Abstract
SUMMARY
In this review we examine the literature related to emerging technologies that will help to reshape the clinical microbiology laboratory. These topics include nucleic acid amplification tests such as isothermal and point-of-care molecular diagnostics, multiplexed panels for syndromic diagnosis, digital PCR, next-generation sequencing, and automation of molecular tests. We also review matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) and electrospray ionization (ESI) mass spectrometry methods and their role in identification of microorganisms. Lastly, we review the shift to liquid-based microbiology and the integration of partial and full laboratory automation that are beginning to impact the clinical microbiology laboratory.
INTRODUCTION
Despite technological advances in laboratory diagnostics, the clinical microbiology laboratory continues to rely heavily on traditional methods, including culture, phenotypic, and biochemical tests, to identify microorganisms present in clinical specimens. This is due, in part, to the complexity and variability of specimens received by the clinical laboratory. The specimen type and test order dictate the processing and culture medium that are used for bacterial and fungal culture, and they also play a role in interpretation of culture results. Much of clinical virology has shifted to tests based on molecular methods due to the increased sensitivity and specificity and reduced turnaround time (TAT) compared with those for viral culture. This shift has also resulted in reduced labor by eliminating time-consuming tasks, including maintenance of permissive host cell lines, repeated microscopic examination, and immunostaining, associated with viral culture. Historically, nucleic acid amplification tests (NAATs) for both viral and bacterial etiologies were largely considered “high-complexity” tests and were limited to molecular laboratories staffed with skilled technologists. Many molecular tests used by clinical laboratories are still developed in-house or utilize analyte-specific reagents (ASRs) and are considered laboratory-developed tests (LDTs). These tests, as well as many U.S. Food and Drug Administration (FDA)-cleared tests, require offline nucleic acid extraction and addition of several reagents to make PCR “master mixes.” The multistep process can make these assays laborious to set up and allow for the introduction of contamination at several steps. Advances in technology such as real-time PCR (RT-PCR), quantitative PCR (qPCR), and automation in the form of sample-to-result instrumentation have alleviated some of these issues. Automation and simplification of molecular assays have led to FDA-cleared assays categorized as “moderate complexity,” which facilitates adoption by smaller laboratories or those less well staffed. Multiplex tests are now available that enable single specimens to be interrogated for the presence of multiple pathogens associated with various clinical syndromes. Digital PCR and next-generation sequencing (NGS) have pushed the landscape of molecular diagnostics further, allowing for analysis of complex, polymicrobial specimens and enabling accurate quantification of organisms present as <0.01% of the microbial consortium in a specimen. For specimens which are still best analyzed using culture, automation of primary processing and plating, coupled with initial culture examination aided by high-resolution optics, has reduced time spent on mundane tasks associated with the initial steps of clinical bacteriology and improved laboratory efficiency. Meanwhile, rapid and accurate identification of these cultured microorganisms is made possible using mass spectrometry (MS).
While these advances aim to improve laboratory performance and efficiency and the quality of patient care, they are not without drawbacks. Higher levels of automation of preanalytic and postanalytic processes can potentially diminish technologist skill sets in those areas through attrition and loss of familiarity with basic skills and concepts, such as the qualitative and quantitative streaking of culture media or appropriate work practices to mitigate the risk of contamination when working with molecular assays. The challenge surrounding education of technologists is to learn new skills while maintaining expertise in classic techniques. The transition from viral culture to largely molecular techniques has been the best documented case study in embracing new technologies. In virology, culture of many viruses is difficult or viruses cannot be grown at all, while other viruses require specialized culture systems that are either not available or too complicated (1). Traditional tube cultures, although comprehensive, fail to isolate viruses in many instances and can take days to weeks to provide a final result. In contrast, molecular assays allow the early detection of pathogens prior to development of an immune response or before a virus can be grown or its antigens detected. This can result, according to Hodinka (1), in “an early and accurate diagnosis that can have a prompt and significant impact on patient care by providing timely treatment that may limit the extent of disease and reduce associated sequelae and by reducing or eliminating unnecessary hospitalization, diagnostic procedures, inappropriate use of antimicrobial agents, and associated costs.” The resulting change has reinvigorated the clinical impact of results and is allowing physicians to make informed decisions regarding therapeutic management rather than empirical guesses (1). With these techniques, turnaround has improved and sensitivity has increased, attributes that few would disagree with. However, the transition to molecular biology has brought viral culture near to extinction in the clinical laboratory. Many trainees in laboratory science are no longer educated in viral cytopathic effect, tissue culture, or reading of viral cultures. In contrast, in areas such as parasitology and mycology, there remains a comparative lack of novel methods for rapid identification of pathogens. In these areas it will be important to maintain the traditional skills of clinical microbiologists until new technologies are more widely available and are fully vetted. Similarly, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS has demonstrated considerable improvement in accuracy, cost effectiveness, and timeliness of bacterial and yeast identification; however, limitations such as the differentiation of Escherichia coli from Shigella spp. and identification of organisms not well represented in commercially available reference libraries have been well documented (2).Therefore, a combination of new technologies and classic techniques is central to the successful accurate identification of all microorganisms encountered in the laboratory. This supports the need to maintain traditional microbiological skills.
In this review, we examine current literature related to emerging technologies that will help to reshape workflow and improve the quality of results provided by the clinical microbiology laboratory.
MOLECULAR METHODS (NUCLEIC ACID BASED)
Molecular methods, including PCR, microarray, and nucleic acid sequencing, have taken a prominent place in the clinical laboratory. These methods provide sensitive and specific identification of microorganisms or genetic polymorphisms through amplification and detection of specific nucleic acid targets. Recent advances in high-density or massively parallel sequencing technologies have removed the limitation of a priori target selection inherent to traditional PCR/probe-based assays and as such have broadened the diagnostic capabilities of these tests. Regardless of methodology, molecular diagnostics have the capability to reduce the time to results and provide more accurate diagnosis. Despite these clear advantages, molecular diagnostic methods are not without drawbacks.
Inherent to all nucleic acid amplification and non-culture-based methods is the lack of a suitable “gold standard” for comparison. Molecular and amplified nucleic acid methods are often more sensitive than the culture methods to which they are being compared. This can be problematic during validation of new molecular tests when specimens are NAAT positive but culture negative. One solution is to use clinical diagnosis as a gold standard, but it can often be difficult to reach a definitive clinical diagnosis when symptoms are nonspecific (e.g., with viral respiratory illness). Alternative methods to validate a new molecular test include the use of well-characterized reference samples or a second validated molecular test which targets a genetic sequence different from the sequence targeted by the test undergoing validation (3–5). For an excellent review of challenges and methods for validation of molecular diagnostic tests, the reader is directed to a review by Burd (3). Still, it is important to recognize that these approaches only confirm the presence of a nucleic acid target and do not prove the presence of a viable organism. In the absence of culture positivity, it is impossible to conclusively rule out nucleic acid (template or amplicon) contamination or the detection of nonviable organisms which are not significant in making a diagnosis. Therefore, interpretation of NAAT-positive, culture-negative results can be challenging even following a rigorous laboratory validation.
In addition to the different chemistries and approaches used by molecular assays, it is also worth considering the variety of platforms on which these assays are designed to run. These platforms can be available as “open” or “closed” systems. Closed-system platforms are designed to run specific assays which are cleared by regulatory agencies, including the U.S. Food and Drug Administration (FDA), the European health, safety, and environmental agency (CE-Mark) and Heath Canada. Examples include many of the “sample-to-result” platforms such as GeneXpert (Cepheid, Sunnyvale, CA), FilmArray (BioFire, Salt Lake City, UT), Tigris (GenProbe, San Diego, CA), and Verigene (Nanosphere, Northbrook, IL). Many of these closed-platform tests can be simplified to gain designation as “moderate complexity,” and as such, the end user has limited ability to modify the assay or result interpretation. Open-system platforms available for real-time and quantitative PCR analysis include the SmartCycler (Cepheid), ABI 7500FastDx (Applied Biosystems), and LightCycler 2.0 (Roche). There are also automated or “sample-to-result” open platforms available, including the BD Max (BD, Sparks, MD) and Abbott m2000 (Abbott, North Chicago, IL). FDA-cleared molecular assays for use on these platforms may be available from the manufacturer of the platform or another diagnostics company; however, the platforms are also suitable for running laboratory-developed tests (LDTs) or “home brew” assays. While the menu of FDA-cleared in vitro diagnostic (IVD) molecular assays continues to expand, the ability of laboratories to develop and validate their own assays is critical to providing high-quality molecular diagnostics for novel or esoteric targets, including those involved in infectious disease. For this reason, open-system platforms will continue to have a prominent place in most clinical laboratories.
With these considerations in mind, we highlight several approaches to nucleic acid detection, including amplification and nonamplification methods for singleplex and multiplex detection of microorganisms.
Singleplex Nucleic Acid Tests
Nucleic acid amplification, including PCR and TMA.
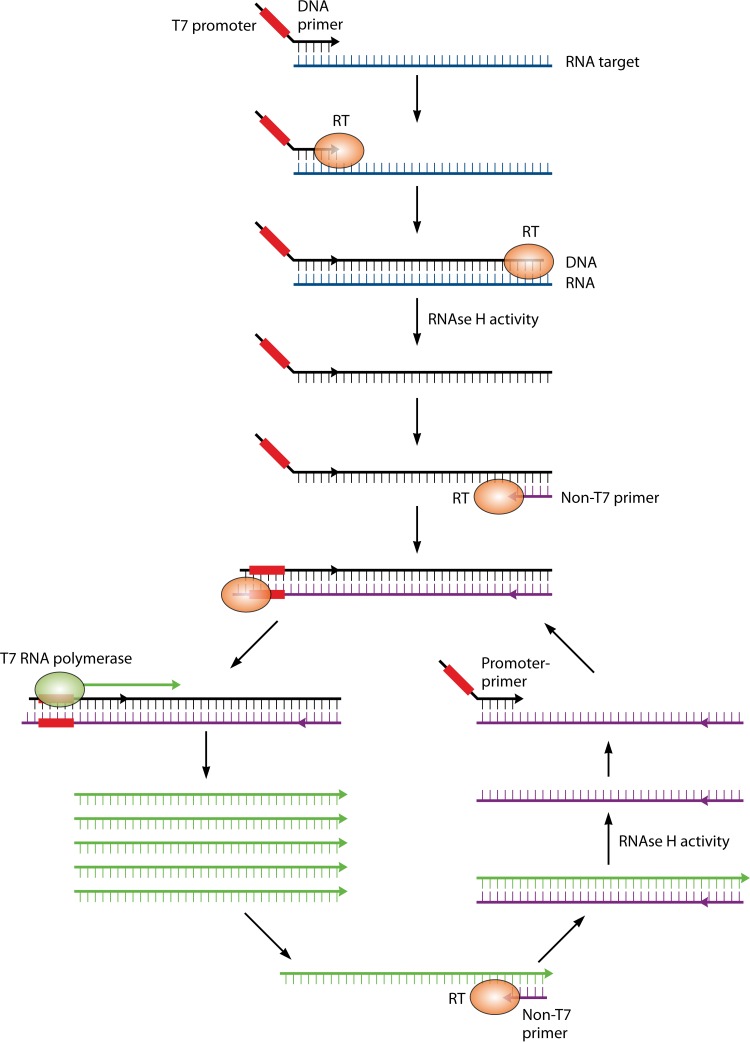
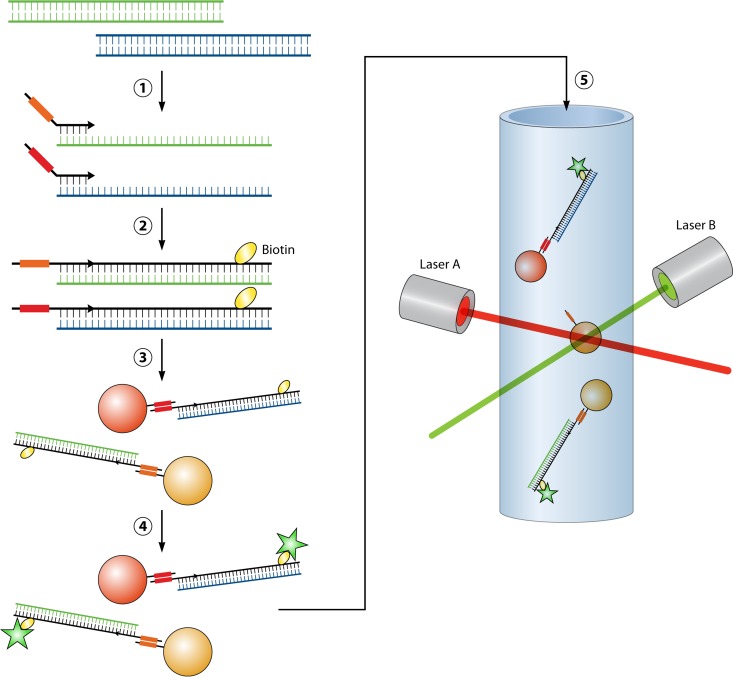
Nucleic acid amplification using thermostable polymerase (PCR) was initially reported in 1988, and this method remains largely unchanged as it forms the backbone of molecular diagnostics in clinical microbiology laboratories today (6). Properties such as high sensitivity and specificity, an extremely low limit of detection (1 to 10 copies of the target), and rapid results have led to proposed changes in the definition of the “gold standard” method for detection and identification of microorganisms in clinical specimens, especially for those that are difficult to culture, including fastidious bacterial or viral pathogens (7–10). While the basic principle of nucleic acid amplification tests (NAATs) has not changed, technologies surrounding this core, including amplification strategy, amplicon detection, multiplexing of reactions, and automation of the entire process into sample-to-result platforms, have provided a large menu of options for the molecular microbiology laboratory to choose from. One such modification is the departure from PCR-based amplification to transcription-mediated amplification (TMA) of a nucleic acid target. This method differs from PCR in that the target sequence is typically an RNA molecule (mRNA or rRNA), which may be present at a high copy number in the cell. Reverse transcriptase and engineered oligonucleotide primers are used to simultaneously generate a cDNA template and incorporate a promoter sequence recognized by a highly efficient, phage-encoded RNA polymerase enzyme. This enzyme enables isothermal synthesis of 100 to 1,000 copies of each starting template cDNA, which are in turn used as the template for subsequent rounds of amplification (11) (Fig. 1). The multicopy nature of the RNA target and ability to amplify beyond a log-linear rate without the need for thermocycling theoretically increase the speed and sensitivity of TMA compared to that of standard PCR. To date, the most widely used molecular assays target a single or few analytes, employing one or few oligonucleotide primer sets (11–13). Using target amplification coupled with fluorescence probe-based detection, these tests provide a mechanism for rapid and sensitive diagnostic tests.
FIG 1.
Transcription-mediated amplification (TMA). The single-stranded RNA target is bound by a cDNA primer engineered to contain a T7 viral RNA polymerase promoter sequence (red box). Reverse transcriptase (RT) extends the DNA primer to form an RNA-cDNA duplex, and the RNA template strand is degraded by RNase H activity. A second primer anneals to the single-stranded cDNA (black) and is extended by RT, which incorporates the T7 promoter into the double-stranded DNA sequence. T7 RNA polymerase recognizes the incorporated T7 promoter sequence and synthesizes 100 to 1,000 copies of single-strand RNA amplicon (green). These amplicons serve both as a target for detection probes and as a single-stranded template for subsequent rounds of amplification using the non-T7 primer to initiate cDNA synthesis by RT.
The majority of molecular tests in use today are qualitative tests. Qualitative tests are best suited for the detection of microorganisms in specimens whose presence, at any level, is associated with a disease state. This includes microorganism that are not regarded as normal flora, as well as any organism isolated from a sterile site. A prime illustration is the use of NAATs for the detection of microorganisms associated with sexually transmitted illnesses, including Neisseria gonorrhoeae, Chlamydia trachomatis, Trichomonas vaginalis, and Mycoplasma genitalium. Culture of these organisms is either impractical or unreliable due to loss of viability during transport, which further decreases the sensitivity of culture methods. NAATs have demonstrated increased sensitivity compared to that of culture methods and dramatically reduced turnaround time for detection of these pathogens (12, 14–16). This enables more rapid, accurate identification of the pathogen(s) responsible for nonspecific symptoms of urethritis or pelvic inflammatory disease and also may aid in limiting the spread of these organisms by identifying asymptomatic carriers. Additionally, the increased sensitivity of NAATs can enable the analysis of specimens obtained by less invasive techniques or of patient-collected specimens, including urine and self-collected vaginal swabs, without affecting the accuracy of the test (12, 13, 17–19). The ability to use these types of specimens can contribute to higher participation in routine screening exams (12, 13, 17–19). Other pathogens commonly identified using qualitative NAATs include respiratory viruses, herpesviruses, Clostridium difficile, Staphylococcus aureus, methicillin-resistant S. aureus (MRSA), Streptococcus pyogenes, Streptococcus agalactiae, Bordetella pertussis, and bacterial pathogens associated with atypical pneumonia. Another use of qualitative tests is to obtain a rapid result for preoperative screening or for infection control purposes. A recent randomized trial compared targeted screening and decolonization of intensive care unit (ICU) patients to a universal decolonization program to reduce the rate of MRSA infection in hospital ICUs (20). While universal decolonization of all patients was associated with the lowest hazard ratio for infection (0.62), targeted screening and decolonization also demonstrated a reduced hazard ratio (0.75). Although screening and targeted decolonization of patients may not be as effective as universal decolonization, studies have demonstrated that sensitive detection of MRSA can significantly reduce the rate of postsurgical infection by identifying those patients who will benefit from preoperative prophylaxis and decolonization (21, 22). As a result, reduced rates of postsurgical infection resulting from molecular screening methods have been shown to reduce the cost of health care to both the hospital and third-party payer (23). Likewise, rapid and accurate detection of MRSA, vancomycin-resistant Enterococcus (VRE), or carbapenem-resistant Enterobacteriaceae (CRE) may aid infection control efforts by identifying those patients requiring contact isolation.
A potential drawback to the use of NAATS is the interpretation of positive results from asymptomatic patients or those who have received appropriate therapy following an initially positive result. While other technologies, including direct microscopy and antigen-based tests, are not immune from this shortcoming, the exquisite sensitivity of the PCR and TMA-based methods used for qualitative NAATs makes these methods most susceptible to potential overreporting of positive results. For these assays, any amount of nucleic acid detected in a specimen is reported as positive, regardless of whether it represents an infectious process due to a live organism, low-level or asymptomatic colonization, or free nucleic acid in the absence of a viable organism. This concern has been highlighted recently by several publications focused on selection of the most appropriate test or algorithm for diagnosis of C. difficile infection (24, 25). Because high rates of asymptomatic carriage of C. difficile are reported among elderly residents in long-term care facilities, it has been proposed that positive NAATs be followed by a direct test for the presence of C. difficile toxins (tcdA and tcdB) to differentiate between carriage and infection (26, 27). Supporting this notion, detection of toxin from patients following a positive NAAT has been correlated with worse prognosis than for patients with a positive NAAT alone (28). Furthermore, NAATs for C. difficile were positive up to 4 weeks following appropriate antibiotic treatment and resolution of symptoms in >50% of patients tested (29). These concerns were addressed directly in a study which demonstrated significantly reduced specificity of molecular modalities when patient symptoms were included as criteria for “gold standard” positive results (30). An excellent review of the diagnostic assays available and difficulties in interpretation of results pertaining to C. difficile has been published (31). Similarly, “pseudo-outbreaks” of Bordetella pertussis have been reported due to the use of NAATs that target the multicopy IS481 chromosomal element (32, 33). In both pseudo-outbreaks, NAAT-positive results could not be confirmed when using an alternative NAAT targeting a single copy genetic target, and 92% to 100% of patients did not meet clinical criteria for pertussis, or were seronegative for antipertussis toxin IgG (32). The effect of these “false-positive” results was the unnecessary prescription of antimicrobial therapy for the patient as well as close contacts and temporary isolation of patients, which constitute a needless financial and social burden on those affected (32). It has been established that NAATs that target the IS481 gene in B. pertussis are capable of detecting <1 organism per sample and that detection at PCR cycle thresholds of >35 has <50% correlation with clinical pertussis disease (33, 34). Therefore, it may be useful to incorporate clinical symptoms and results of other testing modalities when defining a positive cycle threshold for molecular tests (34, 35).
In both examples, positive results may have been due to persistence of nucleic acid or nonviable organism in the specimen. This reinforces the point that molecular assays should be interpreted in the context of clinical presentation and should not be used as a test of cure.
LAMP and HDA technologies.
To maximize the benefits of molecular testing, developers of diagnostics have begun to focus on technologies that employ both simplified technology and simplified specimen preparation in an attempt to bring molecular assays closer to the patient. These technologies have the potential to further reduce TAT, which may positively impact patient care and reduce the overall cost of health care. Isothermal amplification methods, including loop-mediated isothermal amplification (LAMP) and helicase-dependent amplification (HDA), effectively remove the need for expensive thermocyclers and technical optimization of cycling conditions. These methods can be coupled to alternative detection technologies (i.e., fluorescent probe-independent detection methods) that eliminate the need sophisticated optics. This further reduces the cost of instrumentation and enables these tests to be used outside today's “molecular laboratory” and closer to the point of care (POC).
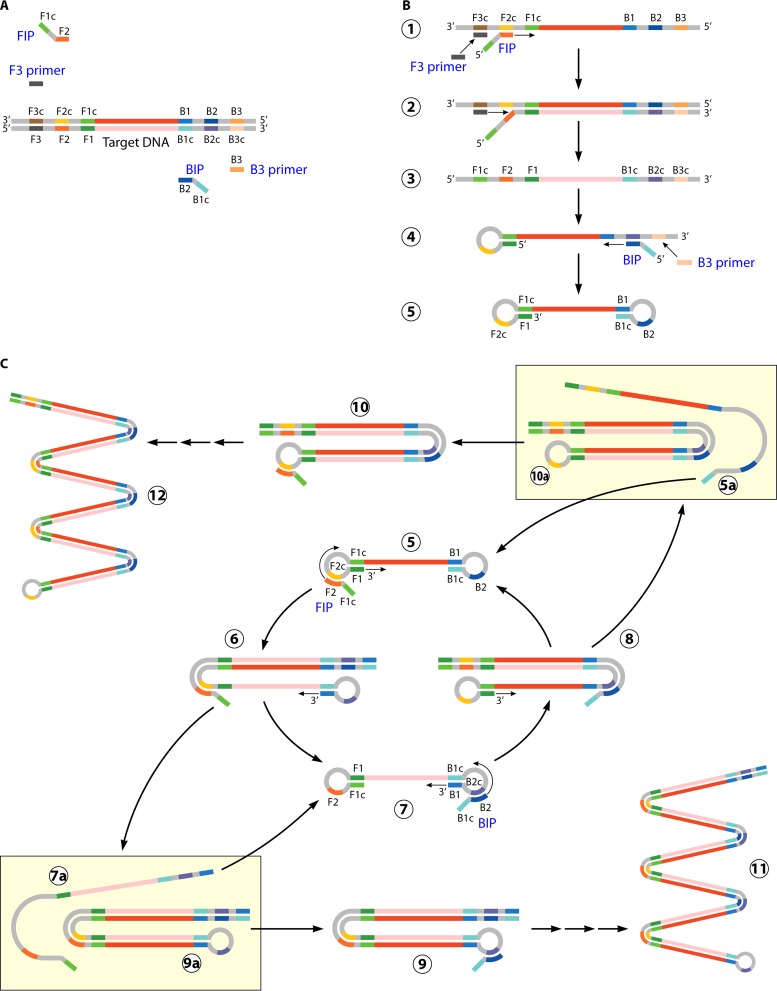
LAMP utilizes 4 primers and 6 recognition (annealing) sites per target to create high levels of amplicon in <60 min. An “inner” set of primers initiates target amplification, while a second, “outer” set of primers initiates a round of replication that displaces the initial product, thus regenerating a single-strand template without the need for heat denaturation (36) (Fig. 2). The use of 6 primers and 4 recognition sites provides specificity higher than that of standard PCRs that utilize only 2 primers. The increased specificity eliminates the need for expensive fluorescence-labeled probes and accompanying optics and allows detection of amplified product based on by-products of DNA replication (37). Pyrophosphate ion, generated by target amplification, can be precipitated by the addition of magnesium ion to the reaction mixture. This enables visual inspection of the reaction tube for turbidity as an indication of a positive result. An increase in the turbidity of the reaction mixture can also be measured in real time using comparatively simple optics to permit the use of LAMP in quantitative assays (38). There are a number of FDA-cleared and laboratory-developed tests that utilize the LAMP technology. FDA-cleared tests utilizing LAMP include those for C. difficile, group A and B Streptococcus, Mycoplasma pneumoniae, and B. pertussis (39, 40). Clinical evaluations of the C. difficile and group A Streptococcus tests have demonstrated sensitivity and specificity similar to those of traditional real-time PCR, though a slight decrease in sensitivity for C. difficile has been noted (39–44). Laboratory-developed and commercially available research-use-only (RUO) tests using LAMP have targeted diverse groups of microorganisms, including Plasmodium spp., Giardia lamblia, Leishmania, Mycobacterium spp., and hepatitis viruses (45–50). Specifically, LAMP-based testing for Plasmodium spp. and Plasmodium falciparum demonstrated >97% sensitivity and >98% specificity compared to nested PCR in patients with parasitemia of ≥1 parasite/μl and was significantly more sensitive than standard microscopy (49, 50). The use of heat-treated whole blood rather than extracted nucleic acid, a simple heat block or water bath to maintain 60 to 65°C for isothermal target amplification, and visual determination of a positive result based on turbidity give LAMP an advantage over traditional PCR methods in resource-limited regions of the world, including many countries where malaria is endemic (51, 52). Further, the use of a pocket warmer (exothermic chemical reaction pouch) to drive LAMP maintained 90.5% sensitivity for detection of Mycobacterium ulcerans compared the same test run using a powered heat block (48). A major limitation of LAMP is the inability to multiplex. This is due to the nonspecific and indirect turbidity-based detection of the amplicon. Still, the noted advantages of inexpensive reagents, simple instrumentation, and “moderate complexity” designation make LAMP technology an emerging player in the field of molecular diagnostics.
FIG 2.
Loop-mediated isothermal amplification (LAMP). (A) LAMP-based amplification requires 4 primers complementary to 6 different regions of the nucleic acid target (F1, F2, F3, B1, B2, and B3). The “inner primers” FIP and BIP each contain two regions complementary to the target sequence; one anneals to the template strand (F2 and B2), and one anneals to the complementary strand (F1c and B1c). The “outer primers” (F3 and B3) are complementary to a single sequence upstream of FIP and BIP, respectively. (B) Replication initiates with annealing and extension of the FIP and BIP “inner primers.” The “outer primers” F3 and B3 anneal upstream of FIP and BIP and are extended, which displaces the strands initiated by the FIP and BIP inner primers. The displaced strands form 5′ loop structures through complementary binding, resulting in a single-strand “dumbbell” structure. (C) The single-strand “dumbbell” serves as the template for subsequent rounds of amplification using the FIP and BIP primers to initiate elongation. Single-stranded template is maintained through formation of loop structures which can be extended to displace newly synthesized double-strand product (C5 through C8). (Adapted from reference 36 with permission from Macmillan Publishers Ltd.)
