiting is less prevalent in astrovirus infection than in rotavirus or calicivirus infection, and HAstV infections also show a longer incubation period. Based on data from adult volunteer studies and outbreaks of gastroenteritis in a child care center, the mean incubation period of HAstV infections was calculated to be 4.5 days (92). In general, HAstV diarrhea is milder than those caused by rotaviruses or noroviruses, and it resolves spontaneously, although in some cases HAstV infections have required hospitalization. Asymptomatic infections have also been described in children and adults (93–96), although HAstV prevalence as asymptomatic pathogens has yet to be characterized. Studies in immunodeficient patients, including HIV-infected individuals, have associated HAstV infections with symptomatic gastroenteritis (97, 98), but a recent report has also shown that classic HAstV infections can also spread systemically and cause severe disseminated lethal infections in highly immunocompromised children (99). Consistent with this idea, recent work on murine AstVs (MuAstVs) has demonstrated distribution of viruses beyond the gastrointestinal tract (i.e., in the spleen, liver, and kidney) in immunodeficient mice (100). Finally, the relationship of HAstV infection to the etiology of intussusception in children has also been studied. Although with less frequency than rotavirus, norovirus, and adenovirus infection, HAstV infection has also been identified as a potential risk factor for intussusception in infected children (101–103).
Correlations between specific serotypes of classic HAstVs, level and duration of viral load, and severity of disease have not been deeply analyzed. In one study, quantification of shedding levels of serotypes 1, 2, 3, 4, and 8 ascertained viral concentrations ranging from 3.4 × 108 to 1 × 1013 per gram of feces and significantly higher titers in the serotype 3-containing feces. In addition, serotype 3 was associated with a more severe gastroenteritis (104). Later on, differences in viral load in fecal samples were further associated with variability in one of the viral nonstructural proteins, the nsP1a/4 protein (60), suggesting that nonstructural proteins may also influence the pathogenicity of the HAstV strain.
Besides classic HAstVs, infections caused by the other two novel types of nonclassic HAstVs (HAstV-MLB and HAstV-VA/HMO) are generally associated with gastroenteritis as well, but their pathogenic role in human health has not been clearly demonstrated. Both types of viruses were almost simultaneously identified in pediatric stool specimens from children with diarrhea (26–29), but their role as etiological agents of gastroenteritis needs to be further clarified.
HAstV-MLB (MLB1, MLB2, and MLB3) strains have so far been detected in children with gastroenteritis in Australia (26), the United States (30), India (27), Mexico (105), Turkey (106), Egypt (107), Bhutan (108), China (109), and Europe (110). However, a case-control study on HAstV-MLB1 and classic HAstV infections in India showed that while classic HAstVs were significantly associated with diarrhea, HAstV-MLB1 was not (111). Furthermore, MLB1 titers in stool did not differ significantly between symptomatic and asymptomatic individuals. Since HAstV-MLB2 viruses have also been isolated in the plasma and nasopharynx of a child with an upper respiratory infection (112, 113), the authors have hypothesized that HAstV-MLB pathogenicity may affect extraenteric tissues and that its presence in stool may simply be a consequence of its mode of transmission. No other pathogen was detected in these samples, suggesting that HAstV-MLB2 was the cause of the patient's fever. Hence, the tropism of selected strains of HAstV-MLB may not be restricted to the gastrointestinal tract, and the role of these viruses in pediatric infections demands further investigation.
HAstV-VA/HMO (VA1, VA2, and VA3 [also known as HMO-C, HMO-A, and HMO-B, respectively] and VA4) have been detected in pediatric gastroenteritis samples in Nigeria, Nepal, and Pakistan (29), the United States (27), Egypt (107), and China (109), but so far they have never been reported in Europe (114). Serological studies in the United States have confirmed that HAstV-HMO-C (VA1) is a highly prevalent human-infectious agent (115). In a recent study, a strain closely related to HAstV-VA1 was isolated in a patient with new-onset celiac disease (114). Finally, similarly to what has been observed for classic HAstVs, extraintestinal dissemination in immunodeficient individuals has also been observed in HAstV-HMO-C (VA1) virus infections, when the agent was detected in neural tissue from an immunocompromised child with encephalitis (116). Future research efforts should assess the extent and severity of diseases associated with novel HAstVs.
Histopathological Changes
Despite the impact of HAstV on human health, knowledge on astrovirus pathogenesis is still limited (117), mainly due to the lack for many years of a good small-animal model and to the fact that most data gathered so far are restricted to classic HAstVs. For humans, there is only one report describing the histopathological examination of small intestinal biopsy specimens in an immunodeficient child with HAstV infection and persistent diarrhea after a bone marrow transplant (118). The child shed HAstV for more than 60 days. The main findings evidenced villous blunting, irregular superficial epithelial cells, and an increase in lamina propria inflammatory cell density. Viral replication was targeted to surface epithelial cells, mostly at the villus tips in the jejunal and duodenal biopsy specimens. Although the patient was immunocompromised, the study concluded that the intestinal inflammatory response was only mild.
Ultrastructural studies in symptomatic animals have been performed in experimentally infected lambs, calves, and turkeys. Experimentally infected lambs showed villus atrophy in the small intestine (84, 119). Astroviruses infected only mature enterocytes (viruses were seen in paracrystalline arrays, along the microvilli, and within lysosomes and autophagic vacuoles) in the villi of the small intestine and macrophages (viruses were seen in lysosomes) in the lamina propria (84, 119). Infected enterocytes could be observed sloughing into the gut, and by 5 days postinfection, the villi appeared normal. Maximum lesions were observed in the jejunum and ileum, without observation of infected cells in the colon (119).
The bovine virus was found to mostly infect the epithelium covering the dome villi of the jejunal and ileal Peyer's patches (120). Both M cells and absorptive enterocyte cells of the dome epithelium contained virus, and microvilli of infected M cells were often severely stunted.
In turkeys, replication of turkey astroviruses (TAstVs) was detected in the upper sectors of the small intestine, but it was later on detected in the large intestine as well, infecting epithelial cells at the basal borders of the villi (121). Infectious TAstV-2 was isolated from the bursa, thymus, spleen, kidney, skeletal muscle, pancreas, and plasma, confirming that a viremic stage occurs during infection (122). However, despite detection of infectious particles and viral antigen in these extraintestinal tissues, in situ hybridization assays indicated that astrovirus replication was confined to the intestines. No clear histological changes were either observed in any of these tissues. Histological analysis of the intestine indicated only minor enteric damage, in the form of mild epithelial necrosis, lamina propria infiltration, minimal villous atrophy, and mild crypt hyperplasia. The lack of major villus atrophy, cell death, and inflammation indicated that astrovirus-induced diarrhea is caused by a mechanism other than damage of the intestinal epithelium or an inflammatory response. The increased activation of the potent immunosuppressive cytokine transforming growth factor β (TGF-β) observed in infected turkeys may partially explain the lack of inflammation (122).
Finally, the recent isolation and characterization of MuAstVs offers new perspectives for the use of the mouse model for the study of many aspects of viral replication in vivo and the histopathological changes induced by the infection (100). Although in the murine model, animals do not suffer diarrhea and different mouse strains show different susceptibilities, MuAstV shedding in wild-type mice for more than 14 days has been observed, and viral RNA has been detected in duodenum, ileum, proximal colon, and mesenteric lymph nodes (100).
Mechanisms of Pathogenesis and Induction of Diarrhea
Since astrovirus-infected intestines show relatively minor histological changes and inflammation, other mechanisms of virus-induced diarrhea have been studied. Among the primary mechanisms described for diarrhea induction by other enteric infections (reviewed in references 123 and 124), major destruction of the intestinal epithelium and inflammatory responses do not seem to play a role in AstV infection. Alternatively, rapid loss of fluids and electrolytes may originate from inhibition of the usual absorptive function of the intestine, activation of secretory processes, or loss of intestinal epithelial barrier permeability.
Studies in the turkey animal model showed that specific maltase activity decreases during infection. Decreased specific maltase activity caused disaccharide maldigestion and malabsorption, followed by osmotic diarrhea affecting intestinal absorption (125). Later on, sodium malabsorption, likely by redistribution of specific sodium transporters, was reported in turkey poults with diarrhea (126). Levels of the sodium hydrogen exchanger NHE2 were found to increase upon infection, and NHE3 was translocated from the membrane to the cytoplasm. In addition, analysis of TAstV-infected enterocytes failed to show evidence of paracellular dilatation, although F-actin rearrangements were identified in the apical regions of cells.
Finally, using HAstV-infected differentiated CaCo-2 cells, F-actin rearrangements have also been observed, together with disruption of the cell barrier and redistribution of occludin (68). Discrepancies with data from AstV infection in turkeys, in which no alterations in tight junctions were observed, could be due to the use of different viruses and/or different cellular models or to differences in the virus infection stage at the time of analysis. Of note, the effect of HAstV infection on epithelial barrier permeability occurs independently of viral replication, suggesting that the capsid protein of HAstVs may act as an enterotoxin (68). Although the study did not report significant amounts of cell death upon HAstV infection at the time of the loss of barrier function and these changes were mainly attributed to the disruption of tight junctions, HAstV-induced enterocyte death may eventually occur at a late stage of the infection. Indeed, other studies using CaCo-2 cells have shown that infection induces cell death by apoptosis (37, 127). In agreement with these hypotheses, the release of virions from infected cells is thought to occur through a nonlytic mechanism (37).
Host Immune Responses and Control of Infection
The observation that AstV infection and gastroenteritis in most species are age dependent led to the assumption that immune protection is acquired after an initial infection. However, the specific immune responses activated upon AstV infection are not yet completely understood (reviewed in reference 128). Epidemiological and clinical studies have demonstrated that the humoral immune response plays a role in restraining infection and disease in humans. Seroprevalence studies show that the vast majority of healthy young adults have antibodies against the most prevalent classic HAstV serotypes (90) and also against some of the novel HAstVs (115) and that the increase in seroprevalence correlates as well with age during childhood (91, 115, 129). Maternal-fetal transmission of antibody occurs, and antibody levels decrease by 6 to 8 months of age (91, 129). Infections induce antibodies in humans of all age groups, which may persist for extended periods of time (90, 129). These findings are in agreement with the observations made during the two clinical studies performed in human volunteers; the severity of infection signs negatively correlated with the presence of anti-HAstV antibodies before the challenge (95, 96). Although seroprevalence studies detected serotype-specific neutralizing antibodies, further studies would be necessary to assess whether cross-reaction and cross-protection may occur between all serotypes. Although mucosal IgA levels in the gastrointestinal tract have been shown to be major immune effector products against gastrointestinal viral infections and IgAs are induced upon AstV infection, it is yet to be elucidated whether they are required for protection (130).
In addition to this indirect evidence that points to a major role of humoral responses, the challenge of small intestinal biopsy specimens from healthy adults in an organ culture system with inactivated HAstVs demonstrated the presence of HAstV-specific CD4+ and CD8+ T cells residing in the normal tissue (131). Thus, it is clear that both humoral and cellular adaptive immune responses are involved in protecting normal healthy adults from reinfections. Finally, besides its role in protection from reinfections, adaptive immunity has also been shown to restrict astroviral replication in primary infections in the murine model (100). Compared to wild-type mice, Rag1−/− mice deficient in B and T cells show significantly higher levels of viral shedding in feces and higher levels of viral genome copies detected in intestinal tissues, mesenteric lymph nodes, spleen, liver, and kidney, showing that dissemination of infection is restricted by B and T cells in wild-type mice.
While it is evident that adaptive immunity is a clear mediator both for protection and for controlling infection and disease, many authors suggest that innate immune responses may also play an important role in controlling and limiting disease in primary infections, especially in young individuals. Indeed, evidence in this regard comes from published in vitro data on human viruses (132–136), as well as from in vivo studies with infected young turkeys (122, 137) and immunodeficient mice (100). Unpublished studies in our laboratory have also explored the interplay between HAstV replication and the interferon (IFN) response in cultured cells, observing that infection of CaCo-2 cells induces little type I IFN (IFN-I) response and that this occurs at late stages of infection.
Data on human viruses have demonstrated that the capsid protein of HAstV acts as an inhibitor of the complement system, which constitutes an important innate response against bacterial and viral infections. Similarly to what has been observed with the IFN response, many pathogens have developed ingenious methods by which to evade destruction by the complement system (138). Purified recombinant HAstV capsid protein is able to inhibit complement-mediated hemolysis through direct interaction with two of the complement regulatory molecules, C1 and MBL (133, 134). Both the classical and lectin activation pathways of complement may be inhibited by HAstV capsid protein. The capsid region responsible for the interaction was narrowed down to a 15-residue sequence located in the highly conserved N-terminal domain of the capsid polyprotein (residues 79 to 108), which is highly conserved among classic HAstVs, feline AstVs (FAstVs), and porcine AstVs (PAstVs) but less conserved in the novel HAstVs and mink, ovine, and avian viruses. The biological role of the interaction of the HAstV capsid protein and the complement system remains to be elucidated. Among possible effects, this mechanism of immune evasion would inhibit the process of inflammatory cell migration to infected tissues, although some authors have additionally speculated that HAstV particles may bind C1q (or MBL) and use C1q receptors expressed on the cell surfaces for cellular entry or initial attachment (139). Besides its role on the pathophysiology of HAstV infection, the discovery of this peptide has inspired novel therapeutic options to treat complement-mediated diseases such as ABO-incompatible blood transfusion reactions, which result in acute intravascular hemolytic disease processes (136).
In the turkey model, in which adaptive immune responses do not seem to be critical for viral clearance (137), infection associated with poult enteritis and mortality syndrome (PEMS) induces functional defects in macrophages. In the context of TAstV infection, macrophages showed decreased phagocytosis and intracytoplasmic bacterial killing, and reduced activities of macrophage-mediated proinflammatory cytokines (interleukin-1 [IL-1] and IL-6) were also detected, leading to an increase in the susceptibility of turkeys to secondary infections (140). Infection also compromised the lymphocyte-mediated immune defenses by reducing lymphoproliferative responses (141) and induced increased levels of active TGF-β in serum (122). Finally, the expression of inducible nitric oxide synthase (iNOS) and the subsequent increase in its immune mediator NO have been shown to play a role in controlling TAstV replication both in vitro and in vivo (137). When stimulated ex vivo with lipopolysaccharide, adherent spleen cells isolated from infected animals produced more NO, suggesting that TAstV infection primes macrophages in vivo and makes them more susceptible to activation upon stimulation. In addition, pretreatment of viruses with NO donor compounds significantly inhibited replication, while treatment of embryonated turkey eggs with specific iNOS inhibitors enhanced virus replication (137). Although treatment of a macrophage cell line with the virus resulted in increased NO activity, the source of NO in vivo was not identified until recently, and it is now known that infected intestinal epithelial cells are the ones responsible for iNOS upregulation upon TAstV infection (142).
In the murine model, although it is clear that adaptive immunity restricts replication, innate immunity contributes to the control of MuAstV replication as well (100). Although infection is asymptomatic, viral replication in intestinal tissues of Stat1−/− mice is significantly higher than that in wild-type mice. Overall, the Jak/Stat1 pathway is required for type I and II IFN signaling, and it has been shown to be critical for resistance to other enteric viruses such as noroviruses (143). Although it has not yet been proved whether iNOS activity is also relevant in controlling the infection in mice as happens in avian species and which cell type would produce it, it has been demonstrated that it plays a role in protecting mice from norovirus infection (143), suggesting that it might play a role against astroviruses as well. On the other hand, however, since NO has also been shown to regulate intestinal mucosal permeability and ion transport, a role of NO as a mediator of diarrhea caused by other viral infections such as rotaviruses has also been demonstrated (144, 145).
TAXONOMY, EVOLUTION, AND EMERGENCE
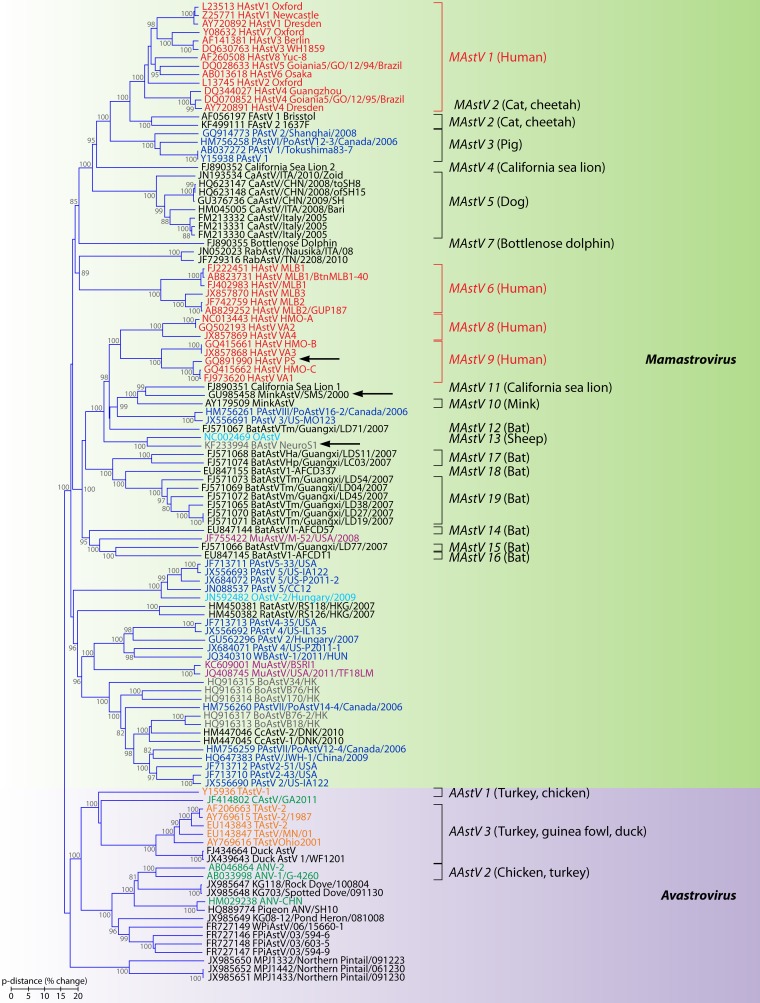
The Astroviridae family was established by the ICTV in 1995, and, consistent with phylogenetic reconstruction, it is now divided in two genera: genus Mamastrovirus (MAstV), including viruses infecting mammals, and genus Avastrovirus (AAstV), including viruses infecting avian species. Since 2008, the number of animal hosts that have been found to be infected with AstVs has quadrupled (comprising at least 30 mammal species and 14 avian species), and classification within the family has gained complexity (146). Classification within each genus was based initially only on the species of the host of origin, but at present genera are divided into “viral species” or “genotypes” on the basis of host range but also on genetic differences in the complete capsid sequence (viruses with differences at the capsid protein level that are higher than approximately 0.30 to 0.35 are regarded as different genotype species). On average, the mean amino acid distance (p-dist) between the two genera is 0.83, while within genera, mean distances are 0.72 and 0.64 for MAstV and AAstV, respectively. The official 2012 ICTV release (http://www.ictvonline.org/virusTaxonomy.asp) recognized 19 species within the MAstV genus and 3 species within the AAstV genus (Fig. 6), but following the standardized criteria for classification, at the time of writing of this review, the two genera could be divided into 33 and 11 genotype species, respectively. Thus, the Astroviridae family currently constitutes a remarkably genetically diverse emerging family with a long evolutionary history and a broad spectrum of host species, and its nomenclature and taxonomy should be discussed, agreed upon, and updated regularly.
