Abstract
SUMMARY
Nontuberculous mycobacteria (NTM) are present in the environment, mainly in water, and are occasionally responsible for opportunistic infections in humans. Despite the fact that NTM are characterized by a moderate pathogenicity, the diseases caused by NTM at various body sites are increasing on a worldwide level. Among over 150 officially recognized NTM species, only two or three dozen are familiar to clinicians, and even to most microbiologists. In this paper, approximately 50 new species described in the last 8 years are reviewed, and their role in human infections is assessed on the basis of reported clinical cases. The small number of reports concerning most of the “new” mycobacterial species is responsible for the widespread conviction that they are very rare. Their role is actually largely underestimated, mainly because they often remain unrecognized and misidentified. Aiming to minimize such bias, emphasis has been placed on more common identification pitfalls. Together with new NTM, new members of the Mycobacterium tuberculosis complex described in the last few years are also an object of the present review.
INTRODUCTION
Mycobacteriology, like every other branch of microbiology, has been characterized in the last 20 years by an extraordinary flourishing of new species consequent to the availability of molecular techniques, in particular genetic sequencing, with discriminative power which would otherwise be unattainable through conventional phenotypic approaches. Authoritative reviews have been published previously, starting with the master paper of Wolinsky in 1979 (1), followed several years later by the work of Wayne and Sramek (2). Two systematic reviews have been published in the last decade (3, 4), along with a significant work oriented toward rapidly growing mycobacteria (5). Since then, however, more than 50 new mycobacterial species have been described (Table 1). The aim of this work is to provide a concise but thorough update to help clinicians and microbiologists in facing the continuous enlargements of the taxonomy of the Mycobacterium genus.
TABLE 1.
Major features of new Mycobacterium speciesa
| Species | Phylogenetic group | Source | Growth | Pigment | Type strain |
|---|---|---|---|---|---|
| M. algericum | MTeC | Animals | Slow | NC | CIP 110121 = DSM 45454 |
| “M. alsiense” | Humans | Slow | SC | ||
| M. arabiense | Environment | Rapid | SC | DSM 45768 = JCM 18538 | |
| M. aromaticivorans | Environment | Rapid | SC | ATCC BAA-1378 = CIP 109274 | |
| M. arosiense | MAC | Humans | Slow | SC | DSM 45069 = ATCC BAA-1401 |
| M. bacteremicum | M. neoaurum | Humans | Rapid | SC | ATCC 25791 |
| “M. barrassiae” | Humans | Rapid | NC | CIP 108545 = CCUG 50398 | |
| M. bouchedurhonense | MAC | Humans | Slow | NC | CCUG 56331 = CIP 109827 = CSUR P34 |
| M. bourgelatii | Animals | Rapid | NC | DSM 45746 = CIP 110557 | |
| M. celeriflavum | Humans | Rapid | SC | DSM 46765 = JCM 18439 | |
| M. crocinum | Environment | Rapid | SC | ATCC BAA-1373 = CIP 109262 | |
| M. engbaekii | MTeC | Humans | Slow | SC | ATCC 27353 = DSM 45694 |
| M. europaeum | MSC | Humans | Slow | SC | DSM 45397 = CCUG 58464 |
| M. fragae | M. celatum | Humans | Slow | NC | INCQS/CMRVS P4051 = DSM 45731 |
| “M. fukienense” | M. chelonae | Humans | Rapid | NC | |
| “M. franklinii” | M. chelonae | Humans | Rapid | NC | ATCC BAA-2149 = DSM 45524 |
| M. heraklionense | MTeC | Humans | Slow | NC | NCTC 13432 = LMG 24735 = CECT 7509 |
| “M. hippocampi” | Animals | Rapid | SC | DSM 45391 = LMG 25372 | |
| M. insubricum | Humans | Slow | NC | DSM 45132 = CIP 109609 | |
| M. iranicum | Humans | Rapid | SC | DSM 45541 = CCUG 62053 = JCM 17461 | |
| M. koreense | M. triviale | Humans | Slow | NC | DSM 45576 = KCTC 19819 |
| M. kumamotonense | MTeC | Humans | Slow | NC | GTC 2729 = JCM 13453 = CCUG 51961 |
| M. kyorinense | M. celatum | Humans | Slow | NC | JCM 15038 = DSM 45166 |
| “M. lepromatosis” | M. leprae | Humans | |||
| M. litorale | Environment | Rapid | SC | CGMCC 4.5724 = JCM 17423 | |
| “M. liflandii” | M. marinum | Animals | Slow | NC | |
| M. llatzerense | Humans and environment | Rapid | NC | CECT 7273 = CCUG 54744 | |
| M. longobardum | MTeC | Humans | Slow | NC | DSM 45394 = CCUG 58460 |
| M. mantenii | M. scrofulaceum | Humans | Slow | SC | NLA000401474 = CIP 109863 = DSM 45255 |
| M. marseillense | MAC | Humans | Slow | NC | CCUG 56325 = CIP 109828 = CSUR P30 |
| M. minnesotense | MTeC | Environment | Slow | SC | DSM 45633 = JCM 17932 = NCCB 100399 |
| M. monacense | Humans | Rapid | SC | DSM 44395 = CIP 109237 | |
| “M. mungi” | MTC | Animals | Slow | NC | |
| M. noviomagense | Humans | Slow | NC | DSM 45145 = CIP 109766 | |
| “M. orygis” | MTC | Animals and humans | Slow | NC | |
| M. pallens | Environment | Rapid | SC | ATCC BAA-1372 = CIP 109268 | |
| M. paraffinicum | M. scrofulaceum | Humans and environment | Slow | SC | ATCC 12670 = DSM 44181 = NCIMB 10420 |
| M. paragordonae | M. gordonae | Humans | Slow | SC | JCM 18565 = KCTC 29126 |
| M. parakoreense | M. triviale | Humans | Slow | NC | DSM 45575 = KCTC 19818 |
| M. paraseoulense | M. scrofulaceum | Humans | Slow | NC | DSM 45000 = KCTC 19145 |
| “M. paraterrae” | MTeC | Humans | Slow | SC/NC | DSM 45127 = KCTC 19556 |
| M. riyadhense | Humans | Slow | NC | CIP 109808 = DSM 45176 | |
| M. rufum | Environment | Rapid | SC | ATCC BAA-1377 = CIP 109273 | |
| M. rutilum | Environment | Rapid | SC | ATCC BAA-1375 = CIP 109271 | |
| M. salmoniphilum | M. chelonae | Animals | Rapid | NC | ATCC 13578 = DSM 43276 |
| M. sediminis | Environment | Rapid | SC | DSM 45643 = KCTC 19999 | |
| M. senuense | MTeC | Humans | Slow | NC | DSM 44999 = KCTC 19147 |
| M. seoulense | M. scrofulaceum | Humans | Slow | SC | DSM 44998 = KCTC 19146 |
| M. setense | M. fortuitum | Humans | Rapid | NC | CIP 109395 = DSM 45070 |
| M. sherrisii | MSC | Humans and environment | Slow | NC/PC | ATCC BAA-832 = DSM 45441 |
| “M. shigaense” | MSC | Humans | Slow | NC | DSM 46748 |
| M. shinjukuense | Humans | Slow | NC | JCM 14233 = CCUG 53584 | |
| “M. simulans” | Humans | Slow | NC | DSM 45395 | |
| “M. sinense” | MTeC | Humans | Slow | NC | |
| M. stomatepiae | MSC | Animals | Slow | NC | DSM 45059 = CIP 109275 = NCIMB 14252 |
| “M. suricattae” | MTC | Animals | Slow | NC | |
| M. timonense | MAC | Humans | Slow | NC | CCUG 56329 = CIP 109830 = CSUR P32 |
| M. vulneris | MAC | Humans | Slow | SC | DSM 45247 = CIP 109859 |
| M. yongonense | MAC | Humans | Slow | NC | DSM 45126 = KCTC 19555 |
MTeC, M. terrae complex; MAC, M. avium complex; MSC, M. simiae complex; MTC, M. tuberculosis complex; NC, nonchromogenic; SC, scotochromogenic; PC, photochromogenic.
Identification of Mycobacteria
Accurate identification of the etiologic agent is critical for diagnosis and management of infectious diseases and for outbreak detection. The identification of nontuberculous mycobacteria (NTM) is still problematic, despite the number of different methodologies explored and used over the last few years.
Identification based on the analysis of biochemical characteristics and cultural traits (6) remained the method of choice for many years. It has now been abandoned, because in addition to relying on tests that are poorly reproducible and unbearably time-consuming, it lacks sufficient discriminative power (7).
The peculiar lipid-rich composition of the cell wall of mycobacteria may be investigated for identification purposes by chromatographic methods. Thin-layer chromatography (8) and gas-liquid chromatography (GLC) (9) have been used mainly for research purposes and have gained very limited use in diagnostic laboratories. High-performance liquid chromatography (HPLC) of cell wall mycolic acids (10) is the method based on lipid analysis which has obtained the largest popularity among reference laboratories, particularly in the United States. Identification springs from comparison of the chromatographic profiles of an unknown species with reference patterns; elution times and relative heights of single peaks are critical. In the last few years, the emergence of new species sharing common HPLC profiles has steadily diminished the discriminatory capacity of this method, which is clearly beginning to become dated. In this review, information on lipid patterns is reported when available, referring only to the HPLC approach. The interpretation of profiles given by the authors of studies was minimized, however, when discriminatory power appeared to be assigned to very minor differences consistent with the variability of the method. A computer-assisted method, Sherlock Microbial ID System (MIDI, Newark, DE), is commercially available for both GLC (11) and HPLC (12); unfortunately, the databases included are largely incomplete. An almost exhaustive HPLC database is available on the Internet (www.MycobacToscana.it/page4.htm) but is nevertheless unsuitable for accurate profile comparison.
In recent years, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry has gained a large reputation among diagnostic laboratories as a rapid and inexpensive tool for the identification of “common,” clinically relevant bacteria. Very good results have also been reported related to the identification of a number of Mycobacterium species (13). Important limitations of the method are represented at the moment by the need for a sample preparation specific for mycobacteria that is not optimized yet and the large incompleteness of databases. It is my opinion that the identification accuracy achievable at present with genetic approaches is out of reach of MALDI-TOF technology.
The molecular approach, using both in-house and commercial systems, is the most frequently used method worldwide for the identification of mycobacterial species.
PCR restriction analysis (PRA) is based on digestion with restriction enzymes of the PCR-amplified 441-bp fragment of the hsp65 gene, encoding the 65-kDa heat shock protein (14). The patterns of fragments produced by the BstEII and HaeIII enzymes (14) are somehow species specific and allow the identification of most clinically significant mycobacteria. A wide PRA pattern database is available online (http://app.chuv.ch/prasite/index.html). The increasing number of shared and ambiguous patterns nowadays makes the system very prone to misidentifications.
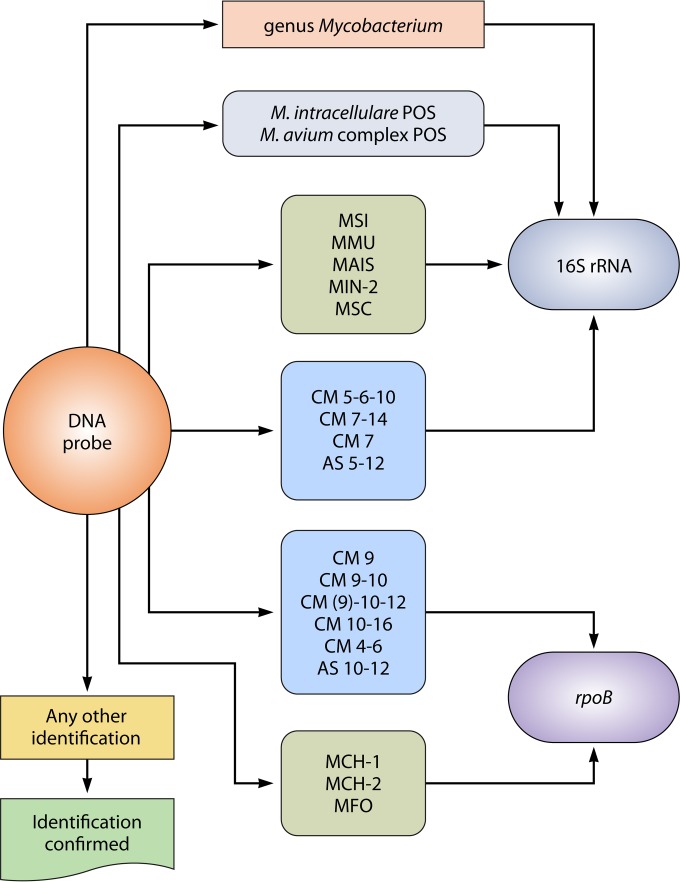
The introduction of commercial DNA probes is the basis for the most important improvement in the identification of mycobacteria in clinical laboratories. Three different commercial systems dominate the market. The AccuProbe system (Hologic Gen-Probe, San Diego, CA), made available nearly 30 years ago, targets the 16S rRNA gene with species-specific probes and does not require amplification. Specific commercial kits identify the most frequently isolated Mycobacterium species, such as Mycobacterium avium and Mycobacterium intracellulare, both individually and at the level of the M. avium complex (MAC) (15). Solid-phase reverse hybridization technology subsequently made the identification of multiple species with a single test possible. The INNO-LiPA Mycobacteria system (Fujirebio Europe, Ghent, Belgium) targets the internal transcribed spacer (ITS1) interposed between the 16S rRNA and 23S rRNA genes; the kit identifies 18 NTM species simultaneously, 2 of them also at the subtype level (16). The GenoType Mycobacterium system (Hain Lifescience, Nehren, Germany) targets the 23S rRNA gene and is available as two separate kits: CM, which identifies 22 of the most frequently isolated species; and AS, which identifies an additional 13 less common species (17). In each of the commercial systems, some probes are not 100% specific, and several cross-reactions, in addition to the ones declared by the producers, are observed (18).
Genetic sequencing of conserved genes is the reference method for the identification of mycobacteria. A number of housekeeping genes were found to be suitable for this purpose, including the ones encoding the 16S rRNA (19), the 65-kDa heat shock protein (hsp65) (20), the RNA polymerase β-subunit (rpoB) (21, 22), and the superoxide dismutase (sodA) (23), as well as several others (24). Sequencing of noncoding regions, such as ITS1 (25), can also be used for species identification.
Antimicrobial Susceptibility of NTM
An excellent review on susceptibility testing and treatment of infections due to NTM was published recently (26), focusing on the most frequently encountered mycobacterial species. In it, as well as in the guidelines of the Clinical and Laboratory Standards Institute (27), the different susceptibility patterns of slow and rapid growers and the testing method are clearly defined. The antimicrobials with potential activity against slow growers include amikacin, ciprofloxacin, clarithromycin, ethambutol, linezolid, moxifloxacin, rifabutin, rifampin, streptomycin, and trimethoprim-sulfamethoxazole. The panel of those recommended for rapid growers consists of amikacin, cefoxitin, ciprofloxacin, clarithromycin, doxycycline, imipenem, linezolid, meropenem, moxifloxacin, trimethoprim-sulfamethoxazole, and, for Mycobacterium chelonae only, tobramycin. Determining the MIC in broth is the method of choice for susceptibility testing of NTM. Unfortunately, these recommendations are often ignored in reports concerning novel mycobacteria. In this review, only susceptibility data for drugs considered appropriate are reported; results for drugs improperly tested were disregarded.
Phylogenetic Remarks
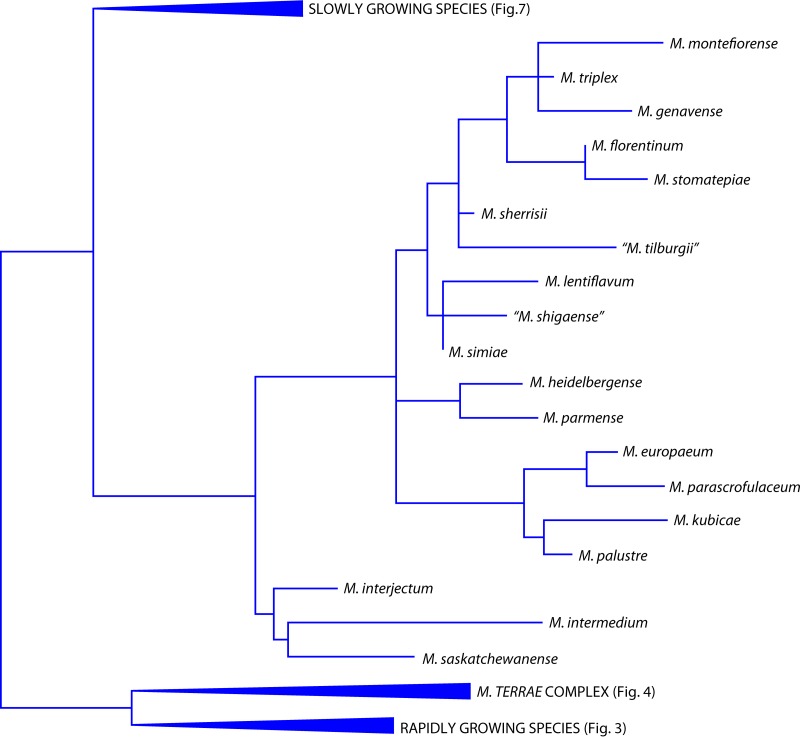
Investigation conducted by comparing genetic sequences of conserved genes allows reconstruction of the evolutionary pathway of mycobacteria. Present data show that rapidly growing species descended directly from ancestral mycobacteria, while slow growers originated from a more recently emerged branch. The 16S rRNA gene is the single gene which best allows the trace of the evolution of the genus Mycobacterium. A particular region, corresponding to helix 18 of the secondary structure of the 16S rRNA, is the site of a significant genetic signature: helix 18 of slow growers is 12 nucleotides longer than that of rapid growers and 2 nucleotides shorter than that of a particular evolutionary branch known as the M. terrae complex, whose members are characterized by an intermediate growth rate. An exception in this scheme is represented by a cluster of species (M. simiae complex) which, although belonging to a recent branch as confirmed by the slow growth, present a short helix 18 like that of the ancestral mycobacteria. A further genetic signature, located right in helix 18, is a unique sequence (Fig. 1) which is shared by the whole M. simiae complex (7). The evolutionary scenario based on the 16S rRNA gene is substantially confirmed by other housekeeping genes, in particular when genes are concatenated to form a single sequence. Furthermore, good agreement exists with preliminary data emerging from whole-genome sequencing.
FIG 1.

Sequence of helix 18 of the 16S rRNA genes (corresponding to Escherichia coli positions 430 to 500) of different Mycobacterium species. Sequences: 1, shared by all species of the M. simiae complex; 2, M. tuberculosis (representative of slowly growing species); 3, M. fortuitum (representative of rapidly growing species); 4, M. arupense (representative of the M. terrae complex). Dots indicate identities, and dashes indicate deletions.