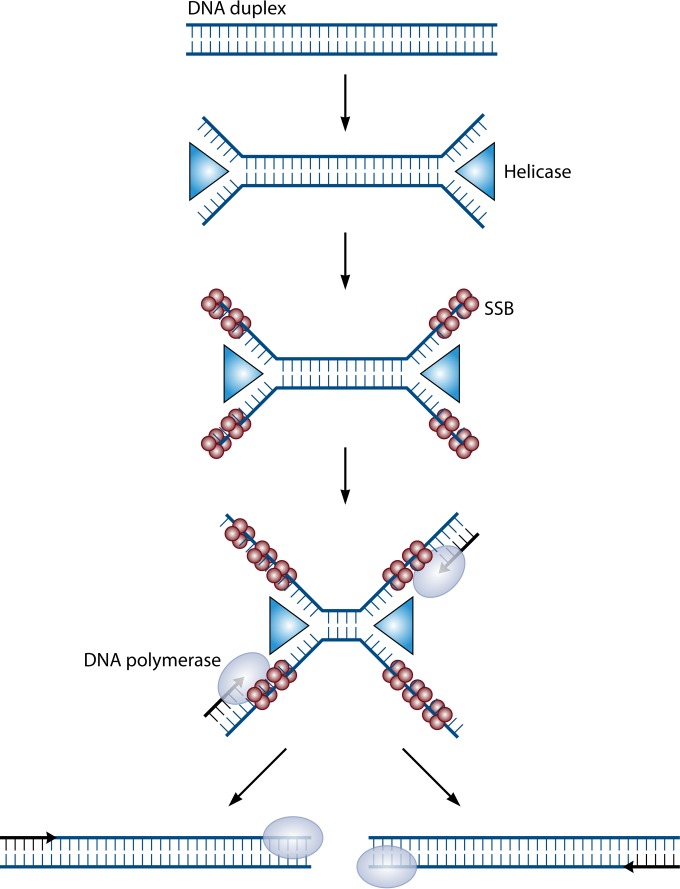
Helicase-dependent amplification (HDA) is another isothermal amplification technology that could be adapted to point-of-care testing. This technology utilizes UvrD (DNA helicase) and MutL enzymes isolated from E. coli and single-strand binding proteins to create and maintain a single-stranded template for primer annealing and subsequent rounds of amplification (53) (Fig. 3). An initial heat-based denaturation is required for optimal efficiency; however, reliance on a single reaction temperature without initial denaturation maintains 40% to 60% efficiency and is adequate to generate sufficient amplicon for endpoint detection assays (53). Like LAMP, the isothermal amplification can be carried out using simple instrumentation in the absence of electricity (54). HDA has been applied to identification of C. difficile, Plasmodium spp., and S. aureus (55, 56). An advantage of HDA is that detection of target can be achieved by incorporation of fluorescein or digoxigenin into the amplicon, followed by capture and visualization of the amplicon as a colored line on an enzyme immunoassay (EIA) lateral-flow strip (56–58). This maintains the ability to utilize these assays without sophisticated instrumentation but also allows the detection of multiple targets in a single reaction. A test developed to detect and differentiate herpes simplex virus 1 (HSV-1) and HSV-2 using this approach has demonstrated 100% sensitivity compared to viral culture, with a limit of detection as low as 5.5 copies per reaction (59). Further, this test could be performed on oral and genital cutaneous or mucocutaneous sources without the need for nucleic acid extraction and could be completed within 75 min.
FIG 3.
Helicase-dependent amplification (HDA). HDA uses the UvrD (helicase) (blue triangles) and MutL (accessory protein required for efficient UvrD loading to DNA) enzymes from E. coli to catalyze temperature-independent creation of a single-stranded DNA template for nucleic acid amplification. The UvrD/MutL complex unwinds double-stranded DNA to form an open complex. Single-strand binding proteins (SSB) (red circles) bind to the denatured strands to prevent association of the complementary strands. Specific primers are designed to anneal to the target sequence, and DNA polymerase (gray oval) extends the primers to the generate target amplicon. This amplicon serves as the template for subsequent rounds of amplification. (Adapted from reference 53 with permission [copyright Wiley-VCH Verlag GmbH & Co. KGaA].)
Automation of NAATs and impact on laboratory workflow and patient care.
Qualitative NAATs vary widely in the level of automation, ranging from largely manual (offline nucleic acid extraction, manual preparation of master mix, and addition of template) to fully automated “sample-to-result” platforms (Table 1). Full automation is typically focused on high-volume or screening tests such as those used for N. gonorrhoeae and C. trachomatis, C. difficile, MRSA, VRE, and HSV. These highly automated sample-to-result platforms decrease technologist hands-on time and may provide more consistency by reducing the risk of cross-contamination of specimens, pipetting error, or other preanalytic errors attributable to human labor. Despite these obvious advantages, there are still impediments to maximizing the value of molecular testing when using these systems. Until recently, the majority of molecular tests have been considered “high complexity” and as such have been confined to molecular laboratories staffed with skilled technologists. This requires that specimens be transported to the laboratory for analysis. For inpatients, the delay resulting from transport of a specimen may not be significant; however, for outpatient clinics, the time between collection of the specimen and receipt by the laboratory may be several hours. This delay due to transport abrogates one of the key advantages of molecular tests, namely, rapid TAT. Additionally, some molecular assays are best suited for batch testing due to multistep processing or efficiency factors related to batching of specimens on automated platforms. Finally, the large capital expenditure for high-capacity fully automated instruments must be considered.
TABLE 1.
Automated molecular platformsa
| Manufacturer | Platform | Technology | Multiplex capabilities | Open or closed system | FDA-cleared tests | Level of automation | Throughput | Turnaround time |
|---|---|---|---|---|---|---|---|---|
| Cepheid | GeneXpert | Real-time PCR with fluorescent probe-based detection | Up to 6 channels for detection of fluorescence | Closed | S. aureus including MRSA (nasal, skin, soft tissue, blood culture), C. difficile including NAP1/027 strain, VRE, influenza viruses A/B, enterovirus (CSF), M. tuberculosis including rifampin resistance, group B Streptococcus (direct or broth enriched), C. trachomatis/N. gonorrhoeae | On-demand random access, sample to result; some assays require dilution of specimen into buffer prior to loading of test cartridge | Variable depending on no. of test modules; Xpert systems available as 1-, 2-, 4-, and 16-module benchtop systems; Infinity systems available with 48 or 80 test modules and include automatic loading and unloading of test cartridges | Most assays complete in approx 1 h; hands-on time <1 min per sample |
| BD Diagnostics | BD Max | Real-time PCR with fluorescent probe-based detection | Up to 6 channels for detection of fluorescence | Open | S. aureus including MRSA (nasal), C. difficile, group B Streptococcus, enteric bacterial panel (Salmonella, Shigella, Campylobacter, stx1/stx2) | Batch, sample to result; specimens inoculated to a sample buffer tube before loading to BD Max; user also must load reagent strip and 24-well PCR cartridge | BD Max capable of batch processing and analyzing up to 24 specimens simultaneously | Approx 2.5-h run time with additional 15–30 min hands-on time for completion of 24 specimens |
| BD Viper | Strand displacement amplification | No | Closed | C. trachomatis, N. gonorrhoeae, HSV-1, HSV-2, T. vaginalis | Continuous batch processing, sample to result; fully automated | BD Viper capable of batch processing 96 samples simultaneously and reporting 184 results within approx 2.5 h, with subsequent batches of 184 results reported within approx 1.5 h; ability to load different assays simultaneously | Approx 2.5-h run time with 10–20 min hands-on time per 96 specimens | |
| Hologic Gen-Probe | Tigris | Transcription-mediated amplification | No | Closed | C. trachomatis/N. gonorrhoeae (available as combination- or single-target tests), T. vaginalis, HPV (14 high-risk types, nondifferentiated), HPV genotyping | Batch, sample to result; user loads collection tube to instrument for processing and analysis; reagents and disposable plastics reloaded manually | Instrument can accommodate 9 racks of 20 samples (180 samples total); additional samples can be loaded and processing initiated prior to completion of the first 180 samples | Approx 4.5 h from test initiation to result reporting |
| Panther | Transcription-mediated amplification | No | Open | C. trachomatis/N. gonorrhoeae (available as combination- or single-target tests), T. vaginalis, HPV (14 high-risk types, nondifferentiated), HPV genotyping | Random-access processing; prioritization of specimens for STAT requests; fully automated extraction, amplification, and detection of target | Random-access loading to a capacity of 120 specimens | Approx 3.5 h for initial result; requires 10 s hands-on time per sample for loading | |
| Nanosphere | Verigene | Multiplex PCR followed by solid-microarray detection using nanoparticle-conjugated probes | Microarray contains up to 400 capture probes | Closed | Blood culture Gram positive (12 Gram-positive genus or species targets and 3 resistance markers), blood culture Gram negative (8 Gram-negative genus or species targets and 6 resistance markers), C. difficile including NAP1/O27 strain, respiratory virus panel (influenza virus A including H1/H3 subtyping, RSV A/B) | On-demand random access; user must load test cartridge along with 2–3 disposable consumables for each test; upon completion of test, user must transfer test cartridge to analyzer to read microarray results | Variable depending on no. of Verigene sample processors; single reader can accommodate up to 32 processors | Tests require 2–2.5 h depending on specific test; each sample requires 1–2 min of hands-on time |
| Great Basin | Portrait | Helicase-dependent amplification or PCR followed by solid-microarray detection based on presence of colorimetric substrate | Array can accommodate up to 64 different target spots, including controls | Closed | C. difficile | On-demand random access; offline sample prepn; on-board extraction, processing and, result analysis within the Portrait processor/analyzer instrument; user must load test card for each test run into Portrait analyzer | Variable depending on no. of Portrait analyzers; each analyzer requires dedicated CPU for data analysis and storage | Test requires dilution, filtration, and heat treatment of specimens prior to loading to test card; total assay run time is 90 min, with an additional 10 min of hands-on time for sample prepn |
| BioFire | FilmArray | Two-stage “nested” PCR; second stage involves parallel singleplex reactions followed by melt analysis | Approx 100 microwells for second-stage target-specific PCR | Closed | Blood culture BCID (8 Gram positive, 11 Gram negative, 5 Candida spp. and 3 antibiotic resistance markers), respiratory panel (17 viral and 3 bacterial targets), gastrointestinal panel (13 bacterial, 5 viral, and 4 parasitic targets) | On-demand random access, Sample to result; user dilutes specimen into provided buffer and loads into test pouch | Variable depending on no. of FilmArray analyzers; each analyzer requires dedicated CPU for data analysis and storage. | Test requires approx 1 h, with 5 min of hands-on time |
| Roche | Cobas AmpliPrep/Cobas TaqMan | Multiplex PCR followed by fluorescent probe-based detection | Up to 4 channels for detection of fluorescence | Open | Qualitative and quantitative assays available; HIV-1, HBV, HCV; additional tests available on Cobas 4800 system include HPV and C. trachomatis/N. gonorrhoeae | Fully automated sample-to-result batch processing; on-board nucleic acid extraction, setup of PCR, amplification, and detection of target | Up to 96 samples can be batch processed | Variable by test, ranging from 4–5 h for full batch of 96 samples |
| Abbot | m2000 (m2000sp and m2000rt) | Multiplex PCR followed by fluorescent probe-based detection | Up to 4 channels for detection of fluorescence | Open | HIV-1, HCV+/− genotyping, HBV, CMV, HPV (high risk), vancomycin resistance careen, influenza viruses A/B and RSV, C. difficile, HSV-1 and -2, group B Streptococcus, EBV, VZV | Fully automated batch processing, including nucleic acid extraction and setup of PCR on m2000sp; user must transfer PCR plate to m2000rt for real-time PCR analysis | Up to 96 samples can be batch processed simultaneously | Variable by test, ranging from 3–4 h run tine with additional 1 h hands-on time for full batch of 96 specimens |
For an exhaustive list of automated molecular platforms and their characteristics, see the October 2013 issue of CAP Today (258).
The trends toward consolidation/centralization of laboratories and bundled care reimbursement structures favors highly automated systems with large-throughput batch processing of specimens to achieve a low cost per test (60). Systems like the m2000 (Abbott), and Cobas AmpliPrep (Roche) feature a two-step system whereby automated nucleic acid extraction is followed by automatic addition of all reagents required for an RT-PCR on one instrument. These instruments can process up to 96 specimens per run; however, prepared specimens must be physically moved to a thermocycler within 30 to 150 min to complete analysis of the specimen. The need for human intervention and a narrow window for transfer of specimens to a thermocycler limit the walkaway capability and present difficulty for laboratories not well staffed on all shifts. Other batch-type platforms such as the BD Max and BD Viper (BD), Tigris (Hologic Gen-Probe), and Cobas AmpliPrep/Cobas TaqMan system (Roche) are true sample-to-result platforms. Most of these platforms are classified as high-complexity molecular assays; however, the BD Max offers FDA-cleared moderate-complexity in vitro diagnostic (IVD) tests as well. These systems incorporate thermocyclers capable of RT-PCR and result reporting along with sample preparation. In addition to simplifying workflow, sample-to-result instruments may also reduce contamination or labeling errors by reducing the number of times that specimens are manipulated by technologists. A major disadvantage of batch platforms is the delay in availability of results compared to on-demand NAATs. In the case of outpatient surgeries, some institutions maintain presurgical clinics scheduled 1 to 2 weeks prior to the scheduled surgery, while in other institutions more than 80% of patients may be admitted on the day of surgery (21). In these cases, a point-of-care or on-demand test may be a better solution to benefit the patient rather than batched molecular assays. For example, real-time on-demand screening for colonization with MRSA or VRE could potentially alter presurgery prophylaxis or infection control measures (20–22, 61).
The advantages of point-of-care (POC) testing have reviewed by Robinson et al. and include a reduction in repeat and unnecessary test orders, a reduced length of stay, and shorter times to appropriate therapy; however, the authors acknowledge the lack of published studies objectively examining quantifiable outcomes related to the use of POC testing (60). There are several on-demand sample-to-result molecular testing platforms, including the GeneXpert (Cepheid, Sunnyvale, CA), Verigene (Nanosphere, Northbrook, IL), Portrait (Great Basin, Salt Lake City, UT), and FilmArray (BioFire, Salt Lake City, UT) (Table 1). Currently, these platforms are best suited to the laboratory; however, movement toward use as point-of-care (POC) tests is being pursued through miniaturization, automation, and simplification of the testing process. Other platforms, including Illumigene (Meridian Bioscience, Cincinnati, OH), and AmpliVue (Quidel Molecular, San Diego, CA) lack automation but have been simplified to potentially enable use as POC or “near-POC” diagnostics. Fully automated on-demand or single-test formats are often significantly more expensive on a per-test basis than batched testing formats; however, the rapid results provided by these systems often enable patient management decisions that can reduce the total cost of care. Many of the studies that demonstrate this principal utilize on-demand molecular tests for the identification of S. aureus and MRSA in positive blood culture broths. One such study compared cohorts of patients with Gram-positive bloodstream infection (BSIs) pre- and postimplementation of an on-demand molecular test that identified S. aureus and differentiated S. aureus from MRSA. The authors reported a 1.6-day reduction in time to optimal antibiotic therapy and a 6.2-day reduction in hospital stay for the cohort of patients tested using the NAAT (62). This also translated to >US$21,000 reduction in the total cost of care for these patients. In contrast, a similar study conducted using a lower-cost-per-test batch-format NAAT to identify S. aureus and MRSA in positive blood culture broths failed to demonstrate such savings (63). Importantly, failure to actively report laboratory values also decreased the benefits of NAAT results in the latter study. Another area of great interest is the use of rapid and accurate molecular assays for the identification of Mycobacterium tuberculosis in patient specimens (64). The recent availability of an FDA-cleared NAAT (Xpert TB/RIF; Cepheid) for the identification of M. tuberculosis, including strains resistant to rifampin, has prompted studies assessing the cost-effectiveness of such a test. The cost of sputum smear as a primary diagnostic test for patients suspected of having active tuberculosis is <7% the cost of Xpert TB/RIF; however, overall savings of US$2,278 per admission could be realized when considering the reduced occupation of isolation rooms for patients with a negative result (65). Other studies have reported up to a 94% decrease in unnecessary antituberculosis treatment and an average 1.5-month reduction in unnecessary therapy as well as a reduction in time in isolation for patients who were smear positive but culture negative for M. tuberculosis when an NAAT was used (66, 67). Importantly, these data were based on a studies conducted in high-prevalence populations (6% to 37% positive for M. tuberculosis). For hospitals and laboratories serving low-prevalence populations, implementation of a more costly molecular test for all smear-positive specimens may increase the overall cost of care for these patients. In these cases, communication between the laboratory and clinician to establish the patient history and risk of M. tuberculosis may be beneficial to reduce unnecessary cost of a molecular test.
In all cases, to reap the greatest benefit from these technologies, the assays must be able to be conducted and results reported in a true “real-time” 24-h-per-day, 7-days-per-week fashion, or the benefit of rapid TAT to patient care will be lost.
Multiplex Nucleic Acid Tests
The combination of multiple primer sets into a single PCR (multiplex PCR) for simultaneous detection of several targets was reported shortly after the initial description of PCR-based amplification methods (68). Multiplex PCR can be very beneficial when testing specimens from patients presenting with nonspecific symptoms attributable to a number of different pathogens. Examples include respiratory specimens from patients with suspected viral illness, stool specimens from patients with enteritis, and positive blood cultures. Approaches to multiplex PCR tests include (i) single reactions containing fluorescently labeled probes for each target, (ii) parallel singleplex reactions conducted in microwell-size vessels in a single run, (iii) traditional microarray-based detection utilizing capture probes immobilized on a solid surface, and (iv) newer liquid-array approaches that involve capture probes immobilized on microbeads which can be sorted using flow cytometry. Each approach has characteristics amenable to different aspects of diagnostic testing, including cost, throughput, automation, and level of multiplex capability.
Multiplex PCR and probe-based detection.
The introduction of platforms equipped with optics capable of excitation and detection of multiple fluorophores in a closed system in real time (real-time PCR [RT-PCR]) made multiplex pathogen detection a simple and viable option for molecular diagnostics in routine clinical laboratories. Laboratory-developed tests (LDTs) have taken advantage of the different probes and platforms in the design of multiplex tests for the detection of a variety of analytes. Only recently have larger multiplex panels begun to be available as FDA-cleared tests for use in clinical diagnostics. These PCR-probe-based tests are typically capable of low-density multiplexing of 4 to 6 unique targets. This limitation is imposed by the number of optical channels and ability to differentiate between fluorescent dyes with similar emission wavelengths. The optics on early platforms, including SmartCycler II (Cepheid, Sunnyvale, CA) and first-generation BD Max (BD, Sparks, MD) were limited to a maximum of 4 channels. Newer platforms, including the GeneXpert (Cepheid), LightCycler 2.0 (Roche, Indianapolis, IN), second-generation BD Max (BD), and ABI 7500 Fast Dx and ABI QuantStudio (ABI, Foster City, CA) are capable of detection in up to 6 different channels. Compared to more recently developed multiplexing technologies, including solid and liquid microarray (discussed below) methods, the ability to multiplex 4 or 6 targets can be a limitation. This is especially true for specimen types in which there are numerous, diverse microorganisms capable of causing similar symptoms or syndromes such as upper respiratory illness, gastroenteritis, or bacterial and fungal sepsis. Despite the limitations in the number of targets that can be detected simultaneously, numerous FDA-cleared tests using these platforms have been favorably evaluated and are applicable in the clinical laboratory.
The SmartCycler II and LightCycler 2.0 are open platforms for RT-PCR. Both require preextraction of nucleic acids to obtain template and manual pipetting of each PCR component or master mix into individual RT-PCR tubes. Multiplex assays using analyte-specific reagents (ASRs) for influenza viruses A and B, respiratory syncytial viruses (RSV) A and B, and HSV-1 and -2 have demonstrated high sensitivities compared to other rapid tests, and results are available days earlier than with viral culture methods (10, 69–72). A recently developed and FDA-cleared test for the detection of bacterial causes of enteritis demonstrated 100% sensitivity and >99% specificity for 5 targets (Salmonella spp., Shigella spp., Campylobacter coli/jejuni, stx1, and stx2) compared to culture and an alternative molecular assay (73). A drawback to this test is the need for offline nucleic acid extraction and the necessity to set up parallel reactions for each specimen to accommodate all 5 assay targets due to limitations of the SmartCycler II optics.
Molecular tests have also been developed for detection of bacterial and fungal pathogens associated with bloodstream infection (BSI) (Table 2). Initial tests were developed using the SmartCycler II or LightCycler 2.0 for low-density multiplexing (63, 74, 75). The SeptiFast assay (Roche) is unique among these tests in that it is intended for use with whole blood specimens prior to broth culture enrichment. This assay has not received FDA clearance for use in the United States. Although run on the LightCycler 2.0, the use of 3 parallel real-time PCRs with different primer/probe combinations and postamplification melt curve analysis expanded the number of bacterial and fungal targets that could be detected using SeptiFast to 20 (75). Importantly, the low number of organisms per ml in direct whole-blood specimens limited the sensitivity to 42% to 79% compared to culture (75–77). The specificity of this test was reported as 95.0 to 97.1% in patients without clinical signs of sepsis but was 74% in symptomatic patients (76). A positive SeptiFast result was confirmed by culture in only 67% of specimens; however, the detected organism was recovered from other clinically relevant samples in approximately half of the discordant cases (75). Together this suggests that a NAAT may be more sensitive than culture in patients with clinical symptoms of sepsis; however, additional studies are needed to correlate positive NAAT results with clinical outcomes. Because of the difficulties in molecular analysis of whole blood, more recent molecular tests have focused on analysis of positive blood cultures. The StaphSR test is performed on positive blood cultures containing Gram-positive cocci. This test is designed to detect and differentiate methicillin-susceptible and -resistant strains of S. aureus (63, 74). Initial studies reported sensitivity and specificity for identification of S. aureus of 96.7% to 99.4% and sensitivity for MRSA of 100% (63, 78); however, subsequent studies report sensitivities as low as 50% depending on the type of SCCmec cassette present in circulating strains (79, 80) (Table 2). Another drawback of this assay is the requirement for offline extraction and manual setup of individual RT-PCRs, which lends to batching of specimens. In the case of positive blood cultures, batching of specimens contributes to delays in reporting of results, which can abrogate the benefit that rapid molecular diagnostics can have for patient care (63). Finally, while S. aureus is of major concern in BSIs, it comprises only about 20% of positive cultures (81). The limited number of fluorophores that can be differentiated in a single reaction using standard RT-PCR platforms prevents the inclusion of additional targets required to make this type of test applicable to the majority of positive blood cultures. For laboratories with larger specimen volumes or limited staffing, the offline processing and manual setup of reactions can complicate assay setup and strain resources and may also be a potential source for cross-contamination of specimens.
TABLE 2.
Comparison of FDA-cleared molecular methods for detection of microorganisms in positive blood culture broths
| Test | Targets | Sensitivity (%) | Specificity (%) | Time to result (h) | Format and setup | References |
|---|---|---|---|---|---|---|
| Verigene BC-GP | 12 Gram-positive genus or species targets and 3 resistance markers (mecA, vanA, vanB) | 92–100 | 98–100 | 2.5 | On-demand, microarray, automated sample processor, manual transfer of array to analyzer | 95–97, 107 |
| Verigene BC-GN | 8 Gram-negative genus or species targets and 6 resistance markers (KPC, NDM, CTX-M, VIM, IMP, OXA) | 81–100 | 98–100 | 2 | On-demand, microarray, automated sample processor, manual transfer of array to analyzer | 101, 102 |
| FilmArray BCID | 8 Gram-positive, 11 Gram-negative, and 5 yeast genus or species targets, 4 resistance markers (mecA, vanA/B, KPC, NDM) | 88–100 | 94–100 | 1 | On-demand, parallel miniaturized singleplex RT-PCR, full sample-to-result capability | 85, 90 |
| GeneOHM StaphSR | S. aureus, MRSA | 50–100 | 98–99 | 2 | Batch, RT-PCR, offline manual sample lysis, extraction, and RT-PCR setup | 63, 78–80 |
| Xpert MRSA/SA Blood Culture | S. aureus, MRSA | 69–100 | 98–100 | 1 | On-demand, RT-PCR, full sample-to-result capability | 259–261 |
| Septifasta | 6 Gram-positive, 8 Gram-negative, and 5 yeast targets and A. fumigatus | 42–79 | 67–97 | 6 | Batch, 1.5–10 ml whole blood; offline extraction and setup of 3 parallel RT-PCRs | 75–77 |
Not cleared by FDA for clinical use. Data are from direct analysis of whole blood.
Miniaturization of singleplex reactions can overcome some of the limitations to traditional PCR-probe-based multiplexing. Conducting singleplex real-time PCR in multiple individual wells enables simultaneous amplification and detection of different targets, but all within a single test device. This can be accomplished using a thermocycler capable of real-time quantitative PCR such as the ABI 7500 FastDx or ABI QuantStudio, which can accommodate 96- or 384-well microplates and can interrogate each well separately. Importantly, these platforms are not sample-to-result platforms, and this approach still requires extraction and manual setup of multiple real-time PCR wells per specimen. In contrast, the FilmArray system (BioFire, Salt Lake City, UT) is a sample-to-result multiplex PCR system contained within a single test pouch. In addition to simplifying workflow, this methodology also enables the assay to be classified as a moderate-complexity IVD test. The clinical specimen is diluted and added directly to a sample port. The specimen then passes through multiple chambers containing reagents for lysis and extraction of nucleic acids from the specimen. Once extracted, the nucleic acids undergo a nested PCR in which the first reaction utilizes degenerate primers to broadly amplify target sequences. Products from the first PCR are then diluted and inoculated into 102 microwells, each of which contains reagents for singleplex amplification and detection of a specific target sequence (82). Each well can be individually interrogated for fluorescence, allowing the use of a single fluorophore for detection of amplicon. Tests using this approach are available or under development for the detection of respiratory viruses, bacteria, and fungi in positive blood cultures and bacterial, viral, and protozoan pathogens in stool (82–85).