FIG 6.
Phylogenetic relationships within the family Astroviridae. The predicted amino acid sequences of the entire capsid polyprotein were aligned using Clustal Omega (309). The phylogenetic tree was generated using the neighbor-joining algorithm (310) implemented in the MEGA6 program (311). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) which are higher than 70 are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances (p-dist) used to infer the phylogenetic tree. All positions containing alignment gaps and missing data were removed only in pairwise sequence comparisons (pairwise deletion option). Genotype species officially recognized by the ICTV are indicated, with hosts of origin shown in parentheses. Human viruses are colored in red, porcine viruses are colored in dark blue, mink viruses are colored in dark gray, bovine viruses are colored in brown, ovine viruses are colored in light blue, bat viruses are colored in black, murine viruses are colored in purple, turkey viruses are colored in orange, and chicken viruses are colored in green. Arrows indicate strains which have been isolated from neurologic tissues. HAstV, human astrovirus; FAstV, feline astrovirus; PAstV, porcine astrovirus; CaAstV, canine astrovirus; RabAstV, rabbit astrovirus; OAstV, ovine astrovirus; BoAstV, bovine astrovirus; MuAstV, murine astrovirus; WBAstV, wild boar astrovirus; CcAstV, deer astrovirus; TAstV, turkey astrovirus; CAstV, chicken astrovirus; ANV, avian nephritis virus; WPIAstV, wild pigeon astrovirus.
In fact, within the Astroviridae family, it is common to find that a single host species, such as humans, is susceptible to infection by divergent astroviruses (Fig. 6). Besides humans, many other animal species, such as pigs (147), bats (15, 148), sea lions (18), sheep (149), cows (150–152), mice (100, 153), mink (154), turkeys (20, 24, 155), chickens (64), ducks (24, 156), and doves (24, 32, 157), can be infected with viruses belonging to different genetic lineages.
For most of the cases, it is still unknown whether these divergent viruses are dispersed globally or whether they are restricted to specific geographic areas. Finally, whether these viruses may show different clinical presentations when infecting the same animal species is yet to be elucidated for most cases. In general, AstV infections are restricted to the gastrointestinal tract, with symptomatic cases occurring predominantly in young individuals, but extraintestinal symptomatic and asymptomatic infections have also been described for both mammal and avian viruses, indicating that the tissue tropism of these viruses may be wide.
Reconstruction of evolutionary events within the Astroviridae family indicates that the separation between mammalian and avian virus lineages occurred 310 million years ago and was followed by the divergence within the mammalian viruses and the occurrence of several cross-species transmissions (158, 159). Besides observational studies, analysis of the tree topology suggests that interspecies transmission may be plausible for these viruses and highlights their zoonotic potential. Indeed, some of the officially recognized AstV species (genotypes MAstV 2, AAstV 1, AAstV 2, and AAstV 3) may infect more than one animal species, indicating that cross-species transmission is frequent, especially in avian viruses. While cross-species infections have been well documented in poultry (160, 161) and have also been suggested in mammals, especially in pigs, cats, and humans (147, 159, 162, 163), it is still unclear whether avian viruses may be able to infect mammal species and vice versa. Interestingly, especially within viruses infecting humans, the distinct evolutionary relationships among the different genotype species that infect humans indicate that several introductions of HAstVs into the human population have occurred (31). As a consequence, the zoonotic potential of these viruses is high, and future nonhuman-to-human transmissions are likely to occur.
Human Astroviruses
Four phylogenetic clades or genotype species of HAstVs have been identified in human stools. Besides the 8 serotypes of classic HAstV (MAstV 1), which are widely recognized as a common cause of nonbacterial gastroenteritis in children, human viruses are found in genotypes MAstV 6, MAstV 8, and MAstV 9 (Fig. 6, red). While classic HAstV (MAstV 1) and HAstV-MLB (MAstV 6) form a monophyletic group together with viruses from cats, pigs, dogs, rabbits, California sea lions, and dolphins, the other two genotypes, MAstV 8 and MAstV 9, are genetically related to each other and to viruses from mink, sheep, California sea lions, bats, cattle, pigs, and mice.
Within genotypes, strains can be grouped in serotypes based on their antigenicity. While all eight classic HAstVs clearly correspond to different serotypes, antigenic differences between MLB1, MLB2, and MLB3 and between VA1, VA2, VA3, and VA4 still need to be investigated. While the eight serotypes of MAstV 1 share 63 to 84% amino acid similarities at the capsid level, similarity among strains within MAstV 6, MAstV 8, and MAstV 9 ranges between 74 and 90%, 78 and 79%, and 75 and 77%, respectively, suggesting that they would likely show different antigenic properties. Finally, subtypes or lineages have also been identified within each serotype, with a cutoff 93 to 95% nucleotide homology to a reference strain needed to be considered a new subtype (164–168). In light of this, HAstV-1 has so far been classified into 6 lineages (1a to 1f), HAstV-2 into 4 lineages (2a to 2d), HAstV-3 into 2 lineages (3a and 3b), and HAstV-4 into 3 lineages (4a to 4c) (166, 169–174).
Although strains are usually classified according to ORF2 variability and a high correlation is detected between serotypes and genotypes, classic HAstVs have also been typed based on ORF1a. Phylogenetic analysis of several partial sequences of ORF1a resulted in only two clearly separated groups, which have been called genogroup A (which comprises serotypes HAstV-1 to HAstV-5 and HAstV-8) and genogroup B (serotypes HAstV-6 and HAstV-7) (86, 175). As suggested, this different phylogenetic clustering may have resulted from recombination events between structural and nonstructural coding regions (86, 175).
As viruses with RNA genomes, nucleotide mutations and recombination events are, among other factors, important in their genome evolution (reviewed in reference 176). The HAstV genome mutation rate has been analyzed, and it generally matches the variation rates for single-stranded positive-sense RNA viruses which evolve rapidly, such as picornaviruses (177). According to this study, variation rates are approximately 3.7 × 10−3 nucleotide substitutions per site per year and 2.8 × 10−3 synonymous changes per site per year. A higher genetic variability in ORF2 than in ORF1a and -1b has been confirmed, suggesting the independent evolution of HAstV genes and even gene regions and pointing to different selection pressures and/or evolutionary constraints for specific genomic regions (158, 178). Overall, negative or purifying selection is a dominant force among all HAstV genes, and only few codon sites, such as those coding for residues located within the capsid protein exposed to the immune pressure or which are involved in cellular recognition, show positive selection and molecular adaptation during evolution (155, 178). Sites which are prone to positive selection are concentrated within the C-terminal half of ORF2, but they are not restricted to it. In ORF1a, clusters of such sites are also located in nonconserved regions around amino acid positions 614 to 624, 775 to 777, and 812 to 817, also close to its C terminus (178). Finally, when comparing HAstVs belonging to different genetic lineages or genotypes, a marked constrained evolution has been observed within both ORF1a and ORF1b in classic HAstVs (MAstV 1) but not in MLB and VA/HMO viruses (MAstV6, MAstV8, and MAstV 9), suggesting that ORFs for nonstructural genes of classic HAstVs may contain RNA secondary structures or cryptic overlapping ORFs that are absent in the same region of the other human viruses (31).
When comparing evolutionary rates among different AstVs, higher rates and an enhanced accumulation of synonymous substitutions, especially in ORF2, have been observed in porcine, ovine, mink, and turkey AstVs than in viruses infecting humans, cats, and chickens (178). The major evolutionary forces acting specifically at synonymous positions are nucleotide composition, translational selection, and codon usage. A study performed in 2007 (159) demonstrated that the codon usage is an operational driving force especially among the expansion of mammalian AstVs after their separation from the avian AstVs and that AstV codon usage points toward the host species codon usage. In addition, this switch in codon usage during diversification of mammalian AstVs was related to their different nucleotide compositions, rather than being caused by translational selection (159).
Finally, recombination events after coinfection of the same host by different viruses are another of the main mechanisms that have played a role in HAstV evolution and in generation of novel strains. Natural recombinants have been commonly found between strains belonging to the same genotype, such as different strains within the same serotype or different serotypes of classic HAstV (MAstV 1) (168, 172, 179–182), but it is still unknown whether recombination may also occur between strains belonging to different genotype species even if they are adapted to infect the same host. So far, recombination events involving human strains have been suggested to have occurred between classic HAstV and a California sea lion AstV (18) and between classic HAstV and PAstV (163). Recombinations between different animal AstVs have also been described (152, 155, 162, 183). Although most recombination breakpoints have been identified upstream of the conserved ORF1b/ORF2 junction region (155, 168, 172, 182), they are also frequent within ORF2 (152, 155, 172, 177), and they may also exist within ORF1a (155, 181) and ORF1b (155, 177, 180).
Nonhuman Astroviruses
Besides HAstVs, many other mamastroviruses show a remarkable genetic diversity, and some of them are of special interest because they have also been isolated both in feces and in extraintestinal tissues. These include mink AstVs (154) and bovine AstVs (BoAstVs) (150–152), which have been isolated in neurologic tissues in association with neurologic diseases, porcine AstVs (PAstVs) (184, 185), which have been isolated in blood, and murine AstVs (MuAstVs), which have been isolated in mesenteric lymph nodes, spleens, livers, and kidneys of wild-type or immunodeficient mice (100).
AstVs infecting mink (genotype MAstV 10) have traditionally been considered agents of the preweaning diarrhea syndrome (14), but recently a novel virus which could be classified as a different genotype (Fig. 6, dark gray) has been isolated in neural tissue from minks suffering from the so-called shaking mink syndrome (SMS) (154). Similarly, among BoAstVs, which are also associated with gastrointestinal infections and may be grouped in 4 genotypes (Fig. 6, brown), the strain called BoAstV-NeuroS1 has been detected in brain tissue in association with neurologic disease (150). Of note, the 3 MAstV species associated with neurologic symptoms (HAstV-PS, mink AstV-SMS, and BoAstV-NeuroS1) are included in the same genetic cluster of astroviruses (Fig. 6).
Pigs have been extensively studied in different parts of the world because they serve as a reservoir for many zoonotic diseases. To date, the ICTV officially recognizes only one PAstV genotype (MAstV 3), but at least five genetic lineages which would eventually correspond to genotypes have been documented (162, 185–188) (Fig. 6, dark blue). PAstVs are widely distributed, and they have been isolated from feces of both diarrheic and healthy pigs in many different parts of the world (187, 189–191). Recently, viremia in healthy pigs infected with lineages 2 and 4 has also been documented, with a viral RNA prevalence in serum of 3.89% (184).
Surprisingly, it has recently been recognized that MuAstVs are commonly found infecting laboratory mice in different countries, including breeding facilities, research institutions, and commercial research mouse facilities (100, 153, 192, 193) (Fig. 6, purple). Although the mice are asymptomatic, the viruses have been isolated in the gastrointestinal tracts and mesenteric lymph nodes of wild-type mice but also in spleens, livers, and kidneys of immunodeficient mice (100, 192).
Among the other MAstVs described to date, large genetic diversity has been described in ovine AstVs (OAstVs) (Fig. 6, light blue) (119, 149) and bat AstVs (BatAstVs) (Fig. 6, black) (15, 148, 194, 195). In most cases, viruses have been mostly isolated from healthy animals, and prevalences are high. The genetic diversity of feline AstVs (FAstVs), canine AstVs (CaAstVs), rabbit AstVs (RabAstVs), rat AstVs, and AstVs infecting sea lions and dolphins has not been studied so extensively, and the clinical impact of their infections seems to be low and has not been well explored yet (11, 16, 17, 196–200).
In contrast, the pathogenicity of Avastroviruses is significant, especially in poultry, with high economic costs and affecting the food industry worldwide. These viruses have been associated with avian diseases, including poult enteritis mortality syndrome (PEMS) in turkeys, chickens, and guinea fowl, mild growth loss, nephritis, the runting-and-stunting syndrome (RSS), and gout in chickens, and fatal hepatitis in ducklings (20, 64, 146, 201, 202). The ICTV officially recognizes 3 species within AAstVs: AAstV 1 (turkey AstV 1), AAstV 2 (avian nephritis virus [ANV]), and AAstV 3 (turkey AstV 2 and duck AstV) (Fig. 6). Among them, TAstV and ANV from chickens are the two most widely studied virus strains, and surveys indicate that these viruses are worldwide distributed. Recently, AstVs infecting pigeons have been described (157), and a novel virus related to ANV has also been isolated and characterized in pigeons with gastroenteritis (22, 32), as well as in healthy chickens (22). Finally, viruses similar to previously known viruses infecting poultry have been also isolated in wild aquatic birds, and wild migratory bird populations have been shown to commonly carry novel and diverse AAstVs, likely playing a role in AAstV ecology (24).
EPIDEMIOLOGY
HAstV Transmission
HAstVs are transmitted essentially through the fecal-oral route, as proven in human volunteer studies (95, 96). However, in these studies, few adult recipient individuals developed gastroenteric disease with virus shedding in feces, probably because of their age.
Food and water may act as vehicles for human enteric virus transmission (203, 204). Several large HAstV outbreaks have been associated with consumption of contaminated food (205–207). In addition, fecally contaminated fomites have been implicated in outbreaks declared in institutions such as day care centers, hospitals, nurseries, schools and military facilities (208, 209). Moreover, waterborne transmission of HAstVs has been suggested as a risk of digestive morbidity for the general population (210), and HAstV was recovered in surface water from an area in which an outbreak of gastroenteritis was concomitantly occurring (211). The environmental transmission of HAstV is supported by the stability of the virus in drinking water (212), freshwater (213), and marine water (213).
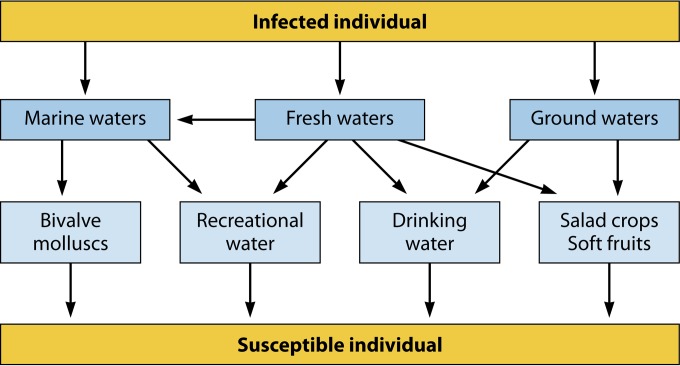
Figure 7 depicts the potential environmental routes of HAstV transmission. HAstV may be shed in very high numbers, reaching up to 1013 genome copies per gram, in the stools of infected individuals (104). Since wastewater treatment practices do not ensure the complete removal of viral pathogens, HAstV present in untreated and treated wastewaters are discharged into the environment (214–217) and may become contaminants of marine water, freshwater, and groundwater (211, 218). Groundwater may also receive viral contamination through the injection of reclaimed wastewater (219).
FIG 7.
Flowchart of potential routes of environmental transmission of human astroviruses (see the text for details).
Among foods susceptible to become contaminated with HAstV at the preharvest stage are bivalve mollusks grown in polluted waters (218) and fresh produce irrigated with contaminated water, such as lettuce, green onion, and other greens, as well as soft fruits such as raspberries and strawberries (220). These products usually undergo little if any processing before consumption and hence are susceptible to act as vehicles for human enteric virus transmission (221).
Surface water and groundwater are employed as sources of drinking water for public consumption, and poor water quality is recognized as a major health threat. The WHO estimates that almost 90% of the global burden of gastroenteritis is attributable to unsafe water and poor sanitation and hygiene, with the pediatric population in developing areas being the most vulnerable group (222). As an example of this situation, HAstV genomes have been detected in tap water in Ghana (223).
Although the risk derived from recreational activities in sewage-polluted waters is much lower than that from drinking contaminated water, an outbreak of gastroenteritis occurred among children and adults who bathed in a Helsinki outdoor wading pool that had norovirus and astrovirus contamination (224). Additionally, a seroprevalence study in the United Kingdom showed that 93% of surfers had HAstV antibodies, while only 23% of age-matched controls had these antibodies (225). Chlorine and other disinfectants are routinely employed for drinking water treatment and are effective for the inactivation of HAstV in water (212, 226).