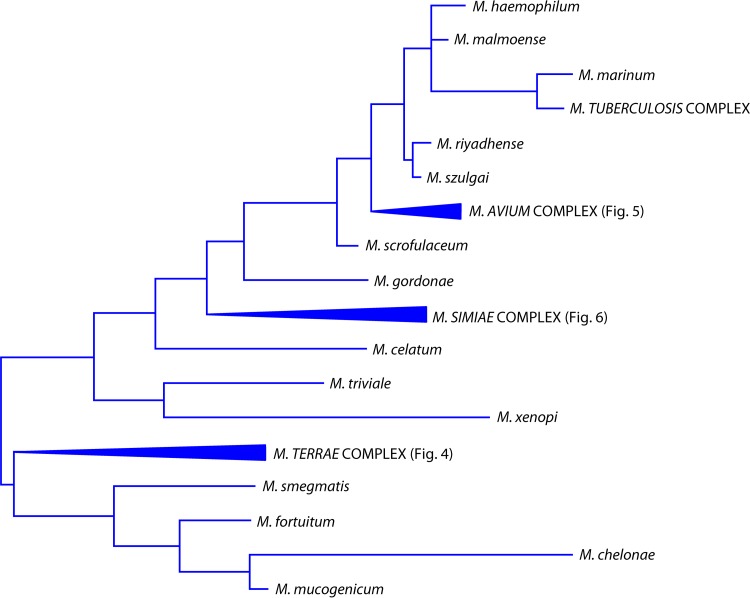
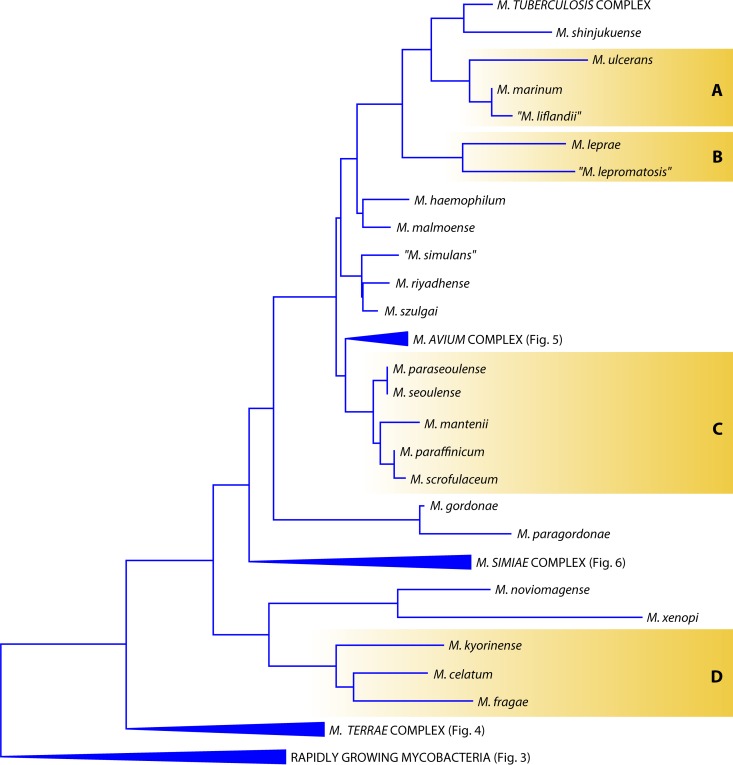
In the Mycobacterium evolutionary tree, species are located both in isolated branches and in clusters (Fig. 2); among the latter, the groups, or complexes, are named after the representative species: M. avium, M. simiae, M. terrae, M. scrofulaceum, M. fortuitum, M. chelonae, and M. smegmatis. The last is also called the thermotolerant, rapidly growing group.
FIG 2.
Phylogenetic tree, based on the 16S rRNA gene, including representative species of the genus Mycobacterium.
Novel Mycobacterium Species
More than 150 Mycobacterium species have been validly published to date, according to the List of Prokaryotic Names with Standing in Nomenclature (http://www.bacterio.net/mycobacterium.html). To these, a number of non-officially recognized species can be added. According to the International Committee on Systematic Bacteriology, species whose descriptions are published in the International Journal of Systematic and Evolutionary Microbiology are automatically validated. The validation of species described elsewhere can be requested at the office of the above-mentioned journal and is subject to evaluation. The status of validated and nonvalidated species is therefore mutable and is in fact ignored even by databases such as GenBank; the decision to include both types of species in this review aims to provide readers with the largest possible coverage. Following a universally accepted custom, the names of nonvalidated species are given in quotation marks. The species have been divided into three groups: rapidly growing NTM, slowly growing NTM, and members of the Mycobacterium tuberculosis complex. Within each group, alphabetical order is used to make searches easier.
RAPIDLY GROWING NTM
Mycobacterium abscessus
The taxonomic position of M. abscessus has been the object of continuous debate. It was described in 1953 (28), downsized to a subspecies of M. chelonae in 1972 (29), and reinstated as a species in 1992 (30). More recently, two previously described species, Mycobacterium bolletii (31) and Mycobacterium massiliense (32), were reclassified as subspecies of M. abscessus and considered a single taxon with the name M. abscessus subsp. bolletii (33), distinct from M. abscessus sensu stricto, which is now officially named M. abscessus subsp. abscessus. More recent data, however, based on whole-genome sequencing, support the splitting of the species M. abscessus into three different subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense (34–36).
In recent years, the involvement of M. abscessus in invasive infections in both immunocompetent and immunocompromised patients has also been emphasized in consideration of problematic treatments (37). Increasing numbers of isolates obtained from patients with cystic fibrosis (38) are also of concern. Most strains are susceptible only to macrolides and amikacin, but mutants resistant to such drugs are not unusual; resistance to macrolides may be either constitutive or inducible. In this respect, the differentiation of M. abscessus subsp. massiliense has clinical relevance; in fact, most of the strains belonging to this subspecies, because of an inactivating deletion in the erm gene, lack inducible resistance, and infections with this organism are characterized by a better prognosis (39–41).
The identification of M. abscessus at the subspecies level is not feasible with available commercial DNA probes and requires the partial genetic sequencing of rpoB and, at times, hsp65 as well.
Mycobacterium arabiense
M. arabiense was described in 2014, along with the closely related species M. sediminis (42), on the basis of a single strain isolated from a sand sample from the Dubai coast.
The species presents the signature of rapid growers (short helix 18) in the 16S rRNA gene but is not clearly related to a specific phylogenetic branch.
Orange-red, smooth, nonphotochromogenic colonies develop in about 3 days at 25 to 37°C, but a scanty growth is also obtained at lower temperatures, down to 5°C.
No isolation from human or animal specimens has been reported so far.
Mycobacterium aromaticivorans
M. aromaticivorans was described in 2009, along with four other new species degrading polycyclic aromatic hydrocarbons (43). The single strain on which the species nova description is based was isolated from the contaminated soil of an oil-gasification company in Hawaii.
The species presents the nucleotide deletion in the 16S rRNA gene that is typical of rapid growers.
Growth on solid media, although rapid, is obtained only from heavy inocula suitable to produce confluent colonies, which are characterized by a mucoid appearance and yellow scotochromogenic pigmentation.
M. aromaticivorans has never been grown so far from human or animal specimens.
Mycobacterium bacteremicum
M. bacteremicum was described in 2010 (44), on the basis of 10 clinical isolates previously considered a variant of Mycobacterium neoaurum.
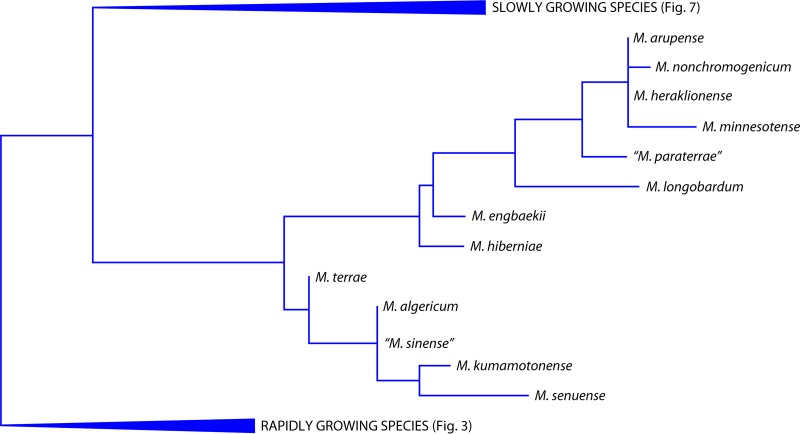
The species is included among the rapid growers (Fig. 3) and is closely related to M. neoaurum. It is characterized by a high level of microheterogeneity in the hsp65 and rpoB genes, resulting in multiple sequevars.
FIG 3.
Phylogenetic tree, based on the 16S rRNA gene, including representative rapidly growing Mycobacterium species (A, M. triviale group; B, M. smegmatis group; C, M. fortuitum complex; and D, M. chelonae complex).
The colonies, which are yellow and scotochromogenic, grow in less than a week on solid media.
Eighty percent of the investigated strains presented either direct or inducible resistance to clarithromycin, while all were susceptible to the remaining drugs included in the panel for testing rapidly growing mycobacteria.
The 10 strains reported so far were isolated from young patients, 2 of whom were immunocompromised. In three cases, the strain grew from blood, and in one case, it grew from a central catheter. No data concerning the treatment of M. bacteremicum are available. In the years preceding the description of the new species, a number of papers reported cases related to an M. neoaurum-like organism. At least some of them can be attributed retrospectively to M. bacteremicum; they were successfully treated with combinations of two or more drugs, which were administered for periods ranging from 3 to 12 weeks (44).
“Mycobacterium barrassiae”
“M. barrassiae” was described in 2006 (45), based on a single clinical strain.
The species presents the genetic signature of rapid growers in the 16S rRNA gene (Fig. 3) and is most closely related to M. moriokaense.
Unpigmented colonies develop in 3 to 6 days at temperatures ranging from 25 to 42°C.
The strain is susceptible in vitro to the whole panel of drugs with potential activity against rapidly growing mycobacteria.
The only strain reported so far was repeatedly isolated from a patient with radiographic evidence of pulmonary disease, which was successfully treated with a 3-month course of amikacin, clarithromycin, and ciprofloxacin.
Mycobacterium bourgelatii
M. bourgelatii was described in 2013, on the basis of three strains isolated from cattle from unrelated herds (46).
The phylogenetic position of M. bourgelatii is confusing. It presents the 12-nucleotide insertion in the 16S rRNA gene that is specific for slowly growing mycobacteria except for those of the M. simiae complex. Nevertheless, M. bourgelatii is most closely related to M. intermedium, a member of the M. simiae complex, and is characterized by rapid growth. Microheterogeneity is present in the investigated housekeeping genes, except for the 16S rRNA gene.
The colonies, which are smooth, shiny, and nonchromogenic, grow in less than a week at temperatures between 25 and 42°C.
Five strains have been reported so far, all of them isolated from bovine lymph nodes.
Mycobacterium celeriflavum
M. celeriflavum was described in 2014 (179), on the basis of six clinical strains isolated in Iran, Turkey, and Italy.
The most closely associated species are Mycobacterium flavescens and Mycobacterium novocastrense (Fig. 3), although this relatedness is not clearly evident in some of the investigated genetic targets (the 16S rRNA gene in particular).
The colonies, which are rough and pale yellow (scotochromogenic), take 5 to 7 days to develop at 37°C. The HPLC profile is characterized by a continuous series of peaks eluting between 3 and 9 min, with the most prominent clustering before the 4th and around the 8th minute. The strains are susceptible to amikacin, clarithromycin, linezolid, quinolones, and doxycycline and resistant to cefoxitin.
Two of the six strains characterized for the description of the new species were considered responsible for pulmonary disease, and the patients were cured with a combination of clarithromycin, rifampin, and ethambutol.
Mycobacterium crocinum
M. crocinum was described in 2009, along with four other new species degrading polycyclic aromatic hydrocarbons (43). The three strains on which the species nova description is based were isolated from the soil on the island of Oahu, HI.
The phylogenetic position of the species is unclear; in fact, it correlates in various genetic regions with mycobacteria belonging to different branches. The three strains share an identical sequence for hsp65, while microheterogeneity exists in the 16S rRNA and rpoB genes.
The colonies grow in less than a week, may be mucoid or dry, and are yellow-orange under both light and dark growth conditions.
M. crocinum has never been grown from human or animal specimens; the only strains reported so far are those that were characterized when the new species was described.
“Mycobacterium franklinii”
“M. franklinii” was described in 2011, on the basis of 26 clinical strains from the United States, most of which were isolated in Pennsylvania (47).
The species has been characterized carefully and is evidently part of the Mycobacterium chelonae-abscessus complex: the 16S rRNA gene is identical to that of M. chelonae, while in other genetic targets, which host moderate levels of microheterogeneity, the species is most closely associated with other members of the complex (Fig. 3). The concatenated sequence of 43 genes places “M. franklinii” in a separate phylogenetic branch halfway between M. chelonae and Mycobacterium immunogenum. The colonies are unpigmented and grow in 2 to 3 days at temperatures ranging from 25 to 37°C.
Differently from M. chelonae, “M. franklinii” presents intermediate or full susceptibility to cefoxitin.
The large majority of the strains were isolated from respiratory specimens from patients with underlying pulmonary disease; only in one case was the strain isolated repeatedly, and none of the patients underwent specific treatment. Two strains closely related to “M. franklinii” were isolated from conduit water in the Netherlands (48).
“Mycobacterium fukienense”
“M. fukienense” was described in 2013 (49), on the basis of five clinical isolates.
The description is very approximate; no information is given about the availability of a type strain in international collections or of sequences in public databases. Only a 300-bp fragment of the rpoB gene can be inferred from a figure in the publication. For different genetic regions, the five investigated strains are reported to present the closest similarity with various species of the M. chelonae-abscessus group. Microheterogeneity is recorded for ITS1.
The species grows in less than a week, forming unpigmented colonies. The GenoType Mycobacterium CM test identifies the strains as M. chelonae.
All the strains were isolated from patients with pulmonary infections which had been misdiagnosed as tuberculosis.
“Mycobacterium hippocampi”
“M. hippocampi” was described in 2011, following isolation from aquarium seahorses with tail rot disease (white cutaneous spots and necrotic tail lesions) (50). Multiple isolations were obtained, but only one of the isolates was characterized.
Mycobacterium flavescens is the species presenting the closest similarity on the basis of the 16S rRNA gene sequence (Fig. 3), while based on the rpoB gene, “M. hippocampi” is quite distant from any other mycobacterium. Scotochromogenic, orange colonies develop at 25°C in 5 days.
The strain is susceptible in vitro to ciprofloxacin, clarithromycin, and rifampin.
Mycobacterium insubricum
M. insubricum was described in 2009, on the basis of five independent clinical strains (51).
M. insubricum clearly differs from any other mycobacterium (Fig. 3), although it presents some similarity, in different genetic regions, to Mycobacterium brumae and Mycobacterium fallax. Genetic microheterogeneity is present in hsp65 and ITS1 but not in the 16S rRNA gene or rpoB.
Smooth, white colonies develop within 7 days at 25 to 37°C. The HPLC profile of mycolic acids is unique but similar to the one of M. brumae, M. fallax, and Mycobacterium triviale.
M. insubricum is susceptible in vitro to the entire panel of antimicrobials recommended for rapidly growing mycobacteria.
All the strains characterized for the description of the new species were isolated from sputum samples, some of them repeatedly; it seems unlikely that the criteria for clinical significance were fulfilled in any of the cases.
Mycobacterium iranicum
M. iranicum was described in 2013, on the basis of eight independent strains isolated from clinical specimens in six different countries (52).
The species occupies a peculiar phylogenetic position among rapid growers (Fig. 3). It is in fact characterized by two 4-nucleotide deletions in the 16S rRNA gene, which are unique within the genus Mycobacterium. The microheterogeneity level is high and is responsible for multiple sequevars in different housekeeping genes.
Mature colonies, which are deep orange, develop on solid media in less than a week at temperatures ranging from 25 to 40°C. The mycolic acid HPLC profile is unique and is characterized by three well-separated peak clusters.
The strains are susceptible in vitro to amikacin, cefoxitin, clarithromycin, ethambutol, minocycline, imipenem, linezolid, and sulfamethoxazole.
M. iranicum was revealed to be pathogenic in two of the eight strains investigated for the description of the new species: it was responsible for a chronic pulmonary disease in an elderly patient and for a cutaneous lesion in a kidney transplant subject. In both cases, treatment with amikacin (combined with ciprofloxacin for the respiratory infection) produced a clear improvement. Two new cases were reported immediately after the description of the species. One case was in the United States, in a woman with bronchiectasis who was treated with tetracyclines, to which the strain was susceptible in vitro (53). The other case was in Iran, involving a severely immunocompromised HIV-positive man with pulmonary disease, who rapidly improved following a 3-month course of amikacin and ciprofloxacin treatment (54).
Mycobacterium litorale
M. litorale was described in 2012, on the basis of a single strain isolated from a soil sample from a seashore in China (55).
Phylogenetically, M. litorale is included among the rapid growers (Fig. 3) and is most closely related to Mycobacterium monacense.
Light yellow colonies develop in 1 to 2 days at 37°C. The HPLC mycolic acid pattern grossly resembles that of MAC strains and is characterized by an early major peak cluster and two late minor clusters.
It is susceptible in vitro to amikacin, ciprofloxacin, doxycycline, linezolid, tobramycin, and sulfamethoxazole but resistant to clarithromycin.
M. litorale has never been grown from human or animal specimens.
Mycobacterium llatzerense
M. llatzerense was described in 2008, based on six strains isolated from water samples (56).
The species harbors the mark of rapid growers in the 16S rRNA gene (Fig. 3) and is closely related to Mycobacterium aubagnense and Mycobacterium mucogenicum. The strains investigated are genetically very homogeneous. Unpigmented colonies grow in 3 to 4 days at temperatures ranging from 22 to 30°C; no growth is observed at 37°C.
The species is susceptible to ciprofloxacin, clarithromycin, and minocycline.
M. llatzerense is misidentified as M. mucogenicum by the GenoType Mycobacterium AS test.
The strains on which the species nova description is based were isolated from hemodialysis water in a hospital in Majorca, Spain, and other reports subsequently documented the isolation of M. llatzerense from tap water (48, 57). Two clinical cases were recently reported, one in the United States, concerning a liver transplant recipient with pulmonary disease (58), and one case of abdominal abscess, in Spain (59).
Mycobacterium monacense
M. monacense was described in 2006, based on four independent clinical isolates (60).
The species occupies a position clearly divergent from the major evolutionary branches of rapidly growing mycobacteria (Fig. 3). The ITS1 genetic region is characterized by a high level of microheterogeneity.
Smooth, yellow, scotochromogenic colonies develop in a week at temperatures ranging from 25 to 45°C. The mycolic acids are characterized by an HPLC pattern grossly resembling that of the M. terrae complex. The species is susceptible in vitro to amikacin, clarithromycin, doxycycline, and ciprofloxacin.
Three of the four strains on which the description of the species is based were isolated from respiratory specimens from unrelated patients; one of the patients underwent a tuberculosis-specific treatment, without any improvement. One case concerned a young boy presenting a fistula consequent to the accidental deep penetration of a screwdriver into the thigh; an in-depth surgical intervention was needed to resolve the problem. A number of cases were reported subsequently: one concerning a hand infection, which was cured with a 6-week course of treatment with clarithromycin and levofloxacin (61), and three related to pulmonary diseases (62–64). Follow-up information is available for only one of the patients, who was treated successfully with amikacin and ciprofloxacin for 12 months.
