Abstract
Dimorphic transitions between yeast-like and filamentous forms occur in many fungi and are often associated with pathogenesis. One of the cues for such a dimorphic switch is the availability of nutrients. Under conditions of nitrogen limitation, fungal cells (such as those of Saccharomyces cerevisiae and Ustilago maydis) switch from budding to pseudohyphal or filamentous growth. Ammonium transporters (AMTs) are responsible for uptake and, in some cases, for sensing the availability of ammonium, a preferred nitrogen source. Homodimer and/or heterodimer formation may be required for regulating the activity of the AMTs. To investigate the potential interactions of Ump1 and Ump2, the AMTs of the maize pathogen U. maydis, we first used the split-ubiquitin system, followed by a modified split-YFP (yellow fluorescent protein) system, to validate the interactions in vivo. This analysis showed the formation of homo- and hetero-oligomers by Ump1 and Ump2. We also demonstrated the interaction of the high-affinity ammonium transporter, Ump2, with the Rho1 GTPase, a central protein in signaling, with roles in controlling polarized growth. This is the first demonstration in eukaryotes of the physical interaction in vivo of an ammonium transporter with the signaling protein Rho1. Moreover, the Ump proteins interact with Rho1 during the growth of cells in low ammonium concentrations, a condition required for the expression of the Umps. Based on these results and the genetic evidence for the interaction of Ump2 with both Rho1 and Rac1, another small GTPase, we propose a model for the role of these interactions in controlling filamentation, a fundamental aspect of development and pathogenesis in U. maydis.
INTRODUCTION
In many fungi, the transition from the yeast-like budding form to a filamentous form is important for growth under different conditions and, importantly, serves as an essential strategy associated with causing disease in hosts. The transition has been demonstrated to be under the control of the two main signal transduction pathways, the mitogen-activated protein kinase (MAPK) and cyclic AMP (cAMP)-dependent protein kinase A (PKA) pathways (1–6), and is often associated with nitrogen availability. Studies with the plant pathogens Colletotrichum lindemuthianum and Magnaporthe grisea have also shown reductions in pathogenicity and in the level of expression of the pathogenesis genes, respectively, due to mutations in genes controlling nitrogen metabolism (7, 8).
An important component linking fungal dimorphism to nitrogen availability is a subset of members of the ammonium transporter (AMT) family of membrane proteins. Such components are essential for sensing and transporting ammonium, a preferred nitrogen source in all organisms. Ammonium transporter proteins have been identified in the major kingdoms of living organisms; representatives are found in eubacteria, archaea, fungi, nematodes, insects, fish, plants, and even humans (9, 10). Like other transporters, ammonium transporters are important components of metabolic processes for cell growth and development.
Regulation of metabolic functions at the membranes is important, because the transporters contained there often help catalyze the first step in a pathway. Transporters are required for membrane stability and are often linked to the signal transduction components, which help the cells communicate with their environment. Multimerization of these membrane proteins might provide a mechanism for specific regulation and membrane structure stability (11, 12).
A variety of studies lend support to the idea that AMTs, too, function as multimers. Paralogous copies of the amt gene are found in the genomes of many organisms. Biochemical studies show that the AMTs in Escherichia coli form trimers, whereas both biochemical and genetic studies reveal that the different Saccharomyces cerevisiae Mep transporters could interact directly; moreover, the Mep2 monomer could interact with itself to form a trimeric complex (4, 13–15). Genetic studies similar to those done with S. cerevisiae, conducted with Aspergillus nidulans, Arabidopsis, and tomato, all indicate interaction among the ammonium transporters (16–18).
Ammonium transporters are also important in the life cycle of the biotrophic phytopathogenic fungus Ustilago maydis. In order to complete its life cycle, U. maydis, the pathogen of maize, requires a switch to dikaryotic hyphae after the successful mating of haploid yeast-like cells (19). In addition, yeast-like cells of U. maydis become elongated and/or filamentous when starved for nitrogen (20, 21). The relevance of nitrogen/nutrient acquisition and sensing for the pathogenicity of U. maydis and its control has been suggested in several studies (22–24). The U. maydis genome encodes two membrane-bound proteins associated with the transport of ammonium, characterized as high- and low-affinity ammonium transporters (20). The high-affinity ammonium transporter, Ump2, in addition to transporting ammonium, also plays a role in sensing the availability of low ammonium levels in the surrounding environment and is required for a filamentous response to low ammonium levels (20). Thus, it would take on the role of “transceptor” attributed to a subset of the AMT family (4, 6, 9, 25).