Poor hygiene practices with improper food handling are usually responsible for postharvest contamination of food products, particularly ready-to-eat foods such as salads, sandwiches, and deli meat that are all consumed either uncooked or lightly cooked. HAstVs cause asymptomatic infections in children and adults (61, 93), and asymptomatic food workers are more often implicated in gastroenteritis outbreaks than symptomatic food handlers (227). It is critical to ascertain the significance of asymptomatic as well as presymptomatic and postsymptomatic HAstV shedding in the transmission of food-borne infections.
Fomites or inanimate surfaces play an important role in the institutional spread of HAstV infections (208, 209, 228). Fomites include serving and chopping utensils, and the ability of the virus to persist dried onto these surfaces long enough and in numbers high enough to represent a risk is a major public health issue. Disinfection practices, as well as improved hygiene measures, particularly in high-risk settings such as hospitals, institutions for the elderly, day care centers, or restaurants, are of major importance in preventing the spread of infections. Antiseptics and disinfectants must be employed for the inactivation of pathogens on skin (notably hands) and fomites, respectively.
Although astroviruses were initially considered to be so species specific that it led to their classification based on the host species (45), concern about the potential zoonotic transmission of astroviruses to humans has recently arisen (146). As discussed above, the advent of pyrosequencing enabled the characterization of previously unrecognized HAstVs, termed nonclassic HAstVs (25). Some of these nonclassic HAstVs have been shown to have common ancestors with recently described rat AstV (11), while others are phylogenetically related to mink and ovine AstVs (29).
As mentioned above, there is evidence of recombination events between human and animal AstVs. Recombination in the variable region of ORF2 has been shown to occur between human and porcine strains of AstV (163). These AstV recombinants were described in areas in which human and porcine strains were in close contact. Since the genetic variability of the HAstV strains appeared to be much higher than that of the porcine strains, it was concluded that zoonotic transmission likely took place from humans to pigs instead of vice versa, which had previously been suggested (158, 163).
High genetic variability, together with the occurrence of interspecific infections and recombination events, makes AstVs serious candidates for emerging zoonotic infections.
Age, Geographic, and Temporal Distribution
HAstVs affect predominantly the pediatric population (117, 164, 229, 230), although infections in elderly people and immunocompromised hosts are also reported (231–236). In addition, astrovirus gastroenteritis may also affect healthy adults (88, 89, 207). The age of children infected with classic HAstV is highly variable, ranging from newborns to over 5 years (33, 164, 167, 237–243); however, infection is more common among those younger than 2.
Classic HAstVs are distributed all over the world and are associated with 2 to 9% of cases of acute, nonbacterial diarrhea in children (reviewed in reference 146), although incidences as high as 61% have been reported (94). Table 2 shows some examples of studies on classic HAstV prevalence in stool conducted in different regions and settings. The mean incidence worldwide is 11%, with 7% and 23% incidences in urban and rural areas, respectively (244). HAstV incidence is usually higher in developing countries, although paradoxically, a low incidence of infection is observed in sub-Saharan Africa (245–247).
TABLE 2.
Studies of human astrovirus prevalence in stool
| Region and country (setting, period of study) | Age (yr) of study population | Astrovirus prevalence (%) | Detection method | Reference |
|---|---|---|---|---|
| North Africa | ||||
| Egypt (rural, 1995–1998) | <2 | 26.7 | EIA | 237 |
| Tunisia (urban and rural 2003–2005) | <5 | 6.0 | EIA | 306 |
| Sub-Saharan Africa | ||||
| Botswana (urban, 2001–2002) | <5 | 2.7 | EIA | 245 |
| Kenya (urban and rural 1999–2005) | <5 | 5.3 | EIA | 246 |
| Ghana (urban, 2005–2006) | <12 | 4.8 | RT-PCR | 247 |
| Asia | ||||
| Thailand (urban, 1985–1987) | <5 | 8.6 | EIA | 239 |
| Japan (urban and rural 1982–1992) | Children of all age groups, mostly <10 | 18 | EIA | 249 |
| China (urban and rural, 1998–2005) | <5 | 5.5 | EIA plus RT-PCR | 307 |
| Vietnam (urban, 2005–2006) | <3 | 13.9 | RT-PCR | 241 |
| India (urban, 2004–2008) | <8 years, adults | 3.1 | RT-PCR | 308 |
| Japan (urban 2008–2009) | Children of all age groups | 1.7 | RT-PCR | 250 |
| North America | ||||
| Mexico (rural, 1992–1995) | <3 | 61 | EIA and RT-PCR | 94 |
| USA (urban, 1993–1994) | <3 | 20 | EIA | 266 |
| USA (urban, 1996–1997) | Children of all age groups, mostly <3 | 6.9 | EIA | 230 |
| Central America | ||||
| Guatemala (rural, 1987–1989) | <3 | 38.6 | EIA | 33 |
| South America | ||||
| Chile (urban, 1985–1987; 1993–1995) | <3 | 23.5 | EIA | 240 |
| Brazil (urban, 1990–1992) | <2 | 5.8 | EIA plus RT-PCR | 265 |
| Argentina (urban, 1995–1998) | <3 | 3.7 | EIA plus RT-PCR | 267 |
| Colombia and Venezuela (urban, 1997–1999) | <5 | 5.0 | EIA | 167 |
| Europe | ||||
| France (urban, 1995–1998) | <3 | 6.0 | EIA | 242 |
| Spain (urban, 1997–2000) | Children of all age groups, Mostly <5 | 4.9 | RT-PCR | 164 |
| Italy (urban, 1999–2000) | <2 | 3.1 | EIA | 264 |
| Germany (urban, 2000) | All ages | 1.2 | EIA | 276 |
| France (urban, 2007) | <5 | 1.8 | EIA | 248 |
| Oceania | ||||
| Australia (urban, 1995) | <5 | 4.2 | Northern (RNA) dot blot | 35 |
A tendency toward a decrease in classic HAstV incidence seems to have occurred in the last decades, which is evidenced by comparing the data from studies conducted in the same area several years apart, e.g., in France (242, 248) or Japan (249, 250). The mean classic HAstV incidence in the 1980s was 22%, while it was 15% in the 1990s and 5% in the first decade of the new millennium (244). However, there are differences in the methods employed and the study populations that make difficult to adequately compare these studies. It is plausible that the decrease in the incidence of classic HAstV infection may be related to the displacement of classic HAstV infections by those with novel HAstV strains such as MLB and VA/HMO, which could be outcompeting classic HAstV. As a matter of fact, recent reports point to a high prevalence of some of these strains (115). The diminution in the number of astrovirus gastroenteritis cases is not observed with other viral agents such as noroviruses, whose incidence has increased in the same time period (251–255).
The classic HAstV distribution follows a seasonal pattern with a higher incidence of infection in temperate regions in the cold-weather period, and although there is a dramatic decline in the number of cases in the warm-weather period, HAstVs continue to circulate throughout the year, and infections do occur in summer too (61, 164, 243, 249, 256, 257). This seasonality may be the consequence of a better stability of the virus at lower temperatures (208, 212). Other viral gastroenteritis agents such as rotaviruses and noroviruses, but not adenoviruses, show a similar seasonal pattern (258–260). In tropical areas, the maximum incidence of HAstV infections tends to occur in the rainy season (33, 94). Nevertheless, some studies have failed to observe a distinct peak of infection throughout the year (243). In addition, other studies have reported a greater burden of astrovirus diarrhea in children in alternate years (164, 167, 243, 257).
A different behavior regarding seasonality seems to occur with nonclassic HAstV strains. In one study (109), novel astrovirus (MLB1, MLB2, and VA/HMO) infections were observed in March, April, May, July, and November. However, more studies are required to elucidate whether nonclassic HAstV strains follow a seasonal distribution pattern.
Molecular Epidemiology of HAstV
In addition to the intrinsic variability caused by the error-prone RdRp (176), the occurrence of recombination events between nonstructural and structural coding regions, sometimes from viruses infecting different host species, adds more diversity and complexity to the AstV molecular typing (18, 146, 163, 175, 181).
In 1984, Kurtz and Lee set the methodology, based on the use of reference sera to recognize the different serotypes (five at the time) of HAstVs (261). Since what can be considered the birth of AstV epidemiology, up to eight serotypes of classic HAstV (HAstV-1 to HAstV-8) have been identified based on their antigenicity. Phylogenetic analysis of the capsid region, particularly of the carboxy-terminal hypervariable domain of the ORF2-encoded structural protein, points to a high correlation between serotypes and genotypes, thus allowing typing of the different AstV isolates through sequence analysis (45, 158, 262, 263). As explained above, sequence diversity in the capsid region allows the recognition of subtypes (165, 166, 168) or lineages (164, 167, 175, 264, 265) within the different genotypes.
Moreover, analysis of the well-conserved partial sequence near the protease motif-coding region enables the establishment of two genogroups, A (with serotypes HAstV-1 to HAstV-5 and HAstV-8) and B (with serotypesHAstV-6 and HAstV-7) (175). Sequence analysis of the hypervariable region in the nsP1a/4-coding region in ORF1a enables the additional differentiation of 12 genotypes, nine within genogroup A and three within genogroup B (86). In order to clearly distinguish them from ORF2-derived serotypes, the ORF1a-derived genotypes are named using roman numerals. While in most cases, a correlation exists between ORF1a genotypes and ORF2 serotypes, some genotypes are shared by more than one serotype, and some serotypes may be found in association with more than one genotype, highlighting the occurrence of recombination events between strains. Selected ORF1a genotypes have been associated with higher levels of viral shedding (86).
Table 3 shows the prevalence of classic HAstV serotypes worldwide. Overall, HAstV-1 is the most frequently isolated type (35, 164, 237, 241, 250, 256, 265–268). However, there are exceptions, such as in one study conducted in Mexico (238) in which HAstV-2 was reported to be the most prevalent. It is not well defined which is the second most frequent type, since differences related to the geographical localization are observed (164, 243, 269). Classic HAstV prevalence may be influenced by the lack of heterotypic immunity, since children in a given location may suffer subsequent infections with different serotypes. This was observed in one study (164) in which most infections with HAstV-1 and HAstV-3 occurred in children younger than 2 years old, while HAstV-4 infections were observed in children older than 4 years of age, thus confirming the apparent lack of cross-protection after infection with one serotype before infections with other types or variants.
TABLE 3.
Prevalence of serotypes (genotypes) of classic human astrovirus
| Region and country (setting, period of study) | Prevalence (%) of serotype: |
Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| North Africa | |||||||||
| Egypt (rural, 1995–1998) | 43.3 | 3.6 | 12.0 | 4.8 | 15.7 | 7.2 | 0 | 12.0 | 237 |
| Asia | |||||||||
| Vietnam (urban, 2005–2006) | 100.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 241 |
| India (urban, 2004–2008) | 6707 | 9.7 | 0 | 0 | 6.5a | 0 | 0 | 16.0 | 308 |
| Japan (urban 2008–2009) | 91.0 | 0 | 9.0 | 0 | 0 | 0 | 0 | 0 | 250 |
| North America | |||||||||
| USA (urban, 1993–1994) | 55.0 | 17.0 | —b | — | — | — | — | — | 266 |
| South America | |||||||||
| Brazil (urban, 1990–1992) | 45.5 | 27.3 | 12.1 | 12.1 | 0 | 3.0 | 0 | 0 | 265 |
| Argentina (urban, 1995–1998) | 41.0 | 13.0 | 13.0 | 25.0 | 8.0 | 0 | 0 | 0 | 267 |
| Europe | |||||||||
| Spain (urban, 1997–2000) | 38.0 | 6.0 | 19.0 | 26.0 | 0 | 0 | 0 | 11.0 | 164 |
| Oceania | |||||||||
| Australia (urban, 1995) | 85.0 | 0 | 0 | 15.0 | 0 | 0 | 0 | 0 | 35 |
ORF1a/ORF2 recombinant strains.
—, there were no data on this genotype in the study.
LABORATORY DIAGNOSTICS
Early Developments: Electron Microscopy and Immunological Assays
Although cell culture propagation of some classic HAstVs is possible with the aid of trypsin (51, 59, 270), virus isolation in cell culture is not applicable for astrovirus diagnosis in fecal specimens. In the past, HAstVs were routinely detected by direct transmission electron microscopy (EM) in negatively stained stool samples (1, 2, 228). This procedure is laborious, time-consuming, and subjective and involves highly experienced personnel. Further limitations were that, as mentioned above, only around 10% of the particles show the characteristic star-like surface structure and that the sensitivity of EM requires concentrations of around 107 particles per gram of stool for visualization of the virus (34). Such levels are usually detected for only 12 to 48 h after the onset of symptoms, with numbers often too low thereafter for EM detection.
The use of antibodies in immune electron microscopy (IEM) or solid-phase immune electron microscopy (SPIEM) enhances the sensitivity of the assay while enabling confirmation of the identity of the viral agent (88, 207, 252, 271). However, the limited availability of antibodies for the different HAstV types hampers the use of these techniques (34).
Enzyme immunoassays (EIAs) were developed for the detection (272, 273) and typing (256, 271, 274) of HAstV. Herrmann and coworkers (273) developed in 1990 an EIA based on the use as capture antibody of a monoclonal antibody (8E7) directed toward the conserved domain of the capsid and a peroxidase-labeled polyclonal antibody as the detection antibody. A further refinement was labeling the detection antibody with biotin (275).
At least two commercial EIAs are available for classic HAstV detection in stool specimens. Although the assays do not enable typing of the isolates, they are useful for rapid detection of the virus when large numbers of samples must be analyzed (167, 276–279). EIA-based techniques have been reported to have the same sensitivity as direct EM observation (228) but obviously are much less time-consuming. In addition, several rapid immunochromatographic tests against astroviruses are commercially available, and some of them have been evaluated (280).
Molecular Assays
The advent of molecular detection techniques, based initially on probe hybridization and later on genome amplification, opened the possibility to develop assays with exquisite sensitivity and specificity for the diagnosis of AstVs in stool. While the sensitivity of probe hybridization has been described to be on the same level as that of EIA (34), reverse transcriptase PCR (RT-PCR) offers thresholds of detection as low as 10 to 100 genome copies per gram of stool (34). Nevertheless, the sensitivity of these molecular assays relies greatly on several variables, such as primer/probe selection, enzymatic amplification reaction conditions, and, last but not least, the efficiency of the target nucleic acid extraction (281). In any case, studies employing EIA and RT-PCR in parallel reported a higher number of positive samples scored by the latter technique as well as a longer period of viral shedding that extended beyond the resolution of symptoms (206, 228).
Conventional RT-PCR methods are still broadly used today in many research laboratories both for screening and for further isolate typing. While some of the most extensively used common-type and type-specific primers for the detection of 8 serotypes of classic HAstV have been previously reviewed (282), primers Mon244/Mon245 (and Mon269/Mon270) described by Noel et al. (274), targeting the conserved 5′ end of ORF2 and allowing typing by sequencing, are some of the most widely used ones. Upon the discovery of novel HAstVs, consensus astrovirus RT-PCR primers targeting ORF1b were also published (15, 29, 30). Although these primers should be able to detect all known human and animal AstVs, so far they have allowed the detection of both classic and novel human AstVs (107, 110). Primers designed to specifically detect HAstV-MLB and HAstV-VA/HMO have also been described (27, 30, 114).
Finally, microarrays have also been shown to be an especially useful option for virus detection and typing. One of the described microarray-based methods is able to distinguish representatives of the eight known serotypes of classic HAstVs (283), and another one is a combimatrix for simultaneous detection of various enteric viral pathogens, including HAstVs (284). Additionally, other, less conventional molecular techniques, such as nucleic acid sequence-based amplification (NASBA), have also been described for HAstVs (285).
A further refinement was the development of quantification techniques such as real-time RT-quantitative PCR (RT-qPCR) assays, which provide high speed, sensitivity, and reproducibility and reduction of contamination risk in the diagnostic of HAstVs, (214, 286–289). Similarly, a real-time reverse transcription loop-mediated isothermal amplification has been developed for rapid and quantitative detection of HAstV-1 (290). Interestingly, some of the published RT-qPCR methods already come in a multiplex format allowing the simultaneous detection of classic HAstVs and other targets such as noroviruses, rotaviruses, sapoviruses, adenoviruses, and/or bocaviruses (291–294). One of these methods even includes up to 19 targets, including bacteria, protozoa, and helminths (295). Interestingly, that assay also includes the detection of two extrinsic controls to monitor extraction and amplification efficiencies (295), which may be highly relevant in surveillance or food virology applications. Commercial monoplex and multiplex real-time RT-qPCR kits are also available, although most of them are especially relevant for the clinical diagnostic setting. Remarkable among them is the FilmArray gastrointestinal panel (BioFire Diagnostics, Inc., USA), which is currently under development and aims to simultaneous detect 23 gastrointestinal pathogens from stool, including HAstVs and 4 other enteric viruses. Regarding novel HAstVs, although not designed to be used in surveillance studies, two RT-qPCR assays targeting HAstV-MLB have been applied for quantification of viral loads in stool and serum (111, 112). If the importance of novel HAstVs in human health is clinically confirmed, molecular methods should be soon updated to incorporate broadly reactive primers/probes to universally detect all HAstVs.
Finally, although classic HAstVs are somewhat fastidious to grow in vitro and novel HAstVs have not yet been propagated in any cell line, the combination of molecular techniques with infectivity assays may help to ultimately analyze the importance that positive results in terms of genome copies may have on public health when present in environmental or food samples. The use of CaCo-2 cells combined with pretreatment of virus inoculums with trypsin remains the gold standard for classic HAstV isolation from clinical and environmental samples (59, 67), and several integrated cell culture (ICC) methods combined with RT-PCR detection have been described (296, 297). Whether these methods allow the isolation of novel HAstVs or whether these viruses show significant differences in cell tropism still remains to be elucidated.