Many studies have evaluated the FilmArray respiratory panel (RP), and the performance in these studies has been reviewed by Babady (86). In general, evaluation of the FilmArray respiratory assay in adult and pediatric populations has demonstrated 80% to 100% agreement with alternative molecular tests, with notable deficiencies in detection of specific adenoviruses (83, 84, 87, 88). This deficiency has been addressed in a more recent version of the assay (version 1.7), which has demonstrated an increase in sensitivity from 43% to 66% to 88% to 91% for detection of 39 clinically relevant adenovirus serotypes (89). Compared to other molecular tests for respiratory viruses, the FilmArray had the highest cost per test, but this was countered by the full sample-to-result capability, highest number of targets detected (n = 20), and fastest total time to result (1 h) (83). In addition to the relatively high per-test cost, a second potential drawback to the use of the FilmArray as a mainstream method for analysis of respiratory specimens is the limited throughput. Each FilmArray is capable of analyzing only a single specimen per run. This can be a significant bottleneck for larger laboratories, which may receive hundreds of respiratory specimens per day in peak respiratory illness season. Therefore, use of the FilmArray with its broadly inclusive panel may be best suited to critically ill or immunocompromised patients rather than for routine testing of all community patients suffering from respiratory symptoms during “influenza season.”
Initial clinical evaluations of the FilmArray BCID blood culture assay demonstrated overall sensitivity of 91% to 99%, including 98.5%, 96.7%, and 100% for 11 Gram-negative, 8 Gram-positive, and 2 yeast targets, respectively, with specificity of 97% to 100% for each of the individual targets on the panel (85, 90) (Table 2). A potential weakness of the assay is the inclusion of a single “Enterococcus spp.” target which is unable to differentiate between E. faecalis and E. faecium. This distinction can be helpful when considering antimicrobial therapy because of differences in susceptibility patterns between the two species. Specifically, resistance to ampicillin and vancomycin are rare in E. faecalis, 1.3%, and 0.5%, respectively, while 82.4% and 9.6% of E. faecium isolates are resistant to ampicillin and vancomycin, respectively (91) A second potential shortcoming is the failure to reliably detect all components present in polymicrobial cultures. Overall, the FilmArray BCID detected all microorganisms present in just 71% of polymicrobial cultures. While many of these were organisms not present on the BCID panel, E. faecalis was missed in two polymicrobial cultures, while E. coli and a viridans group Streptococcus spp. were missed in two other polymicrobial cultures (85). Finally, while the assay includes a total of 24 genus or species targets commonly associated with bloodstream infection, up to 8% of blood cultures contain organisms not present on the BCID panel (85, 90). Therefore, a primary Gram stain of all positive blood culture broths as well as routine culture of broths which are both positive and negative by BCID is prudent before finalizing results.
Microarray methods.
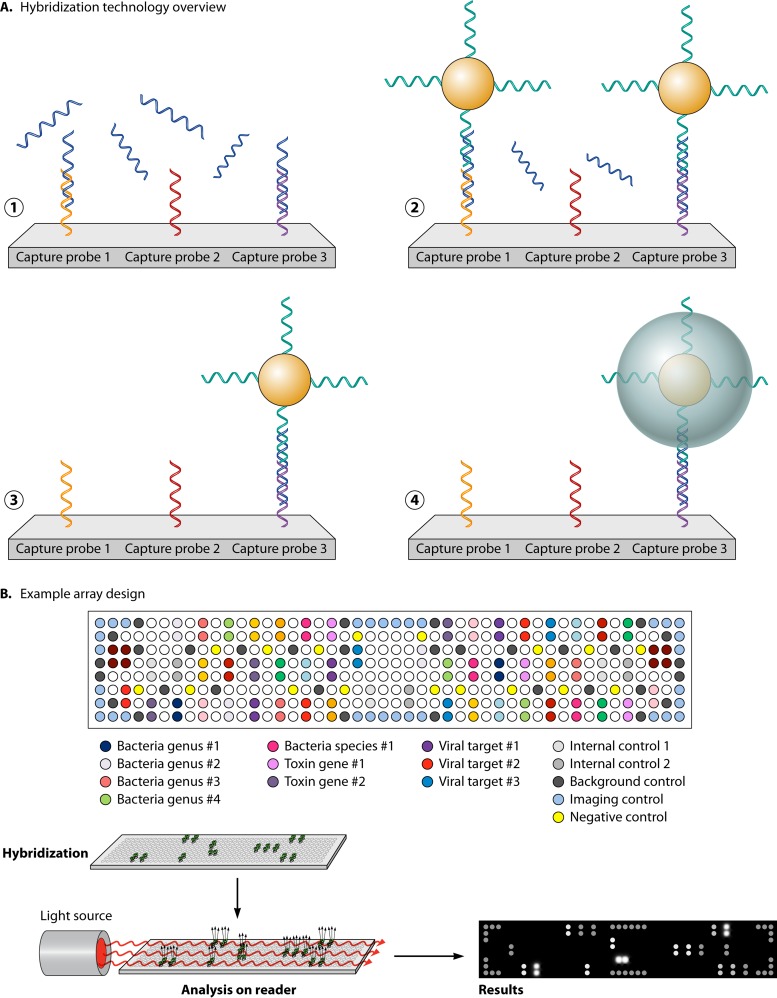
Several approaches have been explored to expand the number of targets detectable in a single multiplex nucleic acid test. Collectively, these are referred to as microarrays. Microarrays can be broadly broken into two classes: solid arrays, which rely on spatial detection of targets arranged on a solid surface, and liquid arrays, which utilize target-specific capture probes conjugated to microspheres which can be detected using flow cytometry. For a thorough review of microarray technologies, the reader is referred to the article by Miller and Tang (92). Microarrays are attractive in diagnostics because they can reduce the cost per target tested and allow simultaneous testing for multiple pathogens associated with similar symptoms.
Traditional microarrays are composed of synthetic oligonucleotides or peptides (capture probes) immobilized on a solid substrate such as a glass slide or nitrocellulose membrane. The number of unique capture probes on a single array can range from 100 on low-density printed arrays to >1 million on in situ-synthesized high-density arrays. The probes on high-density arrays are typically shorter (20 to 25 nucleotides [nt]) and are designed to have target redundancy to increase the specificity of target detection (92). Because of the large number of probes, these arrays are most commonly used for whole-genome expression profiling or for other genome-wide comparisons such as mutations or deletions. Low-density arrays consist of longer probes, typically 50 to 800 nucleotides in length, which may be chemically synthesized or created as amplicons by PCR. The use of PCR amplicons and liquid spotting of probes makes this type of array comparatively inexpensive to manufacture. The relatively long length of amplicon probes increases target sensitivity because several polymorphisms can be tolerated during hybridization steps; however, this can also results in decreased specificity for the target (92, 93). Therefore, each probe is typically spotted in replicate on a single array to increase test specificity (92). Each synthesized oligonucleotide or amplicon probe corresponds to a single gene and is spotted or printed to the array solid surface. Inexpensive manufacturing and high sensitivity make low-density printed arrays a reasonable choice for diagnostic tests designed for use in clinical microbiology laboratories.
The commercially available and FDA-cleared Verigene system (Nanosphere, Northbrook, IL) (Fig. 4) has offers microarray-based tests for identification of respiratory viruses (RV+), C. difficile (CDF), blood cultures containing Gram-positive bacteria (BC-GP) or Gram-negative bacteria (BC-GN), and identification of genetic variants, including Factor V Leiden and CYP450 2C19 *2 and *3, which impact patients with coagulation disorders (94–102).
FIG 4.
Verigene solid-phase microarray. (A) Single-stranded, target-specific capture probes are arrayed spatially and immobilized onto the surface of a glass slide. The nucleic acid target (PCR amplicon or extracted nucleic acid) is denatured and applied to the glass slide. If present, the target nucleic acid will anneal to the complementary capture probe. Gold microspheres coated with single-stranded nucleic acid complementary to a different region of the target sequence are added and anneal to the capture probe-target sequence hybrid to form a “sandwich” nucleic acid structure. The array is washed to remove unbound nucleic acid and gold microparticles. Application of colloidal silver increases the size of the bound microspheres to increase the sensitivity of detection. (B) Target-specific capture probes, along with internal controls, are spotted in triplicate to different locations on the glass slide to ensure consistency of the annealing and hybridization steps and increase accuracy of results. Target detection is accomplished using a light source shown across the plane of the array. If present, bound silver microspheres diffract the light, which is then detected by an optical camera in the array reader.
The RV+ test simultaneously tests specimens submitted in viral transport medium for influenza viruses A and B, including subtypes H1, H3, and 2009 H1N1, and RSV A/B. Clinical evaluations have reported sensitivities of 96.6% to 100% for influenza virus A, 96.8% to 100% for influenza virus B, and 89.8% to 91.7% for RSV, with specificities of >96.5% for all targets (100, 103, 104), which in one comparative study was superior to results for a traditional RT-PCR test (103).
A clinical evaluation of the CDF assay demonstrated 98.7% sensitivity and 87.5% specificity for detection of toxigenic C. difficile based on the presence of tcdA and/or tcdB, the primary toxin-encoding genes present in toxigenic strains of C. difficile (94). In addition, the CDF assay contains capture probes for detection of the Δ117 deletion in tcdC, which encodes the repressor of tcdA and -B expression, and genes encoding binary toxin (cdtA and cdtB) (94). Strains with the Δ117 deletion produce up to 23-fold more toxin than wild-type strains (105). Additionally, the Δ117 deletion and the presence of binary toxin are characteristic of strains of the O27/NAP1 ribotype, which has been associated with more severe disease (105, 106).
Direct detection of microorganisms from positive blood cultures is an area of great interest because of the potential benefits of rapid identification to patient care, antimicrobial stewardship, and health care cost. Clinical performance of the BC-GP assay has been evaluated in several studies, including large multicenter efforts encompassing all commercially available blood culture systems. These studies have reported sensitivities of >96% for most of the 12 identification targets on the panel; however, lower sensitivities of 67.0% to 94.8% were reported for E. faecalis and E. faecium, and false-positive Streptococcus pneumoniae results were reported for isolates of Streptococcus mitis/Streptococcus oralis (95–97, 107–109). Importantly, the sensitivity of BC-GP for detection of mecA in methicillin-resistant S. aureus (MRSA) in these studies was >99%. Additionally, the performance does not appear to be affected by the type of blood culture broth used (i.e., aerobic versus anaerobic, pediatric versus adult, containing charcoal or resin, etc.). Literature regarding the performance of the more recently FDA-cleared BC-GN test is currently limited to studies including a small number of prospectively collected clinical specimens or studies using primarily simulated specimens (101, 102). In a multicenter evaluation of 104 clinical specimens, the overall sensitivity of BC-GN was 91%, with sensitivities of 67% to 100% for the 9 Gram-negative genus or species targets (101). In a larger study of 397 blood cultures (75% simulated specimens), sensitivity was >98% for 7 of the 8 targets present on the FDA-cleared panel. The single target demonstrating poor performance was Klebsiella pneumoniae, which was reported to be 86.1% sensitive (102). Interestingly, all of the specimens with false-negative results were phenotypically identified as Klebsiella pneumoniae; however, 16S rRNA gene sequence analysis identified these isolates as K. variicola. A distinguishing characteristic of the BC-GN compared to the FilmArray BCID is the inclusion of the resistance markers blaCTX-M, blaIMP, blaVIM, and blaOXA in addition to blaKPC and blaNDM, which are present in both assays. The sensitivity of BC-GN for these 6 genetic markers of antibiotic resistance is reported to be 100% compared to sequence analysis of the strains (102). Importantly, additional studies to demonstrate phenotypic correlation with detection of these markers are needed.
A potential strength of solid-array technology is the ability to correctly identify multiple targets in the same specimen; however, in studies involving the BC-GP and BC-GN, all targets in a polymicrobial culture were correctly identified in only 60.0% to 81.3% of specimens (96, 97, 101, 102). This limitation is similar to that observed with the FilmArray BCID (discussed above). Additionally, unlike the FilmArray, the Verigene blood culture assays are restricted to Gram-positive or Gram-negative targets. Selection of the correct test depends on accurate reading of the primary Gram stain. These limitations again underscore the importance of primary Gram staining as well as routine culture of all positive blood culture broths prior to finalizing the culture.
Liquid-array technology, typified by the xTAG assays (Luminex, Toronto, Canada), involves an initial multiplexed PCR step, followed by target-specific primer extension that incorporates a unique nucleic acid “tag” and biotin label into each target amplicon. Tagged amplicons are then incubated with microbeads of various fluorescent potential, each type coated with a unique antitag sequence. Amplified target sequences with incorporated tags complementary to those on a specific bead will hybridize. Finally, a streptavidin-fluorophore conjugate is added and hybridizes to biotin-labeled amplicons immobilized on the beads. Detection of a target is accomplished using two lasers that interrogate each bead for (i) the presence of a captured amplicon as indicated by streptavidin-fluorophore and ii) the identity of amplicon as indicated by fluorescence of the bead specific for each antitag (Fig. 5) (110). The xTAG test for agents of gastroenteritis (xTAG GPP) includes targets for 15 bacterial, viral, and protozoan pathogens associated with gastroenteritis. Few clinical evaluations of the assay have been published, but initial reports demonstrate sensitivity and specificity ranging from 82 to 100% depending on the comparator used as gold standard (111, 112). A larger number of studies have evaluated the xTAG assay for respiratory pathogens (xTAG RVP), which detects 12 to 19 viruses (FDA-cleared versus CE-Mark targets) associated with respiratory illness. These studies have found 92 to 100% agreement of xTAG RVP with other molecular platforms and sensitivities of 91 to 100% with specificities of >99% for individual targets on the panel (87, 113, 114).
FIG 5.
xTAG liquid-phase microarray. Target sequences (blue and green) are amplified using multiplex PCR. Following amplification, a second set of target-specific primers containing “universal tag sequences” (orange and red boxes) unique to each target primer are used for a primer extension reaction. During primer extension, a biotin label is also incorporated into the amplicon. Labeled amplicons are then incubated with polystyrene microbeads. Microbeads are uniquely colored, allowing differentiation of up to 100 different types of microbeads by the analyzer. Each color bead is also coated with a single-strand nucleic acid probe complementary to one of the universal tag sequences (antitag). Amplicons labeled with universal tag sequences will hybridize to the microbeads containing the antitag. Additionally, a streptavidin-fluorophore conjugate (green star) is added and hybridizes to biotin-labeled amplicons immobilized on the beads. Following hybridization steps, beads are analyzed using a cell sorter equipped with two lasers. The first detects the presence of the fluorophore conjugated to biotin, indicating the presence of an amplicon bound to a specific microbead. The second laser interrogates the bead to determine which dye is present, thereby identifying the specific target amplicon present. The center bead in step 5 lacks amplicon and thus would be negative for the biotin-fluorophore signal. This bead would not be analyzed by the second laser.
Impact of large multiplexed panels on laboratory workflow and patient care.
Multiplexed molecular panels containing up to 20 targets or more can simplify ordering for the physician and simplify workflow in the laboratory by consolidating what were previously individual tests into a single “complex panel” for patients with respiratory illness, gastroenteritis, or positive blood cultures. An obvious benefit of these large multiplex molecular tests is the ability to detect numerous pathogens in a specimen without having to rely on different methodologies, including culture, molecular, EIA, or direct staining procedures as appropriate for the various pathogens that may be present in a single specimen. Especially in the case of fully automated platforms, this can ease the burden on the laboratory and reduce the dependence on experienced technologists for such tasks as identification of protozoan pathogens in a trichrome stain. Large multiplex panels also simplify test ordering for physicians, who may miss a diagnosis because of failure to order the correct test. For example, the diversity of targets on the Luminex GPP test enabled detection of a pathogen that would have been missed in up to 65% of specimens because the appropriate routine test to detect these pathogens was not ordered (111). An additional potential benefit is the ability to detect multiple pathogens simultaneously. Up to 10% of stool specimens may be positive for multiple targets which can be an indication of coinfection; however, these results must be interpreted with caution, since the presence of nucleic acid does not always correlate with clinical illness (111). Asymptomatic carriage of C. difficile, which can be as high as 15 to 20%, asymptomatic shedding of adenoviruses, or residual nucleic acid in the absence of viable organisms following treatment are potential sources of false-positive results (115–117). Other considerations include the pretest probability for a given pathogen and the cost per test, which is often higher for densely multiplexed and fully automated tests. During peak respiratory illness season, use of a batched molecular test for influenza viruses A and B for the majority of clinic patients may be more economical than a large on-demand multiplexed panel.
Identification of the organism present in positive blood culture broths using multiplexed molecular assays has been the focus of several recent publications because of the potential to dramatically impact patient care and reduce the total cost of care for patients suffering from bloodstream infections. Studies using NAATs or fluorescent in situ hybridization (FISH) for rapid identification of 2 to 4 targets in a positive blood culture broth have demonstrated significant reductions in the time on suboptimal antimicrobial therapy, length of hospital or ICU stay, and overall cost of care for patients infected with S. aureus, MRSA, Enterococcus, or Candida species (62, 118–120). While these outcomes are impressive, each test is limited to a relatively small number of microorganism targets, making each applicable to only 5% to 50% of cultures having a Gram stain consistent with specific test targets (81). Larger multiplex panels containing 12 or more targets are more broadly applicable across all blood cultures. For example, the Verigene BC-GP test (12 Gram-positive identification targets) effectively identified the bacterium present in 92.5% of cultures containing Gram-positive organisms, and the FilmArray BCID test (8 Gram-positive, 11 Gram-negative, and 5 Candida targets) accommodated >90% of the microorganisms present in all positive blood cultures (85, 97).
The level of automation of multiplexed tests and the level of complexity (high complexity versus moderate complexity) are features that broadly divide these tests and must be considered when choosing the most appropriate test for a laboratory. Tests that require “offline” extraction of nucleic acids and manual pipetting to set up the PCR(s) are designated high-complexity tests and as a result may not be suitable for most laboratories with restricted staffing or expertise. Alternatively, sample-to-result platforms typically gain approval as moderate-complexity tests which can be adopted by laboratories which lack staff with appropriate training/certification or which are not designated “high-complexity” laboratories by CLIA. This may be an important factor for many laboratories when selecting a molecular platform that best suits their needs. Another factor that needs to be considered is the per-test cost. As discussed above, the cost per test may be reduced for batch-type platforms compared to sample-to-results tests; however, the turnaround time for reporting of results will suffer. Evaluations of total time to result, throughput, and cost of the xTAG, FilmArray, and Verigene have reported a total turnaround time of 7 to 8 h for xTAG, with up to 21 samples reported in this run time (87). The extended TAT for xTAG is a result of the requirement for offline extraction and manual setup, which require 3 to 5 steps and over 1 h of hands-on time. In contrast, the FilmArray and Verigene are true sample-to-result platforms that provide a reportable result in 1 to 2 h, with <5 min of hands-on time. As discussed above, a rapid TAT is essential to maximizing the clinical benefits for identification of microorganisms in blood culture but may not be as critical for other specimens types, such as stool specimens, received from outpatient clinics. The major drawback to both sample-to-result platforms is limited throughput, which is only 4 to 8 tests per 8-h shift (87). Therefore, the benefit of a rapid, on-demand result must be weighed against specimen throughput capabilities. These decisions may be affected by season (e.g., respiratory illness season) or by patient population (inpatient versus outpatient), so it is important for the clinical laboratory to fully assess the needs of its specific institution or clinics when deciding on a platform for multiplex testing of clinical specimens.
Digital PCR
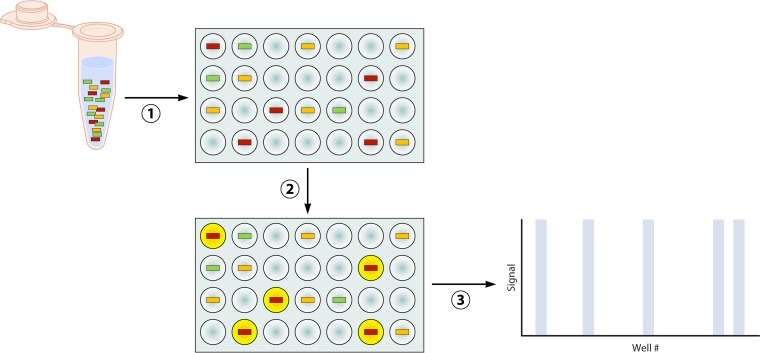
Quantitation of nucleic acid in a specimen using quantitative RT-PCR (qPCR) has become an essential task for clinical and molecular microbiology laboratories. Disease progression, prognosis, selection of antivirals, and response to therapy have been linked to the initial viral load or changes in load observed during continuous monitoring for HIV, cytomegalovirus (CMV), Epstein-Barr virus (EBV), human herpesvirus 6 (HHV-6), and BK virus (121–126). Likewise, identification and quantification of genetic markers of malignancy, including mutations, rearrangements, and expression of dysfunctional microRNAs (miRNAs), can aid in management of these patients (127–130). Standard qPCR is dependent on the measurement of increasing fluorescent signal generated during each cycle of RT-PCR. Quantitation is achieved through establishment of a signal threshold, generally the cycle at which the fluorescent signal is ≥10 times the standard deviation of the background noise, and creation of a standard curve using specimens with a known quantity of template (131). Although qPCR is widely used, is suffers from some significant drawbacks. First, the signal threshold and standard curve are test and instrument specific and must be calibrated regularly to ensure accuracy. Second, different RT-PCR platforms, probe types, and calibration standards can all affect the cycle threshold obtained and thus impact the quantitation of template in the original specimen. Finally, quantitation is accurate only along the log-linear portion of the calibration curve, where each PCR cycle represents a true doubling of amplicon (131). For these reasons, there is often poor correlation and high coefficients of variation (CVs) when specimens are tested on different instruments or by different laboratories. The intralaboratory CV has been reported to be as high as 246.8% in split-specimen surveys (132), and CVs of 20 to 70% have been reported even when assays are run on the same instrument by skilled technologists (131, 133–135). The CV is often greatest in these cases when analyzing specimens with the template near the lower limit of quantification, where amplification is transitioning from early exponential to log-linear phase. These shortcomings of qPCR highlight the analogue nature of quantitation made using calculations based on amplification curves. Digital PCR (dPCR) aims to eliminate amplification curve-based calculations by enumerating the actual number of templates in a specimen. This is achieved by dilution and segregation of the specimen into thousands of miniaturized parallel RT-PCR mixtures. Each reaction mixture will contain either one or zero copies of template. Following massively parallel RT-PCR, the number of wells with endpoint positivity for an amplification product is a direct measure of the copy number of template present in the specimen (Fig. 6) (136, 137). Because the actual number of copies is determined by the number of positive reactions, there is no need to construct a calibration or standard curve for comparison. Additionally, a low or high copy number of template can be accurately quantified because accurate quantification is not limited to the log-linear phase of the PCR. For example, Hindson et al. demonstrated a 37% to 86% decrease in CV when comparing dPCR to qPCR in both water and serum matrix (138).
FIG 6.
Digital PCR. A nucleic acid template containing target sequences (colored boxes) in the original sample is diluted into individual microwells (plate PCR, pictured) or picoliter droplets (emulsion PCR) such that each well or droplet contains one or zero copies of the target sequence. Following partitioning of the specimen, endpoint PCR is carried out and amplicon is detected using fluorescent dyes or probes. Each well will be either positive or negative for fluorescent signal depending on the presence of the target sequence and resulting amplicon (yellow circles correspond to blue bars on the graph). The number of wells or droplets positive for fluorescent signal (yellow circles) directly corresponds to the number of specific target sequences (red boxes) present in the original sample.
Digital PCR was originally developed in 1992 using the same primers and probes utilized in traditional RT-PCR (139). Since that time, advances in microfluidics have enabled automation and miniaturization using two different approaches for segregation of template prior to RT-PCRs. The first involves a silicon microfluidics chip containing up to 20,000 individual wells into which the substrate is diluted. The QuantStudio 3D digital PCR system manufactured by Life Technologies (Carlsbad, CA) is capable of thermocycling and analysis of 24 chips simultaneously, allowing quantitation of multiple specimens per run (137). A potential limitation of this approach is the comparatively limited analytical measurement or dynamic range that can be achieved. Assuming 20,000 individual wells, the maximum achievable dynamic range would be 4 log10 (1 to 20,000 copies). The second method overcomes this potential limitation through the use of emulsion PCR. This method utilizes an emulsion combining oil, aqueous specimen, and all necessary components for RT-PCR. The emulsion is then divided into up to 10 million picoliter droplets, each containing a maximum of one copy of template (137). Following PCR, each droplet is interrogated for fluorescent signal using flow cytometry to determine the number of droplets containing amplicon. Emulsion dPCR systems are manufactured by both Bio-Rad Laboratories (Hercules, CA) and RainDance (Billerica, MA).
Applications of dPCR encompass accurate quantification of a target sequence; however, the addition of multiplexing expands the utility to identification and quantification of rare alleles or minor variant species present in a specimen. Ma et al. demonstrated the ability of dPCR to detect a variant sequence when diluted 1:1,024 with wild-type sequence in a specimen. This was in comparison to traditional qPCR, which could detect a variant sequence to a dilution of only 1:256 (140). Because of the novelty of dPCR and relatively recent availability of commercially manufactured systems, there are few large clinical evaluations of this technology. The majority of studies have focused on the detection and quantification of mutations in oncogenes. This includes the quantification of free tumor DNA (tDNA) or microRNAs (miRNAs) from plasma specimens as an indicator of some types of carcinomas and a method to simultaneously screen for and quantify 4 common mutations in the KRAS gene in serum specimens (127, 141–143). Digital PCR has also been used in infectious disease for detection of S. aureus and C. trachomatis (144, 145) and has demonstrated the ability to accurately quantify HIV and hepatitis C virus (HCV) loads over a wide dynamic range (146, 147). A very recent publication has demonstrated the utility of dPCR to differentiate between HHV-6 reactivation and chromosomally integrated HHV-6 (ciHHV-6) in pre- and posttransplant patients (148). Differentiation is based on accurate determination of the ratio of HHV-6 to a eukaryotic cell marker, which is less precise using standard qPCR. This distinction is critical for posttransplant patients with a high HHV-6 load, since ciHHV-6 is often not associated with disease (126). Despite the current paucity of clinical studies, based on these examples it is easy to speculate that the primary strength of dPCR in infectious disease is quantification of viral load, including identification of minor species in a specimen, such as drug-resistant HIV quasispecies.
The limitations of dPCR are similar to those encountered with traditional qPCR. The ability to identify multiple or poorly conserved single nucleotide polymorphisms (SNPs) in a specimen is complicated by the necessity for specific primers and probes for each SNP of interest. Therefore, only well-characterized mutations can be identified and quantified. Additionally, multiplexing of dPCR will be limited by the instrument optics and number of fluorophores available for assay design. Currently, dPCR platforms range in multiplexing capability from 2 to 5 colors, which limits the ability of dPCR to evaluate complex genotypes or simultaneously detect multiple markers of antimicrobial resistance. For these applications, next-generation sequencing is likely the technology of choice.
Nucleic Acid Sequencing Methods
Sanger sequencing.