Mycobacterium pallens
M. pallens was described in 2009, along with four other new species degrading polycyclic aromatic hydrocarbons (43), on the basis of a single strain isolated from the soil on the island of Oahu, HI.
The phylogenetic position of the species is unclear, as it is closely related to different rapidly growing mycobacteria depending on which genes are investigated.
The colonies grow in less than a week, are flat and dry with an undulated border, and turn pale orange once exposed to light.
M. pallens has never been grown from human or animal specimens; the only isolation reported so far is the one on which the description of the species is based.
Mycobacterium rufum
M. rufum was described in 2009, along with four other new species degrading polycyclic aromatic hydrocarbons (43). The single strain on which the species nova description is based was isolated from the contaminated soil of an oil-gasification company in Hawaii.
The phylogenetic position of the species is unclear, as it is most closely related to different rapidly growing mycobacteria depending on which genes are investigated.
The colonies grow within 7 days and are large, raised, and dry, with an orange pigmentation in light or darkness. The optimum growth is obtained at 28°C.
M. rufum has never been grown from human or animal specimens; the only isolation reported so far is the one on which the description of the species is based.
Mycobacterium rutilum
M. rutilum was described in 2009, along with four other new species degrading polycyclic aromatic hydrocarbons (43). The three strains on which the species nova description is based were isolated from the soil of an urban park in Honolulu, HI.
The phylogenetic position of the species, which is characterized by wide microheterogeneity in all the investigated housekeeping genes, is unclear, as it is most closely related to different rapidly growing mycobacteria depending on the considered gene.
The colonies are pigmented deeply orange after exposure to light, have an undulated transparent border, and may be mucoid or dry. Their growth takes less than a week at temperatures ranging from 28 to 45°C.
M. rutilum has never been grown from human or animal specimens; the only isolations reported so far are the ones on which the description of the species is based.
Mycobacterium salmoniphilum
M. salmoniphilum was first described in 1960 (65), but the new species was not validated. It was revived in 2007, on the basis of 11 strains isolated from salmonid fish (66).
The species is phylogenetically located among rapid growers within the M. chelonae complex (Fig. 3). Very high levels of genetic microheterogeneity are present.
Smooth, cream-colored, shiny colonies grow after 4 to 6 days at temperatures ranging from 20 to 30°C, but not at 37°C. The HPLC mycolic acid profile, characterized by two late-emerging peak clusters, is similar to that of M. chelonae.
M. salmoniphilum is pathogenic for salmonid fish (67–71), with the disease affecting predominantly the kidneys. It has also been isolated from tap water (48) but never from human specimens.
Mycobacterium sediminis
M. sediminis was described in 2014 (42), on the basis of a single strain isolated from a sediment sample collected in the South China Sea.
The phylogenetic position is controversial, and the present structure of the genome has been hypothesized to originate from a large lateral gene transfer event.
The colonies develop in about 1 week at 25 to 37°C, but they can grow more slowly at temperatures as low as 5°C. They are orange-red, rough, and not photochromogenic.
No isolation from human or animal specimens has been reported so far.
Mycobacterium setense
M. setense was described in 2008, on the basis of a single clinical isolate (72).
The species occupies a phylogenetic position among the rapid growers within the Mycobacterium fortuitum complex (Fig. 3) and is more closely related, in the 16S rRNA gene, to Mycobacterium houstonense and Mycobacterium senegalense.
The colonies are smooth and nonpigmented, do not produce aerial hyphae, and grow in 2 to 4 days at 25 to 37°C. The HPLC mycolic acid pattern presents similarities to those of M. fortuitum and other related species.
The strain is susceptible in vitro to quinolones, cefoxitin, and amikacin.
M. setense is misidentified as M. fortuitum by the INNO-LiPA Mycobacteria test.
The strain from which the description of the new species originated was isolated from a patient with a posttraumatic lesion of the foot complicated by osteitis and tenosynovitis. The initial treatment with clarithromycin for 1 month was ineffective; the patient was subsequently cured with a 1-month course of treatment with amikacin and levofloxacin. A further case of bone infection was reported for a recipient of an osseous graft (73), which was cured with a 4-month treatment with ciprofloxacin. More recently, three infections due to M. setense were reported in Iran (74): in a man with fever and inflammation following a knee joint replacement, an HIV-positive young woman with a subcutaneous abscess consequent to an accidental injury, and a renal transplant recipient under immunosuppressive therapy and with chronic bronchitis. The first two patients were successfully treated with amikacin, and the outcome for the third is unknown.
SLOWLY GROWING NTM
Mycobacterium algericum
M. algericum was described in 2011, based on a single strain isolated from the lung lesion of a goat (75).
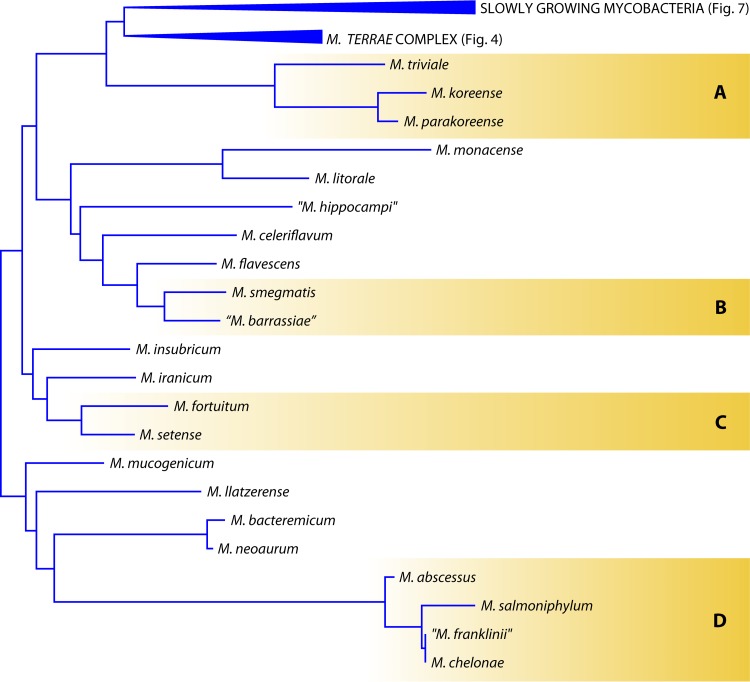
The species belongs to the Mycobacterium terrae complex (Fig. 4), as demonstrated by the 14-nucleotide insertion in helix 18 of the 16S rRNA gene.
FIG 4.
Phylogenetic tree, based on the 16S rRNA gene, for the species belonging to the M. terrae complex.
The colonies, which are smooth and unpigmented, develop in more than a week at temperatures between 25 and 40°C. The HPLC profile of mycolic acids is characterized by two separate peak clusters and does not allow for discrimination between the species within the complex (76).
Only one other strain, isolated from a fish (77), has been reported so far.
“Mycobacterium alsiense”
“M. alsiense” was described in 2007 (78), on the basis of two clinical strains isolated from unrelated patients.
The phylogenetic position of “M. alsiense” is not clear, as it groups with Mycobacterium gordonae and Mycobacterium asiaticum based on the 16S rRNA gene but shows similarities with other Mycobacterium species in various genetic regions.
Yellowish scotochromogenic and smooth colonies grow slowly at 25 to 36°C. The mycolic acid HPLC profile is characterized by a single, late-emerging peak cluster that is different enough from other species presenting a comparable peak arrangement.
The only strain tested so far proved to be susceptible to clarithromycin, ethambutol, rifampin, and streptomycin.
In the GenoType Mycobacterium CM commercial assay, “M. alsiense” presents the same hybridization pattern as that shown by the species M. scrofulaceum, M. parascrofulaceum, and M. paraffinicum.
The only two strains reported so far (78) were isolated from respiratory specimens from elderly immunocompetent subjects. Both strains were considered responsible for pulmonary disease, and the patients were treated with rifampin (combined with ethambutol in one case and with clarithromycin in the other), with a clear improvement of the radiological picture.
Mycobacterium arosiense
M. arosiense was described in 2008 (79), on the basis of a single clinical strain.
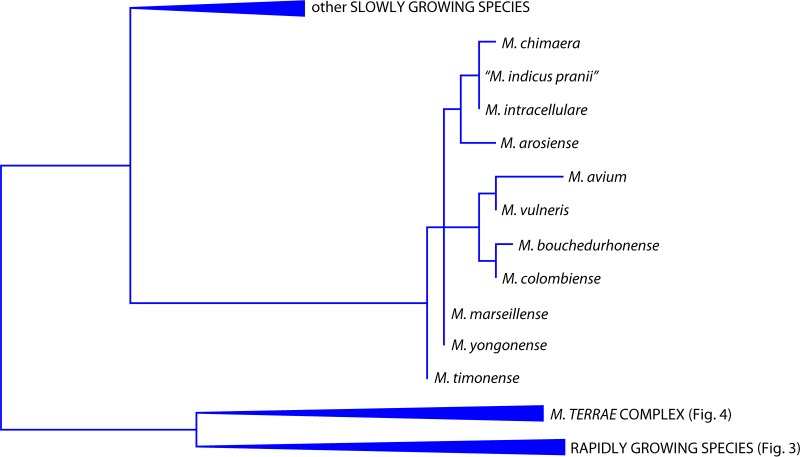
The species belongs to the MAC and is genetically most closely related to M. intracellulare (Fig. 5).
FIG 5.
Phylogenetic tree, based on the 16S rRNA gene, for the species belonging to the M. avium complex.
The colonies are scotochromogenic, yellow, and smooth; they grow in two or more weeks at temperatures ranging from 36 to 42°C, with the latter being the optimum. This species shares the typical HPLC profile of mycolic acids with the other species of the MAC, presenting an early major peak cluster and two late minor ones.
All the strains isolated so far were susceptible in vitro to clarithromycin and amikacin.
The observation that M. arosiense belongs to the MAC is confirmed by the hybridization patterns presented with commercial DNA probes. All of them assign M. arosiense to the MAC: the AccuProbe and GenoType Mycobacterium CM tests misidentify it as M. intracellulare, while the INNO-LiPA Mycobacteria test includes it in the M. avium-intracellulare-scrofulaceum (MAIS) group and recognizes it as being different from M. avium, M. intracellulare, and Mycobacterium scrofulaceum (18).
The strain from which the description of the new species originated was responsible for disseminated osteomyelitis in a 7-year-old child with underlying gamma interferon receptor alpha-1 deficiency (79). Other clinical strains have been reported, including five from respiratory specimens and two from urine (80). At least one of the cases fulfilled the American Thoracic Society (ATS) criteria for clinical significance (81); this case concerned an elderly patient presenting with pulmonary infection and a history of Hodgkin's lymphoma, splenectomy, and gastrectomy, who was successfully treated with clarithromycin, rifampin, and ethambutol. The involvement of M. arosiense in human infections is probably underestimated because of the large use of DNA probe identification systems which systematically misidentify this species as M. intracellulare.
Mycobacterium bouchedurhonense
M. bouchedurhonense was described in 2009, along with two other new species belonging to the MAC, on the basis of a single strain isolated from a sputum specimen (82).
Its inclusion in the MAC (Fig. 5) is supported by the close genetic similarity restricted to species belonging to the complex.
The colonies are rough and unpigmented and grow on solid media in about 2 weeks at temperatures between 30 and 45°C.
M. bouchedurhonense is misidentified as M. intracellulare by the GenoType Mycobacterium CM test; hybridization with other commercial DNA probes has not been determined.
No clinical information is available regarding the patient from whom the only reported strain so far was isolated.
Mycobacterium engbaekii
The M. engbaekii species was first proposed in 1972 (83), but it was only officially described in 2013 (76), based on seven clinical isolates.
The species presents the typical signature of the M. terrae complex (Fig. 4) in the 16S rRNA gene: a 14-nucleotide insertion in helix 18. Microheterogeneity is present in the hsp65 and rpoB genes but not in the 16S rRNA gene. The colonies grow on solid media in about 10 days at 25 to 37°C, are rough, and develop pink pigmentation following exposure to light. The HPLC profile of mycolic acids supports the phylogenetic position, in that M. engbaekii is characterized by the traits common to all of the species belonging to the M. terrae complex.
M. engbaekii is susceptible in vitro to amikacin, clarithromycin, ethambutol, linezolid, and rifabutin and is resistant to doxycycline and sulfamethoxazole.
About 50 strains of M. engbaekii have been characterized so far, most of which were isolated from human specimens. No case meeting the ATS criteria for clinical relevance has been reported.
Mycobacterium europaeum
M. europaeum was described in 2011, on the basis of five independent strains isolated in different European countries (84).
The species belongs to the Mycobacterium simiae complex (Fig. 6), as shown by the short helix 18 characterized by the specific sequence.
FIG 6.
Phylogenetic tree, based on the 16S rRNA gene, for the species belonging to the M. simiae complex.
The colonies are smooth and deep yellow in both dark and light and grow in 2 to 3 weeks at temperatures between 30 and 37°C. By HPLC, the mycolic acid profile grossly resembles that of the species belonging to the MAC. The sequences of housekeeping genes other than the 16S rRNA gene are characterized by microheterogeneity with multiple sequevars.
M. europaeum is resistant in vitro to quinolones and is susceptible to the other drugs included in the panel designed for slowly growing mycobacteria.
Four clinically significant cases due to M. europaeum have been documented. Two of the strains characterized on the occasion of the description of this new species were responsible for cavitary pneumonia or cervical lymphadenitis. Two further cases of lung disease have been reported, concerning an HIV-positive subject and a patient with cystic fibrosis. The former was cured with amikacin, while the latter underwent an antituberculosis regimen (85).
Mycobacterium fragae
M. fragae was described in 2013, on the basis of a single clinical strain (86).
The species is part of the phylogenetic branch occupied by the slowly growing species Mycobacterium branderi, Mycobacterium celatum, and Mycobacterium kyorinense (Fig. 7).
FIG 7.
Phylogenetic tree, based on the 16S rRNA gene, including representative slow-growing Mycobacterium species (A, M. marinum group; B, M. leprae group; C, M. scrofulaceum group; and D, M. celatum group).
The colonies are smooth and unpigmented and grow in 3 to 4 weeks at temperatures ranging from 30 to 37°C. M. fragae possesses a unique HPLC pattern of mycolic acids and is susceptible in vitro to the whole panel of antimicrobials potentially active against slowly growing species.
The only strain of M. fragae isolated so far was grown repeatedly from a patient with pulmonary disease. The patient underwent three pharmacological cycles, two based on antituberculosis drugs and the third, which was effective, with amikacin, clarithromycin, and ethambutol (unpublished data).
Mycobacterium heraklionense
M. heraklionense was described in 2013, on the basis of 23 strains isolated from clinical specimens in Greece, Italy, and India (76).
The species presents the typical 14-nucleotide insertion in the 16S rRNA gene, considered the signature of the species included in the M. terrae complex (Fig. 4). Wide heterogeneity is present in the sequences of the hsp65 and rpoB genes.
The colonies develop on solid media in 5 to 12 days at 25 to 37°C and are smooth and unpigmented. The HPLC profile of mycolic acids supports the phylogenetic location. It is in fact not substantially different from that of the other species belonging to the M. terrae complex.
M. heraklionense is susceptible in vitro to clarithromycin and is resistant to quinolones, rifampin, sulfamethoxazole, and doxycycline.
Around 20 strains of M. heraklionense have been investigated so far and were isolated from various clinical specimens. The only infection meeting the criteria for clinical relevance concerns a case of tenosynovitis in an immunocompetent patient (87).
“Mycobacterium indicus pranii”
“M. indicus pranii” (sic), although known since the 1970s, was described in 2009 (88). It includes a single strain, originally named “Mycobacterium W,” which was chosen from a collection of NTM for its ability to evoke cell-mediated immune responses against M. leprae (89).
The strain presents, in the large majority of housekeeping genes, complete identity with M. intracellulare (Fig. 5), of which it should probably be considered a subspecies. Surprisingly, M. intracellulare is not included among the species with which the recently determined complete genome of “M. indicus pranii” has been compared (90).
The colonies are unpigmented and smooth and develop on solid media in 6 to 8 days at temperatures between 25 and 45°C. The strain shares antigens with M. tuberculosis and M. leprae.
Because of its immunomodulatory activity, it has been used to treat various human diseases, including tuberculosis (91), either alone or in combination with chemotherapy. “M. indicus pranii” is also used as an immunotherapeutic vaccine against leprosy (92).
Mycobacterium koreense
M. koreense was described in 2012, on the basis of three strains: a clinical isolate and two reference strains (ATCC 23290 and ATCC 23291) previously assigned to the species M. triviale (93).
M. koreense presents the short helix 18 in the 16S rRNA gene that is typical of rapid growers (Fig. 3). It is therefore not part of the M. terrae complex as stated in the species nova description. None of the housekeeping genes investigated is exempt from microheterogeneity.
Nonchromogenic colonies, which may be rough or smooth, grow in 1 to 2 weeks at temperatures ranging from 25 to 37°C. The HPLC profile is distinguishable from that of M. triviale only because of the presence of a minor early peak cluster.
Among the main antimycobacterial drugs, only quinolones are fully active in vitro.
No clinical information is available about the patient from whose sputum M. koreense was isolated.
Mycobacterium kumamotonense
M. kumamotonense was described in 2006, on the basis of a single strain isolated from a sputum sample (94).
M. kumamotonense presents the 14-nucleotide insertion distinctive of the M. terrae complex in the 16S rRNA gene (Fig. 4) and is most closely related to the M. terrae species. A large degree of sequence variability is present in all of the investigated housekeeping genes (76).
Colonies become visible after 7 to 14 days of incubation at 25 to 42°C. The mycolic acid profile in HPLC is similar to that of other members of the M. terrae complex but often presents with the peaks of the first cluster arranged in an unusual rising order (76).
The species is susceptible in vitro to amikacin, clarithromycin, rifampin, and quinolones.
No clinical information is available about the patient from whose sputum the original strain of M. kumamotonense was isolated. One case of generalized lymphadenopathy in an HIV-positive patient has been reported (95); the patient was treated with an antituberculosis regimen for 18 months. The same paper also reported the misidentification of the strain as a member of the M. tuberculosis complex by commercial probes (AccuProbe and INNO-LiPA Mycobacteria); such a finding has not been confirmed with other strains. One isolate from fish has also been reported (77).
M. kumamotonense, although described on the basis of a single strain, has proven to be very common. Within the M. terrae complex, it is, along with M. arupense, the species most frequently isolated, while M. terrae, with which it was confused before the description of the new species, is very rare (76).
Mycobacterium kyorinense
M. kyorinense was described in 2009, on the basis of three strains isolated from independent patients (96).
M. kyorinense occupies a position in the same phylogenetic branch as M. branderi, M. celatum, and M. fragae (Fig. 7). In various genetic regions, an unexpected identity of its sequence was reported with M. celatum type 2, one of the three subtypes of this species (97, 98). Subsequent studies demonstrated, however, that M. celatum type 2 and M. kyorinense are the same species (99). Like M. celatum, M. kyorinense is characterized by two copies of the 16S rRNA gene (100), which is an exceptional condition among the slow growers, as they typically harbor only one copy. Microheterogeneity is absent in M. kyorinense sequences.
The colonies, which are smooth and nonchromogenic, grow at 28 to 42°C in more than 4 weeks.