Next-generation technologies, including whole-genome sequencing and viral metagenomics, may rapidly generate sequence-based data that will be determinant for optimizing the currently used primers and will also help to identify additional genomic regions for analysis. Furthermore, whole-genome sequencing will provide genetic information that may also correlate with virulence factors and may help us to better understand the basis of viral pathogenesis, and metagenomics will be useful to identify new emerging strains.
PREVENTION AND TREATMENT
The prevention of HAstV infections is essentially based on the control of the transmission routes and on the prevention and control of disease at the host level. The control of the transmission routes includes virus detection and inactivation in water and food and disinfection of contaminated fomites. Methods for the detection and quantification of HAstVs are available (see above), but the threshold of acceptable genome copy numbers in water and food matrices has yet to be defined not only for HAstVs but for other gastroenteritis agents as well. Although the survival of classic HAstV in drinking water is high, disinfection treatments of 2 h with1 mg/ml of free chlorine is quite effective (212). However, genogroup B is somewhat more resistant to chlorine disinfection than genogroup A (215), suggesting that differences in environmental persistence may exist between strains. Astrovirus survival and inactivation in food matrices have not been extensively studied, probably because only a few HAstV food-borne outbreaks have been described (207, 298) and because efforts have been dedicated to the application of emerging technologies for the inactivation of other viral agents, such as norovirus and hepatitis A virus (299, 300). Regarding disinfection of contaminated fomites, 90% alcohol has been proved useful (301). Unfortunately, no information on inactivation is yet available for nonclassic HAstVs.
Prevention of disease development in the host includes vaccination and potentiation of the natural defenses such as the gut microbiota. No vaccines have been developed for HAstVs despite several descriptions of virus-like particle (VLP) production in different systems (302, 303). This lack of commercial interest in vaccine production may be due to the low clinical impact of astrovirus infection in healthy patients and to the need for a multivalent vaccine to cover all circulating serotypes and strains or at least the most prevalent ones, since apparently there is not heterologous protection (90). Some authors have speculated that probiotics, which may interfere with the biological cycle of enteric virus at many different steps, may be used as a measure to prevent and/or treat intestinal viral infections (304, 305). Additionally, the antiviral activity of some synthetic flavonoids on astrovirus replication has been described (226).
The usual therapy applied to patients (young children or adults) suffering from serious gastroenteritis consists of oral or intravenous fluid replacement to avoid dehydration. Intravenous treatment with IgG for immunocompromised hosts with severe or persistent diarrhea has also been proposed, although the efficacy of this treatment has yet to be established in large-scale studies (231).
CONCLUSIONS
In the last few years, the global vision of AstV diversity has dramatically changed. Although they were traditionally regarded as agents of acute gastroenteritis in children, it is now known that the viruses may spread systemically, especially in immunocompromised individuals. Viremia has been documented in some human cases, pigs, and turkeys, and AstVs have been isolated in neurologic tissue in association with neurologic disorders in humans, cattle, and mink. The emergence of such a number of different viruses infecting so many different species indicates the possibility of zoonotic transmission of viruses to humans. In this regard, the AstV diversity found in avian species, pigs, and bats is remarkable, with these species frequently being associated with the zoonotic transmission and the emergence of new strains of influenza viruses and coronaviruses. In fact, the novel HAstV-VA/HMO viruses (genotypes MAstV 8 and MAstV 9) are phylogenetically closer to the bat isolates than to the classic HAstVs (MAstV 1), and some of these novel viruses have been associated with extraintestinal manifestations. All these concerns highlight the need to improve broadly reactive diagnostic techniques allowing the detection of all HAstVs. A better understanding of the AstV replicative cycle, particularly of the novel viruses, and of their role in clinical disease and their mechanisms of pathogenesis is required to prevent and treat potential new infections.
ACKNOWLEDGMENTS
This work was supported in part by grants 2009SGR00024, XRB-Biotechnology Reference Network (Generalitat de Catalunya; www.gencat.cat/agaur), and FRI-2011 (Institute of Nutrition and Food Safety, University of Barcelona).
Biographies

Albert Bosch (Full Professor) has over 35 years of experience in virology and has published over 150 peer-reviewed articles on enteric viruses in scientific journals as well as complete books. He has been member of numerous scientific committees, notably the scientific committee of the Spanish Food Safety Authority and the CEN committee of the EU on standardization of molecular methods for virus detection in foods; he also belongs to the scientific panel on food- and waterborne diseases of the European Centre for Disease Control and Prevention (ECDC). He acted as chairman of the Astrovirus Study Group of the International Committee on Taxonomy of Viruses (ICTV). He is president of the International Society for Food and Environmental Virology and the Spanish Society for Virology. He was the president of the water microbiology section of the Spanish Society for Microbiology and is a member of the steering committee of the Health-Related Water Microbiology Group of the International Water Association.

Rosa M. Pintó (Associate Professor) has over 25 years of experience in the molecular virology of enteric viruses, particularly hepatitis A virus and human astroviruses, and has published over 100 peer-reviewed articles on these issues. She has been the leader of several projects on the molecular biology of hepatitis A virus, with special emphasis on the biotechnological applications of the generated basic research and particularly on the development of diagnostic molecular procedures. This expertise has led to her membership in the scientific committee of the Spanish Food Safety Authority (AESAN) as well as in the CEN committee of the EU on standardization of molecular methods for virus detection in foods.

Susana Guix graduated from the University of Barcelona in 1997 with a degree in biology. After graduation, she performed her Ph.D. work on human astroviruses at the Department of Microbiology of the University of Barcelona. She performed the first studies on the molecular epidemiology of astroviruses in Spain and characterized the apoptotic response in infected cells. In 2004, she received a Fulbright postdoctoral fellowship and performed her postdoctoral work at Baylor College of Medicine in Houston, TX, working on human noroviruses. For the past 6 years, she has worked as a Lecturer Professor at the Department of Microbiology of the University of Barcelona. She was a member of the Astrovirus Study Group of the International Committee on Taxonomy of Viruses (ICTV) from 2005 to 2012. Her current research interests on astroviruses and noroviruses include their molecular epidemiology, the study of the role of the viral nonstructural proteins, and the characterization of the innate immune responses activated upon infection.
REFERENCES
- 1.Appleton H, Higgins PG. 1975. Viruses and gastroenteritis in infants. Lancet i:1297. (Letter.). [DOI] [PubMed] [Google Scholar]
- 2.Madeley CR, Cosgrove BP. 1975. 28 nm particles in faeces in infantile gastroenteritis. Lancet 6:451–452 [DOI] [PubMed] [Google Scholar]
- 3.Madeley CR. 1989. Epidemiology of gut viruses, p 5–15 In Farthing CF. (ed), Viruses and the gut. Smith, Kline & French Laboratories Ltd., Welwyn Garden City, Hertfordshire, United Kingdom [Google Scholar]
- 4.Monroe SS, Jiang B, Stine SE, Koopmans M, Glass RI. 1993. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J. Virol. 67:3611–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monroe SS, Carter MJ, Herrmann JE, Kurtz JB, Matsui SM. 1995. Astroviridae, p 364–384 In Murphy FA, Bishop DHL, Ghabrail SA, Jarvis AW, Martielli GP, Mayo MA, Summers MD. (ed), Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, Vienna, Austria [Google Scholar]
- 6.Hoshino Y, Zimmer JF, Moise NS, Scott FW. 1981. Detection of astroviruses in feces of a cat with diarrhea. Arch. Virol. 70:373–376. 10.1007/BF01320252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woode GN, Bridger JC. 1978. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 11:441–452. 10.1099/00222615-11-4-441 [DOI] [PubMed] [Google Scholar]
- 8.Tzipori S, Menzies JD, Gray EW. 1981. Detection of astrovirus in the faeces of red deer. Vet. Rec. 108:286. 10.1136/vr.108.13.286 [DOI] [PubMed] [Google Scholar]
- 9.Castro TX, Cubel Garcia RC, Costa EM, Leal RM, Xavier MP, Leite JP. 2013. Molecular characterisation of calicivirus and astrovirus in puppies with enteritis. Vet. Rec. 172:557. 10.1136/vr.101566 [DOI] [PubMed] [Google Scholar]
- 10.Kjeldsberg E, Hem A. 1985. Detection of astroviruses in gut contents of nude and normal mice. Arch. Virol. 84:135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu DKW, Chin AWH, Smith GJ, Chan K-H, Guan Y, Peiris JSM, Poon LLM. 2010. Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. J. Gen. Virol. 91:2457–2462. 10.1099/vir.0.022764-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bridger JC. 1980. Detection by electron microscopy of caliciviruses, astroviruses and rotavirus-like particles in the faeces of piglets with diarrhoea. Vet. Rec. 107:532–533 [PubMed] [Google Scholar]
- 13.Jonassen CM, Jonassen TT, Sveen TM, Grinde B. 2003. Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res. 91:195–201. 10.1016/S0168-1702(02)00269-1 [DOI] [PubMed] [Google Scholar]
- 14.Englund L, Chriel M, Dietz HH, Hedlund KO. 2002. Astrovirus epidemiologically linked to pre-weaning diarrhoea in mink. Vet. Microbiol. 85:1–11. 10.1016/S0378-1135(01)00472-2 [DOI] [PubMed] [Google Scholar]
- 15.Chu DKW, Poon LLM, Guan Y, Peiris JSM. 2008. Novel astroviruses in insectivorous bats. J. Virol. 82:9107–9114. 10.1128/JVI.00857-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkins A, Wellehan JFX, Jr, Childress AL, Archer LL, Fraser WA, Citino SB. 2009. Characterization of an outbreak of astroviral diarrhea in a group of cheetahs (Acinonyx jubatus). Vet. Microbiol. 136:160–165. 10.1016/j.vetmic.2008.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martella V, Moschidou P, Pinto P, Catella C, Desario C, Larocca V, Circella E, Banyai K, Lavazza A, Magistrali C, Decaro N, Buonavoglia C. 2011. Astroviruses in rabbits. Emerg. Infect. Dis. 17:2287–2293. 10.3201/eid1712.110967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivera R, Nollens HH, Venn-Watson S, Gulland FMD, Wellehan JFX., Jr 2010. Characterization of phylogenetically diverse astroviruses of marine mammals. J. Gen. Virol. 91:166–173. 10.1099/vir.0.015222-0 [DOI] [PubMed] [Google Scholar]
- 19.Koci MD, Seal BS, Schultz-Cherry S. 2000. Molecular characterization of an avian astrovirus. J. Virol. 74:6173–6177. 10.1128/JVI.74.13.6173-6177.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantin-Jackwood MJ, Strother KO, Mundt E, Zsak L, Day JM, Spackman E. 2011. Molecular characterization of avian astroviruses. Arch. Virol. 156:235–244. 10.1007/s00705-010-0849-z [DOI] [PubMed] [Google Scholar]
- 21.Asplin FD. 1965. Duck hepatitis: vaccination against two serological types. Vet. Rec. 77:1529–1530 [PubMed] [Google Scholar]
- 22.Zhao W, Zhu AL, Yu Y, Yuan CL, Zhu CX, Yang ZB, Cui L, Hua XG. 2011. Complete sequence and genetic characterization of pigeon avian nephritis virus, a member of the family Astroviridae. Arch. Virol. 156:1559–1565. 10.1007/s00705-011-1034-8 [DOI] [PubMed] [Google Scholar]
- 23.Canelli E, Cordioli P, Barbieri I, Catella A, Pennelli D, Ceruti R, Moreno A, Lavazza A. 2012. Astroviruses as causative agents of poultry enteritis: genetic characterization and longitudinal studies on field conditions. Avian Dis. 56:173–182. 10.1637/9831-061311-Reg.1 [DOI] [PubMed] [Google Scholar]
- 24.Chu DKW, Leung CYH, Perera HKK, Ng EM, Gilbert M, Joyner PH, Grioni A, Ades G, Guan Y, Peiris JSM, Poon LLM. 2012. A novel group of avian astroviruses in wild aquatic birds. J. Virol. 86:13772–13778. 10.1128/JVI.02105-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 4:e1000011. 10.1371/journal.ppat.1000011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finkbeiner SR, Kirkwood CD, Wang D. 2008. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 5:117. 10.1186/1743-422X-5-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, Kang G, Wang D. 2009. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol. J. 6:161. 10.1186/1743-422X-6-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, Virgin HW, Anderson LJ, Vinje J, Wang D, Tong S. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 83:10836–10839. 10.1128/JVI.00998-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, Zaidi SZ, Delwart E. 2009. Multiple novel astrovirus species in human stool. J. Gen. Virol. 90:2965–2972. 10.1099/vir.0.014449-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkbeiner SR, Le BM, Holtz LR, Storch GA, Wang D. 2009. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg. Infect. Dis. 15:441–444. 10.3201/1503.081213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H, Holtz LR, Bauer I, Franz CJ, Zhao G, Bodhidatta L, Shrestha SK, Kang G, Wang D. 2013. Comparison of novel MLB-clade, VA-clade and classic human astroviruses highlights constrained evolution of the classic human astrovirus nonstructural genes. Virology. 436:8–14. 10.1016/j.virol.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao W, Zhu AL, Yuan CL, Yu Y, Zhu CX, Lan DL, Yang ZB, Cui L, Hua XG. 2011. Detection of astrovirus infection in pigeons (Columbia livia) during an outbreak of diarrhoea. Avian Pathol. 40:361–365. 10.1080/03079457.2011.587792 [DOI] [PubMed] [Google Scholar]
- 33.Cruz JR, Bartlett AV, Herrmann JE, Caceres P, Blacklow NR, Cano F. 1992. Astrovirus-associated diarrhea among Guatemalan ambulatory rural children. J. Clin. Microbiol. 30:1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glass RI, Noel J, Mitchell D, Herrmann JE, Blacklow NR, Pickering LK, Dennehy P, Ruiz-Palacios G, de Guerrero ML, Monroe SS. 1996. The changing epidemiology of astrovirus-associated gastroenteritis: a review. Arch. Virol. Suppl. 12:287–300 [DOI] [PubMed] [Google Scholar]
- 35.Palombo EA, Bishop RF. 1996. Annual incidence, serotype distribution, and genetic diversity of human astrovirus isolates from hospitalized children in Melbourne, Australia. J. Clin. Microbiol. 34:1750–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter JE, Mitchell DK. 2003. Astrovirus infection in children. Curr. Opin. Infect. Dis. 16:247–253. 10.1097/00001432-200306000-00011 [DOI] [PubMed] [Google Scholar]
- 37.Mendez E, Salas-Ocampo E, Arias CF. 2004. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J. Virol. 78:8601–8608. 10.1128/JVI.78.16.8601-8608.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendez E, Fernandez-Luna T, Lopez S, Mendez-Toss M, Arias CF. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J. Virol. 76:7996–8002. 10.1128/JVI.76.16.7996-8002.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishna NK. 2005. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 18:17–26. 10.1089/vim.2005.18.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong J, Dong L, Méndez E, Tao Y. 2011. Crystal structure of the human astrovirus capsid spike. Proc. Natl. Acad. Sci. U. S. A. 108:12681–12686. 10.1073/pnas.1104834108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Risco C, Carrascosa JL, Pedregosa AM, Humphrey CD, Sanchez-Fauquier A. 1995. Ultrastructure of human astrovirus serotype 2. J. Gen. Virol. 76:2075–2080. 10.1099/0022-1317-76-8-2075 [DOI] [PubMed] [Google Scholar]