Originally described in 1976, DNA sequencing has undergone significant modifications, culminating in massive parallel or “next-generation” whole-genome sequencing (WGS) methods which are gaining favor for use in clinical microbiology laboratories. Allan Maxam and Walter Gilbert first achieved reliable DNA sequencing using a method that involved radioactive labeling of the DNA and chemical treatment to break DNA into small fragments. Fragments from each of four parallel reaction mixtures (one for each nucleotide) were electrophoresed side by side, with visualization using X-ray film autoradiography. Banding patterns in each lane corresponded to radiolabeled DNA fragments containing one of four radiolabeled nucleotides, from which the sequence could be inferred. Maxam-Gilbert sequencing quickly fell out of favor due to its technical complexity and use of hazardous chemicals, which complicated scale-up and prevented its use in standard molecular biology kits. Chain termination sequencing, or Sanger sequencing, improved upon the Maxam-Gilbert method by using dideoxynucleoside triphosphates (ddNTPs) as DNA elongation terminators. Sequencing reaction mixtures were again divided into four parallel vessels, each containing one of four ddNTPs (ddATP, ddGTP, ddCTP, or ddTTP) along with an excess of standard dNTPs. The resulting reaction mixtures contained DNA fragments of different lengths representing each size fragment produced by termination following inclusion of the given ddNTP. Fragments from the four reaction mixtures could then be separated by gel or capillary electrophoresis with a resolution of one nucleotide. The ability to radioactively or fluorescently label each ddNTP enabled detection of fragments and reading of sequence data by automated sequencing instruments. These advances in automation and analysis made nucleic acid sequencing a realistic option for diagnostics; however, several technical challenges still prevented the widespread use of Sanger sequencing in the clinical laboratory. Poor quality in the first 15 to 40 bases of the sequence and deteriorating quality of sequencing data after 700 to 900 bases limit its applicability to relatively short DNA fragments. Additionally, Sanger sequencing reactions are limited to sequence analysis of a single amplicon per reaction. This prevents the analysis of complex specimens such as sputum or abscess which contain multiple organisms (149).
NGS.
Next-generation sequencing (NGS) refers to a high-throughput sequencing method that parallelizes the sequencing process, producing thousands or millions of sequences at once. Intentionally broad, next-generation sequencing encompasses several different sequencing technologies that have been adapted to high-throughput, low-cost sequencing. A thorough review and comparison of these methods has been published elsewhere (150), but we summarize the key differences and applications of the major NGS approaches (Table 3).
TABLE 3.
Comparison of nucleic acid sequencing methodsa
| Characteristic | Ion Torrent | 454 Sequencing | Sanger sequencing | SOLiD |
|---|---|---|---|---|
| Sequencing chemistry | Ion semiconductor sequencing | Pyrosequencing | Terminator sequencing | Ligation-based sequencing |
| Amplification approach | Emulsion PCR | Emulsion PCR | Liquid-phase reaction | Emulsion PCR |
| Mb/run | 100–400 | 400–700 | 0.001 (1,000 bp) | 150,000 |
| Time/run | 1.5 h | 7–10 h | 3 h | 7–9 days |
| Read length (bp) | 200 | 400 | 800–1,500 | 35 × 75 |
| Reads/run | ∼1,000,000 | ∼1,000,000 | Not applicable | 700,000,000–1 billion |
| Sequence accuracy (%) | 98.4–98.9 | 99.51–99.96 | 99.999 | 99.94–99.99 |
| Cost (US$) per: | ||||
| Run | ∼500–700 | 6,000–8,000 | 100.00 | 4,000 |
| Mb | <5.00 | 10.00–15.00 | 2,400.00 | 0.04 |
| Instrument | 50,000 | 500,000 | 100,000 | 595,000 |
Pyrosequencing, licensed by 454 Life Sciences and later purchased by Roche, was the first next-generation sequencing method commercially marketed. Pyrosequencing employs a “sequence-by-synthesis” approach, meaning that it generates sequence data during DNA synthesis rather than analyzing nucleic acid amplicons postsynthesis as is the case with Sanger sequencing (151, 152) (Fig. 7). Amplified or chromosomal target nucleic acid is fragmented, and synthetic nucleic acid adaptors are enzymatically ligated to each end of the product. One adaptor serves as an adaptor for hybridization of the nucleic acid product to a microbead, and the other serves as a sequencing primer. Following a PCR to amplify the target sequence, microbeads coated with amplicon are segregated into microwells. Each well contains all the reagents required for sequencing, including DNA polymerase, luciferase, ATP sulfurylase, and apyrase. Each of the four dNTPs is individually added and washed away from the wells in repeating cycles. When a complementary dNTP is added, it is incorporated by DNA polymerase, with the concomitant release of pyrophosphate as a by-product of DNA synthesis. ATP sulfurylase converts the released pyrophosphate to ATP, which is used to drive luciferase activity, resulting in the production of light (153). Sequence data are generated by monitoring the microwell reactions for a pulse of light following addition of each dNTP. Since each microwell contains a single microbead harboring a unique region of chromosomal DNA, parallel sequencing of hundreds of regions of the chromosome achieves high sequence coverage in a single run. Additionally, because sequencing reactions are carried out in picoliter-volume reaction wells, this technology is capable of sequencing 400 to 600 megabases of DNA per 10-h run at a price per base up to 100-fold lower than that for Sanger sequencing (154) (Table 3). Pyrosequencing was initially capable of generating accurate reads of approximately 100 bases, with the limiting factor related to decreasing efficiency of apyrase in degrading unincorporated nucleotides in each successive cycle (155). Replacement of apyrase with thorough washing to remove unused nucleotides can extend the effective read length to approximately 400 bases. This is still a relatively short read in comparison to that with the Sanger method, but it is significantly longer than those of other NGS methods (155). An extended read length can be advantageous when attempting rapid whole-genome sequencing (WGS), especially when coupled with the speed of pyrosequencing technology and sophisticated software capable of assembling short individual reads into a confluent genome sequence. The overall accuracy of the sequence data generated is 99.51% to 99.96% (156, 157). A potential drawback to pyrosequencing is the inability to generate reliable sequences of homopolymers of >4 bases in length (156). In a study assessing the accuracy of sequences generated by pyrosequencing, 39% of errors were attributable to homopolymer sequences (156).
FIG 7.
Next generation sequencing by synthesis. Next generation sequencing by 454 (pyrosequencing) and Ion Torrent (semiconductor sequencing) utilize similar techniques to generate sequence information. In an initial step, genomic or amplified DNA to be sequenced is fragmented and single-strand overhangs are enzymatically removed. Synthetic nucleic acid adaptors (red and green) are ligated to each end of the target nucleic acid fragment. The modified target is then denatured and incubated with microbeads coated with a single-stranded capture probe (red) complementary to one of the adaptors (red). Hybridization immobilizes the target onto the surface of the bead, and beads are then partitioned into oil emulsion droplets containing reagents required for PCR. The PCR amplifies the target sequence, resulting in a single bead coated with thousands of identical copies of the target sequence. Following PCR, the beads are partitioned into microwells for sequencing. Each well contains the reagents required for sequencing with the exception of nucleotides. For both pyrosequencing and semiconductor sequencing, wells are washed with each of the four nucleoside bases in sequential order. 5a, in pyrosequencing, addition of a complementary nucleoside results in the release of pyrophosphate (PPi). The PPi is converted to ATP by ATP sulfurylase in the presence of adenosine 5′ phosphosulfate (APS), which is used to drive light production by luciferase. 5b, in semiconductor sequencing, release of H+ upon addition of a complementary nucleoside results in a change in pH, which is measured by a semiconductor in the bottom on the sequencing well. In both cases, the intensity of the signal (light or pH change) is proportional to the number of nucleotides incorporated. Therefore, addition of two consecutive nucleotides (e.g., GG) will generate a signal approximately twice the intensity of that generated by a single nucleoside insertion.
Semiconductor sequencing, typified by the Ion Torrent system (ABI), is a similar “sequence-by synthesis” technology. Parallel sequencing reactions are carried out in 1.2 million microwells on the surface of a low-cost semiconductor chip (158). Each picoliter well contains template and DNA polymerase, to which each of the four nucleosides is added in sequential order, however; Ion Torrent sequencing differs from pyrosequencing in that it uses production of hydrogen as the sole marker for determining the sequence (Fig. 7) (158). Release of hydrogen ions following incorporation of a complementary nucleotide is detected by a miniaturized ion sensor integrated into each reaction well. This technology is capable of generating up to 25 Mb of sequence data in a single run with a 2-h run time (158). Independence from the use of multiple enzymes, sensitive optics, or modified nucleotides dramatically reduces the cost of reagents and equipment compared to those with Sanger or other NGS methods. The reported cost of an Ion Torrent instrument is approximately US$50,000, excluding sample preparation equipment and a server for data analysis (159). The reported accuracy of semiconductor sequencing systems, including Ion Torrent, ranges from 98.4% to 98.9% (158, 160) (Table 3). The major limitations of this system are that it has difficulty in enumerating long repeats (homopolymers of >6 nt in length) and has a read length of 50 to 100 nt, which is relatively a short compared to that of Sanger sequencing or pyrosequencing (158).
Applications of pyrosequencing and semiconductor sequencing include whole-genome sequencing (WGS), amplicon sequencing, transcriptome sequencing, and metagenomics. The strength of pyrosequencing for WGS was demonstrated by Margulies et al., who sequenced the entire genome of M. genitalium (580,096 bp) with >99.9% accuracy and 96% genome coverage in a single run (157). More impressively, pyrosequencing was utilized to sequence the entire 6-gigabase human genome with 7.4× coverage in just 2 months (154). Similarly, the whole genomes of Escherichia coli and Vibrio fischeri were sequenced with 96.8 to 99.9% coverage with 98.9% accuracy in a single run using Ion Torrent (158). While the sequencing and assembly of an entire genome in days to months are remarkable, the most immediate use of NGS in clinical microbiology is likely amplicon sequencing. Amplicon sequencing is targeted to full sequencing of one or more genetic loci concurrently. This method is valuable when identification of multiple mutations or SNPs in a genetic locus is required to predict oncogenic potential or antimicrobial resistance. In addition to detection of multiple SNPs in a single locus, parallel sequencing offers the ability to generate sequences for multiple loci simultaneously. Next-generation sequencing is among the molecular technologies that can be applied to the identification of mycobacteria, including the prediction of resistance to antituberculosis therapies (64). Determination of resistance to first-line antituberculosis drugs (rifampin [RIF], isoniazid [INH], pyrazinamide [PZA], and ethambutol [EMB]) requires the analysis of several SNPs contained on 5 different genes (161). SNPs associated with resistance to rifampin are relatively conserved, with 3 mutations accounting for up to 75% of resistance (161). In this instance, routine probe-based amplification tests can be up to 98% sensitive (162). However, SNPs resulting in resistance to other first line antituberculosis drugs are considerably less conserved, rendering detection by a limited number of probes impractical. Pyrosequencing has been exploited for the simultaneous detection of resistance mutations in multiple genes to rapidly identify multidrug-resistant (MDR) strains of M. tuberculosis (163, 164). Resistance to rifampin, isoniazid, and fluoroquinolones was determined using 4 sequencing primers to identify multiple point mutations in rpoB, katG, and gyrA, with sensitivities of 96.7%, 63.8%, and 70%, respectively. The specificity of the pyrosequencing reaction was reported to be 97.3% to 100% (163). Variable sensitivity for predicting susceptibility to the 3 drugs reflects the lack of knowledge regarding the mutations and mechanisms which contribute to a resistant phenotype. This limitation is inherent to all molecular testing strategies and will be overcome only through continued research to characterize mutations conferring resistance and development of more complete reference libraries for sequence comparison.
Analogous to the use of NGS methods to sequence multiple targets in a single organism is the utility of NGS to simultaneously sequence and identify multiple organisms in a single specimen. Many of these studies, known as metagenomics, have been conducted to characterize complex bacterial communities in environmental specimens. Clinically, NGS has been used characterize the microbial community present in the airways of patients with cystic fibrosis (CF) using sputum specimens (149, 165). An advantage of NGS is the detection of nonculturable or fastidious organisms that may be outcompeted and overlooked in routine CF cultures. In a cohort of 66 sputum specimens from CF patients, NGS identified 122 different microbial species, compared to only 18 identified by culture (149). In an analytic study, organisms representing as little as 0.25% of the total nucleic acid template in a specimen were reproducibly identified (149). This ability to better define the microbiological components of the CF lung could aid in a better understanding of the associated illness and inform therapeutic strategies. A potential drawback to this type of metagenomic study is the semiquantitative nature NGS, which prevents an accurate assessment of the proportion of each organism present at a single point or changes in the composition of microorganisms in serial specimens. Similarly, the presence of nucleic acid is not necessarily indicative of a viable organism and may represent residual nucleic acid from flora or exogenous sources entering the upper respiratory tract.
A final application of pyrosequencing and semiconductor sequencing is in epidemiological investigation of outbreaks. Most notably, Mellmann et al. used the Ion Torrent NGS to identify and characterize a novel strain of enterohemorrhagic E. coli (EHEC) responsible for a large outbreak in Germany in 2011 (166). Whole-genome sequencing of 4 isolates from geographically distinct cities along with relevant historical reference strains was conducted. Sequencing and analysis of the strains were completed in 2 to 3 days and enabled near-real-time phylogenetic linkage of these strains (166). Investigators were also able to propose a likely evolutionary pathway linking the outbreak strain to an earlier progenitor strain identified 20 years earlier. In a smaller study, investigators were able to examine 33 multidrug-resistant isolates of E. coli obtained from patients in a neonatal intensive care unit using Ion Torrent NGS (167). The authors reported a 5-day turnaround and a cost of US$300 per isolate for whole-genome sequencing. Sequencing resulted in 88% to 89% genome coverage, which was sufficient to link all strains phylogenetically and identify them as most closely related to multiresistant strains of the ST-131 multilocus sequence type (MLST). While approximately twice the cost of traditional strain typing using pulse-field gel electrophoresis (PFGE) or MLST, NGS provided additional useful information, including the specific identification of the blaCTX-M-15 extended-spectrum beta-lactamase (ESBL) gene and the presence of other genes and point mutations associated with resistance to several classes of antimicrobials (167).
The term “ultradeep sequencing” (UDS) refers to amplicon sequencing designed to allow mutations to be detected at extremely low levels in a population. Initial PCR amplification of a genetic region of interest followed by segregation of each amplicon into a separate reaction well allows sequencing and identification of rare sequence variants. For example, ultradeep sequencing has been successfully used to detect HIV quasispecies and the emergence of resistant subpopulations. Analysis of blood samples from HIV-infected patients using pyrosequencing identified strains with mutations in the viral reverse transcriptase gene at levels of <0.1% of the total viral population (168, 169). Similarly, Ion Torrent sequencing was utilized to identify the emergence of mutations conferring resistance to nonnucleoside reverse transcriptase inhibitors (NNRTIs) and protease inhibitors (PIs) at a level of <1% of the total population through an average of 13,700× coverage of the gag-pol loci, though it was noted that coverage decreased significantly in a homopolymeric region containing five consecutive guanine residues (170). This is again in comparison to routine Sanger methods, which demonstrate a limit of detection of approximately 20 to 35% of the population (171). Accurate sequence data with error rates of <0.05% are easily achieved due to the high number of parallel reads, which provide highly redundant coverage of the target sequence (168, 169). Pretherapy resistance testing has been recommended to identify quasispecies with mutations known to confer resistance to antiretrovirals and is also recommended following a rise in HIV load attributed to therapy failure (172). Early detection of mutant alleles present at a low frequency is key in selection of antiretroviral therapy, since discontinuation of a specific antiviral can result in reversion of mutant populations to a susceptible, pretherapy genotype (168, 173). Future clinical applications of pyrosequencing include transcriptome sequencing, which aims to efficiently create RNA profiles and examine the effects of mRNA transcript expression. The majority of research using NGS for transcriptome analysis has involved the basic sciences; however, recent studies have utilized this method for comparison of mRNA expression in normal and malignant cell populations and for discovery of latent or cryptic viruses whose presence and expression may be associated with malignancies (174–176).
Other NGS platforms such as Illumina and SOLiD are capable of generating 1.5 to 4.0 Gb of data per single run at a cost of less than $0.10 per kilobase, which is significantly less expensive than Sanger or other NGS methods (177, 178). Illumina (Solexa) sequencing is based on reversible dye terminators. DNA molecules are first attached to primers on a slide and amplified so that local clonal colonies are formed. Four types of reversible terminator bases are added, and nonincorporated nucleotides are washed away. A camera takes images of the fluorescently labeled nucleotides, and then the dye along with the terminal 3′ blocker is chemically removed from the DNA, allowing the next cycle. In contrast, SOLiD (supported oligonucleotide ligation and detection) is a method of sequencing by ligation. A target-specific sequencing primer is used to initiate sequencing by the sequential addition of octamer probes, each containing 2 specific nucleotides at the 5′ terminus followed by 6 degenerate nucleotides. Each of the 16 possible combinations of two nucleotides is represented, and octamers are fluorescently labeled with one of 4 fluorophores. The 16 octamers are then grouped into 4 sets (each containing one each of the 4 fluorophores) and are added sequentially to the sequencing reaction mixture for 7 full cycles of the 4 groups. Fluorescence is measured after addition of each 4-member group of probes, and the 2-base sequence is determined by the fluorophore detected. Gaps in the sequence corresponding to the 6 degenerate nucleotides in each probe are filled in by repeating the reaction using additional sequencing primers, each offset by one nucleotide (n − 1, n − 2, etc.) from the initial primer (177). This results in short reads (26 nucleotides); however, the sequencing error rate is reduced to 0.001 because each nucleotide in the template is read twice (177). The disadvantage of this technology is turnaround time. The run time for a single sequencing reaction is 2.5 to 6 days, resulting in turnaround time for a full genome sequence of up to 2 weeks (178). Because of the large amount of sequence data generated per run, low cost per base sequenced, and extended TAT, these platforms are currently best suited to whole-genome sequencing projects rather than rapid identification of microorganisms or SNP polymorphisms in a clinical laboratory. Most recently, Illumina has begun offering full genome sequencing through it reference laboratory at a reported cost of $4,000.00 per genome.
MASS SPECTROMETRY METHODS
Mass spectrometry (MS) methods, including gas chromatography-MS (GC-MS), liquid chromatography-MS (LC-MS), and high-resolution tandem MS (LC-MS/MS), have increasingly been exploited by clinical chemistry laboratories for monitoring of drug and hormone levels in blood and urine specimens (179). Only recently have mass spectrometry methods been broadly applied to the identification of bacteria and other microorganisms in the clinical microbiology laboratory. Among these methods are electrospray ionization (ESI)-MS, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS, and ion trap-based identification technologies. Each MS approach has unique strengths and weaknesses which must be considered when implementing MS for routine use in a clinical laboratory (Table 4).
TABLE 4.
Characteristics of MALDI-TOF MS and ESI-MS systems used in clinical microbiology
| Characteristic | MALDI-TOF MS | ESI-MS |
|---|---|---|
| Platform(s) | Bruker Biotyper, Vitek MS | PLEX-ID |
| Analytes | Cultured whole organisms, including bacteria, yeast, filamentous fungi; extracted protein preparations. | Nucleic acid amplicons; can be applied to bacteria, fungi, viruses, noncultivable microorganisms present in clinical specimens |
| Preanalytic steps | Transfer of organism to target plate, overlay with matrix material; identification may be improved by formic acid extraction of proteins | Multiplexed PCR-based amplification of target sequence to be analyzed; may use specific or broad range primers. |
| Carrier/matrix | Analyte embedded in weak acid matrix material such as alpha-cyano-4 hydroxycinnamic acid or 2,5-dihydroxybenzoic acid | Analyte dissolved in organic aqueous phase carrier |
| Ionization method | Excitation by laser catalyzes charge transfer from matrix to analyte, desorbs ions from solid phase on target plate | Analyte in liquid phase is exposed to a high voltage and passed through a capillary tube, which generates a spray of charged ions |
| Detection | Mass analyzer detects time of flight of each ion species in the specimen | Mass analyzer detects time of flight of each ion species in the specimen |
| Output | Spectral profile based on mass-to-charge ratio of all proteins present in a specimen | Exact mol wt of nucleic acid amplicon present in specimen |
| Basis for identification | Comparison of analyte spectral profile to commercially available or laboratory developed reference spectral library | Weight of amplicon is used to calculate exact no. of each of the 4 nucleotides (A, T, C, G) present in amplicon; nucleic acid composition is compared to reference library |
| Direct specimen analysis | No; requires cultured microorganism. | Yes; initial PCR can be performed directly on clinical specimens, including upper respiratory tract specimens, blood, sterile fluids |
| Limit of detection | 105-106 CFU | 40 genome equivalents |
Matrix-Assisted Laser Desorption Ionization–Time of Flight MS
A major factor enabling the application of MS to the identification of bacteria and other microorganisms was the advent of nonfragmenting or “soft ionization” techniques, including MALDI-TOF MS (Fig. 8), which facilitates the analysis of large macromolecules, including nucleic acids and proteins (180, 181). Various analytes, including whole bacteria, are transferred to the surface of a metal plate and embedded in an acidic matrix material such as alpha-cyano-4-hydroxycinnamic acid (HCCA or CHCA) or 2,5-dihydroxybenzoic acid (DBA). Excitation by a nitrogen laser catalyzes charge transfer from the matrix to the analyte and causes desorption of the newly ionized particles. The resulting ions are then accelerated through a vacuum tube which separates ions based upon their mass-to-charge (m/z) ratio. Detection of ions by a mass analyzer at the distal end of the tube results in creation of a mass spectral profile in which the m/z of each ion in the sample is plotted on the x axis and relative abundance plotted on the y axis. This spectral profile is unique to each analyte. Analysis of an analyte typically involves several hundred independent laser shots to create a consensus spectral profile and establish a low background threshold (180, 181).
FIG 8.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). (A) Preparation of samples for analysis by MALDI-TOF MS can be either by direct transfer of a bacterial colony to the sample plate using a sterile implement (green spots) or as a liquid supernatant following an extraction procedure (blue spots). In either method, the analyte is allowed to dry before being overlaid with a weak acid matrix material. (B) Analysis of the analyte begins with exposure to a laser, which ionizes and desorbs analyte form the sample plate. The created ions are accelerated through the time of flight vacuum tube by application of an electrostatic field until they reach the MS detector. Ions with a larger mass-to-charge ratio (m/z) will take longer to traverse the time of flight tube than ions with a smaller m/z. An MS profile is created with the m/z of each ion species plotted on the x axis and the relative abundance of each m/z ion species on the y axis. This MS profile is compared to a reference spectral library of defined spectra to establish a “best-match” identification of the isolate being analyzed. (Reprinted from reference 2 with permission.)
MALDI-TOF MS has been used in clinical laboratories in Europe for nearly a decade and has been more recently adopted in the United States (182). This method is readily adaptable to the direct analysis of bacterial and fungal isolates, requires inexpensive reagents, and is capable of returning an identification result in approximately 15 to 30 s once the sample is loaded into the MALDI-TOF MS. These attributes make integration of MALDI-TOF MS an attractive option for the microbiology laboratory. The performance and clinical utility of MALDI-TOF have been thoroughly reviewed by Clark et al. (2). In brief, the majority of clinical evaluations have been conducted using one of two commercially available systems, the Bruker Biotyper (Bruker Daltonik, Bremen Germany) and the Vitek MS (bioMérieux, Marcy l'Etoile, France). Large studies have demonstrated similar overall performance of these two systems for the identification of bacteria routinely encountered in the laboratory; however, minor differences have been noted within specific groups of organisms. In head-to-head comparison studies, the Bruker Biotyper provided a larger proportion of “high-confidence” identifications among Gram-negative nonfermenters (97.0% versus 89.5%), while the Vitek MS was superior for identification of anaerobes and viridans group streptococci (183–186). In addition, several multicenter studies have evaluated the ability of the Vitek MS to identify specific subgroups of microorganisms, including Gram-positive isolates (187), fastidious bacterial pathogens (188), Gram-negative non-Enterobacteriaceae (189), and anaerobic bacteria (190). Regardless of system, identification rates for routine clinical isolates using MALDI-TOF MS range from 90% to 95% compared to 16S rRNA gene sequencing and can be completed within minutes, with a substantial savings in cost per identification (191–193).
The clinical application of MALDI-TOF MS has traditionally been restricted to identification of isolated pure colonies following culture on solid medium. This is the result of two technical limitations inherent to the current technology. The first of these is related to the limit of detection. Unlike amplified molecular methods, MALDI-TOF MS relies on analysis of whole-cell or extracted protein specimens. Generation of an adequate spectral profile requires that sufficient material be deposited onto a target for analysis. Studies to determine the minimal amount of cellular material or extracted protein required for MALDI-TOF MS analysis have found a minimum of 1.5 × 105 CFU to be required for reproducible and accurate identification of microorganisms (194, 195). The second limitation is the inability of current software to deconvolute or separate multiple spectra collected simultaneously, as would occur during analysis of mixed or polymicrobial cultures. Simply put, the combined spectra of two organisms will not match any single organism spectrum in the reference library and will result in low or unacceptable confidence scores. This has been observed in the analysis of polymicrobial specimens, including blood cultures containing more than one organism, and in the direct analysis of urine specimens, which are often polymicrobial (194, 196, 197). Combined, these limitations generally restrict the use of MALDI-TOF MS from primary clinical specimens, which typically do not have sufficient cellular material for analysis and, depending on the source, are often polymicrobial in nature.
Despite the limitations discussed above, direct-from-specimen identification has been attempted using positive blood cultures and urine specimens (194, 196, 198–201). Direct analysis of positive blood cultures has been aided by standardized protocols using commercially available (research-use-only) kits and centrifugation-based methods for isolation of bacteria and yeasts from broth culture (196, 198). Using these protocols, the organism present in 85% to 98% of monomicrobial blood cultures was correctly identified in as little as 30 min (196, 198–200). An alternative protocol using filtration-based isolation of microorganisms from positive blood cultures demonstrated approximately 80% success in identification of bacteria and yeast from 225 monomicrobial blood cultures (197). These advantages in laboratory turnaround time, coupled with the initiation of an antimicrobial stewardship program, impacted patient care through significantly shorter times to optimal antimicrobial therapy and reduced 30-day mortality for patients with bloodstream infection (202, 203). Direct analysis of urine has also been attempted using MALDI-TOF MS. Preprocessing steps include low-speed centrifugation to remove leukocytes followed by high-speed centrifugation to pellet any bacteria present. Urine specimens containing >105 CFU/ml of a single species could be reproducibly and correctly identified for 92% to 95% of specimens containing a predominant Gram-negative organism and 75% to 93% containing a predominant Gram-positive organism (194, 201). An alternative method for isolation of microorganisms based on diafiltration of urine has also been evaluated. This method lowered the limit of detection for reliable identification of bacteria by 10-fold compared to previously reported centrifugation- or filter paper-based isolation methods; however, the sensitivity of this method was reported to be 67% (195). In general, for all studies involving direct analysis of urine specimens, those containing <105 CFU/ml of a single organism or those with >1 organism present in similar quantity either failed to generate adequate spectral profiles for analysis or returned low-confidence identifications (194, 201). Combined, these limitations can account for 10% to 15% of specimens which would not be acceptable for identification using MALDI-TOF MS. However, identification of the likely pathogen in up to 90% of urine specimens within minutes may aid in guiding empirical therapy for those pathogens that have predictable resistance or susceptibility to antibiotics commonly used to treat urinary tract infections.