The species is susceptible in vitro to amikacin, clarithromycin, streptomycin, and quinolones but is resistant to rifampin.
The three strains of M. kyorinense characterized for the species nova description were responsible for pneumonia in nonimmunocompromised patients. A review of seven Japanese cases (100), in five patients with lung disease and the others with lymph nodal and joint infection, reports satisfactory outcomes for five of six patients treated with a combination of clarithromycin and quinolones; in some of them, this treatment had been preceded by a standard antituberculosis regimen which proved ineffective. Further cases, again in Japan (101–104), concerned two elderly patients who died of respiratory failure and a case of pulmonary infection successfully treated with clarithromycin and levofloxacin. In Brazil, M. kyorinense was isolated from a subject with extensive fibrotic lesions of the lung, who died despite antitubercular treatment (105). The cases imputed to M. celatum (type 2) before the recognition of M. kyorinense should be added to these.
“Mycobacterium lepromatosis”
“M. lepromatosis” was described in 2008 (106), on the basis of molecular investigations conducted on human tissues from patients with leprosy.
Although most closely related to Mycobacterium leprae, it is clearly distinct from it. The two species, in fact, present similarities of <98% in the 16S rRNA gene and close to 94% and 93% in rpoB and hsp65, respectively (Fig. 7). The phylogenetic relatedness between the two species is confirmed by the C+G content, which is lower than that in other mycobacteria, by a unique insertion (19 bp long in “M. lepromatosis” and 16 bp in M. leprae) in the 16S rRNA gene, and by genomic reduction and degradation.
Like M. leprae, it is uncultivable on artificial media.
“M. lepromatosis” is responsible for a form of diffuse lepromatous leprosy predominantly seen in Mexico (107) and causing substantial mortality when not treated. The disease is characterized by massive mycobacterial invasion and proliferation into the endothelium, with vascular occlusion. Following the recognition of the novel species, other cases have been reported in Singapore (108) and Canada (109).
“Mycobacterium liflandii”
“M. liflandii” was characterized in 2004, on the basis of multiple isolates (110), and the current name was introduced 1 year later (111). Recent data based on whole-genome sequencing suggest that “M. liflandii” may be an ecotype of M. ulcerans, the causative agent of Buruli ulcer (112).
The species belongs to the same phylogenetic branch as Mycobacterium marinum and Mycobacterium ulcerans (Fig. 7). M. marinum is considered the progenitor from which M. ulcerans and “M. liflandii” evolved, following the loss of multiple genomic regions (113).
The growth of this species is fastidious, and colonies, which are rough and unpigmented, take more than 1 month to become visible at 28°C. Like M. ulcerans, “M. liflandii” possesses one plasmid coding for a cytopathogenic toxin (mycolactone).
“M. liflandii” was responsible for cutaneous lesions with a high death rate in amphibians. Epidemics in colonies of laboratory frogs have been reported (114, 115), while there is no report of human infections.
Mycobacterium longobardum
M. longobardum was described in 2013, on the basis of seven independent strains isolated from clinical specimens in Italy (76).
The species harbors the 14-nucleotide insertion in helix 18 of the 16S rRNA gene that is typical of the species included in the M. terrae complex (Fig. 4). A moderate level of microheterogeneity is reported only for the rpoB gene.
Rough and unpigmented colonies develop on solid media in 7 to 14 days at 25 to 37°C. The mycolic acid profile by HPLC is indistinguishable from that of other species belonging to the M. terrae complex.
The species is susceptible in vitro to sulfamethoxazole and clarithromycin and is resistant to quinolones, linezolid, and streptomycin.
The seven strains of M. longobardum characterized for the description of the species were isolated from various types of clinical specimens, but none of the strains were clinically relevant. Very recently, a case of elbow osteomyelitis was reported in which M. longobardum was isolated from a biopsy specimen; the patient was cured with clarithromycin and ethambutol (116).
Mycobacterium mantenii
M. mantenii was described in 2009, based on four independent clinical strains plus one strain of environmental origin (117).
M. mantenii shares an intermediate-length helix 18 in the 16S rRNA gene (3-bp deletion in comparison to other slow growers) with M. scrofulaceum (Fig. 7). The clinical strains present homogeneous sequences in the 16S rRNA gene, ITS1, hsp65, and rpoB, while the environmental one bears minor differences in those sequences.
Colonies, which are smooth and yellow (scotochromogenic), grow on solid media in 4 weeks at 25 to 37°C. The HPLC mycolic acid pattern is not distinguishable from that of M. scrofulaceum and the MAC.
The species is susceptible in vitro to rifamycins and clarithromycin only.
M. mantenii is identified by the INNO-LiPA Mycobacteria test as a member of the MAIS group but different from M. avium, M. intracellulare, and M. scrofulaceum, while it is misidentified as M. intracellulare by the GenoType Mycobacterium CM test (18).
M. mantenii has been isolated from cervical lymph nodes of two otherwise healthy children: one was successfully treated with rifampin and clarithromycin for 3 months, and the other was cured without antibiotic therapy following lymph node drainage. Two other strains, both considered clinically nonsignificant, were isolated from respiratory specimens. The environmental strain was grown from a water sample of the Zambesi River. One isolation from a fish has also been reported (118).
Mycobacterium marseillense
M. marseillense was described in 2009, along with two other new species belonging to the MAC, based on two strains isolated from respiratory specimens from unrelated patients (82).
It is a member of the MAC (Fig. 5) and is closely related to other species of the complex. The 16S rRNA gene sequence does not allow an unequivocal identification of the species because of its identity with M. yongonense; in other housekeeping genes, some microheterogeneity is present.
The colonies are rough and unpigmented and grow on solid media in about 2 weeks at temperatures between 30 and 45°C.
M. marseillense is misidentified as M. intracellulare by the GenoType Mycobacterium CM test; its hybridization with other commercial DNA probes has not been determined.
No information is available about the patients from whose clinical specimens the two strains characterized on occasion of the species nova description were isolated. One case of pulmonary disease, characterized by bilateral bronchiectasis and multiple nodules, was recently reported. The patient remained intermittently culture positive for M. marseillense for over 5 years, and in this period, he underwent three therapeutic cycles, initially with antituberculosis drugs and finally with ethambutol, rifampin, and azithromycin. Despite the negativization of the cultures, the progression of pulmonary involvement was radiologically evident at follow-up (119). Another case of pulmonary disease with multiple isolations of M. marseillense has been reported for a patient with lupus erythematosus (120). Two strains have been isolated from respiratory specimens of cystic fibrosis patients (121).
Mycobacterium minnesotense
M. minnesotense was described in 2013, on the basis of 13 strains isolated from a sphagnum peat (122).
The species bears the signature of members of the M. terrae complex (Fig. 4) in helix 18 of the 16S rRNA gene; the most closely related species is M. arupense, with which it shares an identical hsp65 PRA pattern. Extensive sequence heterogeneity is present in various housekeeping genes, including the 16S rRNA gene.
Smooth, pink-orange, photochromogenic colonies develop on solid media in 7 to 10 days at 27 to 34°C but not at 37°C. The mycolic acid pattern determined by HPLC does not differ significantly from that of other species included in the M. terrae complex.
M. minnesotense has never been grown from human or animal specimens.
Mycobacterium noviomagense
M. noviomagense was described in 2009, on the basis of 17 strains isolated in the Netherlands (123).
The phylogenetic position of this very homogeneous species is unclear, because the most closely related species vary according to different genetic regions (Fig. 7).
Nonchromogenic colonies grow at 37°C but not at 45°C, in about 4 weeks. The HPLC mycolic acid pattern is not distinguishable from that of M. xenopi.
In vitro antimicrobial testing revealed susceptibility to streptomycin, amikacin, ciprofloxacin, and clarithromycin and resistance to rifampin.
All the strains investigated so far were isolated from respiratory specimens in a circumscribed geographic area of the Netherlands; none of them fulfilled the criteria for clinical significance.
Mycobacterium paraffinicum
M. paraffinicum was first described in 1956 (124), but subsequently, its species status was considered illegitimate. The species was definitively revived in 2009 (125), on the basis of six strains.
M. paraffinicum is most closely related to M. scrofulaceum (Fig. 7), with which it shares a helix 18 in the 16S rRNA gene which is three nucleotides shorter than that of other slow growers.
The colonies are yellow and waxy, may be smooth or wrinkled, and grow in more than a week at 22 to 35°C. HPLC of mycolic acids reveals a pattern that is not distinguishable from those of M. scrofulaceum and MAC members.
The strains are susceptible in vitro to rifabutin, linezolid, clarithromycin, and amikacin.
The INNO-LiPA Mycobacteria test misidentifies the species as M. intracellulare type 2 (corresponding to Mycobacterium chimaera), while it is misassigned to the MAC by the AccuProbe test.
The original strain was isolated from samples of oil-field soil. Other strains were grown from respiratory specimens, but no clinical information is available regarding the patients. One pseudo-outbreak involving 21 patients was reported in a hospital where the ice machine was then detected as the source (126). Very recently, a case of symptomatic pulmonary infection was described (127).
Mycobacterium paragordonae
M. paragordonae was described in 2014, on the basis of a single strain (128).
The species is related to M. gordonae (Fig. 7), with which it shares 99% similarity in the 16S rRNA gene.
The colonies, which are orange (scotochromogenic) and smooth, grow at 25 to 30°C but not at 37°C, in more than a week.
The strain was isolated from the sputum of a patient with an unspecified symptomatic pulmonary infection.
Mycobacterium parakoreense
M. parakoreense was described in 2013, based on a single strain (129).
M. parakoreense harbors the signature of rapid growers in helix 18 of the 16S rRNA gene and occupies a phylogenetic position close to that of M. triviale (Fig. 3).
The colonies are rough, are yellow or unpigmented in both dark and light, and grow at 37°C in four or more weeks. The HPLC profile of mycolic acids is hardly distinguishable from that of M. triviale.
The strain is susceptible in vitro to amikacin, clarithromycin, and rifampin.
The only strain isolated so far was grown from the sputum of a patient with an unspecified symptomatic pulmonary infection.
Mycobacterium paraseoulense
M. paraseoulense was described in 2010, based on a single strain (130).
Like Mycobacterium seoulense, with which M. paraseoulense shares an identical 16S rRNA gene sequence, the species occupies a phylogenetic position close to that of M. scrofulaceum (Fig. 7), from which it differs, however, by a two-nucleotide insertion in helix 18.
Orange scotochromogenic colonies, which are smooth and occasionally rough, grow in three or more weeks at 25 to 37°C. The profile of mycolic acids is characterized by two major peak clusters, similar to the HPLC profile of M. gordonae chromatotype 2 (131).
The GenoType Mycobacterium CM test misidentifies M. paraseoulense as M. scrofulaceum (unpublished data).
The sputum of a patient with an unspecified symptomatic pulmonary infection was the source of the only strain isolated so far.
“Mycobacterium paraterrae”
“M. paraterrae” was described in 2010, based on a single strain (132).
The phylogenetic position of “M. paraterrae” is unclear: in the 16S rRNA gene, it shows the genetic signature of M. terrae complex members (Fig. 4), but it is located on distant branches in the trees built from the sequences of other housekeeping genes.
Mature colonies develop in 4 weeks at 25 to 37°C on solid media; they are orange, scotochromogenic, and prevalently smooth. The HPLC pattern of mycolic acids is clearly different from the peak arrangement typical of the species of the M. terrae complex.
The only strain isolated was obtained from the sputum of a patient with an unspecified symptomatic pulmonary infection.
Mycobacterium riyadhense
M. riyadhense was described in 2009, on the basis of a single clinical strain (133).
M. riyadhense is closely related to Mycobacterium szulgai (Fig. 7) in the 16S rRNA gene and ITS1, while it is distant from every other known species when additional genetic regions are considered. Like M. tuberculosis, it possesses the region of difference 1.
The colonies, which are rough and unpigmented, develop on solid media in more than 3 weeks at temperatures ranging from 25 to 36°C. The HPLC profile of mycolic acids grossly resembles that of M. tuberculosis.
The strain is susceptible in vitro to ciprofloxacin, clarithromycin, ethambutol, rifampin, and streptomycin.
M. riyadhense is misidentified as a member of the M. tuberculosis complex by the GenoType Mycobacterium CM test.
The strain on which the description of the new species is based was responsible for posttraumatic infection at the level of the left maxillary sinus. The patient improved clinically and radiologically following a 9-month course of antituberculosis treatment. Recently, two cases of cavitary pulmonary disease were reported (134): the first patient was cured with a 10-month antituberculosis regimen; the second, despite a 1-year treatment with a combination of clarithromycin and ciprofloxacin, had a relapse which was resolved by the addition of antituberculosis drugs to the treatment. A further case of tuberculosis-like pneumonia has been reported in Korea (135) and was cured with a standard antituberculosis regimen. The large number of case reports, probably underestimated due to the misidentification as M. tuberculosis, clearly supports the pathogenic role of M. riyadhense.
Mycobacterium senuense
M. senuense was described in 2008, on the basis of a single strain (136).
The species is located within the M. terrae complex (Fig. 4) and is more closely related in the 16S rRNA gene to Mycobacterium arupense.
Tiny, nonchromogenic colonies develop in 4 weeks at both 25 and 37°C. The HPLC profile of mycolic acids is not different from that of other members of the M. terrae complex.
The only known strain was isolated from the sputum of a patient with an unspecified symptomatic pulmonary infection.
Mycobacterium seoulense
M. seoulense was described in 2007, on the basis of a single strain (137).
Like M. paraseoulense, with which M. seoulense shares an identical 16S rRNA gene sequence, the species occupies a phylogenetic position close to that of M. scrofulaceum (Fig. 7), from which it differs, however, by a two-nucleotide insertion in helix 18.
Smooth, scotochromogenic, orange colonies grow on solid media in three or more weeks at temperatures ranging from 25 to 37°C. The HPLC mycolic acid pattern grossly resembles that of MAC species.
The only strain isolated so far was grown repeatedly from the sputum of a patient with an unspecified symptomatic pulmonary infection.
Mycobacterium sherrisii
Although a number of clinical cases involving M. sherrisii have been reported since 2004, this new species was described in 2011 (138), on the basis of 11 strains collected in different countries.
M. sherrisii is characterized in the 16S rRNA gene by a short helix 18 bearing the specific sequence unique to the members of the M. simiae complex (Fig. 6). Moderate levels of microheterogeneity are present in the hsp65 and rpoB genes.
Mature colonies develop after 2 to 3 weeks of incubation on solid media at 25 to 37°C; different strains display either nonchromogenic or photochromogenic (yellow) colonies. HPLC of the cell wall mycolic acids presents a profile with three late peak clusters also shared by other species belonging to the M. simiae complex.
The strains are resistant to the majority of antimycobacterial drugs recommended against slowly growing mycobacteria, with the exception of clarithromycin and rifabutin.
The GenoType Mycobacterium AS and INNO-LiPA Mycobacteria commercial identification methods misidentify M. sherrisii as M. simiae.
A large number of infections due to M. sherrisii have been reported in the literature, most of them concerning patients from various African countries (139–145). The proportion of HIV-positive subjects (139–147) is also large. The clinical pictures differ in that the species is prevalently disseminated, or both pulmonary and disseminated, in HIV-infected patients, whereas it is mainly pulmonary in other patients. The majority of patients were treated with a combination of one macrolide, one rifamycin, and ethambutol, which in most cases proved to be effective.
“Mycobacterium shigaense”
“M. shigaense” was described in 2012, on the basis of a single clinical strain (148).
From a phylogenetic point of view, the species is included in the M. simiae complex (Fig. 6) due to the presence of the pathognomonic signature consisting of a specific sequence within the short helix 18 of the 16S rRNA gene.
Scotochromogenic yellow colonies grow in culture at 25 to 37°C in around 2 weeks. HPLC of cell wall mycolic acids reveals a profile with the three late peak clusters presented by most species of the M. simiae complex.
The strain was susceptible in vitro to amikacin, clarithromycin, moxifloxacin, and rifampin.
The GenoType Mycobacterium AS test produces a pattern (hybridization bands 6, 8, and 16) not included with those provided by the system (unpublished data).
“M. shigaense” was isolated from a skin biopsy specimen from a patient treated for Hodgkin's disease and presenting disseminated cutaneous nodules. The patient died of a non-Hodgkin's T-cell lymphoma with severe immunodeficiency. A new case of cutaneous infection was subsequently reported for an immunocompetent woman (149). The patient was cured following a 6-month course of treatment with rifampin, moxifloxacin, and clarithromycin.
Mycobacterium shinjukuense
M. shinjukuense was described in 2011, on the basis of seven independent strains (150).
The species is very homogeneous, but its phylogenetic position is unclear, since the relatedness with the M. tuberculosis complex displayed in the 16S rRNA gene (Fig. 7) is not confirmed in other genetic targets.
The colonies are unpigmented and develop in around 3 weeks at 30 to 37°C. The mycolic acid HPLC pattern is characterized by a large cluster of late-eluting peaks grossly resembling those of M. malmoense. M. shinjukuense produces false-positive results with the Amplified Mycobacterium Tuberculosis Direct commercial amplification system (Gen-Probe), which is specific for M. tuberculosis. With the GenoType Mycobacterium CM and AS tests, it is misidentified as Mycobacterium kansasii (AS pattern 10-12).
The strains on which the description of the new species is based and, more recently, two others (151, 152) were isolated in Japan from respiratory specimens from independent patients. Three of the described cases fulfilled the ATS diagnostic criteria for clinical significance; one of the patients was cured with antituberculosis treatment.
“Mycobacterium simulans”
Only one report exists about “M. simulans” (153), and the species has not been officially described.
The strain presents the closest similarity to M. riyadhense (Fig. 7), and likewise, it is misidentified as a member of the M. tuberculosis complex by the GenoType Mycobacterium CM test.
Rough, nonpigmented colonies grow at 37°C in two or more weeks. By HPLC, the cell wall mycolic acids give a pattern characterized by a single, late-emerging peak cluster grossly resembling that specific for M. tuberculosis.
The strain was susceptible in vitro to amikacin, clarithromycin, linezolid, rifabutin, and quinolones.
The only strain of “M. simulans” isolated so far was responsible for severe cavitary pulmonary disease, which was successfully treated with the combination of amikacin, clarithromycin, moxifloxacin, and ethionamide.
“Mycobacterium sinense”
“M. sinense” was described in 2013, on the basis of a single strain (154).
The strain carries the 14-nucleotide insertion in helix 18 of the 16S rRNA gene that is characteristic of the M. terrae complex (Fig. 4).
The colonies, which are unpigmented and smooth, develop in three or more weeks at 28 to 37°C. The paper describing “M. sinense” emphasizes its multidrug resistance; it was actually only tested for drugs specific to M. tuberculosis and showed resistance in vitro to amikacin, ethambutol, rifampin, quinolones, and streptomycin.
The strain was isolated repeatedly from the sputum of a patient with severe lung infiltrates; the infection was resolved after prolonged treatment with second-line antituberculosis drugs.
Mycobacterium stomatepiae
M. stomatepiae was described in 2007, on the basis of three strains isolated from fish (155).
The strains carry the M. simiae complex signature in the 16S rRNA gene, i.e., a short helix 18 presenting the complex-specific sequence (Fig. 6).