- 42.Dryden KA, Tihova M, Nowotny N, Matsui SM, Mendez E, Yeager M. 2012. Immature and mature human astrovirus: structure, conformational changes, and similarities to hepatitis E virus. J. Mol. Biol. 422:650–658. 10.1016/j.jmb.2012.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xing L, Li TC, Mayazaki N, Simon MN, Wall JS, Moore M, Wang CY, Takeda N, Wakita T, Miyamura T, Cheng RH. 2010. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 285:33175–33183. 10.1074/jbc.M110.106336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bass DM, Qiu S. 2000. Proteolytic processing of the Astrovirus capsid. J. Virol. 74:1810–1814. 10.1128/JVI.74.4.1810-1814.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosch A, Méndez E, Krishna NK, Pantin-Jackwood M, Schultz-Cherry S, Monroe SS, Guix S. 2011. Astroviridae, p 953–960 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy, 9th ed. Elsevier, Oxford, United Kingdom [Google Scholar]
- 46.Fuentes C, Bosch A, Pinto RM, Guix S. 2012. Identification of human astrovirus genome-linked protein (VPg) essential for virus infectivity. J. Virol. 86:10070–10078. 10.1128/JVI.00797-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willcocks MM, Carter MJ. 1993. Identification and sequence determination of the capsid protein gene of human astrovirus serotype 1. FEMS Microbiol. Lett. 114:1–7. 10.1111/j.1574-6968.1993.tb06542.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Firth A, Atkins J. 2010. Candidates in astroviruses, seadornaviruses, cytorhabdoviruses, and coronaviruses for +1 frame overlapping genes accessed by leaky scanning. Virol. J. 7:17. 10.1186/1743-422X-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geigenmuller U, Ginzton NH, Matsui SM. 1997. Construction of a genome-length cDNA clone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J. Virol. 71:1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velazquez-Moctezuma R, Banos-Lara MDR, Acevedo Y, Mendez E. 2012. Alternative cell lines to improve the rescue of infectious human astrovirus from a cDNA clone. J. Virol. Methods 179:295–302. 10.1016/j.jviromet.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 51.Monroe SS, Stine SE, Gorelkin L, Herrmann JE, Blacklow NR, Glass RI. 1991. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J. Virol. 65:641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Racaniello V. 2007. Picornaviridae: the viruses and their replication, p 795–838 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 53.Lindenbach BD TH, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 54.Green K. 2007. Caliciviridae: the noroviruses, p 949–979 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 55.Monceyron C, Grinde B, Jonassen TO. 1997. Molecular characterisation of the 3′-end of the astrovirus genome. Arch. Virol. 142:699–706. 10.1007/s007050050112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jonassen CM, Jonassen TO, Grinde B. 1998. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen.Virol. 79:715–718 [DOI] [PubMed] [Google Scholar]
- 57.Wang QH, Kakizawa J, Wen LY, Shimizu M, Nishio O, Fang ZY, Ushijima H. 2001. Genetic analisys of the capsid region of astroviruses. J. Med. Virol. 64:245–255. 10.1002/jmv.1043 [DOI] [PubMed] [Google Scholar]
- 58.Robertson MP, Igel H, Baertsch R, Haussler D, Ares M, Jr, Scott WG. 2005. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 3:e5. 10.1371/journal.pbio.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willcocks MM, Ashton N, Kurtz JB, Cubitt WD, Carter MJ. 1994. Cell culture adaptation of astrovirus involves a deletion. J. Virol. 68:6057–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guix S, Caballero S, Bosch A, Pinto RM. 2005. Human astrovirus C-terminal nsP1a protein is involved in RNA replication. Virology 333:124–131. 10.1016/j.virol.2004.12.023 [DOI] [PubMed] [Google Scholar]
- 61.Mendez E, Arias CF. 2007. Astroviruses, p 981–1000 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 62.Jiang B, Monroe SS, Koonin EV, Stine SE, Glass RI. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. U. S. A. 90:10539–10543. 10.1073/pnas.90.22.10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marczinke B, Bloys AJ, Brown TD, Willcocks MM, Carter MJ, Brierley I. 1994. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J. Virol. 68:5588–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang KI, Icard AH, Linnemann E, Sellers HS, Mundt E. 2012. Determination of the full length sequence of a chicken astrovirus suggests a different replication mechanism. Virus Genes 44:45–50. 10.1007/s11262-011-0663-z [DOI] [PubMed] [Google Scholar]
- 65.Fu Y, Pan M, Wang X, Xu Y, Xie X, Knowles NJ, Yang H, Zhang D. 2009. Complete sequence of a duck astrovirus associated with fatal hepatitis in ducklings. J. Gen. Virol. 90:1104–1108. 10.1099/vir.0.008599-0 [DOI] [PubMed] [Google Scholar]
- 66.Neill JD. 2002. The subgenomic RNA of feline calicivirus is packaged into viral particles during infection. Virus Res. 87:89–93. 10.1016/S0168-1702(02)00086-2 [DOI] [PubMed] [Google Scholar]
- 67.Brinker JP, Blacklow NR, Herrmann JE. 2000. Human astrovirus isolation and propagation in multiple cell lines. Arch. Virol. 145:1847–1856. 10.1007/s007050070060 [DOI] [PubMed] [Google Scholar]
- 68.Moser LA, Carter M, Schultz-Cherry S. 2007. Astrovirus increases epithelial barrier permeability independently of viral replication. J. Virol. 81:11937–11945. 10.1128/JVI.00942-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mendez E, Munoz-Yanez C, Sanchez-San Martin C, Aguirre-Crespo G, Banos-Lara MD, Gutierrez M, Espinosa R, Acevedo Y, Arias CF, Lopez S. 2014. Characterization of human astrovirus cell entry. J. Virol. 88:2452–2460. 10.1128/JVI.02908-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Donelli G, Superti F, Tinari A, Marziano ML. 1992. Mechanism of astrovirus entry into Graham 293 cells. J. Med. Virol. 38:271–277. 10.1002/jmv.1890380408 [DOI] [PubMed] [Google Scholar]
- 71.Moser LA, Schultz-Cherry S. 2008. Suppression of astrovirus replication by an ERK1/2 inhibitor. J. Virol. 82:7475–7482. 10.1128/JVI.02193-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tange S, Zhou Y, Nagakui-Noguchi Y, Imai T, Nakanishi A. 2013. Initiation of human astrovirus type 1 infection was blocked by inhibitors of phosphoinositide 3-kinase. Virol. J. 10:153. 10.1186/1743-422X-10-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorne LG, Goodfellow IG. 2014. Norovirus gene expression and replication. J. Gen. Virol. 95:278–291. 10.1099/vir.0.059634-0 [DOI] [PubMed] [Google Scholar]
- 74.Speroni S, Rohayem J, Nenci S, Bonivento D, Robel I, Barthel J, Luzhkov VB, Coutard B, Canard B, Mattevi A. 2009. Structural and biochemical analysis of human pathogenic astrovirus serine protease at 2.0 A resolution. J. Mol. Biol. 387:1137–1152. 10.1016/j.jmb.2009.02.044 [DOI] [PubMed] [Google Scholar]
- 75.Geigenmuller U, Chew T, Ginzton N, Matsui SM. 2002. Processing of nonstructural protein 1a of human astrovirus. J. Virol. 76:2003–2008. 10.1128/JVI.76.4.2003-2008.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibson CA, Chen J, Monroe SA, Denison MR. 1998. Expression and processing of nonstructural proteins of the human astroviruses. Adv. Exp. Med. Biol. 440:387–391 [DOI] [PubMed] [Google Scholar]
- 77.Kiang D, Matsui SM. 2002. Proteolytic processing of a human astrovirus nonstructural protein. J. Gen. Virol. 83:25–34 [DOI] [PubMed] [Google Scholar]
- 78.Mendez E, Salas-Ocampo MP, Munguia ME, Arias CF. 2003. Protein products of the open reading frames encoding nonstructural proteins of human astrovirus serotype 8. J. Virol. 77:11378–11384. 10.1128/JVI.77.21.11378-11384.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willcocks MM, Boxall AS, Carter MJ. 1999. Processing and intracellular location of human astrovirus non-structural proteins. J. Gen.Virol. 80:2607–2611 [DOI] [PubMed] [Google Scholar]
- 80.Al-Mutairy B, Walter JE, Pothen A, Mitchell DK. 2005. Genome prediction of putative genome-linked viral protein (VPg) of astroviruses. Virus Genes 31:21–30. 10.1007/s11262-004-2196-1 [DOI] [PubMed] [Google Scholar]
- 81.Fuentes C, Guix S, Bosch A, Pinto RM. 2011. The C-terminal nsP1a protein of human astrovirus is a phosphoprotein that interacts with the viral polymerase. J. Virol. 85:4470–4479. 10.1128/JVI.01515-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guix S, Caballero S, Bosch A, Pinto RM. 2004. C-terminal nsP1a protein of human astrovirus colocalizes with the endoplasmic reticulum and viral RNA. J. Virol. 78:13627–13636. 10.1128/JVI.78.24.13627-13636.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mendez E, Aguirre-Crespo G, Zavala G, Arias CF. 2007. Association of the astrovirus structural protein VP90 with membranes plays a role in virus morphogenesis. J. Virol. 81:10649–10658. 10.1128/JVI.00785-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gray EW, Angus KW, Snodgrass DR. 1980. Ultrastructure of the small intestine in astrovirus-infected lambs. J. Gen. Virol. 49:71–82. 10.1099/0022-1317-49-1-71 [DOI] [PubMed] [Google Scholar]
- 85.Jang SY, Jeong WH, Kim MS, Lee YM, Lee JI, Lee GC, Paik SY, Koh GP, Kim JM, Lee CH. 2010. Detection of replicating negative-sense RNAs in CaCo-2 cells infected with human astrovirus. Arch. Virol. 155:1383–1389. 10.1007/s00705-010-0718-9 [DOI] [PubMed] [Google Scholar]
- 86.Guix S, Caballero S, Fuentes C, Bosch A, Pinto RM. 2008. Genetic analysis of the hypervariable region of the human astrovirus nsP1a coding region: design of a new RFLP typing method. J. Med. Virol. 80:306–315. 10.1002/jmv.21058 [DOI] [PubMed] [Google Scholar]
- 87.Banos-Lara MR, Mendez E. 2010. Role of individual caspases induced by astrovirus on the processing of its structural protein and its release from the cell through a non-lytic mechanism. Virology 401:322–332. 10.1016/j.virol.2010.02.028 [DOI] [PubMed] [Google Scholar]
- 88.Belliot G, Laveran H, Monroe SS. 1997. Outbreak of gastroenteritis in military recruits associated with serotype 3 astrovirus infection. J. Med. Virol. 51:101–106. [DOI] [PubMed] [Google Scholar]
- 89.Pager CT, Steele AD. 2002. Astrovirus-associated diarrhea in South African adults. Clin. Infect. Dis. 35:1452–1453. 10.1086/344456 [DOI] [PubMed] [Google Scholar]
- 90.Koopmans MP, Bijen MH, Monroe SS, Vinje J. 1998. Age-stratified seroprevalence of neutralizing antibodies to astrovirus types 1 to 7 in humans in The Netherlands. Clin. Diagn. Lab. Immunol. 5:33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kriston S, Willcocks MM, Carter MJ, Cubitt WD. 1996. Seroprevalence of astrovirus types 1 and 6 in London, determined using recombinant virus antigen. Epidemiol. Infect. 117:159–164. 10.1017/S0950268800001266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee R, Lessler J, Lee R, Rudolph K, Reich N, Perl T, Cummings D. 2013. Incubation periods of viral gastroenteritis: a systematic review. BMC Infect. Dis. 13:446. 10.1186/1471-2334-13-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mendez-Toss M, Griffin DD, Calva J, Contreras JF, Puerto FI, Mota F, Guiscafre H, Cedillo R, Munoz O, Herrera I, Lopez S, Arias CF. 2004. Prevalence and genetic diversity of human astroviruses in Mexican children with symptomatic and asymptomatic infections. J. Clin. Microbiol. 42:151–157. 10.1128/JCM.42.1.151-157.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maldonado Y, Cantwell M, Old M, Hill D, Sanchez ML, Logan L, Millan-Velasco F, Valdespino JL, Sepulveda J, Matsui S. 1998. Population-based prevalence of symptomatic and asymptomatic astrovirus infection in rural Mayan infants. J. Infect. Dis. 178:334–339. 10.1086/515625 [DOI] [PubMed] [Google Scholar]
- 95.Midthun K, Greenberg HB, Kurtz JB, Gary GW, Lin FY, Kapikian AZ. 1993. Characterization and seroepidemiology of a type 5 astrovirus associated with an outbreak of gastroenteritis in Marin County, California. J. Clin. Microbiol. 31:955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurtz JB, Lee TW, Craig JW, Reed SE. 1979. Astrovirus infection in volunteers. J. Med. Virol. 3:221–230. 10.1002/jmv.1890030308 [DOI] [PubMed] [Google Scholar]
- 97.González GG, Pujol FH, Liprandi F, Deibis L, Ludert JE. 1998. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J. Med. Virol. 55:288–292. [DOI] [PubMed] [Google Scholar]
- 98.Grohmann GS, Glass RI, Pereira HG, Monroe SS, Hightower AW, Weber R, Bryan RT. 1993. Enteric viruses and diarrhea in HIV-infected patients. N. Engl. J. Med. 329:14–20. 10.1056/NEJM199307013290103 [DOI] [PubMed] [Google Scholar]
- 99.Wunderli W, Meerbach A, Gungor T, Berger C, Greiner O, Caduff R, Trkola A, Bossart W, Gerlach D, Schibler M, Cordey S, McKee TA, Van Belle S, Kaiser L, Tapparel C. 2011. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One 6:e27483. 10.1371/journal.pone.0027483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yokoyama CC, Loh J, Zhao G, Stappenbeck TS, Wang D, Huang HV, Virgin HW, Thackray LB. 2012. Adaptive immunity restricts replication of novel murine astroviruses. J. Virol. 86:12262–12270. 10.1128/JVI.02018-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aminu M, Ameh EA, Geyer A, Esona MD, Taylor MB, Steele AD. 2009. Role of astrovirus in intussusception in Nigerian infants. J. Trop. Pediatr. 55:192–194. 10.1093/tropej/fmn101 [DOI] [PubMed] [Google Scholar]
- 102.Jakab F, Peterfai J, Verebely T, Meleg E, Banyai K, Mitchell DK, Szucs G. 2007. Human astrovirus infection associated with childhood intussusception. Pediatr. Int. 49:103–105. 10.1111/j.1442-200X.2007.02293.x [DOI] [PubMed] [Google Scholar]
- 103.Lee YW, Yang SI, Kim JM, Kim JY. 2013. Clinical features and role of viral isolates from stool samples of intussuception in children. Pediatr. Gastroenterol. Hepatol. Nutr. 16:162–170. 10.5223/pghn.2013.16.3.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Caballero S, Guix S, El-Senousy WM, Calico I, Pinto RM, Bosch A. 2003. Persistent gastroenteritis in children infected with astrovirus: association with serotype-3 strains. J. Med. Virol. 71:245–250. 10.1002/jmv.10476 [DOI] [PubMed] [Google Scholar]
- 105.Banyai K, Meleg E, Moschidou P, Martella V. 2010. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg. Infect. Dis. 16:169 (Author reply, 16:169–170.) 10.3201/eid1601.091563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mitui MT, Bozdayi G, Matsumoto T, Dalgic B, Nishizono A, Ahmed K. 2013. Complete genome sequence of an MLB2 astrovirus from a Turkish child with diarrhea. Genome Announc. 1(4):e00619-13. 10.1128/genomeA.00619-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ahmed SF, Sebeny PJ, Klena JD, Pimentel G, Mansour A, Naguib AM, Bruton J, Young SY, Holtz LR, Wang D. 2011. Novel astroviruses in children, Egypt. Emerg. Infect. Dis. 17:2391–2393. 10.3201/eid1712.110909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matsumoto T, Wangchuk S, Tshering K, Yahiro T, Zangmo S, Dorji T, Mitui MT, Nishizono A, Ahmed K. 2013. Complete genome sequences of two astrovirus MLB1 strains from Bhutanese children with diarrhea. Genome Announc. 1(4):e00485-13. 10.1128/genomeA.00485-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y, Li Y, Jin Y, Li DD, Li X, Duan ZJ. 2013. Recently identified novel human astroviruses in children with diarrhea, China. Emerg. Infect. Dis. 19:1333–1335. 10.3201/eid1908.121863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Medici MC, Tummolo F, Calderaro A, Elia G, Banyai K. 2014. MLB1 astrovirus in children with gastroenteritis, Italy. Emerg. Infect. Dis. 20:169–170. 10.3201/eid2001.131259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holtz LR, Bauer IK, Rajendran P, Kang G, Wang D. 2011. Astrovirus MLB1 is not associated with diarrhea in a cohort of Indian children. PLoS One 6:e28647. 10.1371/journal.pone.0028647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Holtz LR, Wylie KM, Sodergren E, Jiang Y, Franz CJ, Weinstock GM, Storch GA, Wang D. 2011. Astrovirus MLB2 viremia in febrile child. Emerg. Infect. Dis. 17:2050–2052. 10.3201/eid1711.110496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. 2012. Sequence analysis of the human virome in febrile and afebrile children. PLoS One 7:e27735. 10.1371/journal.pone.0027735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smits SL, van Leeuwen M, van der Eijk AA, Fraaij PL, Escher JC, Simon JH, Osterhaus AD. 2010. Human astrovirus infection in a patient with new-onset celiac disease. J. Clin. Microbiol. 48:3416–3418. 10.1128/JCM.01164-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burbelo PD, Ching KH, Esper F, Iadarola MJ, Delwart E, Lipkin WI, Kapoor A. 2011. Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PLoS One 6:e22576. 10.1371/journal.pone.0022576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg. Infect. Dis. 16:918–925. 10.3201/eid1606.091536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moser LA, Schultz-Cherry S. 2005. Pathogenesis of astrovirus infection. Viral Immunol. 18:4–10. 10.1089/vim.2005.18.4 [DOI] [PubMed] [Google Scholar]