MALDI-TOF MS has more recently been expanded to include analysis of yeasts, filamentous fungi, and mycobacteria (204–212). Obstacles to the direct identification of fungi, such as a more robust cell wall, were overcome using simple ethanol, formic acid, and acetonitrile extraction steps prior to analysis. Identification of yeasts using these methods demonstrated species-level identification for 92.5% to 98.2% of isolates (211, 213–215). Notably, the use of a formic acid overlay onto the isolate following spotting to the target plate increased the identification rate by approximately 7% for isolates analyzed using the Vitek MS (final identification rate, 97.4%) and by 60% when using the Bruker Biotyper (final identification rate, 84.6%) (212). Additionally, it was necessary to reduce the score threshold for “species-level identification” to 1.7 when using the Bruker Biotyper to increase the proportion of species level identifications from 57% to 85%. It is important to note that these data reflect the analysis of primarily Candida sp. yeasts and that identification of other yeast-like isolates (Aurobasidium, Cryptococcus, Trichosporon, Pichia, and Geotrichum) was less successful, resulting in identification of only 41.2% to 61.9% of isolates (213, 215). This shortcoming was attributable to a lack of representation of these isolates in the spectral reference library and could be corrected by addition of spectra corresponding to type strains of these genera (215, 216). Similar results have been obtained for analysis of routinely encountered filamentous fungi, of which 87% to 89% were correctly identified using MALDI-TOF MS (206, 207). Again, the main limiting factor was representation of species in the reference library. Creation of supplemental libraries has been demonstrated to improve the identification rate, as typified by a recent study reporting correct identification of 97.8% of dermatophyte isolates provided that the species analyzed were well represented in the reference library (205).
Identification of mycobacteria, including both M. tuberculosis complex (MTBC) and nontuberculous mycobacteria (NTM), has also been reported and requires additional preprocessing methods to achieve optimal identification scores and accuracy (208, 209, 217). These steps typically include inactivation using heat- and/or bead-based cell disruption followed by ethanol and formic acid/acetonitrile extraction of the isolate. Early studies demonstrated the ability to generate species-specific spectral profiles that could be used to differentiate closely related strains within both the M. tuberculosis complex and the M. avium-M. intracellulare complex and between closely relates species of rapid grow in mycobacteria, including M. abscessus and M. chelonae (210, 218). In a large study, Lotz et al. demonstrated 97% accuracy of MALDI-TOF MS for identification of 311 isolates cultured on solid medium (219). All of these studies were based on nonstandardized reference libraries developed in-house by individual laboratories or groups. More recently, investigators have evaluated MALDI-TOF using commercially available reference libraries. One such study compared the identification of mycobacteria using 88 isolates grown on solid medium versus direct identification from broth culture. The authors reported a higher identification rate when isolates were identified from broth culture than when they were identified from solid medium (98.8% versus 89.8%, respectively); however, the accuracies of identification from solid and broth culture were similar (92.5% to 95.4%) (208). A similar study examined 178 isolates cultured on two types of solid medium or in broth and reported an overall identification rate of 97.3%, with 93.8% correctly identified to the species level (209). A third study compared two commercially available MALDI-TOF platforms (Bruker MALDI Biotyper and bioMérieux Vitek MS) using 198 clinical isolates cultured on solid medium. The authors reported species-level identification for 94.9% of isolates using the Vitek MS compared to only 79.3% using the MALDI Biotyper. However, reducing the score threshold from 2.0 to 1.7 increased the identification rate for the MALDI Biotyper to 93.9% with only a minimal impact on the accuracy of identification (217). A potential weakness of these three studies was the use of archived clinical isolates or type strains to seed culture medium. This does not account for materials present in clinical specimens, such as other microorganisms, mucus, or other matrix material, that could potentially interfere with the MALDI-TOF analysis. More recent publications have focused mainly on refining and optimizing extraction method and evaluating clinical specimens (217, 220). In summary, these data suggest that MALDI-TOF MS has the potential to accurately identify a wide variety of mycobacteria to the species level from solid or broth culture. Identification directly from broth culture can reduce the time to identification by several days to weeks compared to that with high-pressure liquid chromatography (HPLC) methods, which typically require subculture of positive broths to obtain an isolate on solid medium.
A very recent area of research is the determination of antimicrobial susceptibility using MALDI-TOF MS. Direct detection of drug-modifying enzymes, including beta-lactamases and aminoglycoside-modifying enzymes, is difficult because of the relatively low expression of these proteins in a cell. Similarly, detection of targets modified by methylation or point mutation, such as ribosomes or gyrase enzymes, is hampered by the relatively small difference in m/z between wild-type and modified proteins. These obstacles have been overcome using one of two approaches. In the case of beta-lactam resistance, hydrolyzed antibiotic can be detected using MALDI-TOF MS following 1 to 3 h of incubation of a test strain in the presence of a given antibiotic (221). The sensitivity of this method may be as high as 100%, and this has been applied to the detection of a number of expanded-spectrum cephalosporins and carbapenemases (221, 222). Importantly, this method lacks the ability to detect strains in which resistance is mediated by alternative mechanisms such as altered penicillin binding proteins, reduced permeability, or active efflux pumps. To overcome this limitation, Sparbier et al. have developed an assay based on the differential detection of isotopically labeled proteins (223). Using this approach, the test strain is incubated in growth medium containing a stable amino acid isotope in addition to a given antibiotic. If the strain is resistant to the antibiotic, it will replicate and the isotope will be incorporated into daughter cells. MALDI-TOF MS analysis of the culture can differentiate between proteins of wild-type mass and those containing the heavier isotope, thereby identifying strains able to grow in the presence of the antibiotic. In addition to being rapid (∼3 h), this method should be broadly applicable to any class of antibiotic or resistance mechanism because it is dependent on growth of the microorganism rather than detection of specific genes or proteins. From a practical standpoint, this method is similar to susceptibility methods using broth microdilution to establish a MIC for a given organism/antibiotic combination. However, processing of each well for each antibiotic tested would be required for MALDI-TOF MS analysis. Unless automated, this process is likely too labor-intensive to be widely applicable in clinical microbiology laboratories as a first-line method for susceptibility testing.
Integration of MALDI-TOF MS into the clinical microbiology laboratory has the potential to significantly reduced turnaround time for identification of bacterial, fungal, and mycobacterial isolates at cost of < US$1 per isolate, including labor (193). A further advantage of MALDI-TOF MS compared to other spectrometry or nucleic acid-based identification techniques is the simplicity of the preanalytic steps, which include transfer of an isolate to the target plate and addition of matrix material. This ease of sample preparation lends well to integration into a total laboratory automation (TLA) line, which can further reduce technologist hands-on time and enable “around-the-clock” identification and reporting of results (see “Liquid Microbiology and Total Laboratory Automation” below). Despite these benefits, there are also shortcomings of MALDI-TOF MS which currently limit its utility. Among these is the inability to identify microorganisms directly from clinical specimens such as swab, wound, or biopsy specimens without the need for culture. This limits the organisms that can be identified to those which are readily recovered on standard laboratory media and precludes the use of MALDI-TOF MS for the identification of viruses in clinical specimens. Additionally, the ability to simultaneously identify multiple microorganisms in a complex specimen has not been reliably demonstrated.
Electrospray Ionization MS
Like MALDI-TOF MS, ESI-MS is a form of soft ionization which lends itself to analysis of larger macromolecules, including proteins and nucleic acids (179, 180). In contrast to MALDI, ESI requires analytes to be dissolved in a liquid-phase carrier (aqueous or polar) for analysis (Table 4). The solute is then passed through a heated capillary and is exposed to a high voltage to generate an aerosol of ions, which are analyzed based on time of flight using a detector similar to that used in MALDI-TOF MS (179, 180). To date, clinical microbiology studies evaluating ESI-MS have focused on the analysis of amplified nucleic acid products using the PLEX-ID platform (Abbott Molecular, Des Plaines, IL). This platform requires offline extraction of total nucleic acid from the clinical specimen, which is used as the template for one or more multiplexed PCRs. Amplification reaction mixtures are typically arranged using multiple wells of a 96-well plate with 2 to 4 primer sets per well, which target a specific group of agents (influenza viruses, food-borne pathogens, biothreat agents, etc.). The amplicon from each PCR is subjected to ESI-MS, and the exact molecular weight of the PCR product is determined. This weight is used to calculate the nucleotide composition (number of A's, T's, C's, and G's) of the amplicon, which is unique. The exact nucleotide composition is then compared to a reference library, thereby generating an identification of the microorganism present in the clinical specimen. The specificity of this method is increased by multilocus analysis of each microorganism targeted by the multiplex PCR step. The use of the nucleotide composition of an amplicon rather than probing for a specific sequence enables the detection of novel strains that may be missed when using standard hybridization probe-based detection methods. As an example, initial identification of the 2009 pandemic H1N1 strain of influenza virus using ESI-MS was reported, whereas this strain was untypeable using routine molecular methods (224). Additionally, because ESI-MS is sufficiently sensitive to detect single nucleotide differences (e.g., SNPs), it has found utility for epidemiology and strain typing (224, 225). Potential advantages of this method compared to protein-based analysis using MALDI-TOF MS include the ability to conduct identifications directly from specimens without subculture, increased sensitivity for low-burden specimens, the ability to identify multiple pathogens in a single specimen (i.e., multiplexing), the ability to identify viral pathogens, and the possibility to identify genetic markers of antibiotic resistance.
ESI-MS has been applied to the identification of bacterial pathogens obtained from a variety of matrices, including food, cell cultures, environmental samples, and positive blood culture broths (226–230). A multiplex PCR panel coupled with ESI-MS was evaluated for the identification of food-borne pathogens and demonstrated 99% to 100% sensitivity for Salmonella and E. coli isolates, including accurate serotyping for 30% of Salmonella isolates (229); however, identification of Shigella isolates was variable, ranging from 100% (S. sonnei) to 42.1% (S. dysenteriae). Direct analysis of food samples containing enteric pathogens was also variable, with a sensitivity of less than 50% for tomatoes and chili powder but 100% for cheese and fish (229). ESI-MS has been also utilized to identify and differentiate species within 14 “biothreat clusters,” which include biothreat agents and closely related species (e.g., Francisella tularensis, F. novicida, and F. philomiragia) (226, 230). Based on a panel of 36 primer pairs and ESI-MS, the assay demonstrated analytic sensitivity as low as 40 genome equivalents and was 100% specific even when specimens contained an excess of a closely related, nonpathogenic organism (230). This methods was also evaluated using simulated specimens, including tissue, food, and environmental samples, and demonstrated similar performance (226).
Clinical studies have most thoroughly evaluated ESI-MS for detection of influenza viruses. Accurate detection and discrimination of subtypes are achieved using up to 9 primer pairs targeting broad (pan-influenza) and conserved (subtype-specific) regions of the influenza virus genome, including the nucleoprotein, matrix protein, nonstructural, and polymerase genes (231). Comparison of ESI-MS to routine molecular methods demonstrated 93.5% to 99.9% agreement, with superior performance in discriminating 2009 pandemic H1N1 strains (224, 231, 232). Two studies have applied ESI-MS to the identification of bacteria and yeasts in positive blood cultures (227, 228). In each study, the concordance of ESI-MS with routine biochemical methods was 94% to 96% for genus-level and 87% to 95% for species-level identification (227, 228). Concordance diminished to 76% for identification of multiple organisms in polymicrobial cultures (228). In a comparison of ESI-MS to MALDI-TOF MS, both methods demonstrated >95% concordance with routine identification methods; however, both methods demonstrated diminished performance when analyzing polymicrobial cultures (227).
A potential advantage of ESI-MS is the ability to identify organisms directly from patient specimens without the need for culture. This can be especially advantageous for the detection and identification of fastidious organisms or organisms present at a very low concentration in a specimen. A prime example of this is the diagnosis of prosthetic joint infections (PJI). Recent data have indicated poor recovery of organisms from joints suspected to be harboring bacteria when routine culture methods are employed (233–235). The authors attribute the poor recovery in standard culture to the comparatively low growth rate of bacteria associated with PJI, encasement of these bacteria in biofilms, and patients' recent receipt of antimicrobial therapy. The use of multiplex PCR for identification of bacteria associated with PJI has demonstrated superiority to culture (78% versus 65% sensitivity); however, the PCR test was negative for 22% of specimens which contained organism not included in the multiplex panel (234). Recently, ESI-MS has been used to analyze synovial fluid from presumed PJI affecting hip and knee joints as well as fluid obtained from explanted prostheses (236, 237). Synovial fluid specimens were analyzed using the Ibis T5000 ESI-MS instrument and a PCR plate containing degenerate primers for the identification of 3,400 bacterial pathogens as well as specific primers for identification of resistance markers, including mecA (methicillin resistance), vanA (vancomycin resistance), and blaKPC (carbapenem resistance). ESI-MS identified a likely pathogen in 100% of cases with a clinical diagnosis of PJI (78.3% culture positive) and also detected one or more organisms in 88% of cases clinically diagnosed as aseptic joint failure (236). Another group reported 77.6% to 82.7% sensitivity when using ESI-MS (culture, 69.7% to 76.7% sensitive) to analyze sonicate fluid obtained from explanted prostheses (237). The largest difference in sensitivity was observed in patients who had received antibiotic therapy within the previous 14 days. In this group, ESI-MS was 85.7% sensitive, compared to 65.7% sensitivity for culture (237). ESI-MS has also been applied to the identification of filamentous fungi in sputum specimens (238). Among organisms claimed by the manufacturer to be identified, the assay correctly identified 100% of isolates to genus level and 92.2% to species level; however, ESI-MS results were only 67% concordant with culture results when considering prospectively tested clinical specimens. Over 95% of isolates not detected using ESI-MS were present in low quantity (<20 colonies) in the clinical specimen. Additionally, multiple organisms were correctly detected by ESI-MS in just 6.8% of polymicrobial cultures, which are common among respiratory specimens (238). While ESI-MS appears to be superior to culture or real-time PCR for some applications, the clinical implications of ESI-MS-positive, culture-negative results and the role of identified microorganisms in respiratory cultures or prosthesis failure remain to be determined, and well-controlled clinical studies are needed.
Based upon data presented in these initial studies, ESI-MS (coupled with multiplexed PCRs) has merit as a viable method for the identification of microorganisms in the clinical laboratory. An advantage of this approach is the ability to identify viral, bacterial, fungal, and noncultivable organisms following successful amplification of nucleic acid targets. Additional advantages may include the ability to identify multiple organisms present in polymicrobial specimens and to identify microorganisms directly from specimens, although current literature fails to support this notion. As with any nucleic acid amplification-based technique, ESI-MS is limited by the primer/multiplex PCR design. The approach taken by several investigators is to design several low-density multiplex reaction mixtures containing primers for various “syndromes” or groups of organisms (e.g., food-borne pathogens or influenza viruses). The performance of ESI-MS is also dependent on a robust reference library to which the ESI-MS results are compared. Organisms which are not represented in the library may not be readily identified; however, closely related species or complexes of organisms can be easily separated based on discrimination of single nucleotide substitutions. A potential drawback compared to MALDI-TOF MS is the requirement for preanalytic steps, including nucleic acid extraction and PCR, which extend turnaround time to 4 to 6 h and lend the method better to batch processing than on-demand analysis of isolates. As routine sequencing methods, including next-generation platforms, become more commonplace, it is reasonable to debate the value obtained from determination of complete sequence versus nucleotide composition of amplicons provided by ESI-MS.
LABORATORY AUTOMATION
Workflow in the clinical microbiology laboratory is often manual and laborious, dependent on skilled technologists performing an array of diverse tasks to accommodate the specific needs of each type of specimen received. Specimens received by the clinical microbiology laboratory undergo multiple manipulations and are often handled by several technologists en route to reporting of a final result. These steps include receipt and accessioning, specimen processing, inoculation of culture media or nucleic acid extraction, analysis using phenotypic, biochemical, nucleic acid, or mass spectrometric methods, susceptibility testing as appropriate, and reporting of results. Inoculation of solid and liquid media for routine bacteriology is the first step in primary processing for bacteriology and is largely a manual process. While similar studies in the United States are lacking, a report from the European Union found that inoculation of specimens into culture medium accounted for up to 24% of technologist time during primary processing of specimens (239). The diversity of specimens received in the laboratory and the ever-expanding test menu has prevented a simple “one size fits all” linear flow of specimens through the laboratory. This has in turn complicated the automation of these processes. For an in depth review of the various systems available for front-end automation of the clinical microbiology lab, the reader is referred to a review by Novak and Marlowe (240). An overview of these technologies, including the relative benefits of each approach, follows.
Initial steps toward automation were limited to specific specimen types (often liquid specimens) and were restricted to automation of only one step in the complete process. Examples include the introduction of continuous-monitoring blood culture systems, which eliminated the task of repeated Gram staining and blind subculture, which were both manual and time-consuming. Early automated plate streaking devices were developed to speed and standardize the inoculation of specimens into culture medium, but these were restricted to liquid samples such as urine or bacterial suspensions that were manually prepared prior to plating. A major leap forward came with the miniaturization and automation of biochemical tests used for routine bacterial identification. Platforms such as the Phoenix (BD),Vitek (bioMérieux), and MicroScan (Siemens) replaced the need to manually inoculate, read, and interpret multiple biochemical test tubes for each isolate requiring identification. This allowed a significant increase in bacterial identifications that could be performed without increasing labor requirements. More recently, fully automated sample-to-result molecular platforms have been introduced. These systems (see previous sections) have simplified molecular testing and provide a method for on-demand testing.
These initial steps toward automation have undoubtedly eased the workload for today's laboratories; however, further automation will be required to meet the needs of a changing health care system. Consolidation of hospitals and centralization of laboratory testing will increase the volume of specimens received by laboratories, and specimens collected at satellite locations may arrive during evening or night shifts when staffing may be reduced. Both of these factors put additional stress on a workforce of skilled laboratory technologists that is declining in size and experience (241, 242). Automation of repetitive tasks and high-level screening of negative cultures will enable current technologists to focus on tasks requiring human intervention and technical expertise.
Automation in Specimen Inoculation
Inoculation of the clinical specimen into appropriate culture medium is one of the initial steps in nonmolecular microbiology workflow. A standardized quantity of the clinical specimen is transferred to one or more pieces of medium using a calibrated pipette or loop and is streaked either qualitatively or quantitatively prior to incubation of the culture(s). Specimens can be of various types and viscosities, including blood culture, urine, sputum, stool, pus, or other bodily fluid. Further, these specimens can arrive in the laboratory in containers of different shapes and sizes. Requirements for different inoculum volumes, streak patterns, specimen characteristics (e.g., viscosities), and containers complicate the automation of what may seem like a simple task. Initial steps toward automation of this process included streak-only instruments like the Isoplater (Vista Technology, Edmonton, Alberta, Canada), which still required the technologist to inoculate appropriate culture medium with the specimen prior to loading plates onto the instrument. The instrument would then streak the specimen over the surface of the plate in a set pattern and return the plates to a “completed” rack. A drawback to streak-only instruments is the requirement for a fair amount of manual intervention in the form of inoculation, loading, and unloading of culture plates.
Further advancements were made with the introduction of automated specimen handlers equipped with liquid handling features that could automatically inoculate plates and, in some cases, remove and recap standardized specimen containers. The Previ-Isola (bioMérieux) is one such system that delivers a standardized volume of liquid specimen to culture plates and then uses a disposable comb to spread the specimen over the surface of the plate in a circular pattern. When tested using surgical swab specimens eluted into liquid phase, 21% of specimens plated using the Previ-Isola generated more isolated colonies than manual plating (244). Greater advantages were noted for urine and preserved stool specimens, in which 57% and 58% of specimens, respectively, plated using the Previ-Isola resulted in higher numbers of isolated colonies than with manual plating (244). Single-use disposable tips and combs reduce the risk of cross-contamination of specimens but add cost and generate more waste than systems that use heat sterilization of metal loops or reusable metal beads. A potential drawback to the Previ-Isola is the requirement for manual vortexing, decapping, and recapping of specimen containers prior to loading to the instrument. This prevents the instrument from being a true walk-away system and introduces the risk of contamination during the time that specimens are open to the environment. Other automated specimen handlers, including the InocuLAB and Innova (BD), incorporate automated uncapping and recapping of specimen containers in addition to plate inoculation and streaking (239, 240). A drawback to the InocuLAB is that automated uncapping and recapping are limited to a single type of container within a processing run, which restricts the utility of this system for primary processing of multiple specimen types (245). In contrast, the Innova can accommodate a wide range of commonly used specimen containers and has optional loop- or pipette-driven specimen inoculation to manage a wider variety of specimen types and viscosities (239, 240). Regardless of system, preliminary studies have shown that automated inoculation and streaking of culture plates alone can result in a savings of up to 30 min of hands-on time per 100 cultures inoculated, provide more reproducible streaking results, and yield more isolated colonies than manual inoculation and streaking (243, 244, 246, 247). Additionally, newer identification systems, including MALDI-TOF MS and next-generation sequencing, are capable of generating organism identification and potentially susceptibility results from single colonies. Therefore, the ability to provide well-isolated colonies is of increasing importance, since it can eliminate the need for “isolation plate” subculture and incubation that can delay routine identification by an additional 16 to 24 h.
Liquid Microbiology and Total Laboratory Automation
Automation of specimen processing and plating is largely limited by the type of specimen submitted. Liquid-phase specimens such as urine, preserved stool, respiratory specimens, cerebrospinal fluid (CSF), and other bodily fluids can be manipulated through the use of pipettes or calibrated inoculation loops; however, liquid specimens represent only approximately half of the specimens submitted to the laboratory for routine bacteriology (239). The remaining specimens are comprised primarily of swabs (up to 35% of specimens submitted) but also include tissues, bone fragments, and other solid-phase specimens (239). To expand the proportion of specimens that can be amenable to automated processing, studies have begun to evaluate a new type of swab collection device which differs from traditional swab collection devices in two ways. First, the head of the swab is a solid bulb which is “flocked.” This arrangement employs short synthetic fibers that protrude perpendicular to the swab shaft rather than longer fibers wound around the swab tip as is the case for traditional swabs (Fig. 9). This arrangement allows for more efficient release of microorganisms when the swab is put into liquid, which is in contrast to traditional swabs, in which microorganisms may otherwise become trapped in the wound fibers. The second difference between traditional and flocked swab collection devices is the use of a standardized volume of nonnutritive transport medium (1 ml) rather than a moistened sponge or gel in the transport tube. Combined with more efficient release of organisms from the flocked swab, the use of liquid transport medium converts swab-collected specimens into liquid-phase specimens that are more amenable to automation. These flocked swab collection devices are available from several manufacturers and distributors, including Copan Diagnostics (Murrieta, CA), Puritan (Guilford, ME), and Millipore (Billerica, MA).
FIG 9.
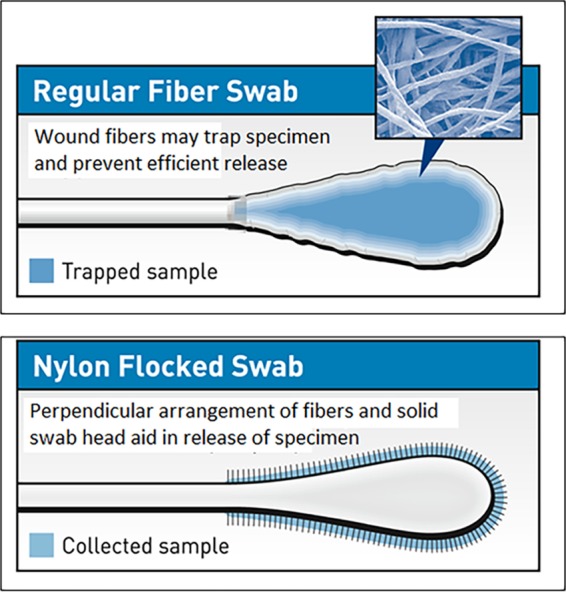
Comparison of wound fiber swabs to flocked swabs. Traditional swabs are constructed by winding fiber strands around the tip of a straight shaft to create a wound fiber bulb for collection of the specimen. Winding of fibers creates a “net” which may entrap microorganisms and prevent efficient release onto solid or liquid culture medium. Flocked swabs are composed of a solid bulbous core at the tip of the swab which is coated with perpendicular fibers. This arrangement allows for more efficient release of microorganisms collected in a specimen onto culture medium.
Analytical comparisons of one of these products, ESwab (Copan) to other frequently used swab collection devices using aerobic, anaerobic, and fastidious organisms have been conducted (248–250). In one such study, suspensions of 10 quality control (QC) strains (5 aerobic and 5 anaerobic) recommended by CLSI document M40-A were sampled using ESwab, BactiSwab (Remel), and CultureSwab (BD) and were held at room temperature or refrigerated for up to 48 h with quantitative sampling at several time points (248). The results demonstrated both maintenance of viability and prevention of overgrowth (<1 log difference between the initial inoculum and 48 h of plating) for ESwab specimens stored at refrigerated temperatures. A similar study demonstrated slightly better recovery of E. coli, S. agalactiae, and Candida albicans using ESwab compared to traditional wound swab when specimens were stored at refrigerated temperatures for 48 h; however, the difference in recovery was <10% (250). Of note, significant growth (>1 log) was observed for Pseudomonas aeruginosa, E. coli, and S. pyogenes after 48 h of storage at room temperature (248, 250). Clinical studies have also been conducted to evaluate the ESwab (251, 252). The total CFU of microorganisms recovered from wound specimens was up to 6 times higher using ESwab (251). In addition, ESwab frequently recovered additional organisms that were not recovered using traditional swabs. This difference was statistically significant for coagulase-negative Staphylococcus spp. and Enterococcus spp. (251). Similarly, the total CFU of MRSA recovered from nasal screening specimens was 3.6 to 9 times higher using ESwab, which could potentially increase the sensitivity of culture-based screening (251, 252). A potential drawback to the use of ESwab collection devices is reported toxicity of the ESwab to various cell lines used in viral culture. It is hypothesized that this toxicity stems from the vegetal coating used on ESwab fibers to aid in preservation of Neisseria spp. and anaerobic bacteria (253).