Smooth and nonchromogenic colonies grow in 3 weeks at 30°C and more slowly at 25°C; no growth is obtained at 37°C. The HPLC pattern is characterized by three peak clusters shared by various species belonging to the M. simiae complex.
No isolation from human specimens has been reported so far.
Mycobacterium timonense
M. timonense was described in 2009, along with two other new species belonging to the MAC, based on two strains isolated from respiratory specimens from unrelated patients (82).
M. timonense clusters with other species of the MAC (Fig. 5) and displays genetic microheterogeneity.
The colonies are rough and unpigmented and grow on solid media in about 2 weeks at temperatures between 30 and 45°C.
The hybridization pattern with commercial DNA probes is not known.
No clinical information is available on the patients from whose specimens the two isolates characterized for the species nova description were isolated. Very recently, two isolations were reported in Latin America: one, in Brazil, from a pulmonary specimen from a cystic fibrosis patient (121), and the other, in Ecuador, from an HIV patient (156).
Mycobacterium vulneris
M. vulneris was described in 2009, based on two independent clinical strains (157).
The species is a member of the MAC (Fig. 5).
Smooth, bright yellow, scotochromogenic colonies develop in more than 3 weeks on solid media. The HPLC pattern of mycolic acids is characterized by a profile with three peak clusters, consistent with that of the other strains included in the MAC.
Among the drugs considered potentially active against slowly growing mycobacteria, only rifamycins and clarithromycin are active in vitro.
The INNO-LiPA Mycobacteria test identifies M. vulneris as a member of the MAIS group that is different from M. avium, M. intracellulare, and M. scrofulaceum. The GenoType Mycobacterium CM test misidentifies the strain as M. intracellulare, while the AccuProbe test correctly includes the species in the MAC.
Of the two strains on which the description of the species is based, the first was isolated from a suppurative wound consequent to a dog bite and treated surgically with repeated debridement; the second was responsible for cervical lymphadenitis in a child, which was successfully excised.
Mycobacterium yongonense
M. yongonense was described in 2013, on the basis of a single clinical strain (158).
M. yongonense is characterized by a 16S rRNA gene sequence that is 100% identical to that of M. marseillense. Both species are members of the MAC (Fig. 5). Interestingly, the rpoB gene sequence is closely related to that of M. parascrofulaceum and is distant from those of the MAC species. To explain such a finding, a lateral gene transfer event has been hypothesized (159). In contrast, a clearly different rpoB sequence that is fully compatible with belonging to the MAC was detected in two strains isolated in Italy. They were identified as M. yongonense on the basis of the consistency of the sequences of seven housekeeping genes (160).
The colonies are mostly nonchromogenic, although some yellow pigmentation may be observed; they develop at 37°C in more than a week. The strain is susceptible in vitro to the majority of the antimicrobials usually tested against slow growers, with the exception of amikacin.
M. yongonense is misidentified as M. intracellulare by the GenoType Mycobacterium CM test.
The strain on which the species nova description is based was isolated from a patient with a pulmonary infection, for which, unfortunately, no clinical information is available. Very recently, two further strains were isolated from respiratory specimens (160). The isolation was considered nonsignificant in one of the patients, while in the other it was responsible for a cavitary disease that improved with a treatment combination of clarithromycin, rifampin, and ethambutol.
NEW MEMBERS OF THE M. TUBERCULOSIS COMPLEX
“Mycobacterium mungi”
“M. mungi” was described in 2010, when it was detected as the cause of an epidemic in mongooses (161).
“M. mungi” is characterized by the identity of more than 99% of its genome with that of M. tuberculosis; in both species, the insertion element IS6110 is present. The identification of the species is possible by spoligotyping of the direct repeat (DR) locus, which is characterized by a unique pattern, with the presence of spacers 1, 2, 4 to 6, 8, 10, 11, 37, 38, and 40 to 43.
Scanty growth of colonies is obtained in 5 to 6 weeks on solid media at 37°C (162).
“M. mungi” is responsible for granulomatous lesions at the level of the nose in mongooses; a few cases of disseminated disease have also been reported. No information is available about its potential pathogenicity in other animals or humans. The organism is probably present in the environment and enters the host through nasal erosions; systemic dissemination is possible via hematogenous and lymphatic spread, and no evidence of aerosol transmission exists.
“Mycobacterium orygis”
“M. orygis” was described as a species or subspecies within the M. tuberculosis complex in 2012, based on 22 isolates selected for the similarity of their IS6110 restriction fragment length polymorphism (RFLP) patterns (163–165).
“M. orygis” is defined by the presence of genomic regions of difference RD1, RD2, RD4, RD5a, RD6, RD13 to RD16, RD701, and RD702; by a C-to-G single nucleotide polymorphism (SNP) at position 551 in mmpL6; and by the deletion of regions RD3, RD5b, RD7 to RD12, RDoryx_1, RDoryx_4, and RDoryx_wag22. The large number of IS6110 elements (from 16 to 22) produces a unique RFLP pattern; the pattern of mycobacterial interspersed repetitive-unit–variable-number tandem repeats (MIRU-VNTR) is equally distinctive. Specific spoligotypes of “M. orygis” do exist and are included in the “Africanum” clade.
The colonies of “M. orygis” are smooth to greasy, domed, and nonchromogenic.
The species is susceptible in vitro to first-line antituberculosis drugs, including pyrazinamide.
The strains are misidentified as M. africanum by the GenoType Mycobacterium MTBC commercial system. The range of hosts includes antelopes in eastern Africa and the Arabian Peninsula, cows and rhesus monkeys in South Asia, and humans (11 of the strains on which the species nova description is based had a human origin). The disease produced in humans is indistinguishable from tuberculosis; also, a case of transmission from a man to a cow has been reported (166).
“Mycobacterium suricattae”
“M. suricattae” was described in 2013, based on three strains isolated from free-living meerkats (167).
From the phylogenetic point of view, the species shares a common ancestor with M. africanum. It is defined by the presence of genomic regions of difference RD1, RD4, and RD12 and by the deletion of RD9. The 16S rRNA gene of “M. suricattae” differs by a SNP from those of all other M. tuberculosis complex members. Twenty-one copies of IS6110 are present in the genome. Spoligotyping does not produce any interpretable pattern because of a wide deletion of the DR locus.
“M. suricattae” has been isolated only from meerkats, in which it is responsible for granulomatous lesions (162).
DISCUSSION OF NEW SPECIES
NTM are found ubiquitously in the environment, where they can be isolated from water and soil; occasionally, they infect humans and animals through aerosols (168) and by contact. In humans, they can be responsible mainly for respiratory diseases but can also have lymph nodal, cutaneous, bone, and joint involvement (169). Disseminated diseases are also possible, especially in immunocompromised patients. The person-to-person transmission of NTM is traditionally excluded, and outbreaks have been reported only for people sharing contact with the same reservoir (170). However, recently, several clusters of cases of respiratory disease due to M. abscessus subsp. massiliense in cystic fibrosis patients suggested the possibility of indirect interhuman transmission (38, 171).
Clinical Significance of NTM
While the isolation of NTM from sterile body sites (tissues or blood) is diagnostic of disease, their isolation from respiratory specimens may also be due to specimen contamination or transient colonization. The diagnostic criteria recommended by the ATS (81) are universally considered the litmus test for clinical significance. Such criteria apply to symptomatic patients with nodular or cavitary disease and with evidence of bronchiectasis and/or multiple small nodules. Nontuberculous pulmonary disease can be diagnosed, following exclusion of other diseases, by the presence of at least two positive sputum cultures or one positive bronchial wash culture.
Geographic Distribution of NTM
Although most NTM species are widespread worldwide (172), for particular species peculiar geographic distributions have been reported (173). It has long been believed that the incidence of NTM in different geographic areas has an inverse relationship with the local burden of tuberculosis. In the last few years, however, the implementation of liquid cultures and the use of molecular probes for NTM differentiation in many countries with a high prevalence of tuberculosis (174) revealed unexpectedly high rates of such organisms in these settings as well.
Description of New Species
In this paper, both validly published and nonofficial species are reviewed. The “validly published” term may lead readers who are not well acquainted with taxonomy to believe that the fulfilment of strict standards is needed to achieve the official recognition of a species. Actually, only recommendations exist, to which a number of consolidated customs are added. Typically, a species nova description consists of the characterization of a combination of phenotypic and genotypic traits followed by a final “description” including the philology of the name of the species and the summary of distinctive features. In the case of mycobacteria, at the phenotypic level, a minimal characterization includes the investigation of biochemical and cultural features and the chemical analysis of cell wall lipids. The genotypic study includes the sequencing of several (usually at least three) housekeeping genes, among which the almost complete 16S rRNA gene is mandatory; their comparison, via alignment, with the sequences of the type strains of the species presenting the highest similarities; and a phylogenetic analysis, preferably based on the concatenation of the determined sequences.
Medical information, in the case of isolation from clinical specimens, and in vitro antimicrobial susceptibility data are optional. No instruction exists concerning the minimal number of strains to be characterized for the description of a new species. It is my firm conviction that some change is needed. Unquestionably, cultural tests remain crucial for the determination of basic characteristics, such as the growth rate, temperature, and pigmentation of colonies. Biochemical tests, in contrast, represent only a historical heritage and are obsolete. Lipid analyses, carried out by multiple approaches, including MALDI-TOF mass spectrometry (94), are sometimes useful. MALDI-TOF analysis of ribosomal proteins should be encouraged as well, in consideration of the increasing diffusion of such approaches among clinical laboratories. Any effort should be made to collect clinical information in cases of strains isolated from humans or animals. Antimicrobial susceptibility testing by determination of MICs of relevant drugs (for slow or rapid growers, and not including unsuited molecules) (27) should be included at least for clinical isolates. The value of DNA-DNA hybridization (DDH) is controversial, although it is still the gold standard for establishing whether or not two closely related strains belong to the same species (175). A robust measure of genomic relatedness, very recently acknowledged by the Committee of Systematic Bacteriology, is now available: average nucleotide identity (ANI). Two strains definitely belong to different species when the ANI between their genomes is lower than 95 to 96% (180). Whole-genome sequencing, a prerequisite for ANI calculation, is becoming widely accessible and handy; DDH, in contrast, still is cumbersome and poorly reproducible.
Does every previously unknown strain represent a new species? This question is very difficult to answer. About half of the species reviewed here were described on the basis of a single strain; statistically, the majority of them have a very low probability of being encountered again. It is the conviction of many taxonomists, including me, that the description of every new bacterial species should be based on “multiple, independent” strains. The meaning of “multiple” is clear: as many as possible and, in any case, at least two. The definition of “independent” is more complex; it seems honest, even if not absolutely true, to consider independent those strains isolated from living organisms or environments without known relatedness.
It is nevertheless clear that reports of single unusual isolates, not aiming for the description of new species, remain welcome. Such publications, as well as the relevant sequences deposited in public databases, may represent opportunities for researchers to become aware that strains with features compatible with those observed by them have been found also elsewhere, thus stimulating collaborative descriptions of “real” new species.
Definitive Identification
Currently, NTM identification relies mostly on the use of commercial DNA probe systems, which give excellent identification results only for a limited number of species. Strains either identified only at the genus Mycobacterium level or incorrectly identified because of cross-reactions of particular probes (Table 2) should be identified by genetic sequencing. The 16S rRNA gene, with few exceptions (Table 3), allows unambiguous identification of the large majority of species and is characterized, in contrast with other genetic targets, by very limited microheterogeneity (Table 4). The sequence of the first 500 bp is generally sufficient; in fact, hypervariable regions A and B, harboring nucleotide polymorphisms suitable for differentiating the large majority of species, are actually included in this region (7).
TABLE 2.
Hybridization results obtained with commercial DNA probes
| Species | Test resulta |
||
|---|---|---|---|
| AccuProbe | INNO-LiPA | GenoType | |
| “M. alsiense” | Not identifiedb | Mycobacterium genusb | SPsP |
| M. arosiense | M. intracellulare | MAIS | M. intracellulare |
| M. bouchedurhonense | NT | NT | M. intracellulare |
| M. llatzerense | NT | Mycobacterium genusb | M. mucogenicum |
| M. mantenii | Not identifiedb | MAIS | M. intracellulare |
| M. paraffinicum | MAC | M. scrofulaceum | SPsPb |
| M. paraseoulense | NT | NT | M. scrofulaceum |
| M. riyadhense | NT | Mycobacterium genusb | M. tuberculosis complex |
| M. sherrisii | NT | M. simiae | M. simiae |
| “M. shigaense” | NT | NT | Bands 6, 8, and 16c |
| M. shinjukuense | NT | NT | M. kansasii |
| “M. simulans” | Not identifiedb | Mycobacterium genusb | M. tuberculosis complex |
| M. vulneris | MACb | MAC− | MAC− |
| M. yongonense | NT | NT | M. intracellulare |
MAIS, belonging to the M. avium-intracellulare-scrofulaceum group but different from M. avium, M. intracellulare, M. chimaera, and M. scrofulaceum; MAC, belonging to the MAC but different from M. avium and M. intracellulare; MAC−, not belonging to the MAC; NT, not tested; SPsP, positive with a probe aiming for M. scrofulaceum, M. parascrofulaceum, and M. paraffinicum.
Correct identification.
The GenoType AS test produces a pattern including bands 6, 8, and 16, which is not interpreted by the system.
TABLE 3.
Species presenting identical sequences in the first 500 bp of the 16S rRNA gene
| Species |
|---|
| M. tuberculosis complex, except for “M. suricattae” |
| M. abscessus, M. chelonae, “M. franklinii” |
| M. alvei, M. setensea |
| “M. barrassiae,” M. moriokaensea |
| M. conceptionense, M. farcinogenes, M. fortuitum, M. houstonense, M. senegalenseb |
| M. gastri, M. kansasii |
| M. marinum, M. ulceransa |
| M. mucogenicum, M. phocaicum |
| M. marseillense, M. yongonense |
| M. murale, M. tokaiense |
| M. paraseoulense, M. seoulense |
| M. peregrinum, M. septicuma |
| M. vaccae, M. vanbaaleniia |
The species can be differentiated by sequencing the whole 16S rRNA gene.
In this group, some species can be differentiated by sequencing the whole gene; M. farcinogenes can be differentiated from the other species because it grows slowly.
TABLE 4.
Genetic microheterogeneity of Mycobacterium speciesa
| Species | No. of sequevars |
No. of strains investigated | |||
|---|---|---|---|---|---|
| 16S rRNA gene | hsp65 | rpoB | ITS1 | ||
| “M. alsiense” | 1 | ND | 1 | 1 | 4 |
| M. arosiense | 1 | 1 | 1 | 1 | 8 |
| M. bacteremicum | 1 | 3 | 7 | ND | 10 |
| M. bourgelatii | 1 | 2 | 3 | ND | 3 |
| M. crocinum | 2 | 1 | 2 | ND | 3 |
| M. celeriflavum | 3 | 3 | 3 | ND | 6 |
| M. engbaekii | 1 | 4 | 4 | ND | 7 |
| M. europaeum | 1 | 2 | 3 | 3 | 4 |
| “M. franklinii” | 1 | 2 | 2 | 2 | 26 |
| M. heraklionense | 1 | 3 | 7 | ND | 23 |
| M. insubricum | 2 | 2 | 1 | 3 | 5 |
| M. iranicum | 2 | 5 | 4 | 3 | 8 |
| M. koreense | 2 | 3 | 1 | ND | 3 |
| M. kumamotonense | 4 | 2 | 8 | ND | 36 |
| M. kyorinense | 1 | 1 | 1 | ND | 3 |
| M. llatzerense | 1 | 1 | 1 | 1 | 6 |
| M. longobardum | 1 | 1 | 2 | ND | 7 |
| M. mantenii | 1 | 2 | 2 | 2 | 5 |
| M. marseillense | 1 | 2 | 2 | 2 | 2 |
| M. minnesotense | 11 | 5 | ND | ND | 13 |
| M. monacense | 1 | 2 | ND | 7 | 4 |
| “M. mungi” | 1 | 1 | 1 | 1 | 8 |
| M. noviomagense | 1 | 1 | 1 | 2 | 18 |
| “M. orygis” | 1 | 1 | 1 | 1 | 22 |
| M. paraffinicum | 1 | 1 | 1 | ND | 6 |
| M. rutilum | 3 | 2 | 2 | ND | 3 |
| M. salmoniphilum | 8 | 6 | 9 | ND | 9 |
| M. sherrisii | 1 | 2 | 1 | 2 | 11 |
| M. shinjukuense | 7 | 1 | 1 | 1 | 7 |
| M. stomatepiae | 1 | 1 | 1 | 1 | 3 |
| M. timonense | 1 | 2 | 1 | 2 | 2 |
| M. vulneris | 1 | 1 | 1 | 1 | 2 |
| M. yongonense | 1 | 1 | 2 | 1 | 3 |
ND, not investigated.
A good-quality sequence is only the starting point toward a careful identification: major hindrances are represented by the high level of similarity characterizing the genus Mycobacterium (3) and by the suboptimal quality of available databases. The skill of the microbiologist plays a pivotal role in deciding whether similar sequences represent sequevars (176) of the same species or distinctive markers of closely related species. Furthermore, the choice of the definitive identification among the entries present in the databases is a real challenge. Strictly controlled databases, both commercial and open access, do exist, but they cover an insufficient number of Mycobacterium species, and this problem may lead to misidentification of strains of novel species not included in the database. This limitation is not present in GenBank and its equivalents (European Molecular Biology Laboratory and DNA Data Bank of Japan), but these are poorly controlled and include large numbers of mislabeled sequences (177), in addition to a huge quantity of duplicates. The latter database, however, remains the best choice because experienced users can select the proper sequence among many incorrect ones in a thorough database but may not be so successful with a database that, although accurate, lacks the appropriate sequence.
A possible identification algorithm combining the use of commercial DNA probes and genetic sequencing is reported in Fig. 8. In this scheme, for identifications emerging from hybridization with DNA probes known to present cross-reactions, confirmation by genetic sequencing is recommended. The most appropriate target is recommended according to different groups of species.
FIG 8.
Identification algorithm combining the use of commercial DNA probes (rounded rectangles and squares) and genetic sequencing (ellipses). For hybridization patterns known to be subject to cross-reactions (rounded rectangles and squares) with different commercial DNA probes (AccuProbe, blue-gray; INNO-LiPA Mycobacteria, gray; GenoType Mycobacterium, light blue), the appropriate sequencing targets are suggested. The hybridization patterns are indicated by the positive probes (probe names are according to the package inserts of different kits).
Epidemiologic Remarks
For the species reviewed here, a clear distinction must be made between the members of the M. tuberculosis complex, and M. lepromatosis as well, and the NTM. “M. mungi,” “M. orygis,” and “M. suricattae,” although isolated from animals, have the potential, like other members of the M. tuberculosis complex that are adapted to nonhuman mammals, of causing tuberculosis in humans, and in the case of “M. orygis,” the organism does indeed cause this disease. In addition, M. lepromatosis is responsible for the most severe forms of leprosy.