- 118.Sebire NJ, Malone M, Shah N, Anderson G, Gaspar HB, Cubitt WD. 2004. Pathology of astrovirus associated diarrhoea in a paediatric bone marrow transplant recipient. J. Clin. Pathol. 57:1001–1003. 10.1136/jcp.2004.017178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Snodgrass DR, Angus KW, Gray EW, Menzies JD, Paul G. 1979. Pathogenesis of diarrhoea caused by astrovirus infections in lambs. Arch. Virol. 60:217–226. 10.1007/BF01317493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Woode GN, Pohlenz JF, Gourley NE, Fagerland JA. 1984. Astrovirus and Breda virus infections of dome cell epithelium of bovine ileum. J. Clin. Microbiol. 19:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Behling-Kelly E, Schultz-Cherry S, Koci M, Kelley L, Larsen D, Brown C. 2002. Localization of astrovirus in experimentally infected turkeys as determined by in situ hybridization. Vet. Pathol. 39:595–598. 10.1354/vp.39-5-595 [DOI] [PubMed] [Google Scholar]
- 122.Koci MD, Moser LA, Kelley LA, Larsen D, Brown CC, Schultz-Cherry S. 2003. Astrovirus induces diarrhea in the absence of inflammation and cell death. J. Virol. 77:11798–11808. 10.1128/JVI.77.21.11798-11808.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hodges K, Gill R. 2010. Infectious diarrhea: cellular and molecular mechanisms. Gut Microbes 1:4–21. 10.4161/gmic.1.1.11036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Viswanathan VK, Hodges K, Hecht G. 2009. Enteric infection meets intestinal function: how bacterial pathogens cause diarrhoea. Nat. Rev. Microbiol. 7:110–119. 10.1038/nrmicro2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Thouvenelle ML, Haynes JS, Sell JL, Reynolds DL. 1995. Astrovirus infection in hatchling turkeys: alterations in intestinal maltase activity. Avian Dis. 39:343–348. 10.2307/1591877 [DOI] [PubMed] [Google Scholar]
- 126.Nighot PK, Moeser A, Ali RA, Blikslager AT, Koci MD. 2010. Astrovirus infection induces sodium malabsorption and redistributes sodium hydrogen exchanger expression. Virology 401:146–154. 10.1016/j.virol.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guix S, Bosch A, Ribes E, Dora Martinez L, Pinto RM. 2004. Apoptosis in astrovirus-infected CaCo-2 cells. Virology 319:249–261. 10.1016/j.virol.2003.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koci MD. 2005. Immunity and resistance to astrovirus infection. Viral Immunol. 18:11–16. 10.1089/vim.2005.18.11 [DOI] [PubMed] [Google Scholar]
- 129.Mitchell DK, Matson DO, Cubitt WD, Jackson LJ, Willcocks MM, Pickering LK, Carter MJ. 1999. Prevalence of antibodies to astrovirus types 1 and 3 in children and adolescents in Norfolk, Virginia. Pediatr. Infect. Dis. J. 18:249–254. 10.1097/00006454-199903000-00008 [DOI] [PubMed] [Google Scholar]
- 130.Blutt SE, Conner ME. 2013. The gastrointestinal frontier: IgA and viruses. Front. Immunol. 4:402. 10.3389/fimmu.2013.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Molberg O, Nilsen EM, Sollid LM, Scott H, Brandtzaeg P, Thorsby E, Lundin KE. 1998. CD4+ T cells with specific reactivity against astrovirus isolated from normal human small intestine. Gastroenterology 114:115–122. 10.1016/S0016-5085(98)70639-0 [DOI] [PubMed] [Google Scholar]
- 132.Bonaparte RS, Hair PS, Banthia D, Marshall DM, Cunnion KM, Krishna NK. 2008. Human astrovirus coat protein inhibits serum complement activation via C1, the first component of the classical pathway. J. Virol. 82:817–827. 10.1128/JVI.01847-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gronemus JQ, Hair PS, Crawford KB, Nyalwidhe JO, Cunnion KM, Krishna NK. 2010. Potent inhibition of the classical pathway of complement by a novel C1q-binding peptide derived from the human astrovirus coat protein. Mol. Immunol. 48:305–313. 10.1016/j.molimm.2010.07.012 [DOI] [PubMed] [Google Scholar]
- 134.Hair PS, Gronemus JQ, Crawford KB, Salvi VP, Cunnion KM, Thielens NM, Arlaud GJ, Rawal N, Krishna NK. 2010. Human astrovirus coat protein binds C1q and MBL and inhibits the classical and lectin pathways of complement activation. Mol. Immunol. 47:792–798. 10.1016/j.molimm.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 135.Krishna NK, Cunnion KM. 2008. Human astrovirus coat protein: a novel C1 inhibitor. Adv. Exp. Med. Biol. 632:237–251. 10.1007/978-0-387-78952-1_17 [DOI] [PubMed] [Google Scholar]
- 136.Mauriello CT, Pallera HK, Sharp JA, Woltmann JL, Jr, Qian S, Hair PS, van der Pol P, van Kooten C, Thielens NM, Lattanzio FA, Cunnion KM, Krishna NK. 2013. A novel peptide inhibitor of classical and lectin complement activation including ABO incompatibility. Mol. Immunol. 53:132–139. 10.1016/j.molimm.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koci MD, Kelley LA, Larsen D, Schultz-Cherry S. 2004. Astrovirus-induced synthesis of nitric oxide contributes to virus control during infection. J. Virol. 78:1564–1574. 10.1128/JVI.78.3.1564-1574.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stoermer KA, Morrison TE. 2011. Complement and viral pathogenesis. Virology 411:362–373. 10.1016/j.virol.2010.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meliopoulos V, Schultz-Cherry S. 2013. Astrovirus pathogenesis, p 65–77 In Schultz-Cherry S. (ed), Astrovirus research: essential ideas, everyday impacts, future directions. Springer, New York, NY [Google Scholar]
- 140.Qureshi MA, Saif YM, Heggen-Peay CL, Edens FW, Havenstein GB. 2001. Induction of functional defects in macrophages by a poult enteritis and mortality syndrome-associated turkey astrovirus. Avian Dis. 45:853–861. 10.2307/1592865 [DOI] [PubMed] [Google Scholar]
- 141.Qureshi MA, Yu M, Saif YM. 2000. A novel “small round virus” inducing poult enteritis and mortality syndrome and associated immune alterations. Avian Dis. 44:275–283. 10.2307/1592540 [DOI] [PubMed] [Google Scholar]
- 142.Meyerhoff RR, Nighot PK, Ali RA, Blikslager AT, Koci MD. 2012. Characterization of turkey inducible nitric oxide synthase and identification of its expression in the intestinal epithelium following astrovirus infection. Comp. Immunol. Microbiol. Infect. Dis. 35:63–69. 10.1016/j.cimid.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578. 10.1126/science.1077905 [DOI] [PubMed] [Google Scholar]
- 144.Borghan MA, Mori Y, El-Mahmoudy A-B, Ito N, Sugiyama M, Takewaki T, Minamoto N. 2007. Induction of nitric oxide synthase by rotavirus enterotoxin NSP4: implication for rotavirus pathogenicity. J. Gen. Virol. 88:2064–2072. 10.1099/vir.0.82618-0 [DOI] [PubMed] [Google Scholar]
- 145.Rodríguez-Díaz J, Banasaz M, Istrate C, Buesa J, Lundgren O, Espinoza F, Sundqvist T, Rottenberg M, Svensson L. 2006. Role of nitric oxide during rotavirus infection. J. Med. Virol. 78:979–985. 10.1002/jmv.20650 [DOI] [PubMed] [Google Scholar]
- 146.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. 2011. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 11:1529–1544. 10.1016/j.meegid.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xiao C-T, Giménez-Lirola LG, Gerber PF, Jiang Y-H, Halbur PG, Opriessnig T. 2013. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J. Gen. Virol. 94:570–582. 10.1099/vir.0.048744-0 [DOI] [PubMed] [Google Scholar]
- 148.Zhu HC, Chu DKW, Liu W, Dong BQ, Zhang SY, Zhang JX, Li LF, Vijaykrishna D, Smith GJD, Chen HL, Poon LLM, Peiris JSM, Guan Y. 2009. Detection of diverse astroviruses from bats in China. J. Gen. Virol. 90:883–887. 10.1099/vir.0.007732-0 [DOI] [PubMed] [Google Scholar]
- 149.Reuter G, Pankovics P, Delwart E, Boros A. 2012. Identification of a novel astrovirus in domestic sheep in Hungary. Arch. Virol. 157:323–327. 10.1007/s00705-011-1151-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, Talbot T, Blanchard P, Rimoldi G, Fahsbender E, Page B, Phan TG, Wang C, Deng X, Pesavento P, Delwart E. 2013. Divergent astrovirus associated with neurologic disease in cattle. Emerg. Infect. Dis. 19:1385–1392. 10.3201/eid1909.130682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Oem JK, An DJ. 2014. Phylogenetic analysis of bovine astrovirus in Korean cattle. Virus Genes 48:372–375. 10.1007/s11262-013-1013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tse H, Chan W-M, Tsoi H-W, Fan RYY, Lau CCY, Lau SKP, Woo PCY, Yuen K-Y. 2011. Rediscovery and genomic characterization of bovine astroviruses. J. Gen. Virol. 92:1888–1898. 10.1099/vir.0.030817-0 [DOI] [PubMed] [Google Scholar]
- 153.Farkas T, Fey B, Keller G, Martella V, Egyed L. 2012. Molecular detection of novel astroviruses in wild and laboratory mice. Virus Genes 45:518–525. 10.1007/s11262-012-0803-0 [DOI] [PubMed] [Google Scholar]
- 154.Blomstrom AL, Widen F, Hammer AS, Belak S, Berg M. 2010. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 48:4392–4396. 10.1128/JCM.01040-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Strain E, Kelley LA, Schultz-Cherry S, Muse SV, Koci MD. 2008. Genomic analysis of closely related astroviruses. J. Virol. 82:5099–5103. 10.1128/JVI.01993-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen L, Xu Q, Zhang R, Li J, Xie Z, Wang Y, Zhu Y, Jiang S. 2012. Complete genome sequence of a duck astrovirus discovered in eastern China. J. Virol. 86:13833–13834. 10.1128/JVI.02637-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kofstad T, Jonassen CM. 2011. Screening of feral and wood pigeons for viruses harbouring a conserved mobile viral element: characterization of novel astroviruses and picornaviruses. PLoS One 6:e25964. 10.1371/journal.pone.0025964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lukashov VV, Goudsmit J. 2002. Evolutionary relationships among Astroviridae. J. Gen. Virol. 83:1397–1405 [DOI] [PubMed] [Google Scholar]
- 159.van Hemert FJ, Berkhout B, Lukashov VV. 2007. Host-related nucleotide composition and codon usage as driving forces in the recent evolution of the Astroviridae. Virology 361:447–454. 10.1016/j.virol.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Biđin M, Biđin Z, Majnarić D, Tišljar M, Lojkić I. 2012. Circulation and phylogenetic relationship of chicken and turkey-origin astroviruses detected in domestic ducks (Anas platyrhynchos domesticus). Avian Pathol. 41:555–562. 10.1080/03079457.2012.733340 [DOI] [PubMed] [Google Scholar]
- 161.Battisti C, Salviato A, Jonassen C, Toffan A, Capua I, Cattoli G. 2012. Genetic characterization of astroviruses detected in guinea fowl (Numida meleagris) reveals a distinct genotype and suggests cross-species transmission between turkey and guinea fowl. Arch. Virol. 157:1329–1337. 10.1007/s00705-012-1311-1 [DOI] [PubMed] [Google Scholar]
- 162.Lan D, Ji W, Shan T, Cui L, Yang Z, Yuan C, Hua X. 2011. Molecular characterization of a porcine astrovirus strain in China. Arch. Virol. 156:1869–1875. 10.1007/s00705-011-1050-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ulloa JC, Gutierrez MF. 2010. Genomic analysis of two ORF2 segments of new porcine astrovirus isolates and their close relationship with human astroviruses. Can. J. Microbiol. 56:569–577. 10.1139/W10-042 [DOI] [PubMed] [Google Scholar]
- 164.Guix S, Caballero S, Villena C, Bartolome R, Latorre C, Rabella N, Simo M, Bosch A, Pinto RM. 2002. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J. Clin. Microbiol. 40:133–139. 10.1128/JCM.40.1.133-139.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jakab F, Walter JE, Berke T, Matson DO, Mitchell DK, Szúcs G. 2003. Molecular characterization and sequence analysis of human astroviruses circulating in Hungary. FEMS Immunol. Med. Microbiol. 39:97–102. 10.1016/S0928-8244(03)00236-0 [DOI] [PubMed] [Google Scholar]
- 166.Liu MQ, Peng JS, Tang L, Zhou Y, Yang BF, Wang YH, Wang B, Zhou DJ, Huang HJ, Ho WZ. 2008. Identification of new subtype of astrovirus type 3 from an infant with diarrhea in Wuhan, China. Virology 375:301–306. 10.1016/j.virol.2008.01.032 [DOI] [PubMed] [Google Scholar]
- 167.Medina SM, Gutierrez MF, Liprandi F, Ludert JE. 2000. Identification and type distribution of astroviruses among children with gastroenteritis in Colombia and Venezuela. J. Clin. Microbiol. 38:3481–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Walter JE, Briggs J, Guerrero ML, Matson DO, Pickering LK, Ruiz-Palacios G, Berke T, Mitchell DK. 2001. Molecular characterization of a novel recombinant strain of human astrovirus associated with gastroenteritis in children. Arch. Virol. 146:2357–2367. 10.1007/s007050170008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Colomba C, Grazia S, Giammanco GM, Saporito L, Scarlata F, Titone L, Arista S. 2006. Viral gastroenteritis in children hospitalised in Sicily, Italy. Eur. J. Clin. Microbiol. Infect. Dis. 25:570–575. 10.1007/s10096-006-0188-x [DOI] [PubMed] [Google Scholar]
- 170.De Grazia S, Platia MA, Rotolo V, Colomba C, Martella V, Giammanco GM. 2011. Surveillance of human astrovirus circulation in Italy 2002-2005: emergence of lineage 2c strains. Clin. Microbiol. Infect. 17:97–101. 10.1111/j.1469-0691.2010.03207.x [DOI] [PubMed] [Google Scholar]
- 171.Gabbay YB, Leite JP, Oliveira DS, Nakamura LS, Nunes MR, Mascarenhas JD, Heinemann MB, Linhares AC. 2007. Molecular epidemiology of astrovirus type 1 in Belem, Brazil, as an agent of infantile gastroenteritis, over a period of 18 years (1982-2000): identification of two possible new lineages. Virus Res. 129:166–174. 10.1016/j.virusres.2007.07.006 [DOI] [PubMed] [Google Scholar]
- 172.Martella V, Medici MC, Terio V, Catella C, Bozzo G, Tummolo F, Calderaro A, Bonura F, Di Franco M, Bányai K, Giammanco GM, De Grazia S. 2013. Lineage diversification and recombination in type-4 human astroviruses. Infect. Genet. Evol. 20:330–335. 10.1016/j.meegid.2013.09.015 [DOI] [PubMed] [Google Scholar]
- 173.Medici MC, Tummolo F, Albonetti V, Abelli LA, Chezzi C, Calderaro A. 2012. Molecular detection and epidemiology of astrovirus, bocavirus, and sapovirus in Italian children admitted to hospital with acute gastroenteritis, 2008-2009. J. Med. Virol. 84:643–650. 10.1002/jmv.23231 [DOI] [PubMed] [Google Scholar]
- 174.Victoria M, Carvalho-Costa FA, Heinemann MB, Leite JPG, Miagostovich MP. 2007. Genotypes and molecular epidemiology of human astroviruses in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Med. Virol. 79:939–944. 10.1002/jmv.20814 [DOI] [PubMed] [Google Scholar]
- 175.Belliot G, Laveran H, Monroe SS. 1997. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch. Virol. 142:1323–1334. 10.1007/s007050050163 [DOI] [PubMed] [Google Scholar]
- 176.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76:159–216. 10.1128/MMBR.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Babkin IV, Tikunov AY, Zhirakovskaia EV, Netesov SV, Tikunova NV. 2012. High evolutionary rate of human astrovirus. Infect. Genet. Evol. 12:435–442. 10.1016/j.meegid.2012.01.019 [DOI] [PubMed] [Google Scholar]
- 178.van Hemert FJ, Lukashov VV, Berkhout B. 2007. Different rates of (non-)synonymous mutations in astrovirus genes; correlation with gene function. Virol. J. 4:25. 10.1186/1743-422X-4-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pativada MS, Chatterjee D, Mariyappa NS, Rajendran K, Bhattacharya MK, Ghosh M, Kobayashi N, Krishnan T. 2011. Emergence of unique variants and inter-genotype recombinants of human astroviruses infecting infants, children and adults in Kolkata, India. Int. J. Mol. Epidemiol. Genet. 2:228–235 [PMC free article] [PubMed] [Google Scholar]