There are currently two manufacturers of commercially available total laboratory automation (TLA) lines for clinical microbiology; Copan (WASP Lab) and BD Kiestra (Kiestra TLA) (Table 5). These TLA systems add connectivity of O2, CO2, and anaerobic incubators to a central processor or work stations through conveyer tracks that move plates to and from the incubators. In all cases inoculated plates are assigned to a specific location in an incubator and can be manually recalled and delivered to the technologist or core processor in <1 min (241). Sophisticated cameras with 5- to 15-megapixel resolution along with various lighting and exposure settings take images of each inoculated plate prior to incubation and can be programmed to retrieve and image the plate at user-defined intervals. The increased sensitivity compared to unaided visual inspection of cultures, as well as more frequent inspection of cultures, can reduce the time to identification of bacteria in clinical specimens. Growth of Gram-negative rods in pure culture could be detected by the BD Kiestra ReadA in as little as 8 h if present at 106 to 108 CFU/ml and within approximately 12 h if present at 102 to 104 CFU/ml (254). Growth of Gram-positive bacteria and Candida spp. was also detected within 7 to 8 h of inoculation if present at higher concentration, although the time to detection was extended if present at lower concentrations. Still, when coupled with MALDI-TOF MS, earlier recognition of culture growth can reduce the time to identification of clinical isolates. Another potential use of serial high-resolution imaging is the earlier detection and measurement of the zone of inhibition for disk-based susceptibility testing, which may aid in earlier reporting of antimicrobial susceptibility results (255). Finally, automated imaging allows technologists to review and compare side-by-side images of complex polymicrobial cultures at various time points to assess microbial growth and determine which colonies may require further investigation. Depending on the TLA system, these colonies may be selected for automated tasks, including restreak, preparation for broth inoculation, or MALDI-TOF identification, or the culture plates may be returned to a manned workstation for manual manipulation. Both manufacturers strive to automate processing, plating, and incubation of specimens submitted for routine culture and also to aid in plate reading and postincubation analysis of cultures through implementation of video microbiology and incorporation of semiautomated or fully automated identification and susceptibility testing systems. Though the core functions and capabilities of these systems are similar, the approaches taken by each to accomplish these tasks are different.
TABLE 5.
Characteristics of total laboratory automation systems for clinical microbiology laboratories
| Characteristic | Copan WASPLab | BD Kiestra TLA | bioMérieux FMLA |
|---|---|---|---|
| Core specimen inoculation module | WASP | InoqulA | Previ-Isola |
| Inoculation conducted in integrated biosafety cabinet | No | Yes | Information unavailable |
| Uninoculated plate capacity (no. of standard plates) | 350 | 720 | 270 |
| Automated decapping and recapping | Yes | Yes | No; specimen containers must be manually decapped prior to loading to Previ-Isola |
| Inoculation method | Automatic, streaking by reusable metal calibrated loops (1 μl, 10 μl, or 30 μl) | Automatic and manual, streaking by rolling-ball method | Automatic, streaking by disposable plastic comb |
| Manual inoculation of nonliquid or nonstandard specimens for automated streaking | No | Yes | Yes |
| Streak pattern | User defined | User defined | Circular pattern |
| Inoculations (no. of standard plates)/h | 180 | 400 | 180 |
| Automated inoculation of broths | Yes, automated on WASP warehouse carousel | Yes, automated on InoqulA | Information unavailable |
| Automated placement of disks | Yes, automated on WASP warehouse carousel | No, manual on ErgonomicA | Information unavailable |
| Automated slide prepn | Yes (staining is manual offline task) | Yes (staining manual at ErgonomicA module) | Information unavailable |
| Automated prepn of MALDI target plates | In development | In development | Information unavailable |
| Commercially available | Yes | Yes | No |
The core of WASPLab is the multifunctional, stand-alone WASP (walk-away specimen processor) unit (Fig. 10). This core unit maintains a relatively small footprint of approximately 75 in. wide by 43 in. deep and is 75 in. high (245). The various functions of the WASP are carried out by two independently operating robots which work in concert to “receive” specimen containers, select appropriate (user-defined) plating media for each specimen type, vortex, decap, and plate the specimen, and then recap the specimen and transfer both the specimen and inoculated medium to a “completed” queue. As specimens are received to the WASP, the existing barcode label is read and dictates the processing and plating program carried out by the WASP. Additional labels are automatically printed and applied to each plate inoculated with the specimen. Specimen plating is carried out using one of three on-board metal inoculating loops (1 μl, 10 μl, and 30 μl), which are heat sterilized between specimens. The volume of the loop used and streak pattern can be selected based on specimen type and are user defined and barcode driven. Solid medium (agar plates) is housed in a rotating carousel with capacity for up to ∼350 standard culture plates arranged in nine individual silos (239, 245). The number of specimens handled per hour is somewhat dependent on the variability of specimens or containers submitted. Batching of one specimen type increases efficiency by reducing the need to change loops or protocols (vortexing or centrifugation of specimen) for different specimen types received in a random order. At peak efficiency, WASP is capable of inoculating 180 plates/h, which is comparable to the case for other automated plate streakers (239). The use of reusable inoculating loops reduces the cost of disposables and waste generated during processing of hundreds or thousands of specimens per day; however, this could be a potential source for cross-contamination. One study assessed the contamination rate using 100 alternating sterile and seeded (105 to 106 CFU/ml E. coli) specimens processed and plated by the WASP and observed zero cross-contamination between specimens (245). Additional features of the WASP include an optional Gram slide preparation module and a “warehouse carousel” that can accommodate broth culture tubes to be inoculated or 4 Kirby-Bauer disk dispensers for automation of agar-based susceptibility testing. The optional MALDI prep module includes a specialized metal probe that is added to the “tool belt” for picking of isolated colonies, a stage to accommodate a MALDI target plate, and a reservoir to hold MALDI matrix material. Inclusion of this module allows a technologist to select a specific colony from an image of the culture plate, which will then be transferred from the culture plate to the MALDI target plate and overlaid with matrix in preparation for identification by MALDI-TOF MS.
FIG 10.
Walk-away specimen processor (WASP). The multifunctional WASP core unit (A) includes two independently operating robotic arms capable of decapping, recapping, and inoculating up to 180 solid agar plates per hour. The core unit is also equipped with a vortex for sample mixing and a “tool belt” which can accommodate three different-size reusable calibrated inoculating loops and a blunt tipped colony-picking instrument which can be accessed by the robotic arm as needed. Culture media are housed in a 9-silo carousel which can accommodate up to 350 standard agar plates. Liquid media for broth culture are housed in a “warehouse carousel” located on the reverse side of the WASP (B). The WASP is also equipped with a barcode label reader capable of reading specimen barcodes and a printer which automatically prints and applies labels to all corresponding plates prior to inoculation. The core WASP unit can be equipped with optional disk dispenser (C) for application of antibiotic disks to inoculated media, a Gram slide prep module for automated preparation of slides (D and E), and a stage for automated transfer of isolated colonies and matrix material to a MALDI target plate for analysis using MALDI-TOF MS (F). Total laboratory automation features the WASP core unit connected via conveyer tracks to smart incubators equipped with high-resolution cameras for imaging of culture plates (G). (Courtesy of Copan Diagnostics.)
Key advantages to the WASP are the relatively small footprint, versatility, and number of functions of the core module. However, taking advantage of all of these functions, including both preincubation (processing, plating, and Gram stain preparation) and postincubation (colony picking, isolation streaking, Kirby-Bauer susceptibility setup, and MALDI prep) tasks is probably impractical without the addition of a second or third WASP unit. Similar to current laboratory workflow, efficiencies in automation may be gained by unidirectional, continuous flow dedicating one WASP for processing and a second for postincubation manipulations. Thus, total automation using the WASPLab may require some physical restructuring of laboratories with “closed” floor plans to accommodate multiple WASP units and accompanying conveyers and incubators.
The second commercially available TLA line for microbiology is the BD Kiestra TLA (Fig. 11). Initial installations of the full BD Kiestra TLA into clinical microbiology laboratories were completed in 2006, and since that time over 40 laboratories have installed this system (241). In contrast to the WASPLab, which relies on a multifunctional core unit coupled to imaging systems and incubators, the BD Kiestra TLA is composed of various task-specific modules which can be configured according to the specific needs of different laboratories. Basic modules are those accommodating tasks including loading and sorting of growth medium plates (SorterA), barcoding of preinoculated media (BarcodeA), and fully automated inoculation of liquid specimens (InoqulA), incubators equipped with high-resolution plate imaging cameras (ReadA), an automated colony picker for preparation of MALDI-TOF MS target plates (MalditofA, in development), and technologist work stations to accommodate more complex manual tasks such as plating of bone, tissue, or other solid specimens (ErgonomicA) (241). Use of the manual interactive (MI) portion of the instrument also ensures specimen traceability through barcoding of inoculated media and automated transfer of plates to the ReadA. The basic modules described above can be combined into small modular workstations such as the InoqulA workstation (comprised of a SorterA, BarcodeA, InoqulA, and ErgonomicA) or Work Cell Automation (WCA), which is composed of an InoqulaA workstation connected to ReadA incubators. Total laboratory automation can be achieved through connection of multiple workstation modules or any configuration and number of basic modules. Like in WASPLab, all modules are connected by a conveyer system which moves specimens and inoculated cultures to the appropriate module automatically according to user-defined protocols or on demand when a technologist selects a given culture for further workup. Plate inoculation using the fully automated section of the InoqulA relies on transfer of liquid specimens by a calibrated pipette, followed by streaking of the specimen using magnetic beads. This differs from WASPLab, which utilizes a reusable metal loop for inoculation and streaking of specimens. Use of a pipette for inoculation of specimens restricts automated inoculation to liquid, nonviscous specimens; however, other specimen types can be manually inoculated to plates for automatic streaking. The ability to simultaneously inoculate multiple plates using magnetic beads allows a throughput of 400 plates/h compared to systems using loop-based inoculation. However, beads must be collected and sterilized prior to reuse, which requires manual intervention and increases the “routine maintenance” associated with the InoqulA (239).
FIG 11.
BD Kiestra total laboratory automation (TLA). The BD Kiestra TLA system is composed of task-specific modules. The stand-alone InoqulA module is capable of automated plate inoculation and streaking of up to 400 plates per hour using the roll bead method (A). Fully automated inoculation, streaking, barcoding, and sorting of inoculated media can be conducted by a combination of the SorterA, BarcodeA, and InoqulA modules, an optional biosafety cabinet for manual plating of nonliquid specimens can also be integrated into front-end workflow (B). Work Cell Automation (WCA) (C) incorporates an ErgonomicA technologist workstation (D) and incubators equipped with high-resolution cameras for imaging culture plates (ReadA). Work Cell Automation modules can be configured into large or small total laboratory automation systems (E) with multiple InoqulA, ErgonomicA, and ReadA modules to accommodate additional specimen volume in medium- to high-throughput microbiology laboratories. (Courtesy of BD Kiestra.)
A major advantage of the BD Kiestra TLA is the open architecture and modular design. This allows laboratories to customize the system to their specific workloads and workflows but also to expand or modify the system if the needs of the laboratory change. This could include the addition of a single SorterA to allow for storage of more plating media or an InoqulA to permit greater throughput. Another advantage is the incorporation of technologist workstations (ErgonomicA) into the work modules and TLA. These workstations are served by a second conveyor track dedicated for retrieval of culture plates from the incubators and delivery to waiting technologists, which enables simple bidirectional flow of plates into and out of the incubators. Automated retrieval and delivery of cultures to technologist workstations accommodate specimens or cultures with unique needs that may not be easily managed by automation. Examples include the processing of nonliquid specimens, inoculation of nonroutine media, and ability to conduct various “offline” tasks such as Gram staining, basic biochemical or latex tests, and subculture of colonies deemed appropriate for further investigation. Incorporation of the BD Kiestra TLA into two large microbiology laboratories has been reported to increase production (number of samples/technologist/day) by 2.0 to 2.6 times versus preautomation capabilities (256, 257).
A third system, the Full Microbiology Lab Automation (FMLA), is currently under development by bioMérieux. While not yet available, this system aims to incorporate the Previ-Isola plate inoculator/streaker with incubators and imaging systems similar to those available with the WASPLab and BD Kiestra TLA systems. Further automation may be available with integration of the Vitek MS MALDI-TOF MS for identification of cultured microbes and integrated susceptibility testing with the Vitek2. A potential advantage to this system is that all components will be developed and manufactured by a single company, and all components will be compatible with the Myla software system. This may aid integration of ID and AST functions, which would need to be purchased separately from other manufacturers and incorporated into the WASPLab and BD Kiestra TLA.
CONCLUSION
The focus of this review was a current look at the literature surrounding emerging technologies in clinical microbiology. Our goal was to provide evidence, supported by peer-reviewed literature, which highlights the applications, performance, advantages, and potential shortcomings of each of the technologies or diagnostic methods discussed. Even at the current time, there are numerous additional technologies under development or with only limited objective supporting literature which are sure to play a role in the future of clinical microbiology. Likewise, investigators continue to push the limits of current technologies, including digital PCR, next-generation sequencing, and MALDI-TOF MS, to broaden their utility in areas including antimicrobial susceptibility testing and identification of oncogenes. The combined efforts of progressive investigators and availability of increasingly sensitive technologies are sure to improve the quality of and add value to the services provided by clinical laboratorians.
ACKNOWLEDGMENTS
Nathan A. Ledeboer serves as a Consultant to Nanosphere, Thermo Fisher Scientific, LabCorp, iCubate, Copan Diagnostics, and BD Diagnostics. He is a Board Member for Evogen. He has received research grants from Meridian, Quidel, IMDx, Cepheid, BD, bioMérieux, Bruker Daltonics, Nanosphere, Seegene, Life Technologies, Prodesse, Great Basin Corp., iCubate, Biohelix, and Bio-Rad. Blake W. Buchan serves as a consultant for GenMark Diagnostics and Nanosphere. He has received honoraria or travel support from BD, Nanosphere, and Quidel Molecular.
Biographies

Blake W. Buchan, Ph.D., D(ABMM), is Assistant Professor of Pathology at the Medical College of Wisconsin and Associate Director of Microbiology at Dynacare Laboratories, Milwaukee, WI. He was awarded a Ph.D. in microbiology from the University of Iowa in 2009 and completed postdoctoral training in clinical microbiology at the Medical College of Wisconsin and Froedtert Hospital. Dr. Buchan has been active in research efforts to evaluate novel diagnostics at various stages of development and has served as principal investigator or co-principal investigator on over 30 research projects in the areas of molecular diagnostics and MALDI-TOF MS. Dr. Buchan serves on the ABMM exam validation committee and is bacteriology special interest group coordinator for the South Central Association for Clinical Microbiology (SCACM).

Nathan A. Ledeboer, Ph.D., D(ABMM) received his B.A. in 2000 and his Ph.D. in microbiology from the University of Iowa in 2005. Following two years of fellowship training in clinical and public health microbiology at Washington University School of Medicine in Saint Louis, MO, he joined the faculty of the Department of Pathology at the Medical College of Wisconsin in Milwaukee, WI. He is currently an Associate Professor and Medical Director of Microbiology, Molecular Diagnostics, Reference Services, and Laboratory Business Development at Froedtert Hospital and Dynacare Laboratories in Milwaukee, WI. His research endeavors, particularly in the area of developing diagnostic tools for infectious diseases, have led to numerous publications in peer-reviewed journals and more than 75 funded research projects. Dr. Ledeboer is currently an editor for the Journal of Clinical Microbiology.
REFERENCES
- 1.Hodinka RL. 2013. Point: is the era of viral culture over in the clinical microbiology laboratory? J. Clin. Microbiol. 51:2–4. 10.1128/JCM.02593-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 26:547–603. 10.1128/CMR.00072-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burd EM. 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 23:550–576. 10.1128/CMR.00074-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valenstein PN. 1990. Evaluating diagnostic tests with imperfect standards. Am. J. Clin. Pathol. 93:252–258 [DOI] [PubMed] [Google Scholar]
- 5.CLSI. 2005. User varification of performance for precisioin and trueness. Approved guideline, 2nd ed. CLSI document EP15-A2. CLSI, Wayne, PA [Google Scholar]
- 6.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491. 10.1126/science.2448875 [DOI] [PubMed] [Google Scholar]
- 7.Mackay IM, Arden KE, Nitsche A. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292–1305. 10.1093/nar/30.6.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monif GR. 1998. A new gold standard for the detection of Chlamydia trachomatis? Infect. Dis. Obstet. Gynecol. 6:44–45. 10.1155/S106474499800009X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455. 10.1086/651706 [DOI] [PubMed] [Google Scholar]
- 10.Ieven M. 2007. Currently used nucleic acid amplification tests for the detection of viruses and atypicals in acute respiratory infections. J. Clin. Virol. 40:259–276. 10.1016/j.jcv.2007.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giachetti C, Linnen JM, Kolk DP, Dockter J, Gillotte-Taylor K, Park M, Ho-Sing-Loy M, McCormick MK, Mimms LT, McDonough SH. 2002. Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J. Clin. Microbiol. 40:2408–2419. 10.1128/JCM.40.7.2408-2419.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wroblewski JK, Manhart LE, Dickey KA, Hudspeth MK, Totten PA. 2006. Comparison of transcription-mediated amplification and PCR assay results for various genital specimen types for detection of Mycoplasma genitalium. J. Clin. Microbiol. 44:3306–3312. 10.1128/JCM.00553-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaydos CA, Quinn TC, Willis D, Weissfeld A, Hook EW, Martin DH, Ferrero DV, Schachter J. 2003. Performance of the APTIMA Combo 2 assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in female urine and endocervical swab specimens. J. Clin. Microbiol. 41:304–309. 10.1128/JCM.41.1.304-309.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin DH, Cammarata C, Van Der Pol B, Jones RB, Quinn TC, Gaydos CA, Crotchfelt K, Schachter J, Moncada J, Jungkind D, Turner B, Peyton C. 2000. Multicenter evaluation of AMPLICOR and automated COBAS AMPLICOR CT/NG tests for Neisseria gonorrhoeae. J. Clin. Microbiol. 38:3544–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Der Pol B, Quinn TC, Gaydos CA, Crotchfelt K, Schachter J, Moncada J, Jungkind D, Martin DH, Turner B, Peyton C, Jones RB. 2000. Multicenter evaluation of the AMPLICOR and automated COBAS AMPLICOR CT/NG tests for detection of Chlamydia trachomatis. J. Clin. Microbiol. 38:1105–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Der Pol B, Liesenfeld O, Williams JA, Taylor SN, Lillis RA, Body BA, Nye M, Eisenhut C, Hook EW., III 2012. Performance of the Cobas CT/NG test compared to the Aptima AC2 and Viper CTQ/GCQ assays for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 50:2244–2249. 10.1128/JCM.06481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mushanski LM, Brandt K, Coffin N, Levett PN, Horsman GB, Rank EL. 2012. Comparison of the BD Viper System with XTR Technology to the Gen-Probe APTIMA COMBO 2 Assay using the TIGRIS DTS system for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens. Sex. Transm. Dis. 39:514–517. 10.1097/OLQ.0b013e31824f2f5b [DOI] [PubMed] [Google Scholar]
- 18.Stewart CM, Schoeman SA, Booth RA, Smith SD, Wilcox MH, Wilson JD. 2012. Assessment of self taken swabs versus clinician taken swab cultures for diagnosing gonorrhoea in women: single centre, diagnostic accuracy study. BMJ 345:e8107. 10.1136/bmj.e8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Der Pol B, Taylor SN, Liesenfeld O, Williams JA, Hook EW., 3rd 2013. Vaginal swabs are the optimal specimen for detection of genital Chlamydia trachomatis or Neisseria gonorrhoeae using the Cobas 4800 CT/NG test. Sex. Transm. Dis. 40:247–250. 10.1097/OLQ.0b013e3182717833 [DOI] [PubMed] [Google Scholar]
- 20.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R. 2013. Targeted versus universal decolonization to prevent ICU infection. N. Engl. J. Med. 368:2255–2265. 10.1056/NEJMoa1207290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pofahl WE, Ramsey KM, Nobles DL, Cochran MK, Goettler C. 2011. Importance of methicillin-resistant Staphylococcus aureus eradication in carriers to prevent postoperative methicillin-resistant Staphylococcus aureus surgical site infection. Am. Surg. 77:27–31 [PubMed] [Google Scholar]
- 22.Jog S, Cunningham R, Cooper S, Wallis M, Marchbank A, Vasco-Knight P, Jenks PJ. 2008. Impact of preoperative screening for meticillin-resistant Staphylococcus aureus by real-time polymerase chain reaction in patients undergoing cardiac surgery. J. Hosp. Infect. 69:124–130. 10.1016/j.jhin.2008.02.008 [DOI] [PubMed] [Google Scholar]
- 23.Lee BY, Wiringa AE, Bailey RR, Goyal V, Tsui B, Lewis GJ, Muder RR, Harrison LH. 2010. The economic effect of screening orthopedic surgery patients preoperatively for methicillin-resistant Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 31:1130–1138. 10.1086/656591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brecher SM, Novak-Weekley SM, Nagy E. 2013. Laboratory diagnosis of Clostridium difficile infections: there is light at the end of the colon. Clin. Infect. Dis. 57:1175–1181. 10.1093/cid/cit424 [DOI] [PubMed] [Google Scholar]
- 25.Peterson LR, Mehta MS, Patel PA, Hacek DM, Harazin M, Nagwekar PP, Thomson RB, Jr, Robicsek A. 2011. Laboratory testing for Clostridium difficile infection: light at the end of the tunnel. Am. J. Clin. Pathol. 136:372–380. 10.1309/AJCPTP5XKRSNXVIL [DOI] [PubMed] [Google Scholar]
- 26.Wilcox MH. 2012. Overcoming barriers to effective recognition and diagnosis of Clostridium difficile infection. Clin. Microbiol. Infect. 18(Suppl 6):S13–S20 [DOI] [PubMed] [Google Scholar]
- 27.Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin. Microbiol. Infect. 15:1053–1066. 10.1111/j.1469-0691.2009.03098.x [DOI] [PubMed] [Google Scholar]
- 28.Longtin Y, Trottier S, Brochu G, Paquet-Bolduc B, Garenc C, Loungnarath V, Beaulieu C, Goulet D, Longtin J. 2013. Impact of the type of diagnostic assay on Clostridium difficile infection and complication rates in a mandatory reporting program. Clin. Infect. Dis. 56:67–73. 10.1093/cid/cis840 [DOI] [PubMed] [Google Scholar]
- 29.Sethi AK, Al-Nassir WN, Nerandzic MM, Bobulsky GS, Donskey CJ. 2010. Persistence of skin contamination and environmental shedding of Clostridium difficile during and after treatment of C. difficile infection. Infect. Control Hosp. Epidemiol. 31:21–27. 10.1086/649016 [DOI] [PubMed] [Google Scholar]
- 30.Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham CA, Dunne WM., Jr 2011. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J. Clin. Microbiol. 49:2887–2893. 10.1128/JCM.00891-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnham CA, Carroll KC. 2013. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin. Microbiol. Rev. 26:604–630. 10.1128/CMR.00016-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.CDC. 2007. Outbreaks of respiratory illness mistakenly attributed to pertussis—New Hampshire, Massachusetts, and Tennessee, 2004-2006. MMWR Morb. Mortal. Wkly. Rep. 56:837–842 [PubMed] [Google Scholar]
- 33.Waters V, Jamieson F, Richardson SE, Finkelstein M, Wormsbecker A, Halperin SA. 2009. Outbreak of atypical pertussis detected by polymerase chain reaction in immunized preschool-aged children. Pediatr. Infect. Dis. J. 28:582–587. 10.1097/INF.0b013e318197fac1 [DOI] [PubMed] [Google Scholar]
- 34.Papenburg J, Fontela P. 2009. What is the significance of a high cycle threshold positive IS481 PCR for Bordetella pertussis? Pediatr. Infect. Dis. J. 28:1143 (Author reply, 28:1143–1144) [DOI] [PubMed] [Google Scholar]
- 35.Guthrie JL, Seah C, Brown S, Tang P, Jamieson F, Drews SJ. 2008. Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J. Clin. Microbiol. 46:3798–3799. 10.1128/JCM.01551-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori Y, Nagamine K, Tomita N, Notomi T. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150–154. 10.1006/bbrc.2001.5921 [DOI] [PubMed] [Google Scholar]
- 38.Mori Y, Kitao M, Tomita N, Notomi T. 2004. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59:145–157. 10.1016/j.jbbm.2003.12.005 [DOI] [PubMed] [Google Scholar]