The NTM can be classified into three major epidemiologic groups (Table 1). Eight species are merely environmental, and it is very unlikely that they will ever be isolated from humans. Five have been isolated so far only from animals (fish and ruminants); however, the occupational risk for workers exposed to such animals cannot be excluded. All of the species remaining have been isolated, prevalently or exclusively, from patients. It is now accepted that different NTM species differ in their pathogenic power (3, 178). Estimates of the pathogenic potentials of the species that are the object of this review is possible only for those for which multiple isolations have been reported. M. engbaekii, M. insubricum, and M. noviomagense have not yet been proven to be pathogenic. The seven new species of the MAC appear to be characterized by the ability to cause disease comparable to that with M. avium or M. intracellulare. Among the four new species of the M. simiae complex, M. sherrisii is often responsible for disease in immunocompromised patients. M. iranicum has frequently been associated with pulmonary disease. M. mantenii, M. monacense, and M. shinjukuense are potential pathogens that are prevalently involved in pulmonary infections. M. bacteremicum, like M. neoaurum, has frequently been detected in catheter-related sepsis. The pathogenic potential of M. kyorinense scores higher than the average for NTM. M. llatzerense is an oligotrophic species fit for survival in waters.
Despite being present in large numbers, the species of the Mycobacterium genus are destined to increase further in the coming years; in fact, isolates not fitting any known species are frequently encountered in reference laboratories.
Biography

Enrico Tortoli, Sc.D., S.M., graduated with a degree in Biology from the University of Florence, with a specialty in Microbiology. He served as a Microbiologist and subsequently was responsible for the Regional Reference Center for Diagnostics of Mycobacteria at the Careggi University Hospital of Florence, Italy, until 2010, when he retired and started collaborating, as Senior Consulting Scientist, with the Emerging Bacterial Pathogens Unit at the San Raffaele Scientific Institute of Milan, Italy. His activity in the field of mycobacteriology dates back to the 1970s and is substantiated by 165 peer-reviewed articles, 14 book chapters, and two editions of a mycobacteriology manual (in Italian). Between 1995 and 2013, he collaborated on the description of 25 new species of mycobacteria. In 2007, at the ASM Congress of Toronto, he received the G. Middlebrook Award for significant contributions in mycobacteriology. He served as President of the European Society of Mycobacteriology for two mandates (2005 to 2010).
REFERENCES
- 1.Wolinsky E. 1979. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 119:107–159 [DOI] [PubMed] [Google Scholar]
- 2.Wayne LG, Sramek HA. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin. Microbiol. Rev. 5:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tortoli E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319–354. 10.1128/CMR.16.2.319-354.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tortoli E. 2006. The new mycobacteria: an update. FEMS Immunol. Med. Microbiol. 48:159–178. 10.1111/j.1574-695X.2006.00123.x [DOI] [PubMed] [Google Scholar]
- 5.Brown-Elliott BA, Wallace RJ., Jr 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716–746. 10.1128/CMR.15.4.716-746.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent PT, Kubica GP. 1985. Public health mycobacteriology. A guide for the level III laboratory. U.S. Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 7.Springer B, Stockman L, Teschner K, Roberts GD, Böttger EC. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan PJ, Heifets M, Ullom BP. 1982. Thin-layer chromatography of lipid antigens as a means of identifying nontuberculous mycobacteria. J. Clin. Microbiol. 15:447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minnikin DE, Minnikin SM, Parlett JM, Goodfellow M, Magnusson M. 1984. Mycolic acid patterns of some species of Mycobacterium. Arch. Microbiol. 139:225–231 [DOI] [PubMed] [Google Scholar]
- 10.Butler WR, Guthertz LS. 2001. Mycolic acid analysis by high-performance liquid chromatography for identification of Mycobacterium species. Clin. Microbiol. Rev. 14:704–726. 10.1128/CMR.14.4.704-726.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smid I, Salfinger M. 1994. Mycobacterial identification by computer-aided gas-liquid chromatography. Diagn. Microbiol. Infect. Dis. 19:81–88. 10.1016/0732-8893(94)90117-1 [DOI] [PubMed] [Google Scholar]
- 12.Kellogg JA, Bankert DA, Withers GS, Sweimler W, Kiehn TE, Pfyffer GE. 2001. Application of the Sherlock Mycobacteria identification system using high-performance liquid chromatography in a clinical laboratory. J. Clin. Microbiol. 39:964–970. 10.1128/JCM.39.3.964-970.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Khéchine A, Couderc C, Flaudrops C, Raoult D, Drancourt M. 2011. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One 6:e24720. 10.1371/journal.pone.0024720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebrun L, Espinasse F, Poveda JD, Vincent-Lévy-Frébault V. 1992. Evaluation of nonradioactive DNA probes for identification of mycobacteria. J. Clin. Microbiol. 30:2476–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tortoli E, Mariottini A, Mazzarelli G. 2003. Evaluation of INNO-LiPA Mycobacteria v2: improved reverse hybridization multiple DNA probe assay for mycobacterial identification. J. Clin. Microbiol. 41:4418–4420. 10.1128/JCM.41.9.4418-4420.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter E, Rusch-Gerdes S, Hillemann D. 2006. Evaluation of the GenoType Mycobacterium assay for identification of mycobacterial species from cultures. J. Clin. Microbiol. 44:1769–1775. 10.1128/JCM.44.5.1769-1775.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tortoli E, Pecorari M, Fabio G, Messinò M, Fabio A. 2010. Commercial DNA-probes for mycobacteria incorrectly identify a number of less frequently encountered species. J. Clin. Microbiol. 48:307–310. 10.1128/JCM.01536-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogall T, Flohr T, Böttger EC. 1990. Differentiation of mycobacterial species by direct sequencing of amplified DNA. J. Gen. Microbiol. 136:1915–1920. 10.1099/00221287-136-9-1915 [DOI] [PubMed] [Google Scholar]
- 20.McNabb A, Eisler D, Adie K, Amos M, Rodrigues M, Stephens G, Black WA, Isaac-Renton J. 2004. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 42:3000–3011. 10.1128/JCM.42.7.3000-3011.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adékambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699–5708. 10.1128/JCM.41.12.5699-5708.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim BJ, Lee SH, Lyu MA, Kim SJ, Bai GH, Kim SJ, Chae GT, Kim EC, Cha CY, Kook YH. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene rpoB. J. Clin. Microbiol. 37:1714–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zolg JW, Philippi-Schulz S. 1994. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J. Clin. Microbiol. 32:2801–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai J, Chen Y, Dean S, Morris JG, Salfinger M, Johnson JA. 2011. Multiple-genome comparison reveals new loci for Mycobacterium species identification. J. Clin. Microbiol. 49:144–153. 10.1128/JCM.00957-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth A, Fisher M, Hamid ME, Michalke S, Ludwig W, Mauch H. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown-Elliott BA, Nash KA, Wallace RJ., Jr 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin. Microbiol. Rev. 25:545–582. 10.1128/CMR.05030-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2011. Susceptibility testing of mycobacteria, nocardiae and other aerobic actinomycetes; approved standard, 2nd ed. M24-A2 CLSI, Wayne, PA: [PubMed] [Google Scholar]
- 28.Moore M, Frerichs JB. 1953. An unusual acid-fast infection of the knee with subcutaneous, abscess-like lesion of the gluteal region. J. Invest. Dermatol. 20:133–169 [DOI] [PubMed] [Google Scholar]
- 29.Kubica GP, Baess I, Gordon RE, Jenkins PA, Kwapinski JBG, McDurmont C, Pattyn SR, Saito H, Silcox V, Stanford JL, Takeya K, Tsukamura M. 1972. A co-operative numerical analysis of rapidly growing mycobacteria. J. Gen. Microbiol. 73:55–70. 10.1099/00221287-73-1-55 [DOI] [PubMed] [Google Scholar]
- 30.Kusunoki S, Ezaki T. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42:240–245. 10.1099/00207713-42-2-240 [DOI] [PubMed] [Google Scholar]
- 31.Adékambi T, Berger P, Raoult D, Drancourt M. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133–143. 10.1099/ijs.0.63969-0 [DOI] [PubMed] [Google Scholar]
- 32.Adékambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, Raoult D, Drancourt M. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493–5501. 10.1128/JCM.42.12.5493-5501.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leao SC, Tortoli E, Euzéby JP, Garcia MJ. 2011. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int. J. Syst. Evol. Microbiol. 61:2311–2313. 10.1099/ijs.0.023770-0 [DOI] [PubMed] [Google Scholar]
- 34.Cho YJ, Yi H, Chun J, Cho SN, Daley CL, Koh WJ, Jae Shin S. 2013. The genome sequence of “Mycobacterium massiliense” strain CIP 108297 suggests the independent taxonomic status of the Mycobacterium abscessus complex at the subspecies level. PLoS One 8:e81560. 10.1371/journal.pone.0081560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sassi M, Drancourt M. 2014. Genome analysis reveals three genomospecies in Mycobacterium abscessus. BMC Genomics 15:359. 10.1186/1471-2164-15-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tettelin H, Davidson RM, Agrawal S, Aitken ML, Shallom S, Hasan NA, Strong M, Nogueira de Moura VC, De Groote MA, Duarte RS, Hine E, Parankush S, Su Q, Daugherty SC, Fraser CM, Brown-Elliott BA, Wallace RJ, Jr, Holland SM, Sampaio EP, Olivier KN, Jackson M, Zelazny AM. 2014. High-level relatedness among Mycobacterium abscessus subsp. massiliense strains from widely separated outbreaks. Emerg. Infect. Dis. 20:364–371. 10.3201/eid2003.131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J. Antimicrob. Chemother. 67:810–818. 10.1093/jac/dkr578 [DOI] [PubMed] [Google Scholar]
- 38.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. 10.1016/S0140-6736(13)60632-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harada T, Akiyama Y, Kurashima A, Nagai H, Tsuyuguchi K, Fujii T, Yano S, Shigeto E, Kuraoka T, Kajiki A, Kobashi Y, Kokubu F, Sato A, Yoshida S, Iwamoto T, Saito H. 2012. Clinical and microbiological differences between Mycobacterium abscessus and Mycobacterium massiliense lung diseases. J. Clin. Microbiol. 50:3556–3561. 10.1128/JCM.01175-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, Park YK, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am. J. Respir. Crit. Care Med. 183:405–410. 10.1164/rccm.201003-0395OC [DOI] [PubMed] [Google Scholar]
- 41.Shallom SJ, Gardina PJ, Myers TG, Sebastian Y, Conville P, Calhoun LB, Tettelin H, Olivier KN, Uzel G, Sampaio EP, Holland SM, Zelazny AM. 2013. New rapid scheme for distinguishing the subspecies of the Mycobacterium abscessus group and identification of Mycobacterium massiliense with inducible clarithromycin resistance. J. Clin. Microbiol. 51:2943–2949. 10.1128/JCM.01132-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang DF, Chen X, Zhang XM, Zhi XY, Yao JC, Jiang Y, Xiong Z, Li WJ. 2013. Mycobacterium sediminis sp. nov. and Mycobacterium arabiense sp. nov., two novel rapidly growing members of the genus Mycobacterium. Int. J. Syst. Evol. Microbiol. 63:4081–4086. 10.1099/ijs.0.050567-0 [DOI] [PubMed] [Google Scholar]
- 43.Hennessee CT, Seo JS, Alvarez AM, Li QX. 2009. Polycyclic aromatic hydrocarbon-degrading species isolated from Hawaiian soils: Mycobacterium crocinum sp. nov., Mycobacterium pallens sp. nov., Mycobacterium rutilum sp. nov., Mycobacterium rufum sp. nov. and Mycobacterium aromaticivorans sp. nov. Int. J. Syst. Evol. Microbiol. 59:378–387. 10.1099/ijs.0.65827-0 [DOI] [PubMed] [Google Scholar]
- 44.Brown-Elliott BA, Wallace RJ, Jr, Petti CA, Mann LB, McGlasson M, Chihara S, Smith GL, Painter P, Hail D, Wilson R, Simmon KE. 2010. Mycobacterium neoaurum and Mycobacterium bacteremicum sp. nov. as causes of mycobacteremia. J. Clin. Microbiol. 48:4377–4385. 10.1128/JCM.00853-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adékambi T, Raoult D, Drancourt M. 2006. Mycobacterium barrassiae sp. nov., a Mycobacterium moriokaense group species associated with chronic pneumonia. J. Clin. Microbiol. 44:3493–3498. 10.1128/JCM.00724-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guerin-Faublee V, Flandrois JP, Pichat C, Boschiroli ML, Lamy B. 2013. Mycobacterium bourgelatii sp. nov., a rapidly growing, non-chromogenic species isolated from the lymph nodes of cattle. Int. J. Syst. Evol. Microbiol. 63:4669–4674. 10.1099/ijs.0.051979-0 [DOI] [PubMed] [Google Scholar]
- 47.Simmon KE, Brown-Elliott BA, Ridge PG, Durtschi JD, Mann LB, Slechta ES, Steigerwalt AG, Moser BD, Whitney AM, Brown JM, Voelkerding KV, McGowan KL, Reilly AF, Kirn TJ, Butler WR, Edelstein PH, Wallace RJ, Jr, Petti CA. 2011. Mycobacterium chelonae-abscessus complex associated with sinopulmonary disease, northeastern U.S.A. Emerg. Infect. Dis. 17:1692–1700. 10.3201/eid1709.101667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Ingen J, Blaak H, de Beer J, de Roda Husman AM, van Soolingen D. 2010. Rapidly growing nontuberculous mycobacteria cultured from home tap and shower water. Appl. Environ. Microbiol. 76:6017–6019. 10.1128/AEM.00843-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang YY, Li YB, Huang MX, Zhao XQ, Zhang LS, Liu WE, Wan KL. 2013. Novel species including Mycobacterium fukienense sp. is found from tuberculosis patients in Fujian Province, China, using phylogenetic analysis of Mycobacterium chelonae/abscessus complex. Biomed. Environ. Sci. 26:894–901. 10.3967/bes2013.018 [DOI] [PubMed] [Google Scholar]
- 50.Balcázar JL, Planas M, Pintado J. 2011. Novel mycobacterium species in seahorses with tail rot. Emerg. Infect. Dis. 17:1770–1772. 10.3201/eid1709.101289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tortoli E, Baruzzo S, Heijdra Y, Klenk HP, Lauria S, Mariottini A, van Ingen J. 2009. Mycobacterium insubricum sp. nov. Int. J. Syst. Evol. Microbiol. 59:1518–1523. 10.1099/ijs.0.003459-0 [DOI] [PubMed] [Google Scholar]
- 52.Shojaei H, Daley C, Gitti Z, Hashemi A, Heidarieh P, Moore ERB, Daei Naser A, Russo C, van Ingen J, Tortoli E. 2013. M. iranicum sp. nov., a rapidly-growing scotochromogenic species isolated from clinical specimens on three different continents. Int. J. Syst. Evol. Microbiol. 63:1383–1389. 10.1099/ijs.0.043562-0 [DOI] [PubMed] [Google Scholar]
- 53.Balakrishnan N, Tortoli E, Engel SL, Breitschwerdt EB. 2013. Isolation of a novel strain of Mycobacterium iranicum from a woman in the United States. J. Clin. Microbiol. 51:705–707. 10.1128/JCM.02560-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hashemi-Shahraki A, Heidarieh P, Azarpira S, Shojaei H, Hashemzadeh M, Tortoli E. 2013. Mycobacterium iranicum infection in HIV-infected patient, Iran. Emerg. Infect. Dis. 19:1696–1697. 10.3201/eid1910.130658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Zhang J, Fang C, Pang H, Fan J. 2012. Mycobacterium litorale sp. nov., a rapidly growing mycobacterium from soil. Int. J. Syst. Evol. Microbiol. 62:1204–1207. 10.1099/ijs.0.033449-0 [DOI] [PubMed] [Google Scholar]
- 56.Gomila M, Ramirez A, Gasco J, Lalucat J. 2008. Mycobacterium llatzerense sp. nov., a facultatively autotrophic, hydrogen-oxidizing bacterium isolated from haemodialysis water. Int. J. Syst. Evol. Microbiol. 58:2769–2773. 10.1099/ijs.0.65857-0 [DOI] [PubMed] [Google Scholar]
- 57.Dubrou S, Konjek J, Macheras E, Welte B, Guidicelli L, Chignon E, Joyeux M, Gaillard JL, Heym B, Tully T, Sapriel G. 2013. Diversity, community composition, and dynamics of nonpigmented and late-pigmenting rapidly growing mycobacteria in an urban tap water production and distribution system. Appl. Environ. Microbiol. 79:5498–5508. 10.1128/AEM.00900-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teixeira L, Avery RK, Iseman M, Arrossi AV, Harrington S, Stephens K, Winans CG. 2013. Mycobacterium llatzerense lung infection in a liver transplant recipient: case report and review of the literature. Am. J. Transplant. 13:2198–2200. 10.1111/ajt.12318 [DOI] [PubMed] [Google Scholar]
- 59.Cárdenas AM, Gomila M, Lalucat J, Edelstein PH. 2014. Mycobacterium llatzerense abdominal abscess. J. Clin. Microbiol. 52:1287–1289. 10.1128/JCM.03525-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reischl U, Melzl H, Kroppenstedt RM, Miethke T, Naumann L, Mariottini A, Mazzarelli G, Tortoli E. 2006. Mycobacterium monacense sp. nov. Int. J. Syst. Evol. Microbiol. 56:2575–2578. 10.1099/ijs.0.64527-0 [DOI] [PubMed] [Google Scholar]