- 180.Pativada M, Bhattacharya R, Krishnan T. 2013. Novel human astrovirus strains showing multiple recombinations within highly conserved ORF1b detected from hospitalized acute watery diarrhea cases in Kolkata, India. Infect. Genet. Evol. 20:284–291. 10.1016/j.meegid.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 181.Wolfaardt M, Kiulia NM, Mwenda JM, Taylor MB. 2011. Evidence of a recombinant wild-type human astrovirus strain from a Kenyan child with gastroenteritis. J. Clin. Microbiol. 49:728–731. 10.1128/JCM.01093-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.De Grazia S, Medici MC, Pinto P, Moschidou P, Tummolo F, Calderaro A, Bonura F, Banyai K, Giammanco GM, Martella V. 2012. Genetic heterogeneity and recombination in human type 2 astroviruses. J. Clin. Microbiol. 50:3760–3764. 10.1128/JCM.02102-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pantin-Jackwood M, Spackman E, Woolcock P. 2006. Phylogenetic analysis of turkey astroviruses reveals evidence of recombination. Virus Genes 32:187–192. 10.1007/s11262-005-6875-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Brnic D, Prpic J, Keros T, Roic B, Staresina V, Jemersic L. 2013. Porcine astrovirus viremia and high genetic variability in pigs on large holdings in Croatia. Infect. Genet. Evol. 14:258–264. 10.1016/j.meegid.2012.12.027 [DOI] [PubMed] [Google Scholar]
- 185.Laurin MA, Dastor M, L'Homme Y. 2011. Detection and genetic characterization of a novel pig astrovirus: relationship to other astroviruses. Arch. Virol. 156:2095–2099. 10.1007/s00705-011-1088-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Machnowska P, Ellerbroek L, Johne R. 2014. Detection and characterization of potentially zoonotic viruses in faeces of pigs at slaughter in Germany. Vet. Microbiol. 168:60–68. 10.1016/j.vetmic.2013.10.018 [DOI] [PubMed] [Google Scholar]
- 187.Reuter G, Pankovics P, Boros A. 2011. Identification of a novel astrovirus in a domestic pig in Hungary. Arch. Virol. 156:125–128. 10.1007/s00705-010-0827-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shan T, Wang C, Tong W, Zheng H, Hua X, Yang S, Guo Y, Zhang W, Tong G. 2012. Complete genome of a novel porcine astrovirus. J. Virol. 86:13820–13821. 10.1128/JVI.02598-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Indik S, Valicek L, Smid B, Dvorakova H, Rodak L. 2006. Isolation and partial characterization of a novel porcine astrovirus. Vet. Microbiol. 117:276–283. 10.1016/j.vetmic.2006.06.020 [DOI] [PubMed] [Google Scholar]
- 190.Luo Z, Roi S, Dastor M, Gallice E, Laurin M-A, L'Homme Y. 2011. Multiple novel and prevalent astroviruses in pigs. Vet. Microbiol. 149:316–323. 10.1016/j.vetmic.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Reuter G, Nemes C, Boros A, Kapusinszky B, Delwart E, Pankovics P. 2012. Astrovirus in wild boars (Sus scrofa) in Hungary. Arch. Virol. 157:1143–1147. 10.1007/s00705-012-1272-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ng TF, Kondov NO, Hayashimoto N, Uchida R, Cha Y, Beyer AI, Wong W, Pesavento PA, Suemizu H, Muench MO, Delwart E. 2013. Identification of an astrovirus commonly infecting laboratory mice in the US and Japan. PLoS One 8:e66937. 10.1371/journal.pone.0066937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart EL. 2011. The fecal viral flora of wild rodents. PLoS Pathog. 7:e1002218. 10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Drexler JF, Corman VM, Wegner T, Tateno AF, Zerbinati RM, Gloza-Rausch F, Seebens A, Muller MA, Drosten C. 2011. Amplification of emerging viruses in a bat colony. Emerg. Infect. Dis. 17:449–456. 10.3201/eid1703.100526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xiao J, Li J, Hu G, Chen Z, Wu Y, Chen Y, Chen Z, Liao Y, Zhou J, Ke X, Ma L, Liu S, Zhou J, Dai Y, Chen H, Yu S, Chen Q. 2011. Isolation and phylogenetic characterization of bat astroviruses in southern China. Arch. Virol. 156:1415–1423. 10.1007/s00705-011-1011-2 [DOI] [PubMed] [Google Scholar]
- 196.Lau SK, Woo PC, Yip CC, Bai R, Wu Y, Tse H, Yuen KY. 2013. Complete genome sequence of a novel feline astrovirus from a domestic cat in Hong Kong. Genome Announc. 1(5):e00708-13. 10.1128/genomeA.00708-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Martella V, Moschidou P, Catella C, Larocca V, Pinto P, Losurdo M, Corrente M, Lorusso E, Banyai K, Decaro N, Lavazza A, Buonavoglia C. 2012. Enteric disease in dogs naturally infected by a novel canine astrovirus. J. Clin. Microbiol. 50:1066–1069. 10.1128/JCM.05018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stenglein MD, Velazquez E, Greenacre C, Wilkes RP, Ruby JG, Lankton JS, Ganem D, Kennedy MA, DeRisi JL. 2012. Complete genome sequence of an astrovirus identified in a domestic rabbit (Oryctolagus cuniculus) with gastroenteritis. Virol. J. 9:216. 10.1186/1743-422X-9-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Toffan A, Jonassen CM, De Battisti C, Schiavon E, Kofstad T, Capua I, Cattoli G. 2009. Genetic characterization of a new astrovirus detected in dogs suffering from diarrhoea. Vet. Microbiol. 139:147–152. 10.1016/j.vetmic.2009.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhu AL, Zhao W, Yin H, Shan TL, Zhu CX, Yang X, Hua XG, Cui L. 2011. Isolation and characterization of canine astrovirus in China. Arch. Virol. 156:1671–1675. 10.1007/s00705-011-1022-z [DOI] [PubMed] [Google Scholar]
- 201.Bulbule NR, Mandakhalikar KD, Kapgate SS, Deshmukh VV, Schat KA, Chawak MM. 2013. Role of chicken astrovirus as a causative agent of gout in commercial broilers in India. Avian Pathol. 42:464–473. 10.1080/03079457.2013.828194 [DOI] [PubMed] [Google Scholar]
- 202.Mor SK, Abin M, Costa G, Durrani A, Jindal N, Goyal SM, Patnayak DP. 2011. The role of type-2 turkey astrovirus in poult enteritis syndrome. Poult. Sci. 90:2747–2752. 10.3382/ps.2011-01617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bosch A. 2007. Human viruses in water, 1st ed. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 204.Koopmans M, Cliver DO, Bosch A. 2008. Foodborne viruses: progress and challenges. ASM Press, Washington, DC [Google Scholar]
- 205.Matsui SM, Greenberg HB. 1994. Medical management of foodborne viral gastroenteritis, p 145–158 In Hui YH, Gorham JR, Murrell KD, Cliver DOE. (ed), Foodborne disease handbook, vol. 2. Diseases caused by viruses, parasites, and fungi. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 206.Mitchell DK, Monroe SS, Jiang X, Matson DO, Glass RI, Pickering LK. 1995. Virologic features of an astrovirus diarrhea outbreak in a day care center revealed by reverse transcriptase-polymerase chain reaction. J. Infect. Dis. 172:1437–1444. 10.1093/infdis/172.6.1437 [DOI] [PubMed] [Google Scholar]
- 207.Oishi I, Yamazaki K, Kimoto T, Minekawa Y, Utagawa E, Yamazaki S, Inouye S, Grohmann GS, Monroe SS, Stine SE, Carcamo C, Ando T, Glass RI. 1994. A large outbreak of acute gastroenteritis associated with astrovirus among students and teachers in Osaka, Japan. J. Infect. Dis. 170:439–443. 10.1093/infdis/170.2.439 [DOI] [PubMed] [Google Scholar]
- 208.Abad FX, Villena C, Guix S, Caballero S, Pinto RM, Bosch A. 2001. Potential role of fomites in the vehicular transmission of human astroviruses. Appl. Environ. Microbiol. 67:3904–3907. 10.1128/AEM.67.9.3904-3907.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gallimore CI, Taylor C, Gennery AR, Cant AJ, Galloway A, Lewis D, Gray JJ. 2005. Use of a heminested reverse transcriptase PCR assay for detection of astrovirus in environmental swabs from an outbreak of gastroenteritis in a pediatric primary immunodeficiency unit. J. Clin. Microbiol. 43:3890–3894. 10.1128/JCM.43.8.3890-3894.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gofti-Laroche L, Gratacap-Cavallier B, Demanse D, Genoulaz O, Seigneurin JM, Zmirou D. 2003. Are waterborne astrovirus implicated in acute digestive morbidity (E.MI.R.A. study)? J. Clin. Virol. 27:74–82. 10.1016/S1386-6532(02)00130-0 [DOI] [PubMed] [Google Scholar]
- 211.Pintó RM, Abad FX, Gajardo R, Bosch A. 1996. Detection of infectious astroviruses in water. Appl. Environ. Microbiol. 62:1811–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Abad FX, Pinto RM, Villena C, Gajardo R, Bosch A. 1997. Astrovirus survival in drinking water. Appl. Environ. Microbiol. 63:3119–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bosch A, Pinto RM, Villena C, Abad FX. 1997. Persistence of human astrovirus in fresh and marine water. Water Sci. Technol. 35:243–247. 10.1016/S0273-1223(97)00266-7 [DOI] [Google Scholar]
- 214.Le Cann P, Ranarijaona S, Monpoeho S, Le Guyader F, Ferre V. 2004. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 155:11–15. 10.1016/j.resmic.2003.09.013 [DOI] [PubMed] [Google Scholar]
- 215.Morsy El-Senousy W, Guix S, Abid I, Pinto RM, Bosch A. 2007. Removal of astrovirus from water and sewage treatment plants, evaluated by a competitive reverse transcription-PCR. Appl. Environ. Microbiol. 73:164–167. 10.1128/AEM.01748-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nadan S, Walter JE, Grabow WO, Mitchell DK, Taylor MB. 2003. Molecular characterization of astroviruses by reverse transcriptase PCR and sequence analysis: comparison of clinical and environmental isolates from South Africa. Appl. Environ. Microbiol. 69:747–753. 10.1128/AEM.69.2.747-753.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Pintó RM, Villena C, Le Guyader F, Guix S, Caballero S, Pommepuy M, Bosch A. 2001. Astrovirus detection in wastewater samples. Water Sci. Technol. 43:73–76 [PubMed] [Google Scholar]
- 218.Le Guyader F, Haugarreau L, Miossec L, Dubois E, Pommepuy M. 2000. Three-year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 66:3241–3248. 10.1128/AEM.66.8.3241-3248.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Asano T, Cotruvo JA. 2004. Groundwater recharge with reclaimed municipal wastewater: health and regulatory considerations. Water Res. 38:1941–1951. 10.1016/j.watres.2004.01.023 [DOI] [PubMed] [Google Scholar]
- 220.Pintó RM, Bosch A. 2008. Rethinking virus detection in food, p 171–188 In Koopmans M, Cliver DO, Bosch A. (ed), Foodborne viruses: progress and challenges. ASM Press, Washington, DC [Google Scholar]
- 221.Kurtz JB, Lee TW. 1987. Astroviruses: human and animal. Ciba Found. Symp. 128:92–107 [DOI] [PubMed] [Google Scholar]
- 222.WHO. 2004. Guidelines for drinking-water quality, 3rd ed, vol 1, p 515 World Health Organization, Geneva, Switzerland [Google Scholar]
- 223.Dongdem JT, Damanka S, Asmah R. 2011. Molecular isolation of human norovirus and astrovirus in tap water by RT-PCR. Int. Res. J. Biochem. Bioinformat. 1:131–138 [Google Scholar]
- 224.Maunula L, Kalso S, von Bonsdorff CH, Ponka A. 2004. Wading pool water contaminated with both noroviruses and astroviruses as the source of a gastroenteritis outbreak. Epidemiol. Infect. 132:737–743. 10.1017/S0950268804002249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Myint S, Manley R, Cubitt D. 1994. Viruses in bathing waters. Lancet 343:1640–1641. 10.1016/S0140-6736(94)93096-1 [DOI] [PubMed] [Google Scholar]
- 226.Superti F, Seganti L, Orsi N, Desideri N, Stein ML, Tinari A, Marziano ML, Donelli G. 1990. In vitro effect of synthetic flavanoids on astrovirus infection. Antiviral Res. 13:201–208. 10.1016/0166-3542(90)90038-9 [DOI] [PubMed] [Google Scholar]
- 227.Todd EC, Greig JD, Bartleson CA, Michaels BS. 2007. Outbreaks where food workers have been implicated in the spread of foodborne disease. 2. Description of outbreaks by size, severity, and settings. J. Food Prot. 70:1975–1993 [DOI] [PubMed] [Google Scholar]
- 228.Cubitt WD, Mitchell DK, Carter MJ, Willcocks MM, Holzel H. 1999. Application of electronmicroscopy, enzyme immunoassay, and RT-PCR to monitor an outbreak of astrovirus type 1 in a paediatric bone marrow transplant unit. J. Med. Virol. 57:313–321. [DOI] [PubMed] [Google Scholar]
- 229.Kirkwood CD, Clark R, Bogdanovic-Sakran N, Bishop RF. 2005. A 5-year study of the prevalence and genetic diversity of human caliciviruses associated with sporadic cases of acute gastroenteritis in young children admitted to hospital in Melbourne, Australia (1998-2002). J. Med. Virol. 77:96–101. 10.1002/jmv.20419 [DOI] [PubMed] [Google Scholar]
- 230.Shastri S, Doane AM, Gonzales J, Upadhyayula U, Bass DM. 1998. Prevalence of astroviruses in a children's hospital. J. Clin. Microbiol. 36:2571–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Bjorkholm M, Celsing F, Runarsson G, Waldenstrom J. 1995. Successful intravenous immunoglobulin therapy for severe and persistent astrovirus gastroenteritis after fludarabine treatment in a patient with Waldenstrom's macroglobulinemia. Int. J. Hematol. 62:117–120. 10.1016/0925-5710(95)00396-A [DOI] [PubMed] [Google Scholar]
- 232.Coppo P, Scieux C, Ferchal F, Clauvel J, Lassoued K. 2000. Astrovirus enteritis in a chronic lymphocytic leukemia patient treated with fludarabine monophosphate. Ann. Hematol. 79:43–45. 10.1007/s002770050008 [DOI] [PubMed] [Google Scholar]
- 233.Cox GJ, Matsui SM, Lo RS, Hinds M, Bowden RA, Hackman RC, Meyer WG, Mori M, Tarr PI, Oshiro LS, Ludert JE, Meyers JD, McDonald GB. 1994. Etiology and outcome of diarrhea after marrow transplantation: a prospective study. Gastroenterology 107:1398–1407. 10.1016/0016-5085(94)90542-8 [DOI] [PubMed] [Google Scholar]
- 234.Cunliffe NA, Glass RI. 1996. Gastrointestinal manifestations of HIV infection. Lancet 348:1037. 10.1016/S0140-6736(05)64970-7 [DOI] [PubMed] [Google Scholar]
- 235.Giordano MO, Martinez LC, Rinaldi D, Espul C, Martinez N, Isa MB, Depetris AR, Medeot SI, Nates SV. 1999. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res. Hum. Retroviruses 15:1427–1432. 10.1089/088922299309937 [DOI] [PubMed] [Google Scholar]
- 236.Gray JJ, Wreghitt TG, Cubitt WD, Elliot PR. 1987. An outbreak of gastroenteritis in a home for the elderly associated with astrovirus type 1 and human calicivirus. J. Med. Virol. 23:377–381. 10.1002/jmv.1890230410 [DOI] [PubMed] [Google Scholar]
- 237.Naficy AB, Rao MR, Holmes JL, Abu-Elyazeed R, Savarino SJ, Wierzba TF, Frenck RW, Monroe SS, Glass RI, Clemens JD. 2000. Astrovirus diarrhea in Egyptian children. J. Infect. Dis. 182:685–690. 10.1086/315763 [DOI] [PubMed] [Google Scholar]
- 238.Guerrero ML, Noel JS, Mitchell DK, Calva JJ, Morrow AL, Martinez J, Rosales G, Velazquez FR, Monroe SS, Glass RI, Pickering LK, Ruiz-Palacios GM. 1998. A prospective study of astrovirus diarrhea of infancy in Mexico City. Pediatr. Infect. Dis. J. 17:723–727. 10.1097/00006454-199808000-00012 [DOI] [PubMed] [Google Scholar]
- 239.Herrmann JE, Taylor DN, Echeverria P, Blacklow NR. 1991. Astroviruses as a cause of gastroenteritis in children. N. Engl. J. Med. 324:1757–1760. 10.1056/NEJM199106203242501 [DOI] [PubMed] [Google Scholar]
- 240.Gaggero A, O'Ryan M, Noel JS, Glass RI, Monroe SS, Mamani N, Prado V, Avendano LF. 1998. Prevalence of astrovirus infection among Chilean children with acute gastroenteritis. J. Clin. Microbiol. 36:3691–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Nguyen TA, Hoang L, Pham le D, Hoang KT, Mizuguchi M, Okitsu S, Ushijima H. 2008. Identification of human astrovirus infections among children with acute gastroenteritis in the Southern Part of Vietnam during 2005-2006. J. Med. Virol. 80:298–305. 10.1002/jmv.21032 [DOI] [PubMed] [Google Scholar]
- 242.Bon F, Fascia P, Dauvergne M, Tenenbaum D, Planson H, Petion AM, Pothier P, Kohli E. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mustafa H, Palombo EA, Bishop RF. 2000. Epidemiology of astrovirus infection in young children hospitalized with acute gastroenteritis in Melbourne, Australia, over a period of four consecutive years, 1995 to 1998. J. Clin. Microbiol. 38:1058–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bosch A, Guix S, Pintó RM. 2013. Epidemiology of human astrovirus, p 1–18 In Schultz-Cherry S. (ed), Astrovirus research: essential ideas, everyday impacts, future directions. Springer, New York, NY [Google Scholar]
- 245.Basu G, Rossouw J, Sebunya TK, Gashe BA, de Beer M, Dewar JB, Steele AD. 2003. Prevalence of rotavirus, adenovirus and astrovirus infection in young children with gastroenteritis in Gaborone, Botswana. East Afr. Med. J. 80:652–655. 10.4314/eamj.v80i12.8783 [DOI] [PubMed] [Google Scholar]
- 246.Kiulia NM, Mwenda JM, Nyachieo A, Nyaundi JK, Steele AD, Taylor MB. 2007. Astrovirus infection in young Kenyan children with diarrhoea. J. Trop. Pediatr. 53:206–209. 10.1093/tropej/fml093 [DOI] [PubMed] [Google Scholar]