- 39.Anderson NW, Buchan BW, Mayne D, Mortensen JE, Mackey TL, Ledeboer NA. 2013. Multicenter clinical evaluation of the illumigene group A Streptococcus DNA amplification assay for detection of group A Streptococcus from pharyngeal swabs. J. Clin. Microbiol. 51:1474–1477. 10.1128/JCM.00176-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gyorke CE, Wang S, Leslie JL, Cohen SH, Solnick JV, Polage CR. 2013. Evaluation of Clostridium difficile fecal load and limit of detection during a prospective comparison of two molecular tests, the illumigene C. difficile and Xpert C. difficile/Epi tests. J. Clin. Microbiol. 51:278–280. 10.1128/JCM.02120-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viala C, Le Monnier A, Maataoui N, Rousseau C, Collignon A, Poilane I. 2012. Comparison of commercial molecular assays for toxigenic Clostridium difficile detection in stools: BD GeneOhm Cdiff, XPert C. difficile and illumigene C. difficile. J. Microbiol. Methods 90:83–85. 10.1016/j.mimet.2012.04.017 [DOI] [PubMed] [Google Scholar]
- 42.Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM. 2012. Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difficile/Epi, and Illumigene C. difficile assays. J. Clin. Microbiol. 50:1331–1335. 10.1128/JCM.06597-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchan BW, Mackey TL, Daly JA, Alger G, Denys GA, Peterson LR, Kehl SC, Ledeboer NA. 2012. Multicenter clinical evaluation of the portrait toxigenic C. difficile assay for detection of toxigenic Clostridium difficile strains in clinical stool specimens. J. Clin. Microbiol. 50:3932–3936. 10.1128/JCM.02083-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henson AM, Carter D, Todd K, Shulman ST, Zheng X. 2013. Detection of Streptococcus pyogenes using illumigene group A Streptococcus assay. J. Clin. Microbiol. 51:4207–4209. 10.1128/JCM.01892-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karani M, Sotiriadou I, Plutzer J, Karanis P. 2014. Bench-scale experiments for the development of a unified loop-mediated isothermal amplification (LAMP) assay for the in vitro diagnosis of Leishmania species' promastigotes. Epidemiol. Infect. 142:1671–1677. 10.1017/S0950268813002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kargar M, Askari A, Doosti A, Ghorbani-Dalini S. 2012. Loop-mediated isothermal amplification assay for rapid detection of hepatitis C virus. Indian J. Virol. 23:18–23. 10.1007/s13337-012-0067-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plutzer J, Karanis P. 2009. Rapid identification of Giardia duodenalis by loop-mediated isothermal amplification (LAMP) from faecal and environmental samples and comparative findings by PCR and real-time PCR methods. Parasitol. Res. 104:1527–1533. 10.1007/s00436-009-1391-3 [DOI] [PubMed] [Google Scholar]
- 48.Ablordey A, Amissah DA, Aboagye IF, Hatano B, Yamazaki T, Sata T, Ishikawa K, Katano H. 2012. Detection of Mycobacterium ulcerans by the loop mediated isothermal amplification method. PLoS Negl. Trop. Diseases. 6:e1590. 10.1371/journal.pntd.0001590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hopkins H, Gonzalez IJ, Polley SD, Angutoko P, Ategeka J, Asiimwe C, Agaba B, Kyabayinze DJ, Sutherland CJ, Perkins MD, Bell D. 2013. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J. Infect. Dis. 208:645–652. 10.1093/infdis/jit184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polley SD, Gonzalez IJ, Mohamed D, Daly R, Bowers K, Watson J, Mewse E, Armstrong M, Gray C, Perkins MD, Bell D, Kanda H, Tomita N, Kubota Y, Mori Y, Chiodini PL, Sutherland CJ. 2013. Clinical evaluation of a loop-mediated amplification kit for diagnosis of imported malaria. J. Infect. Dis. 208:637–644. 10.1093/infdis/jit183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, Abeyewickreme W, Tangpukdee N, Yuen KY, Guan Y, Looareesuwan S, Peiris JS. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chem. 52:303–306. 10.1373/clinchem.2005.057901 [DOI] [PubMed] [Google Scholar]
- 52.Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T. 2007. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J. Clin. Microbiol. 45:2521–2528. 10.1128/JCM.02117-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vincent M, Xu Y, Kong H. 2004. Helicase-dependent isothermal DNA amplification. EMBO Rep. 5:795–800. 10.1038/sj.embor.7400200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang S, Do J, Mahalanabis M, Fan A, Zhao L, Jepeal L, Singh SK, Klapperich CM. 2013. Low cost extraction and isothermal amplification of DNA for infectious diarrhea diagnosis. PLoS One 8:e60059. 10.1371/journal.pone.0060059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frech GC, Munns D, Jenison RD, Hicke BJ. 2012. Direct detection of nasal Staphylococcus aureus carriage via helicase-dependent isothermal amplification and chip hybridization. BMC Res. Notes 5:430. 10.1186/1756-0500-5-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Kumar N, Gopalakrishnan A, Ginocchio C, Manji R, Bythrow M, Lemieux B, Kong H. 2013. Detection and species identification of malaria parasites by isothermal tHDA amplification directly from human blood without sample preparation. J. Mol. Diagn. 15:634–641. 10.1016/j.jmoldx.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller NS, Yen-Lieberman B, Poulter MD, Tang YW, Granato PA. 2012. Comparative clinical evaluation of the IsoAmp HSV Assay with ELVIS HSV culture/ID/typing test system for the detection of herpes simplex virus in genital and oral lesions. J. Clin. Virol. 54:355–358. 10.1016/j.jcv.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Artiushin S, Tong Y, Timoney J, Lemieux B, Schlegel A, Kong H. 2011. Thermophilic helicase-dependent DNA amplification using the IsoAmp SE experimental kit for rapid detection of Streptococcus equi subspecies equi in clinical samples. J. Vet. Diagn. Invest. 23:909–914. 10.1177/1040638711416968 [DOI] [PubMed] [Google Scholar]
- 59.Kim HJ, Tong Y, Tang W, Quimson L, Cope VA, Pan X, Motre A, Kong R, Hong J, Kohn D, Miller NS, Poulter MD, Kong H, Tang YW, Yen-Lieberman B. 2011. A rapid and simple isothermal nucleic acid amplification test for detection of herpes simplex virus types 1 and 2. J. Clin. Virol. 50:26–30. 10.1016/j.jcv.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson A, Marcon M, Mortensen JE, McCarter YS, LaRocco M, Peterson LR, Thomson RB., Jr 1999. Controversies affecting the future practice of clinical microbiology. J. Clin. Microbiol. 37:883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho C, Lau A, Cimon K, Farrah K, Gardam M. 2013. Screening, isolation, and decolonization strategies for vancomycin-resistant enterococci or extended spectrum beta-lactamase-producing organisms: a systematic review of the clinical evidence and health services impact. CADTH Technol. Overviews 3:e3202. [PMC free article] [PubMed] [Google Scholar]
- 62.Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA. 2010. An antimicrobial stewardship program's impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin. Infect. Dis. 51:1074–1080. 10.1086/656623 [DOI] [PubMed] [Google Scholar]
- 63.Frye AM, Baker CA, Rustvold DL, Heath KA, Hunt J, Leggett JE, Oethinger M. 2012. Clinical impact of a real-time PCR assay for rapid identification of staphylococcal bacteremia. J. Clin. Microbiol. 50:127–133. 10.1128/JCM.06169-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo RF, Banaei N. 2013. Molecular approaches and biomarkers for detection of Mycobacterium tuberculosis. Clin. Lab. Med. 33:553–566. 10.1016/j.cll.2013.03.012 [DOI] [PubMed] [Google Scholar]
- 65.Millman AJ, Dowdy DW, Miller CR, Brownell R, Metcalfe JZ, Cattamanchi A, Davis JL. 2013. Rapid molecular testing for TB to guide respiratory isolation in the U.S.: a cost-benefit analysis. PLoS One 8:e79669. 10.1371/journal.pone.0079669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marks SM, Cronin W, Venkatappa T, Maltas G, Chon S, Sharnprapai S, Gaeddert M, Tapia J, Dorman SE, Etkind S, Crosby C, Blumberg HM, Bernardo J. 2013. The health-system benefits and cost-effectiveness of using Mycobacterium tuberculosis direct nucleic acid amplification testing to diagnose tuberculosis disease in the United States. Clin. Infect. Dis. 57:532–542. 10.1093/cid/cit336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis JL, Kawamura LM, Chaisson LH, Grinsdale J, Benhammou J, Ho C, Babst A, Banouvong H, Metcalfe JZ, Pandori M, Hopewell PC, Cattamanchi A. 2014. Impact of GeneXpert MTB/RIF on patients and tuberculosis programs in a low-burden setting: a hypothetical trial. Am. J. Respir. Crit. Care Med. 189:1551–1559. 10.1164/rccm.201311-1974OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. 1988. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 16:11141–11156. 10.1093/nar/16.23.11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Landry ML, Cohen S, Ferguson D. 2000. Impact of sample type on rapid detection of influenza virus A by cytospin-enhanced immunofluorescence and membrane enzyme-linked immunosorbent assay. J. Clin. Microbiol. 38:429–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boivin G, Cote S, Dery P, De Serres G, Bergeron MG. 2004. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J. Clin. Microbiol. 42:45–51. 10.1128/JCM.42.1.45-51.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sails AD, Saunders D, Airs S, Roberts D, Eltringham G, Magee JG. 2009. Evaluation of the Cepheid respiratory syncytial virus and influenza virus A/B real-time PCR analyte specific reagent. J. Virol. Methods 162:88–90. 10.1016/j.jviromet.2009.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buelow DR, Bankowski MJ, Fofana D, Gu Z, Pounds S, Hayden RT. 2013. Comparison of two multiplexed PCR assays for the detection of HSV-1, HSV-2, and VZV with extracted and unextracted cutaneous and mucosal specimens. J. Clin. Virol. 58:84–88. 10.1016/j.jcv.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 73.Buchan BW, Olson WJ, Pezewski M, Marcon MJ, Novicki T, Uphoff TS, Chandramohan L, Revell P, Ledeboer NA. 2013. Clinical evaluation of a real-time PCR assay for identification of Salmonella, Shigella, Campylobacter (Campylobacter jejuni and C. coli), and Shiga toxin-producing Escherichia coli isolates in stool specimens. J. Clin. Microbiol. 51:4001–4007. 10.1128/JCM.02056-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snyder JW, Munier GK, Heckman SA, Camp P, Overman TL. 2009. Failure of the BD GeneOhm StaphSR assay for direct detection of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates in positive blood cultures collected in the United States. J. Clin. Microbiol. 47:3747–3748. 10.1128/JCM.01391-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Westh H, Lisby G, Breysse F, Boddinghaus B, Chomarat M, Gant V, Goglio A, Raglio A, Schuster H, Stuber F, Wissing H, Hoeft A. 2009. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin. Microbiol. Infect. 15:544–551. 10.1111/j.1469-0691.2009.02736.x [DOI] [PubMed] [Google Scholar]
- 76.Bloos F, Hinder F, Becker K, Sachse S, Mekontso Dessap A, Straube E, Cattoir V, Brun-Buisson C, Reinhart K, Peters G, Bauer M. 2010. A multicenter trial to compare blood culture with polymerase chain reaction in severe human sepsis. Intensive Care Med. 36:241–247. 10.1007/s00134-009-1705-z [DOI] [PubMed] [Google Scholar]
- 77.Casalta JP, Gouriet F, Roux V, Thuny F, Habib G, Raoult D. 2009. Evaluation of the LightCycler SeptiFast test in the rapid etiologic diagnostic of infectious endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 28:569–573. 10.1007/s10096-008-0672-6 [DOI] [PubMed] [Google Scholar]
- 78.Stamper PD, Cai M, Howard T, Speser S, Carroll KC. 2007. Clinical validation of the molecular BD GeneOhm StaphSR assay for direct detection of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in positive blood cultures. J. Clin. Microbiol. 45:2191–2196. 10.1128/JCM.00552-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bartels MD, Boye K, Rohde SM, Larsen AR, Torfs H, Bouchy P, Skov R, Westh H. 2009. A common variant of staphylococcal cassette chromosome mec type IVa in isolates from Copenhagen, Denmark, is not detected by the BD GeneOhm methicillin-resistant Staphylococcus aureus assay. J. Clin. Microbiol. 47:1524–1527. 10.1128/JCM.02153-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas L, van Hal S, O'Sullivan M, Kyme P, Iredell J. 2008. Failure of the BD GeneOhm StaphS/R assay for identification of Australian methicillin-resistant Staphylococcus aureus strains: duplex assays as the “gold standard” in settings of unknown SCCmec epidemiology. J. Clin. Microbiol. 46:4116–4117. 10.1128/JCM.01146-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- 82.Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DH, Ririe KM. 2011. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 6:e26047. 10.1371/journal.pone.0026047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Wesenbeeck L, Meeuws H, Van Immerseel A, Ispas G, Schmidt K, Houspie L, Van Ranst M, Stuyver L. 2013. Comparison of the FilmArray RP, Verigene RV+, and Prodesse ProFLU+/FAST+ multiplex platforms for detection of influenza viruses in clinical samples from the 2011-2012 influenza season in Belgium. J. Clin. Microbiol. 51:2977–2985. 10.1128/JCM.00911-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pierce VM, Elkan M, Leet M, McGowan KL, Hodinka RL. 2012. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J. Clin. Microbiol. 50:364–371. 10.1128/JCM.05996-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altun O, Almuhayawi M, Ullberg M, Ozenci V. 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J. Clin. Microbiol. 51:4130–4136. 10.1128/JCM.01835-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Babady NE. 2013. The FilmArray respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev. Mol. Diagn. 13:779–788. 10.1586/14737159.2013.848794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Popowitch EB, O'Neill SS, Miller MB. 2013. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J. Clin. Microbiol. 51:1528–1533. 10.1128/JCM.03368-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Piralla A, Lunghi G, Percivalle E, Vigano C, Nasta T, Pugni L, Mosca F, Stronati M, Torresani E, Baldanti F. 2014. FilmArray respiratory panel performance in respiratory samples from neonatal care units. Diagn. Microbiol. Infect. Dis. 79:183–186. 10.1016/j.diagmicrobio.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Doern CD, Lacey D, Huang R, Haag C. 2013. Evaluation and implementation of FilmArray version 1.7 for improved detection of adenovirus respiratory tract infection. J. Clin. Microbiol. 51:4036–4039. 10.1128/JCM.02546-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Thatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA. 2012. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn. Microbiol. Infect. Dis. 74:349–355. 10.1016/j.diagmicrobio.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Protonotariou E, Dimitroulia E, Pournaras S, Pitiriga V, Sofianou D, Tsakris A. 2010. Trends in antimicrobial resistance of clinical isolates of Enterococcus faecalis and Enterococcus faecium in Greece between 2002 and 2007. J. Hosp. Infect. 75:225–227. 10.1016/j.jhin.2009.12.007 [DOI] [PubMed] [Google Scholar]
- 92.Miller MB, Tang YW. 2009. Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 22:611–633. 10.1128/CMR.00019-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hager J. 2006. Making and using spotted DNA microarrays in an academic core laboratory. Methods Enzymol. 410:135–168. 10.1016/S0076-6879(06)10007-5 [DOI] [PubMed] [Google Scholar]
- 94.Carroll KC, Buchan BW, Tan S, Stamper PD, Riebe KM, Pancholi P, Kelly C, Rao A, Fader R, Cavagnolo R, Watson W, Goering RV, Trevino EA, Weissfeld AS, Ledeboer NA. 2013. Multicenter evaluation of the Verigene Clostridium difficile nucleic acid assay. J. Clin. Microbiol. 51:4120–4125. 10.1128/JCM.01690-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS. 2013. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J. Clin. Microbiol. 51:2072–2076. 10.1128/JCM.00831-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Samuel LP, Tibbetts RJ, Agotesku A, Fey M, Hensley R, Meier FA. 2013. Evaluation of a microarray-based assay for rapid identification of Gram-positive organisms and resistance markers in positive blood cultures. J. Clin. Microbiol. 51:1188–1192. 10.1128/JCM.02982-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buchan BW, Ginocchio CC, Manii R, Cavagnolo R, Pancholi P, Swyers L, Thomson RB, Jr, Anderson C, Kaul K, Ledeboer NA. 2013. Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS medicine. 10:e1001478. 10.1371/journal.pmed.1001478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buchan BW, Peterson JF, Cogbill CH, Anderson DK, Ledford JS, White MN, Quigley NB, Jannetto PJ, Ledeboer NA. 2011. Evaluation of a microarray-based genotyping assay for the rapid detection of cytochrome P450 2C19 *2 and *3 polymorphisms from whole blood using nanoparticle probes. Am. J. Clin. Pathol. 136:604–608. 10.1309/AJCPCPU9Q2IRNYXC [DOI] [PubMed] [Google Scholar]
- 99.Lefferts JA, Jannetto P, Tsongalis GJ. 2009. Evaluation of the Nanosphere Verigene system and the Verigene F5/F2/MTHFR nucleic acid tests. Exp. Mol. Pathol. 87:105–108. 10.1016/j.yexmp.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 100.Jannetto PJ, Buchan BW, Vaughan KA, Ledford JS, Anderson DK, Henley DC, Quigley NB, Ledeboer NA. 2010. Real-time detection of influenza A, influenza B, and respiratory syncytial virus A and B in respiratory specimens by use of nanoparticle probes. J. Clin. Microbiol. 48:3997–4002. 10.1128/JCM.01118-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sullivan KV, Deburger B, Roundtree SS, Ventrola CA, Blecker-Shelly DL, Mortensen JE. 23 April 2014. Rapid detection of inpatient Gram-negative bacteremia; extended-spectrum beta-lactamases and carbapenemase resistance determinants with the Verigene BC-GN test: a multicenter evaluation. J. Clin. Microbiol. 10.1128/JCM.00737-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tojo M, Fujita T, Ainoda Y, Nagamatsu M, Hayakawa K, Mezaki K, Sakurai A, Masui Y, Yazaki H, Takahashi H, Miyoshi-Akiyama T, Totsuka K, Kirikae T, Ohmagari N. 2014. Evaluation of an automated rapid diagnostic assay for detection of gram-negative bacteria and their drug-resistance genes in positive blood cultures. PLoS One 9:e94064. 10.1371/journal.pone.0094064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alby K, Popowitch EB, Miller MB. 2013. Comparative evaluation of the Nanosphere Verigene RV+ assay and the Simplexa Flu A/B & RSV kit for detection of influenza and respiratory syncytial viruses. J. Clin. Microbiol. 51:352–353. 10.1128/JCM.02504-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boku S, Naito T, Murai K, Tanei M, Inui A, Nisimura H, Isonuma H, Takahashi H, Kikuchi K. 2013. Near point-of-care administration by the attending physician of the rapid influenza antigen detection immunochromatography test and the fully automated respiratory virus nucleic acid test: contribution to patient management. Diagn. Microbiol. Infect. Dis. 76:445–449. 10.1016/j.diagmicrobio.2013.04.029 [DOI] [PubMed] [Google Scholar]
- 105.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. 10.1016/S0140-6736(05)67420-X [DOI] [PubMed] [Google Scholar]
- 106.See I, Mu Y, Cohen J, Beldavs ZG, Winston LG, Dumyati G, Holzbauer S, Dunn J, Farley MM, Lyons C, Johnston H, Phipps E, Perlmutter R, Anderson L, Gerding DN, Lessa FC. 2014. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin. Infect. Dis. 58:1394–1400. 10.1093/cid/ciu125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mestas J, Polanco CM, Felsenstein S, Dien Bard J. 2014. Performance of the Verigene gram-positive blood culture assay for direct detection of Gram-positive organisms and resistance markers in a pediatric hospital. J. Clin. Microbiol. 52:283–287. 10.1128/JCM.02322-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beal SG, Ciurca J, Smith G, John J, Lee F, Doern CD, Gander RM. 2013. Evaluation of the nanosphere verigene gram-positive blood culture assay with the VersaTREK blood culture system and assessment of possible impact on selected patients. J. Clin. Microbiol. 51:3988–3992. 10.1128/JCM.01889-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sullivan KV, Turner NN, Roundtree SS, Young S, Brock-Haag CA, Lacey D, Abuzaid S, Blecker-Shelly DL, Doern CD. 2013. Rapid detection of Gram-positive organisms by use of the Verigene Gram-positive blood culture nucleic acid test and the BacT/Alert Pediatric FAN system in a multicenter pediatric evaluation. J. Clin. Microbiol. 51:3579–3584. 10.1128/JCM.01224-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Merante F, Yaghoubian S, Janeczko R. 2007. Principles of the xTAG respiratory viral panel assay (RVP Assay). J. Clin. Virol. 40(Suppl 1):S31–S35. 10.1016/S1386-6532(07)70007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. 2013. Performance of the xTAG(R) gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J. Microbiol. Biotechnol. 23:1041–1045. 10.4014/jmb.1212.12042 [DOI] [PubMed] [Google Scholar]
- 112.Mengelle C, Mansuy JM, Prere MF, Grouteau E, Claudet I, Kamar N, Huynh A, Plat G, Benard M, Marty N, Valentin A, Berry A, Izopet J. 2013. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clin. Microbiol. Infect. 19:E458–E4565. 10.1111/1469-0691.12255 [DOI] [PubMed] [Google Scholar]
- 113.Pabbaraju K, Tokaryk KL, Wong S, Fox JD. 2008. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46:3056–3062. 10.1128/JCM.00878-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Anderson TP, Werno AM, Barratt K, Mahagamasekera P, Murdoch DR, Jennings LC. 2013. Comparison of four multiplex PCR assays for the detection of viral pathogens in respiratory specimens. J. Virol. Methods 191:118–121. 10.1016/j.jviromet.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anonymous. 2004. Adenovirus. Am. J. Transplant. 4(Suppl 10):101–104. 10.1111/j.1600-6135.2004.00733.x [DOI] [PubMed] [Google Scholar]
- 116.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. 1998. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 351:633–636. 10.1016/S0140-6736(97)08062-8 [DOI] [PubMed] [Google Scholar]
- 117.Alasmari F, Seiler SM, Hink T, Burnham CA, Dubberke ER. 2014. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin. Infect. Dis. 59:216–222. 10.1093/cid/ciu258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Forrest GN, Mehta S, Weekes E, Lincalis DP, Johnson JK, Venezia RA. 2006. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J. Antimicrob. Chemother. 58:154–158. 10.1093/jac/dkl146 [DOI] [PubMed] [Google Scholar]
- 119.Forrest GN, Mankes K, Jabra-Rizk MA, Weekes E, Johnson JK, Lincalis DP, Venezia RA. 2006. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J. Clin. Microbiol. 44:3381–3383. 10.1128/JCM.00751-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Forrest GN, Roghmann MC, Toombs LS, Johnson JK, Weekes E, Lincalis DP, Venezia RA. 2008. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob. Agents Chemother. 52:3558–3563. 10.1128/AAC.00283-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Spector SA, Hsia K, Crager M, Pilcher M, Cabral S, Stempien MJ. 1999. Cytomegalovirus (CMV) DNA load is an independent predictor of CMV disease and survival in advanced AIDS. J. Virol. 73:7027–7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Emery VC, Sabin CA, Cope AV, Gor D, Hassan-Walker AF, Griffiths PD. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032–2036. 10.1016/S0140-6736(00)02350-3 [DOI] [PubMed] [Google Scholar]
- 123.Bogdanovic G, Priftakis P, Giraud G, Kuzniar M, Ferraldeschi R, Kokhaei P, Mellstedt H, Remberger M, Ljungman P, Winiarski J, Dalianis T. 2004. Association between a high BK virus load in urine samples of patients with graft-versus-host disease and development of hemorrhagic cystitis after hematopoietic stem cell transplantation. J. Clin. Microbiol. 42:5394–5396. 10.1128/JCM.42.11.5394-5396.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Erard V, Kim HW, Corey L, Limaye A, Huang ML, Myerson D, Davis C, Boeckh M. 2005. BK DNA viral load in plasma: evidence for an association with hemorrhagic cystitis in allogeneic hematopoietic cell transplant recipients. Blood 106:1130–1132. 10.1182/blood-2004-12-4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Korenromp EL, Williams BG, Schmid GP, Dye C. 2009. Clinical prognostic value of RNA viral load and CD4 cell counts during untreated HIV-1 infection—a quantitative review. PLoS One 4:e5950. 10.1371/journal.pone.0005950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee SO, Brown RA, Razonable RR. 2012. Chromosomally integrated human herpesvirus-6 in transplant recipients. Transplant Infect. Dis. 14:346–354. 10.1111/j.1399-3062.2011.00715.x [DOI] [PubMed] [Google Scholar]
- 127.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 128.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 105:10513–10518. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van't Veer LJ, Bernards R. 2008. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature 452:564–570. 10.1038/nature06915 [DOI] [PubMed] [Google Scholar]
- 130.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P. 2008. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J. Clin. Oncol. 26:374–379. 10.1200/JCO.2007.12.5906 [DOI] [PubMed] [Google Scholar]
- 131.Wong ML, Medrano JF. 2005. Real-time PCR for mRNA quantitation. Biotechniques 39:75–85. 10.2144/05391RV01 [DOI] [PubMed] [Google Scholar]
- 132.Versalovic J, Carroll K, Funke G, Jorgensen JH, Landry ML, Warnock DW. (ed). 2011. Manual of clinical microbiology, 10th ed, vol 2 ASM Press, Washington, DC [Google Scholar]
- 133.Boyle DL, Rosengren S, Bugbee W, Kavanaugh A, Firestein GS. 2003. Quantitative biomarker analysis of synovial gene expression by real-time PCR. Arthritis Res. Ther. 5:R352–R360. 10.1186/ar1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lai KK, Cook L, Wendt S, Corey L, Jerome KR. 2003. Evaluation of real-time PCR versus PCR with liquid-phase hybridization for detection of enterovirus RNA in cerebrospinal fluid. J. Clin. Microbiol. 41:3133–3141. 10.1128/JCM.41.7.3133-3141.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 42:1142–1148. 10.1128/JCM.42.3.1142-1148.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sedlak RH, Jerome KR. 2013. Viral diagnostics in the era of digital polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 75:1–4. 10.1016/j.diagmicrobio.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baker M. 2012. Digital PCR hits its stride. Nat. Methods 9:541–544. 10.1038/nmeth.2027 [DOI] [Google Scholar]
- 138.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. 2013. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10:1003–1005. 10.1038/nmeth.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. 1992. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 13:444–449 [PubMed] [Google Scholar]
- 140.Ma J, Li N, Guarnera M, Jiang F. 2013. Quantification of plasma miRNAs by digital PCR for cancer diagnosis. Biomarker Insights 8:127–136. 10.4137/BMI.S13154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ludwig JA, Weinstein JN. 2005. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 5:845–856. 10.1038/nrc1739 [DOI] [PubMed] [Google Scholar]
- 142.Sawyers CL. 2008. The cancer biomarker problem. Nature 452:548–552. 10.1038/nature06913 [DOI] [PubMed] [Google Scholar]
- 143.Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, Atochin I, Link DR, Griffiths AD, Pallier K, Blons H, Bouche O, Landi B, Hutchison JB, Laurent-Puig P. 2013. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 59:1722–1731. 10.1373/clinchem.2013.206359 [DOI] [PubMed] [Google Scholar]