- 61.Taieb A, Ikeguchi R, Yu VL, Rihs JD, Sharma M, Wolfe J, Wollstein R. 2008. Mycobacterium monacense: a mycobacterial pathogen that causes infection of the hand. J. Hand Surg. 33:94–96. 10.1016/j.jhsa.2007.10.016 [DOI] [PubMed] [Google Scholar]
- 62.Shojaei H, Hashemi A, Heidarieh P, Hosseini N, Daei Naser A. 2012. Chronic pulmonary disease due to Mycobacterium monacense infection: the first case from Iran. Ann. Lab. Med. 32:87–90. 10.3343/alm.2012.32.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Therese KL, Gayathri R, Thiruppathi K, Madhavan HN. 2011. First report of isolation of Mycobacterium monacense from sputum specimen in India. Lung India 28:124–126. 10.4103/0970-2113.80326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hogardt M, Schreff AM, Naumann L, Reischl U, Sing A. 2008. Mycobacterium monacense in a patient with a pulmonary tumor. Jpn. J. Infect. Dis. 61:77–78 [PubMed] [Google Scholar]
- 65.Ross A. 1960. Mycobacterium salmoniphilum sp. nov. from salmonid fishes. Am. Rev. Respir. Dis. 18:241–250 [DOI] [PubMed] [Google Scholar]
- 66.Whipps CM, Butler WR, Pourahmad F, Watral VG, Kent ML. 2007. Molecular systematics support the revival of Mycobacterium salmoniphilum (ex Ross 1960) sp. nov., nom. rev., a species closely related to Mycobacterium chelonae. Int. J. Syst. Evol. Microbiol. 57:2525–2531. 10.1099/ijs.0.64841-0 [DOI] [PubMed] [Google Scholar]
- 67.Aro L, Correa K, Martinez A, Ildefonso R, Yanez JM. 2014. Characterization of Mycobacterium salmoniphilum as causal agent of mycobacteriosis in Atlantic salmon, Salmo salar l. from a freshwater recirculation system. J. Fish Dis. 37:341–348. 10.1111/jfd.12108 [DOI] [PubMed] [Google Scholar]
- 68.Berg V, Zerihun MA, Jorgensen A, Lie E, Dale OB, Skaare JU, Lyche JL. 2013. High prevalence of infections and pathological changes in burbot (Lota lota) from a polluted lake (Lake Mjosa, Norway). Chemosphere 90:1711–1718. 10.1016/j.chemosphere.2012.10.017 [DOI] [PubMed] [Google Scholar]
- 69.Righetti M, Favaro L, Antuofermo E, Caffara M, Nuvoli S, Scanzio T, Prearo M. 2014. Mycobacterium salmoniphilum infection in a farmed Russian sturgeon, Acipenser gueldenstaedtii (Brandt & Ratzeburg). J. Fish Dis. 37:671–674. 10.1111/jfd.12143 [DOI] [PubMed] [Google Scholar]
- 70.Zerihun MA, Berg V, Lyche JL, Colquhoun DJ, Poppe TT. 2011. Mycobacterium salmoniphilum infection in burbot Lota lota. Dis. Aquat. Org. 95:57–64. 10.3354/dao02347 [DOI] [PubMed] [Google Scholar]
- 71.Zerihun MA, Nilsen H, Hodneland S, Colquhoun DJ. 2011. Mycobacterium salmoniphilum infection in farmed Atlantic salmon, Salmo salar L. J. Fish Dis. 34:769–781. 10.1111/j.1365-2761.2011.01293.x [DOI] [PubMed] [Google Scholar]
- 72.Lamy B, Marchandin H, Hamitouche K, Laurent F. 2008. Mycobacterium setense sp. nov., a Mycobacterium fortuitum-group organism isolated from a patient with soft tissue infection and osteitis. Int. J. Syst. Evol. Microbiol. 58:486–490. 10.1099/ijs.0.65222-0 [DOI] [PubMed] [Google Scholar]
- 73.Toro A, Adekambi T, Cheynet F, Fournier PE, Drancourt M. 2008. Mycobacterium setense infection in humans. Emerg. Infect. Dis. 14:1330–1332. 10.3201/eid1408.080179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shojaei H, Hashemi A, Heidarieh P, Feizabadi MM, Ataei B, Daei NA. 2011. First report on isolation and molecular characterization of clinical Mycobacterium setense isolates in Asia. Jpn. J. Infect. Dis. 64:234–236 [PubMed] [Google Scholar]
- 75.Sahraoui N, Ballif M, Zelleg S, Yousfi N, Ritter C, Friedel U, Amstutz B, Yala D, Boulahbal F, Guetarni D, Zinsstag J, Keller PM. 2011. Mycobacterium algericum sp. nov., a novel rapidly growing species related to Mycobacterium terrae complex and associated with goat lung lesions. Int. J. Syst. Evol. Microbiol. 61:1870–1874. 10.1099/ijs.0.024851-0 [DOI] [PubMed] [Google Scholar]
- 76.Tortoli E, Gitti Z, Klenk HP, Lauria S, Mannino R, Mantegani P, Mariottini A, Neonakis I. 2013. Survey of 150 strains belonging to the Mycobacterium terrae complex and description of Mycobacterium engbaekii sp. nov., Mycobacterium heraklionense sp. nov. and Mycobacterium longobardum sp. nov. Int. J. Syst. Evol. Microbiol. 63:401–411. 10.1099/ijs.0.038737-0 [DOI] [PubMed] [Google Scholar]
- 77.Mrlik V, Slany M, Kubecka J, Seda J, Necas A, Babak V, Slana I, Kriz P, Pavlik I. 2012. A low prevalence of mycobacteria in freshwater fish from water reservoirs, ponds and farms. J. Fish Dis. 35:497–504. 10.1111/j.1365-2761.2012.01369.x [DOI] [PubMed] [Google Scholar]
- 78.Richter E, Tortoli E, Fischer A, Hendricks O, Engel R, Hillemann D, Schubert S, Kristiansen JE. 2007. Mycobacterium alsiense, a novel, slowly growing species isolated from two patients with pulmonary disease. J. Clin. Microbiol. 45:3837–3839. 10.1128/JCM.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bang D, Herlin T, Stegger M, Andersen Å Torkko BP, Tortoli E, Thomsen BV. 2008. Mycobacterium arosiense sp. nov., a slowly growing, scotochromogenic species causing osteomyelitis in an immunocompromised child. Int. J. Syst. Evol. Microbiol. 58:2398–2402. 10.1099/ijs.0.65503-0 [DOI] [PubMed] [Google Scholar]
- 80.Tortoli E, Adriani B, Baruzzo S, Degl'Innocenti R, Galanti I, Lauria S, Mariottini A, Pascarella M. 2009. Pulmonary disease due to Mycobacterium arosiense, an easily misidentified pathogenic novel mycobacterium. J. Clin. Microbiol. 47:1947–1949. 10.1128/JCM.02449-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburg R, Huit G, Iademarco MF, Iseman M, Olivier K, Rouss S, von Reyn CF, Wallace RJ, Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America 2007. An official ATS/IDSA statement: diagnosis, treatment and prevention of nontuberculous mycobacterial disease. Am. J. Respir. Crit. Care Med. 175:367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- 82.Ben Salah I, Cayrou C, Raoult D, Drancourt M. 2009. Mycobacterium marseillense sp. nov., Mycobacterium timonense sp. nov. and Mycobacterium bouchedurhonense sp. nov., novel species in the Mycobacterium avium complex. Int. J. Syst. Evol. Microbiol. 59:2803–2808. 10.1099/ijs.0.010637-0 [DOI] [PubMed] [Google Scholar]
- 83.Korsak T, Boisvert H. 1972. Mycobactéries a pigment rose. Ann. Inst. Pasteur 122:31–41 [PubMed] [Google Scholar]
- 84.Tortoli E, Böttger EC, Fabio A, Falsen E, Gitti Z, Grottola A, Klenk HP, Mannino R, Mariottini A, Messinò M, Pecorari M, Rumpianesi F. 2011. Mycobacterium europaeum sp. nov., a scotochromogenic species related to the Mycobacterium simiae complex. Int. J. Syst. Evol. Microbiol. 61:1606–1611. 10.1099/ijs.0.025601-0 [DOI] [PubMed] [Google Scholar]
- 85.Pourahmad F, Shojaei H, Heidarieh P, Khosravi A, Hashemi A. 2012. Report of two cases of Mycobacterium europaeum from Iran. Jpn. J. Infect. Dis. 65:539–541. 10.7883/yoken.65.539 [DOI] [PubMed] [Google Scholar]
- 86.Ramos JP, Campos CE, Caldas PC, Ferreira NV, Boas da Silva MV, Redner P, Campelo CL, Vale SF, Barroso EC, Medeiros RF, Montes FC, Galvão TC, Tortoli E. 2013. Mycobacterium fragae sp. nov., a non-chromogenic species isolated from human respiratory specimens. Int. J. Syst. Evol. Microbiol. 63:2583–2587. 10.1099/ijs.0.046862-0 [DOI] [PubMed] [Google Scholar]
- 87.Abedalthagafi M, Rosenberg O, Miller S. 2014. First report of tenosynovitis in a immunocompetent person caused by Mycobacterium heraklionense. J. Med. Microbiol. Case Rep. 10.1099/jmmcr.0.002071 [DOI] [Google Scholar]
- 88.Saini V, Raghuvanshi S, Talwar GP, Ahmed N, Khurana JP, Hasnain SE, Tyagi AK. 2009. Polyphasic taxonomic analysis establishes Mycobacterium indicus pranii as a distinct species. PLoS One 4:e6263. 10.1371/journal.pone.0006263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Talwar GP, Zaheer SA, Mukherjee R, Walia R, Misra RS, Sharma AK, Kar HK, Mukherjee A, Parida SK, Suresh NR, Nair SK, Pandey RM. 1990. Immunotherapeutic effects of a vaccine based on a saprophytic cultivable mycobacterium, Mycobacterium W in multibacillary leprosy patients. Ind. Vaccine 8:121–129 [DOI] [PubMed] [Google Scholar]
- 90.Saini V, Raghuvanshi S, Khurana JP, Ahmed N, Hasnain SE, Tyagi AK, Tyagi AK. 2012. Massive gene acquisitions in Mycobacterium indicus pranii provide a perspective on mycobacterial evolution. Nucleic Acids Res. 40:10832–10850. 10.1093/nar/gks793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Faujdar J, Gupta P, Natrajan M, Das R, Chauhan DS, Katoch VM, Gupta UD. 2011. Mycobacterium indicus pranii as stand-alone or adjunct immunotherapeutic in treatment of experimental animal tuberculosis. Indian J. Med. Res. 134:696–703. 10.4103/0971-5916.90999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nath I. 1998. A vaccine for leprosy. Nat. Med. 4:548–550. 10.1038/nm0598-548 [DOI] [PubMed] [Google Scholar]
- 93.Kim BJ, Jeong J, Lee SH, Kim SR, Yu HK, Park JG, Kim KJ, Kook YH, Kim BJ. 2012. Mycobacterium koreense sp. nov., a slowly growing non-chromogenic species closely related to Mycobacterium triviale. Int. J. Syst. Evol. Microbiol. 62:1289–1295. 10.1099/ijs.0.033274-0 [DOI] [PubMed] [Google Scholar]
- 94.Masaki T, Ohkusu K, Hata H, Fujiwara N, Iihara H, Yamada-Noda M, Nhung PH, Asano Y, Kawamura Y, Ezaki T. 2006. Mycobacterium kumamotonense sp. nov. recovered from clinical specimen and the first isolation report of Mycobacterium arupense in Japan: novel slowly growing, nonchromogenic clinical isolates related to Mycobacterium terrae complex. Microbiol. Immunol. 50:889–897. 10.1111/j.1348-0421.2006.tb03865.x [DOI] [PubMed] [Google Scholar]
- 95.Rodriguez-Aranda A, Jimenez MS, Yubero J, Chaves F, Rubio-Garcia R, Palenque E, Garcia MJ, Menendez MC. 2010. Misidentification of Mycobacterium kumamotonense as M. tuberculosis. Emerg. Infect. Dis. 16:1178–1180. 10.3201/eid1607.091913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Okazaki M, Ohkusu K, Hata H, Ohnishi H, Sugahara K, Kawamura C, Fujiwara N, Matsumoto S, Nishiuchi Y, Toyoda K, Saito H, Yonetani S, Fukugawa Y, Yamamoto M, Wada H, Sejimo A, Ebina A, Goto H, Ezaki T, Watanabe T. 2009. Mycobacterium kyorinense sp. nov., a novel, slow-growing species, related to Mycobacterium celatum, isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 59:1336–1341. 10.1099/ijs.0.000760-0 [DOI] [PubMed] [Google Scholar]
- 97.Bull TJ, Shanson DC, Archard LC, Yates MD, Hamid ME, Minnikin DE. 1995. A new group (type 3) of Mycobacterium celatum isolated from AIDS patients in the London area. Int. J. Syst. Bacteriol. 45:861–862. 10.1099/00207713-45-4-861 [DOI] [PubMed] [Google Scholar]
- 98.Butler WR, O'Connor SP, Yakrus MA, Smithwick RW, Plikaytis BB, Moss CW, Floyd MM, Woodley CL, Kilburn JO, Vadney FS, Gross WM. 1993. Mycobacterium celatum sp. nov. Int. J. Syst. Bacteriol. 43:539–548. 10.1099/00207713-43-3-539 [DOI] [PubMed] [Google Scholar]
- 99.Ohnishi H, Tortoli E, Ramos JP, Kazumi Y, Yonetani S, Matsushima S, Ohtsuka K, Watanabe T. 23 March 2014. Proposal of reclassification of Mycobacterium celatum type 2 as Mycobacterium kyorinense. Ann. Microbiol. 10.1007/s13213-014-0860-9 [DOI] [Google Scholar]
- 100.Ohnishi H, Yonetani S, Matsushima S, Wada H, Takeshita K, Kuramochi D, Caldas PC, Campos CE, da Costa BP, Ramos JP, Mikura S, Narisawa E, Fujita A, Funayama Y, Kobashi Y, Sakakibara Y, Ishiyama Y, Takakura S, Goto H, Watanabe T. 2013. Mycobacterium kyorinense infection. Emerg. Infect. Dis. 19:508–510. 10.3201/eid1903.12-0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kobashi Y, Mouri K, Obase Y, Kato S, Oka M. 2012. Pulmonary Mycobacterium kyorinense disease showed clinical improvement following combined therapy with clarithromycin and levofloxacin. Intern. Med. 51:1923–1926. 10.2169/internalmedicine.51.6431 [DOI] [PubMed] [Google Scholar]
- 102.Terada Y, Takeshita K, Baba R, Hiraoka R, Suzuki Y. 2012. Case report; a case of Mycobacterium kyorinense pulmonary infection. Nihon Naika Gakkai Zasshi 101:2301–2303. 10.2169/naika.101.2301 [DOI] [PubMed] [Google Scholar]
- 103.Wada H, Yamamoto M, Okazaki M, Watanabe T, Goto H. 2009. Isolation of Mycobacterium kyorinense in a patient with respiratory failure. Ann. Intern. Med. 150:568–570. 10.7326/0003-4819-150-8-200904210-00018 [DOI] [PubMed] [Google Scholar]
- 104.Sakakibara Y, Kishimoto K, Kojima K, Fujie T, Inase N. 2014. Fatal nontuberculous mycobacterial lung disease caused by Mycobacterium kyorinense: a case report with five years of follow-up. Kekkaku 89:509–513 [PubMed] [Google Scholar]
- 105.Campos CE, Caldas PC, Ohnishi H, Watanabe T, Ohstuka K, Matsushima S, Ferreira NV, da Silva MV, Redner P, de Carvalho LD, Medeiros RF, Abbud Filho JA, Montes FC, Galvão TC, Ramos JP. 2012. First isolation of Mycobacterium kyorinense from clinical specimens in Brazil. J. Clin. Microbiol. 50:2477–2478. 10.1128/JCM.00023-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Han XY, Seo YH, Sizer KC, Schoberle T, May GS, Spencer JS, Li W, Nair RG. 2008. A new Mycobacterium species causing diffuse lepromatous leprosy. Am. J. Clin. Pathol. 130:856–864. 10.1309/AJCPP72FJZZRRVMM [DOI] [PubMed] [Google Scholar]
- 107.Han XY, Sizer KC, Velarde-Felix JS, Frias-Castro LO, Vargas-Ocampo F. 2012. The leprosy agents Mycobacterium lepromatosis and Mycobacterium leprae in Mexico. Int. J. Dermatol. 51:952–959. 10.1111/j.1365-4632.2011.05414.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han XY, Sizer KC, Tan HH. 2012. Identification of the leprosy agent Mycobacterium lepromatosis in Singapore. J. Drugs Dermatol. 11:168–172 [PubMed] [Google Scholar]
- 109.Jessamine PG, Desjardins M, Gillis T, Scollard D, Jamieson F, Broukhanski G, Chedore P, McCarthy A. 2012. Leprosy-like illness in a patient with Mycobacterium lepromatosis from Ontario, Canada. J. Drugs Dermatol. 11:229–233 [PubMed] [Google Scholar]
- 110.Trott KA, Stacy BA, Lifland BD, Diggs HE, Harland RM, Khokha MK, Grammer TC, Parker JM. 2004. Characterization of a Mycobacterium ulcerans-like infection in a colony of African tropical clawed frogs (Xenopus tropicalis). Comp. Med. 54:309–317 [PubMed] [Google Scholar]
- 111.Mve-Obiang A, Lee RE, Umstot ES, Trott KA, Grammer TC, Parker JM, Ranger BS, Grainger R, Mahrous EA, Small PL. 2005. A newly discovered mycobacterial pathogen isolated from laboratory colonies of Xenopus species with lethal infections produces a novel form of mycolactone, the Mycobacterium ulcerans macrolide toxin. Infect. Immun. 73:3307–3312. 10.1128/IAI.73.6.3307-3312.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tobias NJ, Doig KD, Medema MH, Chen H, Haring V, Moore R, Seemann T, Stinear TP. 2013. Complete genome sequence of the frog pathogen Mycobacterium ulcerans ecovar liflandii. J. Bacteriol. 195:556–564. 10.1128/JB.02132-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Doig KD, Holt KE, Fyfe JA, Lavender CJ, Eddyani M, Portaels F, Yeboah-Manu D, Pluschke G, Seemann T, Stinear TP. 2012. On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer. BMC Genomics 13:258. 10.1186/1471-2164-13-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fremont-Rahl JJ, Ek C, Williamson HR, Small PL, Fox JG, Muthupalani S. 2011. Mycobacterium liflandii outbreak in a research colony of Xenopus (Silurana) tropicalis frogs. Vet. Pathol. 48:856–867. 10.1177/0300985810388520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Suykerbuyk P, Vleminckx K, Pasmans F, Stragier P, Ablordey A, Tran HT, Hermans K, Fleetwood M, Meyers WM, Portaels F. 2007. Mycobacterium liflandii infection in European colony of Silurana tropicalis. Emerg. Infect. Dis. 13:743–746. 10.3201/eid1305.060625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hong SK, Sung JY, Lee HJ, Oh MD, Park SS, Kim EC. 2013. First case of Mycobacterium longobardum infection. Ann. Lab. Med. 33:356–359. 10.3343/alm.2013.33.5.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Ingen J, Lindeboom JA, Hartwig NG, de Zwaan R, Tortoli E, Dekhuijzen RP, Boeree MJ, van Soolingen D. 2009. Mycobacterium mantenii sp. nov., a pathogenic, slowly growing, scotochromogenic mycobacterium. Int. J. Syst. Evol. Microbiol. 59:2784–2787. 10.1099/ijs.0.010405-0 [DOI] [PubMed] [Google Scholar]
- 118.Slany M, Jezek P, Fiserova V, Bodnarova M, Stork J, Havelkova M, Kalat F, Pavlik I. 2012. Mycobacterium marinum infections in humans and tracing of its possible environmental sources. Can. J. Microbiol. 58:39–44. 10.1139/w11-104 [DOI] [PubMed] [Google Scholar]
- 119.Grottola A, Roversi P, Fabio A, Antenora F, Apice A, Tagliazucchi S, Gennari W, Fregni Serpini G, Rumpianesi F, Fabbri LM, Magnani R, Pecorari M. Mycobacterium marseillense: prolonged pulmonary disease in an immunocompetent patient, Italy. Emerg. Infect. Dis., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim SY, Yoo H, Jeong BH, Jeon K, Ha YE, Huh HJ, Ki CS, Lee NY, Shin SJ, Koh WJ. 2014. First case of nontuberculous mycobacterial lung disease caused by Mycobacterium marseillense in a patient with systemic lupus erythematosus. Diagn. Microbiol. Infect. Dis. 79:355–357. 10.1016/j.diagmicrobio.2014.03.019 [DOI] [PubMed] [Google Scholar]