- 247.Reither K, Ignatius R, Weitzel T, Seidu-Korkor A, Anyidoho L, Saad E, Djie-Maletz A, Ziniel P, Amoo-Sakyi F, Danikuu F, Danour S, Otchwemah R, Schreier E, Bienzle U, Stark K, Mockenhaupt F. 2007. Acute childhood diarrhoea in northern Ghana: epidemiological, clinical and microbiological characteristics. BMC Infect. Dis. 7:104. 10.1186/1471-2334-7-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tran A, Talmud D, Lejeune B, Jovenin N, Renois F, Payan C, Leveque N, Andreoletti L. 2010. Prevalence of rotavirus, adenovirus, norovirus, and astrovirus infections and coinfections among hospitalized children in Northern France. J. Clin. Microbiol. 48:1943–1946. 10.1128/JCM.02181-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Utagawa ET, Nishizawa S, Sekine S, Hayashi Y, Ishihara Y, Oishi I, Iwasaki A, Yamashita I, Miyamura K, Yamazaki S. 1994. Astrovirus as a cause of gastroenteritis in Japan. J. Clin. Microbiol. 32:1841–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chan-it W, Thongprachum A, Okitsu S, Mizuguchi M, Ushijima H. 2010. Epidemiology and molecular characterization of sapovirus and astrovirus in Japan, 2008-2009. Jpn. J. Infect. Dis. 63:302–303 [PubMed] [Google Scholar]
- 251.Martínez A, Torner N, Broner S, Bartolomé R, Guix S, de Simón M, Godoy P, Moreno A, Company M, Balanyà PJ, Sala MR, Torra R, Ferrús G, Parrón I, Barrabeig I, Domínguez A. 2013. Norovirus: a growing cause of gastroenteritis in Catalonia (Spain)? J. Food Prot. 76:1810–1816. 10.4315/0362-028X.JFP-12-544 [DOI] [PubMed] [Google Scholar]
- 252.Tanaka H, Kisielius JJ, Ueda M, Glass RI, Joazeiro PP. 1994. Intrafamilial outbreak of astrovirus gastroenteritis in Sao Paulo, Brazil. J. Diarrhoeal Dis. Res. 12:219–221 [PubMed] [Google Scholar]
- 253.Desai R, Hall AJ, Lopman BA, Shimshoni Y, Rennick M, Efron N, Matias Y, Patel MM, Parashar UD. 2012. Norovirus disease surveillance using google internet query share data. Clin. Infect. Dis. 55:e75–78. 10.1093/cid/cis579 [DOI] [PubMed] [Google Scholar]
- 254.Phillips G, Tam CC, Conti S, Rodrigues LC, Brown D, Iturriza-Gomara M, Gray J, Lopman B. 2010. Community incidence of norovirus-associated infectious intestinal disease in England: improved estimates using viral load for norovirus diagnosis. Am. J. Epidemiol. 171:1014–1022. 10.1093/aje/kwq021 [DOI] [PubMed] [Google Scholar]
- 255.Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, Van Duynhoven Y, Gray J, Iturriza M, Bottiger B, Falkenhorst G, Johnsen C, von Bonsdorff CH, Maunula L, Kuusi M, Pothier P, Gallay A, Schreier E, Hohne M, Koch J, Szucs G, Reuter G, Krisztalovics K, Lynch M, McKeown P, Foley B, Coughlan S, Ruggeri FM, Di Bartolo I, Vainio K, Isakbaeva E, Poljsak-Prijatelj M, Grom AH, Mijovski JZ, Bosch A, Buesa J, Fauquier AS, Hernandez-Pezzi G, Hedlund KO, Koopmans M. 2008. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the foodborne viruses in Europe network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 46:2959–2965. 10.1128/JCM.00499-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lee TW, Kurtz JB. 1994. Prevalence of human astrovirus serotypes in the Oxford region 1976-92, with evidence for two new serotypes. Epidemiol. Infect. 112:187–193. 10.1017/S0950268800057551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Steele AD, Basetse HR, Blacklow NR, Herrmann JE. 1998. Astrovirus infection in South Africa: a pilot study. Ann. Trop. Paediatr. 18:315–319 [DOI] [PubMed] [Google Scholar]
- 258.Dey SK, Ushijima H, Phathammavong O, Chanit W, Okitsu S, Mizuguchi M, Ota Y. 2010. Seasonal trend and serotype distribution of rotavirus infection in Japan, 1981-2008. Pediatr. Infect. Dis. J. 29:166–167. 10.1097/INF.0b013e3181b79460 [DOI] [PubMed] [Google Scholar]
- 259.Lopman B, Zambon M, Brown DW. 2008. The evolution of norovirus, the “gastric flu.” PLoS Med. 5:e42. 10.1371/journal.pmed.0050042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Parashar UD, Glass RI. 2003. Viral causes of gastroenteritis, p 9–21 In Desselberger U, Gray J. (ed), Perspectives in medical virology, vol 9 Elsevier, Amsterdam, The Netherlands: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kurtz JB, Lee TW. 1984. Human astrovirus serotypes. Lancet ii:1405. [DOI] [PubMed] [Google Scholar]
- 262.Méndez-Toss M, Romero-Guido P, Munguía ME, Méndez E, Arias CF. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891–2897 [DOI] [PubMed] [Google Scholar]
- 263.Silva PA, Cardoso DD, Schreier E. 2006. Molecular characterization of human astroviruses isolated in Brazil, including the complete sequences of astrovirus genotypes 4 and 5. Arch. Virol. 151:1405–1417. 10.1007/s00705-005-0704-9 [DOI] [PubMed] [Google Scholar]
- 264.De Grazia S, Giammanco GM, Colomba C, Cascio A, Arista S. 2004. Molecular epidemiology of astrovirus infection in Italian children with gastroenteritis. Clin. Microbiol. Infect. 10:1025–1029. 10.1111/j.1469-0691.2004.00995.x [DOI] [PubMed] [Google Scholar]
- 265.Gabbay YB, Linhares AC, Cavalcante-Pepino EL, Nakamura LS, Oliveira DS, da Silva LD, Mascarenhas JD, Oliveira CS, Monteiro TA, Leite JP. 2007. Prevalence of human astrovirus genotypes associated with acute gastroenteritis among children in Belem, Brazil. J. Med. Virol. 79:530–538. 10.1002/jmv.20813 [DOI] [PubMed] [Google Scholar]
- 266.Mitchell DK, Matson DO, Jiang X, Berke T, Monroe SS, Carter MJ, Willcocks MM, Pickering LK. 1999. Molecular epidemiology of childhood astrovirus infection in child care centers. J. Infect. Dis. 180:514–517. 10.1086/314863 [DOI] [PubMed] [Google Scholar]
- 267.Espul C, Martinez N, Noel JS, Cuello H, Abrile C, Grucci S, Glass R, Berke T, Matson DO. 2004. Prevalence and characterization of astroviruses in Argentinean children with acute gastroenteritis. J. Med. Virol. 72:75–82. 10.1002/jmv.10537 [DOI] [PubMed] [Google Scholar]
- 268.Noel J, Mansoor A, Thaker U, Herrmann J, Perron-Henry D, Cubitt WD. 1994. Identification of adenoviruses in faeces from patients with diarrhoea at the Hospitals for Sick Children, London, 1989-1992. J. Med. Virol. 43:84–90. 10.1002/jmv.1890430116 [DOI] [PubMed] [Google Scholar]
- 269.Unicomb LE, Banu NN, Azim T, Islam A, Bardhan PK, Faruque AS, Hall A, Moe CL, Noel JS, Monroe SS, Albert MJ, Glass RI. 1998. Astrovirus infection in association with acute, persistent and nosocomial diarrhea in Bangladesh. Pediatr. Infect. Dis. J. 17:611–614. 10.1097/00006454-199807000-00007 [DOI] [PubMed] [Google Scholar]
- 270.Lee TW, Kurtz JB. 1981. Serial propagation of astrovirus in tissue culture with the aid of trypsin. J. Gen. Virol. 57:421–424. 10.1099/0022-1317-57-2-421 [DOI] [PubMed] [Google Scholar]
- 271.Noel J, Cubitt D. 1994. Identification of astrovirus serotypes from children treated at the Hospitals for Sick Children, London 1981-93. Epidemiol. Infect. 113:153–159. 10.1017/S0950268800051578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Herrmann JE, Nowak NA, Blacklow NR. 1985. Detection of Norwalk virus in stools by enzyme immunoassay. J. Med. Virol. 17:127–133. 10.1002/jmv.1890170205 [DOI] [PubMed] [Google Scholar]
- 273.Herrmann JE, Nowak NA, Perron-Henry DM, Hudson RW, Cubitt WD, Blacklow NR. 1990. Diagnosis of astrovirus gastroenteritis by antigen detection with monoclonal antibodies. J. Infect. Dis. 161:226–229. 10.1093/infdis/161.2.226 [DOI] [PubMed] [Google Scholar]
- 274.Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Moe CL, Allen JR, Monroe SS, Gary HE, Jr, Humphrey CD, Herrmann JE, Blacklow NR, Carcamo C, Koch M, Kim KH. 1991. Detection of astrovirus in pediatric stool samples by immunoassay and RNA probe. J. Clin. Microbiol. 29:2390–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Putzker M, Sauer H, Kirchner G, Keksel O, Malic A. 2000. Community acquired diarrhea—the incidence of Astrovirus infections in Germany. Clin. Lab. 46:269–273 [PubMed] [Google Scholar]
- 277.McIver CJ, Palombo EA, Doultree JC, Mustafa H, Marshall JA, Rawlinson WD. 2000. Detection of astrovirus gastroenteritis in children. J. Virol. Methods 84:99–105. 10.1016/S0166-0934(99)00133-0 [DOI] [PubMed] [Google Scholar]
- 278.Rovida F, Campanini G, Sarasini A, Adzasehoun KMG, Piralla A, Baldanti F. 2013. Comparison of immunologic and molecular assays for the diagnosis of gastrointestinal viral infections. Diagn. Microbiol. Infect. Dis. 75:110–111. 10.1016/j.diagmicrobio.2012.09.016 [DOI] [PubMed] [Google Scholar]
- 279.Huhulescu S, Kiss R, Brettlecker M, Cerny RJ, Hess C, Wewalka G, Allerberger F. 2009. Etiology of acute gastroenteritis in three sentinel general practices, Austria 2007. Infection 37:103–108. 10.1007/s15010-008-8106-z [DOI] [PubMed] [Google Scholar]
- 280.Khamrin P, Dey SK, Chan-it W, Thongprachum A, Satou K, Okitsu S, Maneekarn N, Ushijima H. 2010. Evaluation of a rapid immunochromatography strip test for detection of astrovirus in stool specimens. J. Trop. Pediatr. 56:129–131. 10.1093/tropej/fmp055 [DOI] [PubMed] [Google Scholar]
- 281.Romalde JL, Ribao C, Luz Vilarino M, Barja JL. 2004. Comparison of different primer sets for the RT-PCR detection of hepatitis A virus and astrovirus in mussel tissues. Water Sci. Technol. 50:131–136 [PubMed] [Google Scholar]
- 282.Guix S, Bosch A, Pinto RM. 2005. Human astrovirus diagnosis and typing: current and future prospects. Lett. Appl. Microbiol. 41:103–105. 10.1111/j.1472-765X.2005.01759.x [DOI] [PubMed] [Google Scholar]
- 283.Brown DW, Gunning KB, Henry DM, Awdeh ZL, Brinker JP, Tzipori S, Herrmann JE. 2008. A DNA oligonucleotide microarray for detecting human astrovirus serotypes. J. Virol. Methods 147:86–92. 10.1016/j.jviromet.2007.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Kim J-M, Kim S, Park Y, Kim H, Min B, Cho J-C, Yang J, Cho Y-H, Ko G. 2012. Simultaneous detection of major enteric viruses using a combimatrix microarray. J. Microbiol. 50:970–977. 10.1007/s12275-012-2228-9 [DOI] [PubMed] [Google Scholar]
- 285.Tai JH, Ewert MS, Belliot G, Glass RI, Monroe SS. 2003. Development of a rapid method using nucleic acid sequence-based amplification for the detection of astrovirus. J. Virol. Methods 110:119–127. 10.1016/S0166-0934(03)00108-3 [DOI] [PubMed] [Google Scholar]
- 286.Royuela E, Negredo A, Sanchez-Fauquier A. 2006. Development of a one step real-time RT-PCR method for sensitive detection of human astrovirus. J. Virol. Methods 133:14–19. 10.1016/j.jviromet.2005.10.012 [DOI] [PubMed] [Google Scholar]
- 287.Dai Y-C, Xu Q-H, Wu X-B, Hu G-F, Tang Y-L, Li J-D, Chen Q, Nie J. 2010. Development of real-time and nested RT-PCR to detect astrovirus and one-year survey of astrovirus in Jiangmen City, China. Arch. Virol. 155:977–982. 10.1007/s00705-010-0664-6 [DOI] [PubMed] [Google Scholar]
- 288.Zhang Z, Mitchell DK, Afflerbach C, Jakab F, Walter J, Zhang YJ, Staat MA, Azimi P, Matson DO. 2006. Quantitation of human astrovirus by real-time reverse-transcription-polymerase chain reaction to examine correlation with clinical illness. J. Virol. Methods 134:190–196. 10.1016/j.jviromet.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 289.Sano D, Pintó RM, Omura T, Bosch A. 2010. Detection of oxidative damages on viral capsid protein for evaluating structural integrity and infectivity of human norovirus. Environ. Sci. Technol. 44:808–812. 10.1021/es9018964 [DOI] [PubMed] [Google Scholar]
- 290.Wei H, Zeng J, Deng C, Zheng C, Zhang X, Ma D, Yi Y. 2013. A novel method of real-time reverse-transcription loop-mediated isothermal amplification developed for rapid and quantitative detection of human astrovirus. J. Virol. Methods 188:126–131. 10.1016/j.jviromet.2012.11.040 [DOI] [PubMed] [Google Scholar]
- 291.Liu Y, Xu Z-Q, Zhang Q, Jin M, Yu J-M, Li J-S, Liu N, Cui S-X, Kong X-Y, Wang H, Li H-Y, Cheng W-X, Ma X-J, Duan Z-J. 2012. Simultaneous detection of seven enteric viruses associated with acute gastroenteritis by a multiplexed luminex-based assay. J. Clin. Microbiol. 50:2384–2389. 10.1128/JCM.06790-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.van Maarseveen NM, Wessels E, de Brouwer CS, Vossen AC, Claas EC. 2010. Diagnosis of viral gastroenteritis by simultaneous detection of adenovirus group F, astrovirus, rotavirus group A, norovirus genogroups I and II, and sapovirus in two internally controlled multiplex real-time PCR assays. J. Clin. Virol. 49:205–210. 10.1016/j.jcv.2010.07.019 [DOI] [PubMed] [Google Scholar]
- 293.Mori K, Hayashi Y, Akiba T, Nagano M, Tanaka T, Hosaka M, Nakama A, Kai A, Saito K, Shirasawa H. 2013. Multiplex real-time PCR assays for the detection of group C rotavirus, astrovirus, and Subgenus F adenovirus in stool specimens. J. Virol. Methods 191:141–147. 10.1016/j.jviromet.2012.10.019 [DOI] [PubMed] [Google Scholar]
- 294.Jiang Y, Fang L, Shi X, Zhang H, Li Y, Lin Y, Qiu Y, Chen Q, Li H, Zhou L, Hu Q. 2014. Simultaneous detection of five enteric viruses associated with gastroenteritis using a PCR assay: a single real-time multiplex reaction and its clinical application J. Clin. Microbiol. 52:1266–1268. 10.1128/JCM.00245-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, Verweij JJ, Taniuchi M, Sobuz SU, Haque R, Haverstick DM, Houpt ER. 2013. A laboratory-developed TaqMan array card for simultaneous detection of 19 enteropathogens. J. Clin. Microbiol. 51:472–480. 10.1128/JCM.02658-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Pinto RM, Diez JM, Bosch A. 1994. Use of the colonic carcinoma cell line CaCo-2 for in vivo amplification and detection of enteric viruses. J. Med. Virol. 44:310–315. 10.1002/jmv.1890440317 [DOI] [PubMed] [Google Scholar]
- 297.Grimm AC, Cashdollar JL, Williams FP, Fout GS. 2004. Development of an astrovirus RT-PCR detection assay for use with conventional, real-time, and integrated cell culture/RT-PCR. Can. J. Microbiol. 50:269–278. 10.1139/w04-012 [DOI] [PubMed] [Google Scholar]
- 298.Le Guyader FS, Le Saux JC, Ambert-Balay K, Krol J, Serais O, Parnaudeau S, Giraudon H, Delmas G, Pommepuy M, Pothier P, Atmar RL. 2008. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J. Clin. Microbiol. 46:4011–4017. 10.1128/JCM.01044-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Kingsley DH. 2013. High pressure processing and its application to the challenge of virus-contaminated foods. Food Environ. Virol. 5:1–12. 10.1007/s12560-012-9094-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Oms-Oliu G, Martín-Belloso O, Soliva-Fortuny R. 2010. Pulsed light treatments for food preservation. A review. Food Bioprocess Technol. 3:13–23. 10.1007/s11947-008-0147-x [DOI] [Google Scholar]
- 301.Kurtz JB, Lee TW, Parsons AJ. 1980. The action of alcohols on rotavirus, astrovirus and enterovirus. J. Hosp. Infect. 1:321–325. 10.1016/0195-6701(80)90008-0 [DOI] [PubMed] [Google Scholar]
- 302.Caballero S, Guix S, Ribes E, Bosch A, Pinto RM. 2004. Structural requirements of astrovirus virus-like particles assembled in insect cells. J. Virol. 78:13285–13292. 10.1128/JVI.78.23.13285-13292.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Dalton RM, Pastrana EP, Sánchez-Fauquier A. 2003. Vaccinia virus recombinant expressing an 87-kilodalton polyprotein that is sufficient to form astrovirus-like particles. J. Virol. 77:9094–9098. 10.1128/JVI.77.16.9094-9098.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Colbère-Garapin F, Martin-Latil S, Blondel B, Mousson L, Pelletier I, Autret A, François A, Niborski V, Grompone G, Catonnet G, van de Moer A. 2007. Prevention and treatment of enteric viral infections: possible benefits of probiotic bacteria. Microbes Infect. 9:1623–1631. 10.1016/j.micinf.2007.09.016 [DOI] [PubMed] [Google Scholar]
- 305.Britton RA, Versalovic J. 2008. Probiotics and gastrointestinal infections. Interdiscip. Perspect. Infect. Dis. 2008:10. 10.1155/2008/290769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Fodha I, Chouikha A, Peenze I, De Beer M, Dewar J, Geyer A, Messaadi F, Trabelsi A, Boujaafar N, Taylor MB, Steele D. 2006. Identification of viral agents causing diarrhea among children in the Eastern Center of Tunisia. J. Med. Virol. 78:1198–1203. 10.1002/jmv.20681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Fang ZY, Sun YP, Ye XH, Wang H, Zhang Q, Duan ZJ, Xi J, Steele D, Glass R. 2006. Astrovirus infection among hospitalized children with acute diarrhea in seven regions of China, 1998-2005. Zhonghua Liu Xing Bing Xue Za Zhi 27:673–676 [PubMed] [Google Scholar]
- 308.Verma H, Chitambar SD, Gopalkrishna V. 2010. Astrovirus associated acute gastroenteritis in western India: predominance of dual serotype strains. Infect. Genet. Evol. 10:575–579. 10.1016/j.meegid.2010.01.008 [DOI] [PubMed] [Google Scholar]
- 309.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 311.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]