- 144.Kelley K, Cosman A, Belgrader P, Chapman B, Sullivan DC. 2013. Detection of methicillin-resistant Staphylococcus aureus by a duplex droplet digital PCR assay. J. Clin. Microbiol. 51:2033–2039. 10.1128/JCM.00196-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Roberts CH, Last A, Molina-Gonzalez S, Cassama E, Butcher R, Nabicassa M, McCarthy E, Burr SE, Mabey DC, Bailey RL, Holland MJ. 2013. Development and evaluation of a next-generation digital PCR diagnostic assay for ocular Chlamydia trachomatis infections. J. Clin. Microbiol. 51:2195–2203. 10.1128/JCM.00622-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943. 10.1371/journal.pone.0055943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF. 2011. Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load. J. Am. Chem. Soc. 133:17705–17712. 10.1021/ja2060116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sedlak RH, Cook L, Huang ML, Magaret A, Zerr DM, Boeckh M, Jerome KR. 2014. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin. Chem. 60:765–772. 10.1373/clinchem.2013.217240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Salipante SJ, Sengupta DJ, Rosenthal C, Costa G, Spangler J, Sims EH, Jacobs MA, Miller SI, Hoogestraat DR, Cookson BT, McCoy C, Matsen FA, Shendure J, Lee CC, Harkins TT, Hoffman NG. 2013. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One 8:e65226. 10.1371/journal.pone.0065226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. 2012. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012:251364. 10.1155/2012/251364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nyren P. 1987. Enzymatic method for continuous monitoring of DNA polymerase activity. Anal. Biochem. 167:235–238. 10.1016/0003-2697(87)90158-8 [DOI] [PubMed] [Google Scholar]
- 152.Hyman ED. 1988. A new method of sequencing DNA. Anal. Biochem. 174:423–436. 10.1016/0003-2697(88)90041-3 [DOI] [PubMed] [Google Scholar]
- 153.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. 1996. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242:84–89. 10.1006/abio.1996.0432 [DOI] [PubMed] [Google Scholar]
- 154.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452:872–876. 10.1038/nature06884 [DOI] [PubMed] [Google Scholar]
- 155.Mashayekhi F, Ronaghi M. 2007. Analysis of read length limiting factors in pyrosequencing chemistry. Anal. Biochem. 363:275–287. 10.1016/j.ab.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. 10.1186/gb-2007-8-7-r143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352. 10.1038/nature10242 [DOI] [PubMed] [Google Scholar]
- 159.Perkel J. 2011. Making contact with sequencing's fourth generation. Biotechniques 50:93–95. 10.2144/000113608 [DOI] [PubMed] [Google Scholar]
- 160.Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW. 2013. Shining a light on dark sequencing: characterising errors in ion torrent PGM data. PLoS Comput. Biol. 9:e1003031. 10.1371/journal.pcbi.1003031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, Hooks DP, Cowan LS, Plikaytis BB, Posey JE. 2011. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55:2032–2041. 10.1128/AAC.01550-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015. 10.1056/NEJMoa0907847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bravo LT, Tuohy MJ, Ang C, Destura RV, Mendoza M, Procop GW, Gordon SM, Hall GS, Shrestha NK. 2009. Pyrosequencing for rapid detection of Mycobacterium tuberculosis resistance to rifampin, isoniazid, and fluoroquinolones. J. Clin. Microbiol. 47:3985–3990. 10.1128/JCM.01229-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jureen P, Engstrand L, Eriksson S, Alderborn A, Krabbe M, Hoffner SE. 2006. Rapid detection of rifampin resistance in Mycobacterium tuberculosis by pyrosequencing technology. J. Clin. Microbiol. 44:1925–1929. 10.1128/JCM.02210-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Maughan H, Wang PW, Diaz Caballero J, Fung P, Gong Y, Donaldson SL, Yuan L, Keshavjee S, Zhang Y, Yau YC, Waters VJ, Tullis DE, Hwang DM, Guttman DS. 2012. Analysis of the cystic fibrosis lung microbiota via serial Illumina sequencing of bacterial 16S rRNA hypervariable regions. PLoS One 7:e45791. 10.1371/journal.pone.0045791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. 10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sherry NL, Porter JL, Seemann T, Watkins A, Stinear TP, Howden BP. 2013. Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J. Clin. Microbiol. 51:1396–1401. 10.1128/JCM.03332-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hedskog C, Mild M, Jernberg J, Sherwood E, Bratt G, Leitner T, Lundeberg J, Andersson B, Albert J. 2010. Dynamics of HIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing. PLoS One 5:e11345. 10.1371/journal.pone.0011345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Larsen BB, Chen L, Maust BS, Kim M, Zhao H, Deng W, Westfall D, Beck I, Frenkel LM, Mullins JI. 2013. Improved detection of rare HIV-1 variants using 454 pyrosequencing. PLoS One 8:e76502. 10.1371/journal.pone.0076502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chang MW, Oliveira G, Yuan J, Okulicz JF, Levy S, Torbett BE. 2013. Rapid deep sequencing of patient-derived HIV with ion semiconductor technology. J. Virol. Methods 189:232–234. 10.1016/j.jviromet.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413. 10.1128/JCM.43.1.406-413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vezinet F, Clotet B, Hammer SM, Johnson VA, Kuritzkes DR, Mellors JW, Pillay D, Yeni PG, Jacobsen DM, Richman DD. 2008. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 47:266–285. 10.1086/589297 [DOI] [PubMed] [Google Scholar]
- 173.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, Hullsiek KH, Balduin M, Jakobsen MR, Geretti AM, Thiebaut R, Ostergaard L, Masquelier B, Johnson JA, Miller MD, Kuritzkes DR. 2011. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 305:1327–1335. 10.1001/jama.2011.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rego N, Bianchi S, Moreno P, Persson H, Kvist A, Pena A, Oppezzo P, Naya H, Rovira C, Dighiero G, Pritsch O. 2012. Search for an aetiological virus candidate in chronic lymphocytic leukaemia by extensive transcriptome analysis. Br. J. Haematol. 157:709–717. 10.1111/j.1365-2141.2012.09116.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lan J, Tai HC, Lee SW, Chen TJ, Huang HY, Li CF. 2014. Deficiency in expression and epigenetic DNA Methylation of ASS1 gene in nasopharyngeal carcinoma: negative prognostic impact and therapeutic relevance. Tumour Biol. 35:161–169. 10.1007/s13277-013-1020-8 [DOI] [PubMed] [Google Scholar]
- 176.Leclerc D, Levesque N, Cao Y, Deng L, Wu Q., Powell J, Sapienza C, Rozen R. 2013. Genes with aberrant expression in murine preneoplastic intestine show epigenetic and expression changes in normal mucosa of colon cancer patients. Cancer Prev. Res. 6:1171–1181. 10.1158/1940-6207.CAPR-13-0198 [DOI] [PubMed] [Google Scholar]
- 177.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Sci. 309:1728–1732. 10.1126/science.1117389 [DOI] [PubMed] [Google Scholar]
- 178.Voelkerding KV, Dames SA, Durtschi JD. 2009. Next-generation sequencing: from basic research to diagnostics. Clin. Chem. 55:641–658. 10.1373/clinchem.2008.112789 [DOI] [PubMed] [Google Scholar]
- 179.Dooley KC. 2003. Tandem mass spectrometry in the clinical chemistry laboratory. Clin. Biochem. 36:471–481. 10.1016/S0009-9120(03)00105-X [DOI] [PubMed] [Google Scholar]
- 180.Sauer S, Kliem M. 2010. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 8:74–82. 10.1038/nrmicro2243 [DOI] [PubMed] [Google Scholar]
- 181.Yates JR, Ruse CI, Nakorchevsky A. 2009. Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 11:49–79. 10.1146/annurev-bioeng-061008-124934 [DOI] [PubMed] [Google Scholar]
- 182.La Scola B. 2011. Intact cell MALDI-TOF mass spectrometry-based approaches for the diagnosis of bloodstream infections. Expert Rev. Mol. Diagn. 11:287–298 [DOI] [PubMed] [Google Scholar]
- 183.Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175. 10.1128/JCM.01881-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jamal WY, Shahin M, Rotimi VO. 2013. Comparison of two matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry methods and API 20AN for identification of clinically relevant anaerobic bacteria. J. Med. Microbiol. 62:540–544. 10.1099/jmm.0.053256-0 [DOI] [PubMed] [Google Scholar]
- 185.Marko DC, Saffert RT, Cunningham SA, Hyman J, Walsh J, Arbefeville S, Howard W, Pruessner J, Safwat N, Cockerill FR, Bossler AD, Patel R, Richter SS. 2012. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for identification of nonfermenting Gram-negative bacilli isolated from cultures from cystic fibrosis patients. J. Clin. Microbiol. 50:2034–2039. 10.1128/JCM.00330-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. 2012. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:1313–1325. 10.1128/JCM.05971-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rychert J, Burnham CA, Bythrow M, Garner OB, Ginocchio CC, Jennemann R, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Sercia L, Westblade LF, Ferraro MJ, Branda JA. 2013. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J. Clin. Microbiol. 51:2225–2231. 10.1128/JCM.00682-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Branda JA, Rychert J, Burnham CA, Bythrow M, Garner OB, Ginocchio CC, Jennemann R, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Sercia LF, Westblade LF, Ferraro MJ. 2014. Multicenter validation of the VITEK MS v2.0 MALDI-TOF mass spectrometry system for the identification of fastidious gram-negative bacteria. Diagn. Microbiol. Infect. Dis. 78:129–131. 10.1016/j.diagmicrobio.2013.08.013 [DOI] [PubMed] [Google Scholar]
- 189.Manji R, Bythrow M, Branda JA, Burnham CA, Ferraro MJ, Garner OB, Jennemann R, Lewinski MA, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Westblade LF, Ginocchio CC. 2014. Multi-center evaluation of the VITEK(R) MS system for mass spectrometric identification of non-Enterobacteriaceae Gram-negative bacilli. Eur. J. Clin. Microbiol. Infect. Dis. 33:337–346. 10.1007/s10096-013-1961-2 [DOI] [PubMed] [Google Scholar]
- 190.Garner O, Mochon A, Branda J, Burnham CA, Bythrow M, Ferraro M, Ginocchio C, Jennemann R, Manji R, Procop GW, Richter S, Rychert J, Sercia L, Westblade L, Lewinski M. 2014. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK(R) MS system. Clin. Microbiol. Infect. 20:335–339. 10.1111/1469-0691.12317 [DOI] [PubMed] [Google Scholar]
- 191.Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6:e16424. 10.1371/journal.pone.0016424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Saffert RT, Cunningham SA, Ihde SM, Jobe KE, Mandrekar J, Patel R. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 49:887–892. 10.1128/JCM.01890-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J. Clin. Microbiol. 50:3301–3308. 10.1128/JCM.01405-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ferreira L, Sanchez-Juanes F, Gonzalez-Avila M, Cembrero-Fucinos D, Herrero-Hernandez A, Gonzalez-Buitrago JM, Munoz-Bellido JL. 2010. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:2110–2115. 10.1128/JCM.02215-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Demarco ML, Burnham CA. 2014. Diafiltration MALDI-TOF mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. Am. J. Clin. Pathol. 141:204–212. 10.1309/AJCPQYW3B6JLKILC [DOI] [PubMed] [Google Scholar]
- 196.Buchan BW, Riebe KM, Ledeboer NA. 2012. Comparison of the MALDI Biotyper system using Sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J. Clin. Microbiol. 50:346–352. 10.1128/JCM.05021-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fothergill A, Kasinathan V, Hyman J, Walsh J, Drake T, Wang YF. 2013. Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization-time of flight mass spectrum analysis with the SARAMIS database. J. Clin. Microbiol. 51:805–809. 10.1128/JCM.02326-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen JH, Ho PL, Kwan GS, She KK, Siu GK, Cheng VC, Yuen KY, Yam WC. 2013. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J. Clin. Microbiol. 51:1733–1739. 10.1128/JCM.03259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Moussaoui W, Jaulhac B, Hoffmann AM, Ludes B, Kostrzewa M, Riegel P, Prevost G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry identifies 90% of bacteria directly from blood culture vials. Clin. Microbiol. Infect. 16:1631–1638. 10.1111/j.1469-0691.2010.03356.x [DOI] [PubMed] [Google Scholar]
- 200.Stevenson LG, Drake SK, Murray PR. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:444–447. 10.1128/JCM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang XH, Zhang G, Fan YY, Yang X, Sui WJ, Lu XX. 2013. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J. Microbiol. Methods 92:231–235. 10.1016/j.mimet.2012.12.016 [DOI] [PubMed] [Google Scholar]
- 202.Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J, Collins CD, Nagel JL. 2013. Impact of rapid organism identification via matrix-assisted laser desorption ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin. Infect. Dis. 57:1237–1245. 10.1093/cid/cit498 [DOI] [PubMed] [Google Scholar]
- 203.Perez KK, Olsen RJ, Musick WL, Cernoch PL, Davis JR, Land GA, Peterson LE, Musser JM. 2013. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch. Pathol. Lab. Med. 137:1247–1254. 10.5858/arpa.2012-0651-OA [DOI] [PubMed] [Google Scholar]
- 204.Buchan BW, Ledeboer NA. 2013. Advances in identification of clinical yeast isolates by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:1359–1366. 10.1128/JCM.03105-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.L'Ollivier C, Cassagne C, Normand AC, Bouchara JP, Contet-Audonneau N, Hendrickx M, Fourquet P, Coulibaly O, Piarroux R, Ranque S. 2013. A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med. Mycol. 51:713–720. 10.3109/13693786.2013.781691 [DOI] [PubMed] [Google Scholar]
- 206.Ranque S, Normand AC, Cassagne C, Murat JB, Bourgeois N, Dalle F, Gari-Toussaint M, Fourquet P, Hendrickx M, Piarroux R. 2014. MALDI-TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses 57:135–140. 10.1111/myc.12115 [DOI] [PubMed] [Google Scholar]
- 207.Cassagne C, Ranque S, Normand AC, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. 2011. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One 6:e28425. 10.1371/journal.pone.0028425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Buchan BW, Riebe KM, Timke M, Kostrzewa M, Ledeboer NA. 2014. Comparison of MALDI-TOF MS with HPLC and nucleic acid sequencing for the identification of Mycobacterium species in cultures using solid medium and broth. Am. J. Clin. Pathol. 141:25–34. 10.1309/AJCPBPUBUDEW2OAG [DOI] [PubMed] [Google Scholar]
- 209.Balada-Llasat JM, Kamboj K, Pancholi P. 2013. Identification of mycobacteria from solid and liquid media by matrix-assisted laser desorption ionization–time of flight mass spectrometry in the clinical laboratory. J. Clin. Microbiol. 51:2875–2879. 10.1128/JCM.00819-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pignone M, Greth KM, Cooper J, Emerson D, Tang J. 2006. Identification of mycobacteria by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry. J. Clin. Microbiol. 44:1963–1970. 10.1128/JCM.01959-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Westblade LF, Jennemann R, Branda JA, Bythrow M, Ferraro MJ, Garner OB, Ginocchio CC, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Burnham CA. 2013. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J. Clin. Microbiol. 51:2267–2272. 10.1128/JCM.00680-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pence MA, McElvania Tekippe E, Wallace MA, Burnham CA. 7 May 2014. Comparison and optimization of two MALDI-TOF MS platforms for the identification of medically relevant yeast species. Eur. J. Clin. Microbiol. Infect. Dis. 10.1007/s10096-014-2115-x [DOI] [PubMed] [Google Scholar]
- 213.Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365. 10.1111/j.1469-0691.2010.03398.x [DOI] [PubMed] [Google Scholar]
- 214.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616. 10.1128/JCM.02381-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917. 10.1128/JCM.00389-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486. 10.1128/JCM.00687-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Mather CA, Rivera SF, Butler-Wu SM. 2014. Comparison of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for identification of mycobacteria using simplified protein extraction protocols. J. Clin. Microbiol. 52:130–138. 10.1128/JCM.01996-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:1790–1794. 10.1128/JCM.02135-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lotz A, Ferroni A, Beretti JL, Dauphin B, Carbonnelle E, Guet-Revillet H, Veziris N, Heym B, Jarlier V, Gaillard JL, Pierre-Audigier C, Frapy E, Berche P, Nassif X, Bille E. 2010. Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:4481–4486. 10.1128/JCM.01397-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Balazova T, Makovcova J, Sedo O, Slany M, Faldyna M, Zdrahal Z. 2014. The influence of culture conditions on the identification of Mycobacterium species by MALDI-TOF MS profiling. FEMS Microbiol. Lett. 353:77–84. 10.1111/1574-6968.12408 [DOI] [PubMed] [Google Scholar]
- 221.Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J. Clin. Microbiol. 50:927–937. 10.1128/JCM.05737-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hrabak J, Studentova V, Walkova R, Zemlickova H, Jakubu V, Chudackova E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerova T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2441–2443. 10.1128/JCM.01002-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sparbier K, Lange C, Jung J, Wieser A, Schubert S, Kostrzewa M. 2013. MALDI Biotyper-based rapid resistance detection by stable-isotope labeling. J. Clin. Microbiol. 51:3741–3748. 10.1128/JCM.01536-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Metzgar D, Baynes D, Myers CA, Kammerer P, Unabia M, Faix DJ, Blair PJ. 2010. Initial identification and characterization of an emerging zoonotic influenza virus prior to pandemic spread. J. Clin. Microbiol. 48:4228–4234. 10.1128/JCM.01336-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sampath R, Russell KL, Massire C, Eshoo MW, Harpin V, Blyn LB, Melton R, Ivy C, Pennella T, Li F, Levene H, Hall TA, Libby B, Fan N, Walcott DJ, Ranken R, Pear M, Schink A, Gutierrez J, Drader J, Moore D, Metzgar D, Addington L, Rothman R, Gaydos CA, Yang S, St George K, Fuschino ME, Dean AB, Stallknecht DE, Goekjian G, Yingst S, Monteville M, Saad MD, Whitehouse CA, Baldwin C, Rudnick KH, Hofstadler SA, Lemon SM, Ecker DJ. 2007. Global surveillance of emerging influenza virus genotypes by mass spectrometry. PLoS One 2:e489. 10.1371/journal.pone.0000489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jacob D, Sauer U, Housley R, Washington C, Sannes-Lowery K, Ecker DJ, Sampath R, Grunow R. 2012. Rapid and high-throughput detection of highly pathogenic bacteria by Ibis PLEX-ID technology. PLoS One 7:e39928. 10.1371/journal.pone.0039928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kaleta EJ, Clark AE, Cherkaoui A, Wysocki VH, Ingram EL, Schrenzel J, Wolk DM. 2011. Comparative analysis of PCR-electrospray ionization/mass spectrometry (MS) and MALDI-TOF/MS for the identification of bacteria and yeast from positive blood culture bottles. Clin. Chem. 57:1057–1067. 10.1373/clinchem.2011.161968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kaleta EJ, Clark AE, Johnson DR, Gamage DC, Wysocki VH, Cherkaoui A, Schrenzel J, Wolk DM. 2011. Use of PCR coupled with electrospray ionization mass spectrometry for rapid identification of bacterial and yeast bloodstream pathogens from blood culture bottles. J. Clin. Microbiol. 49:345–353. 10.1128/JCM.00936-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pierce SE, Bell RL, Hellberg RS, Cheng CM, Chen KS, Williams-Hill DM, Martin WB, Allard MW. 2012. Detection and identification of Salmonella enterica, Escherichia coli, and Shigella spp. via PCR-electrospray ionization mass spectrometry: isolate testing and analysis of food samples. Appl. Environ. Microbiol. 78:8403–8411. 10.1128/AEM.02272-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sampath R, Mulholland N, Blyn LB, Massire C, Whitehouse CA, Waybright N, Harter C, Bogan J, Miranda MS, Smith D, Baldwin C, Wolcott M, Norwood D, Kreft R, Frinder M, Lovari R, Yasuda I, Matthews H, Toleno D, Housley R, Duncan D, Li F, Warren R, Eshoo MW, Hall TA, Hofstadler SA, Ecker DJ. 2012. Comprehensive biothreat cluster identification by PCR/electrospray-ionization mass spectrometry. PLoS One 7:e36528. 10.1371/journal.pone.0036528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mengelle C, Mansuy JM, Da Silva I, Guerin JL, Izopet J. 2013. Evaluation of a polymerase chain reaction-electrospray ionization time-of-flight mass spectrometry for the detection and subtyping of influenza viruses in respiratory specimens. J. Clin. Virol. 57:222–226. 10.1016/j.jcv.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tang YW, Lowery KS, Valsamakis A, Schaefer VC, Chappell JD, White-Abell J, Quinn CD, Li H, Washington CA, Cromwell J, Giamanco CM, Forman M, Holden J, Rothman RE, Parker ML, Ortenberg EV, Zhang L, Lin YL, Gaydos CA. 2013. Clinical accuracy of a PLEX-ID flu device for simultaneous detection and identification of influenza viruses A and B. J. Clin. Microbiol. 51:40–45. 10.1128/JCM.01978-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 357:654–663. 10.1056/NEJMoa061588 [DOI] [PubMed] [Google Scholar]
- 234.Achermann Y, Vogt M, Leunig M, Wust J, Trampuz A. 2010. Improved diagnosis of periprosthetic joint infection by multiplex PCR of sonication fluid from removed implants. J. Clin. Microbiol. 48:1208–1214. 10.1128/JCM.00006-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Butler-Wu SM, Burns EM, Pottinger PS, Magaret AS, Rakeman JL, Matsen FA, 3rd, Cookson BT. 2011. Optimization of periprosthetic culture for diagnosis of Propionibacterium acnes prosthetic joint infection. J. Clin. Microbiol. 49:2490–2495. 10.1128/JCM.00450-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Jacovides CL, Kreft R, Adeli B, Hozack B, Ehrlich GD, Parvizi J. 2012. Successful identification of pathogens by polymerase chain reaction (PCR)-based electron spray ionization time-of-flight mass spectrometry (ESI-TOF-MS) in culture-negative periprosthetic joint infection. J. Bone Joint Surg. Am. 94:2247–2254. 10.2106/JBJS.L.00210 [DOI] [PubMed] [Google Scholar]
- 237.Greenwood-Quaintance KE, Uhl JR, Hanssen AD, Sampath R, Mandrekar JN, Patel R. 2014. Diagnosis of prosthetic joint infection by use of PCR-electrospray ionization mass spectrometry. J. Clin. Microbiol. 52:642–649. 10.1128/JCM.03217-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Simner PJ, Uhl JR, Hall L, Weber MM, Walchak RC, Buckwalter S, Wengenack NL. 2013. Broad-range direct detection and identification of fungi by use of the PLEX-ID PCR-electrospray ionization mass spectrometry system. J. Clin. Microbiol. 51:1699–1706. 10.1128/JCM.03282-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Greub G, Prod'hom G. 2011. Automation in clinical bacteriology: what system to choose? Clin. Microbiol. Infect. 17:655–660. 10.1111/j.1469-0691.2011.03513.x [DOI] [PubMed] [Google Scholar]
- 240.Novak SM, Marlowe EM. 2013. Automation in the clinical microbiology laboratory. Clin. Lab. Med. 33:567–588. 10.1016/j.cll.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 241.Bourbeau PP, Ledeboer NA. 2013. Automation in clinical microbiology. J. Clin. Microbiol. 51:1658–1665. 10.1128/JCM.00301-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Burnham CA, Dunne WM, Jr, Greub G, Novak SM, Patel R. 2013. Automation in the clinical microbiology laboratory. Clin. Chem. 59:1696–1702. 10.1373/clinchem.2012.201038 [DOI] [PubMed] [Google Scholar]
- 243. Reference deleted.
- 244.Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S. 2012. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J. Clin. Microbiol. 50:2732–2736. 10.1128/JCM.05501-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bourbeau PP, Swartz BL. 2009. First evaluation of the WASP, a new automated microbiology plating instrument. J. Clin. Microbiol. 47:1101–1106. 10.1128/JCM.01963-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Glasson JH, Guthrie LH, Nielsen DJ, Bethell FA. 2008. Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J. Clin. Microbiol. 46:1281–1284. 10.1128/JCM.01687-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Tilton RC, Ryan RW. 1978. Evaluation of an automated agar plate streaker. J. Clin. Microbiol. 7:298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Van Horn KG, Audette CD, Sebeck D, Tucker KA. 2008. Comparison of the Copan ESwab system with two Amies agar swab transport systems for maintenance of microorganism viability. J. Clin. Microbiol. 46:1655–1658. 10.1128/JCM.02047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Rivers CA, Schwebke JR. 2008. Viability of Trichomonas vaginalis in Copan universal transport medium and eSwab transport medium. J. Clin. Microbiol. 46:3134–3155. 10.1128/JCM.00841-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Nys S, Vijgen S, Magerman K, Cartuyvels R. 2010. Comparison of Copan eSwab with the Copan Venturi Transystem for the quantitative survival of Escherichia coli, Streptococcus agalactiae and Candida albicans. Eur. J. Clin. Microbiol. Infect. Dis. 29:453–456. 10.1007/s10096-010-0883-5 [DOI] [PubMed] [Google Scholar]
- 251.Saegeman V, Flamaing J, Muller J, Peetermans WE, Stuyck J, Verhaegen J. 2011. Clinical evaluation of the Copan ESwab for methicillin-resistant Staphylococcus aureus detection and culture of wounds. Eur. J. Clin. Microbiol. Infect. Dis. 30:943–349. 10.1007/s10096-011-1178-1 [DOI] [PubMed] [Google Scholar]
- 252.Smismans A, Verhaegen J, Schuermans A, Frans J. 2009. Evaluation of the Copan ESwab transport system for the detection of methicillin-resistant Staphylococcus aureus: a laboratory and clinical study. Diagn. Microbiol. Infect. Dis. 65:108–111. 10.1016/j.diagmicrobio.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 253.Indevuyst C, Beuselinck K, Lagrou K. 2012. eSwab flocked swabs unfit for viral culture. J. Clin. Virol. 55:282–283. 10.1016/j.jcv.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 254.Mutters NT, Hodiamont CJ, de Jong MD, Overmeijer HP, van den Boogaard M, Visser CE. 2014. Performance of Kiestra total laboratory automation combined with MS in clinical microbiology practice. Ann. Lab. Med. 34:111–117. 10.3343/alm.2014.34.2.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Matthews S, Deutekom J. 2011. The future of diagnostic bacteriology. Clin. Microbiol. Infect. 17:651–654. 10.1111/j.1469-0691.2011.03512.x [DOI] [PubMed] [Google Scholar]
- 256.Bentley N, Farrington M, Doughton R, Pearce D. 2011. Automating the bacteriology laboratory, abstr P1792. Abstr. Eur. Congr. Clin. Microbiol. Infect. Dis., Milan, Italy [Google Scholar]
- 257.Humphrey G, Malone C, Gough H, Awadel-Kariem M. 2011. Experience with KIESTRA's Total Lab Automation solutiion to meet teh challenge of universal MRSA screening for Lister Hospital, a large UK distract general hospital, abstr P1793. Abstr. Eur. Congr. Clin. Microbiol. Infect. Dis., Milan, Italy [Google Scholar]
- 258.Ford A. 2013. Molecular testing platforms a land of plenty. CAP Today 27:22–42 [Google Scholar]
- 259.Laurent C, Bogaerts P, Schoevaerdts D, Denis O, Deplano A, Swine C, Struelens MJ, Glupczynski Y. 2010. Evaluation of the Xpert MRSA assay for rapid detection of methicillin-resistant Staphylococcus aureus from nares swabs of geriatric hospitalized patients and failure to detect a specific SCCmec type IV variant. Eur. J. Clin. Microbiol. Infect. Dis. 29:995–1002. 10.1007/s10096-010-0958-3 [DOI] [PubMed] [Google Scholar]
- 260.Wolk DM, Struelens MJ, Pancholi P, Davis T, Della-Latta P, Fuller D, Picton E, Dickenson R, Denis O, Johnson D, Chapin K. 2009. Rapid detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in wound specimens and blood cultures: multicenter preclinical evaluation of the Cepheid Xpert MRSA/SA skin and soft tissue and blood culture assays. J. Clin. Microbiol. 47:823–826. 10.1128/JCM.01884-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Spencer DH, Sellenriek P, Burnham CA. 2011. Validation and implementation of the GeneXpert MRSA/SA blood culture assay in a pediatric setting. Am. J. Clin. Pathol. 136:690–694. 10.1309/AJCP07UGYOKBVVNC [DOI] [PubMed] [Google Scholar]
- 262.Glenn TC. 2011. Field guide to next-generation DNA sequencers. Mol. Ecol. Resources 11:759–769. 10.1111/j.1755-0998.2011.03024.x [DOI] [PubMed] [Google Scholar]