- 121.Cândido PH, Nunes LD, Marques EA, Folescu TW, Coelho FS, de Moura VC, Da Silva MG, Gomes KM, Lourenco MC, Aguiar FS, Chitolina F, Armstrong DT, Leão SC, Neves FP, Mello FC, Duarte RS. 2014. Multidrug-resistant nontuberculous mycobacteria isolated from cystic fibrosis patients. J. Clin. Microbiol. 52:2990–2997. 10.1128/JCM.00549-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hannigan JD, Krivogorsky B, Fordice D, Welch JB, Dahl JL. 2013. Mycobacterium minnesotense sp. nov., a photochromogenic bacterium isolated from sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 63:124–128. 10.1099/ijs.0.037291-0 [DOI] [PubMed] [Google Scholar]
- 123.van Ingen J, Boeree MJ, de Lange WC, de Haas PE, van der Zanden AG, Mijs W, Rigouts L, Dekhuijzen PN, van Soolingen D. 2009. Mycobacterium noviomagense sp. nov.; clinical relevance evaluated in 17 patients. Int. J. Syst. Evol. Microbiol. 59:845–849. 10.1099/ijs.0.001511-0 [DOI] [PubMed] [Google Scholar]
- 124.Chase HH, Davis JB, Raymond RL. 1956. Mycobacterium paraffinicum n. sp., a bacterium isolated from soil. Appl. Microbiol. 4:310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Toney NC, Adekambi T, Toney S, Yakrus M, Butler WR. 2010. Revival and emended description of “Mycobacterium paraffinicum” (Davis, Chase and Raymond 1956) as Mycobacterium paraffinicum (ex Davis, Chase and Raymond 1956) sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 60:2307–2313. 10.1099/ijs.0.016972-0 [DOI] [PubMed] [Google Scholar]
- 126.Wang SH, Pancholi P, Stevenson K, Yakrus MA, Butler WR, Schlesinger LS, Mangino JE. 2009. Pseudo-outbreak of “Mycobacterium paraffinicum” infection and/or colonization in a tertiary care medical center. Infect. Control Hosp. Epidemiol. 30:848–853. 10.1086/599071 [DOI] [PubMed] [Google Scholar]
- 127.Chan AW, Kabbani S, Staton G, Kraft CS. 2014. Mycobacterium paraffinicum causing symptomatic pulmonary infection. J. Clin. Microbiol. 52:1281–1283. 10.1128/JCM.03107-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim BJ, Hong SH, Kook YH, Kim BJ. 2014. Mycobacterium paragordonae sp. nov., a novel slowly growing scoto-chromogenic species closely related to Mycobacterium gordonae. Int. J. Syst. Evol. Microbiol. 64:39–45. 10.1099/ijs.0.051540-0 [DOI] [PubMed] [Google Scholar]
- 129.Kim BJ, Hong SH, Yu HK, Park YG, Jeong J, Lee SH, Kim SR, Kim K, Kook YH, Kim BJ. 2013. Mycobacterium parakoreense sp. nov., a slowly growing non-chromogenic species related to Mycobacterium koreense, isolated from a human clinical specimen. Int. J. Syst. Evol. Microbiol. 69:2254–2259. 10.1099/ijs.0.045070-0 [DOI] [PubMed] [Google Scholar]
- 130.Lee HK, Lee SA, Lee IK, Yu HK, Park JG, Hyun JW, Kim K, Kook JH, Kim BJ. 2010. Mycobacterium paraseoulense sp. nov., a slowly growing, scotochromogenic species related genetically to Mycobacterium seoulense. Int. J. Syst. Evol. Microbiol. 60:439–443. 10.1099/ijs.0.012054-0 [DOI] [PubMed] [Google Scholar]
- 131.Cage GD. 1992. High-performance liquid chromatography patterns of Mycobacterium gordonae mycolic acids. J. Clin. Microbiol. 30:2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee H, Lee SA, Lee IK, Yu HK, Park YG, Jeong J, Lee SH, Kim SR, Hyun JW, Kim K, Kook YH, Kim BJ. 2010. Mycobacterium paraterrae sp. nov. recovered from a clinical specimen: novel chromogenic slow growing mycobacteria related to Mycobacterium terrae complex. Microbiol. Immunol. 54:46–53. 10.1111/j.1348-0421.2009.00184.x [DOI] [PubMed] [Google Scholar]
- 133.van Ingen J, Al Hajoj SAM, Boere M, Al Rabiah F, Enaimi M, de Zwaan R, Tortoli E, Dekhuijzen R, van Soolingen D. 2009. Mycobacterium riyadhense sp. nov.; a non-tuberculous species identified as Mycobacterium tuberculosis by a commercial line-probe assay. Int. J. Syst. Evol. Microbiol. 59:1049–1053. 10.1099/ijs.0.005629-0 [DOI] [PubMed] [Google Scholar]
- 134.Godreuil S, Marchandin H, Michon AL, Ponsada M, Chyderiotis G, Brisou P, Bhat A, Panteix G. 2012. Mycobacterium riyadhense pulmonary infection, France and Bahrain. Emerg. Infect. Dis. 18:176–178. 10.3201/eid1801.110751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Choi JI, Lim JH, Kim SR, Lee SH, Park JS, Seo KW, Jeon JB, Jeong J. 2012. Lung infection caused by Mycobacterium riyadhense confused with Mycobacterium tuberculosis: the first case in Korea. Ann. Lab. Med. 32:298–303. 10.3343/alm.2012.32.4.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mun HS, Park JH, Kim H, Yu HK, Park YG, Cha CY, Kook YH, Kim BJ. 2008. Mycobacterium senuense sp. nov., a slowly growing, non-chromogenic species closely related to the Mycobacterium terrae complex. Int. J. Syst. Evol. Microbiol. 58:641–646. 10.1099/ijs.0.65374-0 [DOI] [PubMed] [Google Scholar]
- 137.Mun HS, Kim HJ, Oh EJ, Kim H, Bai GH, Yu HK, Park YG, Cha CY, Kook YH, Kim BJ. 2007. Mycobacterium seoulense sp. nov., a slowly growing scotochromogenic species. Int. J. Syst. Evol. Microbiol. 57:594–599. 10.1099/ijs.0.64744-0 [DOI] [PubMed] [Google Scholar]
- 138.van Ingen J, Tortoli E, Selvarangan R, Coyle MB, Crump JA, Morrissey AB, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2011. Mycobacterium sherrisii sp. nov.; a slow growing nonchromogenic species. Int. J. Syst. Evol. Microbiol. 61:1293–1298. 10.1099/ijs.0.024752-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Crump JA, van Ingen J, Morrissey AB, Boeree MJ, Mavura DR, Swai B, Thielman NM, Bartlett JA, Grossman H, Maro VP, van Soolingen D. 2009. Invasive disease caused by nontuberculous mycobacteria, Tanzania. Emerg. Infect. Dis. 15:53–55. 10.3201/eid1501.081093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gamperli A, Bosshard PP, Sigrist T, Brandli O, Wildermuth S, Weber R, Mueller NJ. 2005. Pulmonary Mycobacterium sherrisii infection in a human immunodeficiency virus type 1-infected patient. J. Clin. Microbiol. 43:4283–4285. 10.1128/JCM.43.8.4283-4285.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ho J, Balm M, Huggan P, Chew N, Venkatachalam I, Archuleta S. 2012. Immune reconstitution inflammatory syndrome associated with disseminated Mycobacterium sherrisii infection. Int. J. STD AIDS 23:369–370. 10.1258/ijsa.2011.011311 [DOI] [PubMed] [Google Scholar]
- 142.Loulergue P, Lamontagne F, Vincent V, Rossier A, Pialoux G. 2007. Mycobacterium sherrisii: a new opportunistic agent in HIV infection? AIDS 21:893–894. 10.1097/QAD.0b013e3280f7750f [DOI] [PubMed] [Google Scholar]
- 143.Tajan J, Espasa M, Sala M, Navarro M, Font B, Gonzalez-Martin J, Segura F. 2013. Disseminated infection by Mycobacterium sherrisii and Histoplasma capsulatum in an African HIV-infected patient. Am. J. Trop. Med. Hyg. 88:914–917. 10.4269/ajtmh.12-0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tortoli E, Galli L, Andebirhan T, Baruzzo S, Chiappini E, de Martino M, Brown-Elliott BA. 2007. The first case of Mycobacterium sherrisii disseminated infection in a child with AIDS. AIDS 21:1496–1497. 10.1097/QAD.0b013e328235a53c [DOI] [PubMed] [Google Scholar]
- 145.Lai C, Lam W, Lee MP. 2014. A case of Mycobacterium sherrisii pneumonia diagnosed by PCR/ESI-MS method. Int. J. Infect. Dis. 5:119–121. 10.1016/j.ijid.2014.03.1391 [DOI] [PubMed] [Google Scholar]
- 146.Lee SM, Myers RA, Singh K, Kansal S. 2013. Disseminated Mycobacterium sherrisii infection in a US-born, HIV-infected patient. J. Int. Assoc. Provid. AIDS Care 12:245–246. 10.1177/2325957413488175 [DOI] [PubMed] [Google Scholar]
- 147.Barrera L, Palmero D, Paul R, Lopez B. 2010. Enfermedad por Mycobacterium simiae y “Mycobacterium sherrisii” en la Argentina. Medicina (Buenos Aires) 70:343–346 [PubMed] [Google Scholar]
- 148.Nakanaga K, Hoshino Y, Wakabayashi M, Fujimoto N, Tortoli E, Makino M, Tanaka T, Ishii N. 2012. Mycobacterium shigaense sp. nov., a novel slowly growing scotochromogenic mycobacterium that produced nodules in an erythroderma patient with severe cellular immunodeficiency and a history of Hodgkin's disease. J. Dermatol. 39:389–396. 10.1111/j.1346-8138.2011.01355.x [DOI] [PubMed] [Google Scholar]
- 149.Cui P, Vissa V, Li W, Zhang X, Lin L, Wang H, Liu X, Wu Q, Zong W. 2013. Cutaneous Mycobacterium shigaense infection in immunocompetent woman, China. Emerg. Infect. Dis. 19:819–820. 10.3201/eid1905.121022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Saito H, Iwamoto T, Ohkusu K, Otsuka Y, Akiyama S, Sato S, Taguchi O, Sueyasu Y, Kawabe Y, Fujimoto H, Ezaki T, Butler R. 2011. Mycobacterium shinjukuense sp. nov., a slowly growing, non-chromogenic species isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 61:1927–1932. 10.1099/ijs.0.025478-0 [DOI] [PubMed] [Google Scholar]
- 151.Futatsugi K, Nishio K, Aida S, Okabayashi K, Nakano Y, Ryuzaki M, Oosone Y, Kazumi Y. 2011. Case report: a case of pulmonary infectious disease due to Mycobacterium shinjukuense. Nihon Naika Gakkai Zasshi 100:3637–3639 [DOI] [PubMed] [Google Scholar]
- 152.Watanabe K, Shinkai M, Yamaguchi N, Shinoda M, Hara Y, Ishigatsubo Y, Kaneko T. 2013. Mycobacterium shinjukuense lung disease that was successfully treated with antituberculous drugs. Intern. Med. 52:2653–2655. 10.2169/internalmedicine.52.1116 [DOI] [PubMed] [Google Scholar]
- 153.Tortoli E, Rogasi PG, Fantoni E, Beltrami C, De Francisci A, Mariottini A. 2010. Infection due to a novel mycobacterium, mimicking multidrug-resistant Mycobacterium tuberculosis. Clin. Microbiol. Infect. 14:1130–1134. 10.1111/j.1469-0691.2009.03063.x [DOI] [PubMed] [Google Scholar]
- 154.Zhang ZY, Sun ZQ, Wang ZL, Hu HR, Wen ZL, Song YZ, Zhao JW, Wang HH, Guo XK, Zhang SL. 2013. Identification and pathogenicity analysis of a novel non-tuberculous mycobacterium clinical isolate with nine-antibiotic resistance. Clin. Microbiol. Infect. 19:91–96. 10.1111/j.1469-0691.2012.03818.x [DOI] [PubMed] [Google Scholar]
- 155.Pourahmad F, Cervellione F, Thompson KD, Taggart JB, Adams A, Richards RH. 2008. Mycobacterium stomatepiae sp. nov., a slowly growing, non-chromogenic species isolated from fish. Int. J. Syst. Evol. Microbiol. 58:2821–2827. 10.1099/ijs.0.2008/001164-0 [DOI] [PubMed] [Google Scholar]
- 156.Zurita J, Ortega-Paredes D, Mora M, Espinel N, Parra H, Febres L, Zurita-Salinas C. 11 May 2014. Characterization of the first report of Mycobacterium timonense infecting an HIV patient in an Ecuadorian hospital. Clin. Microbiol. Infect. 10.1111/1469-0691.12675 [DOI] [PubMed] [Google Scholar]
- 157.van Ingen J, Boeree MJ, Koesters K, Wieland A, Tortoli E, Dekhuijizen PNR, van Soolingen D. 2009. Proposal to elevate Mycobacterium avium complex sequevar MAC-Q to Mycobacterium vulneris sp. nov. Int. J. Syst. Evol. Microbiol. 59:2277–2282. 10.1099/ijs.0.008854-0 [DOI] [PubMed] [Google Scholar]
- 158.Kim BJ, Math RK, Jeon CO, Yu HK, Park YG, Kook YH, Kim BJ. 2013. Mycobacterium yongonense sp. nov., a slow-growing non-chromogenic species closely related to Mycobacterium intracellulare. Int. J. Syst. Evol. Microbiol. 63:192–199. 10.1099/ijs.0.037465-0 [DOI] [PubMed] [Google Scholar]
- 159.Kim BJ, Hong SH, Kook YH, Kim BJ. 2013. Molecular evidence of lateral gene transfer in rpoB gene of Mycobacterium yongonense strains via multilocus sequence analysis. PLoS One 8:e51846. 10.1371/journal.pone.0051846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tortoli E, Mariottini A, Pierotti P, Simonetti TM, Rossolini GM. 2013. Mycobacterium yongonense in pulmonary disease, Italy. Emerg. Infect. Dis. 19:1902–1904. 10.3201/eid1911.130911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Alexander KA, Laver PN, Michel AL, Williams M, van Helden PD, Warren RM, Gey van Pittius NC. 2010. Novel Mycobacterium tuberculosis complex pathogen, M. mungi. Emerg. Infect. Dis. 16:1296–1299. 10.3201/eid1608.100314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alexander KA, Pleydell E, Williams MC, Lane EP, Nyange JF, Michel AL. 2002. Mycobacterium tuberculosis: an emerging disease of free-ranging wildlife. Emerg. Infect. Dis. 8:598–601. 10.3201/eid0806.010358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gey van Pittius NC, van Helden PD, Warren RM. 2012. Characterization of Mycobacterium orygis. Emerg. Infect. Dis. 18:1708–1709. 10.3201/eid1810.120569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, Brosch R, van Soolingen D. 2012. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg. Infect. Dis. 18:653–655. 10.3201/eid1804.110888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van Ingen J, Brosch R, van Soolingen D. 2013. Characterization of Mycobacterium orygis. Emerg. Infect. Dis. 19:521–522. 10.3201/eid1903.121005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dawson KL, Bell A, Kawakami RP, Coley K, Yates G, Collins DM. 2012. Transmission of Mycobacterium orygis (M. tuberculosis complex species) from a tuberculosis patient to a dairy cow in New Zealand. J. Clin. Microbiol. 50:3136–3138. 10.1128/JCM.01652-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Parsons SD, Drewe JA, Gey van Pittius NC, Warren RM, van Helden PD. 2013. Novel cause of tuberculosis in meerkats, South Africa. Emerg. Infect. Dis. 19:2004–2007. 10.3201/eid1912.130268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Falkinham JO., III 2013. Ecology of nontuberculous mycobacteria—where do human infections come from? Semin. Respir. Crit. Care Med. 34:95–102. 10.1055/s-0033-1333568 [DOI] [PubMed] [Google Scholar]
- 169.Tortoli E. 2009. Clinical manifestations of nontuberculous mycobacteria infections. Clin. Microbiol. Infect. 15:906–910. 10.1111/j.1469-0691.2009.03014.x [DOI] [PubMed] [Google Scholar]
- 170.Leao SC, Tortoli E, Viana-Niero C, Ueki SY, Lima KV, Lopes ML, Yubero J, Menendez MC, Garcia MJ. 2009. Characterization of mycobacteria from a major Brazilian outbreak suggests a revision of the taxonomic status of members of the Mycobacterium chelonae-abscessus group. J. Clin. Microbiol. 47:2691–2698. 10.1128/JCM.00808-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Aitken ML, Limaye A, Pottinger P, Whimbey E, Goss CH, Tonelli MR, Cangelosi GA, Dirac MA, Olivier KN, Brown-Elliott BA, McNulty S, Wallace RJ., Jr 2012. Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am. J. Respir. Crit. Care Med. 185:231–232. 10.1164/ajrccm.185.2.231 [DOI] [PubMed] [Google Scholar]
- 172.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, Chimara E, Churchyard G, Cias R, Dasa R, Daley CL, Dekhuijzen PN, Domingo D, Drobniewski F, Esteban J, Fauville-Dufaux M, Folkvardsen DB, Gibbons N, Gomez-Mampaso E, Gonzalez R, Hoffmann H, Hsueh PR, Indra A, Jagielski T, Jamieson F, Jankovic M, Jong E, Keane J, Koh WJ, Lange B, Leao S, Macedo R, Mannsaker T, Marras TK, Maugein J, Milburn HJ, Mlinko T, Morcillo N, Morimoto K, Papaventsis D, Palenque E, Paez-Pena M, Piersimoni C, Polanova M, Rastogi N, Richter E, Ruiz-Serrano MJ, Silva A, da Silva MP, Simsek H, van Soolingen D, Szabo N, Thomson R, Fernandez MT, Tortoli E, Totten SE, Tyrrell G, Vasankari T, Villar M, Walkiewicz R, Winthrop K, Wagner D. 2013. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: a NTM-NET collaborative study. Eur. Respir. J. 42:1604–1613. 10.1183/09031936.00149212 [DOI] [PubMed] [Google Scholar]
- 173.Falkinham JO., III 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jing H, Wang H, Wang Y, Deng Y, Li X, Liu Z, Graviss EA, Ma X. 2012. Prevalence of nontuberculous mycobacteria infection, China, 2004–2009. Emerg. Infect. Dis. 18:527–528. 10.3201/eid1803.110175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wayne LG. 1988. International Committee on Systematic Bacteriology: report of the ad hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 268:433–434 [DOI] [PubMed] [Google Scholar]
- 176.Kirschner P, Böttger EC. 1992. Microheterogeneity within rRNA of Mycobacterium gordonae. J. Clin. Microbiol. 30:1049–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Turenne CY, Tschetter L, Wolfe J, Kabani A. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637–3648. 10.1128/JCM.39.10.3638-3648.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.van Ingen J, Bendien SA, de Lange WC, Hoefsloot W, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2009. Clinical relevance of non-tuberculous mycobacteria isolated in the Nijmegen-Arnhem region, The Netherlands. Thorax 64:502–506. 10.1136/thx.2008.110957 [DOI] [PubMed] [Google Scholar]
- 179.Shahraki AH, Çavuşoğlu C, Borroni E, Heidarieh P, Koksalan KO, Cabibbe AM, Hashemzadeh M, Mariottini A, Mostafavi E, Cittaro D, Feizabadi MM, Lazarevic D, Yaghmaei F, Molinari GL, Camaggi A, Tortoli E. Mycobacterium celeriflavum sp. nov., a scotochromogenic rapid grower isolated from clinical specimens. Int. J. Syst. Evol. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 180.Kim M, Oh HS, Park SC, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64:346–351. 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]