We were interested in determining the interacting partners of the U. maydis AMTs that would regulate or affect the functionality of these proteins. Our investigation of the U. maydis ammonium permeases provides evidence of homo- as well as heterodimer formation. In the present study, we also present in vivo support for interactions of Ump2 with the signaling protein Rho1, which belongs to the conserved family of Rho/Rac GTPase proteins. We had found preliminary evidence (26) of this interaction via yeast two-hybrid experiments to determine the interacting partners of the U. maydis Rho1 proteins; these experiments identified the high-affinity ammonium transporter Ump2 from a cDNA library. This interaction between Rho1 and Ump2 was also confirmed using yeast-based expression and coimmunoprecipitation (26). To explore this potentially novel mechanism of AMT interaction with a protein involved in cellular morphogenesis, we examined in vivo interactions in U. maydis, as well as genetic connections between the proteins. Our study confirms these findings and provides, along with genetic evidence, the first in vivo evidence for the physical interaction of ammonium transporters with a Rho1 homologue, an important signaling component in eukaryotes.
MATERIALS AND METHODS
Strains and growth conditions.
The yeast reporter strain utilized in this study is S. cerevisiae NMY32 [MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ ade2::(lexAop)8-ADE2 GAL4; a gift from J. Heitman, Duke University] provided in the DUALmembrane kit (Dualsystems Biotech, Zurich, Switzerland). The U. maydis strains used in this study are listed in Table 1. E. coli strains DH5α (Bethesda Research Laboratories) and TOP10 (Invitrogen, Carlsbad, CA) were employed for cloning and open reading frame (ORF) amplification requirements.
TABLE 1.
U. maydis strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| ½ WT | a1b1 | 40 |
| Δump2 a1 | a1b1 ump2::hygR | This study |
| rho1crg a1 | a1b1 rho1::Pcrg-rho1-cbxR | 26 |
| rho1crg Δump2 a1 | a1b1 rho1::Pcrg-rho1-cbxR ump2::hygR | This study |
| ½ pUM | a1b1/pUM | 26 |
| ½ pUM-Rho1 | a1b1/pUM-rho1 | 26 |
| ump2otef | a1b1 Potef-ump2-cbxR | This study |
| ump2otef pUM-Rho1 | a1b1 Potef-ump2-cbxR pUM Rho1 | This study |
| rac1otef | Potef-rac1-cbxR | 31 |
| Δump2 rac1otef | a1b1 ump2::hygR Potef-rac1-cbxR | This study |
| Δump2 rac1otef pCM-Ump2 | a1b1 ump2::hygR Potef-rac1-cbxR pCM-ump2-G418R | This study |
| rho1crg rac1otef a1 | a1b1 rho1::Pcrg-rho1-hygR Potef-rac1-cbxR | 26 |
U. maydis cells were grown at 25 to 28°C in YEPS medium (1% yeast extract, 2% each of peptone and sucrose, with or without 1.5% agar) and synthetic low-ammonium dextrose (SLAD) medium [0.17% yeast nitrogen base (YNB) without amino acids, 2% dextrose, 50 μM (NH4)2SO4] as described previously (20). In general, YP medium (1% yeast extract and 2% peptone) or synthetic low ammonium (SLA) medium (0.17% YNB and ammonium sulfate adjusted to a final concentration of 50 µM) were used, with additional supplementations as follows. The Pcrg1 promoter was induced by supplementation of the YP or SLA medium with 2% arabinose (i.e., YPA or SLAA) or was suppressed by supplementation with 2% dextrose (YPD or SLAD). For the split-ubiquitin assay, the yeast cells were grown on SD medium (0.17% YNB and 1× amino acid dropout solution) supplemented with dextrose. The yeast cells were kept at 30°C for 3 to 4 days. Strain NMY32 was maintained on YPAD plates (1% yeast extract, 2% peptone, 2% dextrose, 0.004% adenine sulfate, and 2% agar).
DUALmembrane screen for the split-ubiquitin assay.
The DUALmembrane screen was performed according to the DUALmembrane kit user manual (Dualsystems Biotech, Zurich, Switzerland). pCCW-STE, which is recommended for use with integral membrane proteins that lack a leader sequence, was used as the bait vector. For each known target protein, the respective cDNA was cloned between the two SfiI sites in the multiple cloning site, in frame with the upstream STE sequence (provided to help increase the expression of bait in yeast) and the downstream Cub-LexA-VP16 cassette. The pDL2xN-STE prey vector was chosen for the study, and each target protein cDNA was cloned between the two SfiI sites in the multiple cloning site. The bait was specifically selected to test for interaction with a particular prey in a directed split-ubiquitin assay. This method was utilized in order to specifically look at the interactions of U. maydis ammonium permeases with themselves and each other and also to confirm the interaction with Rho1 detected by Pham et al. (26). Specifically, ump1, ump2, and rho1 genes from U. maydis were each cloned into the bait and the prey vector at the SfiI sites following the cloning of the cDNA fragments into the pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA).
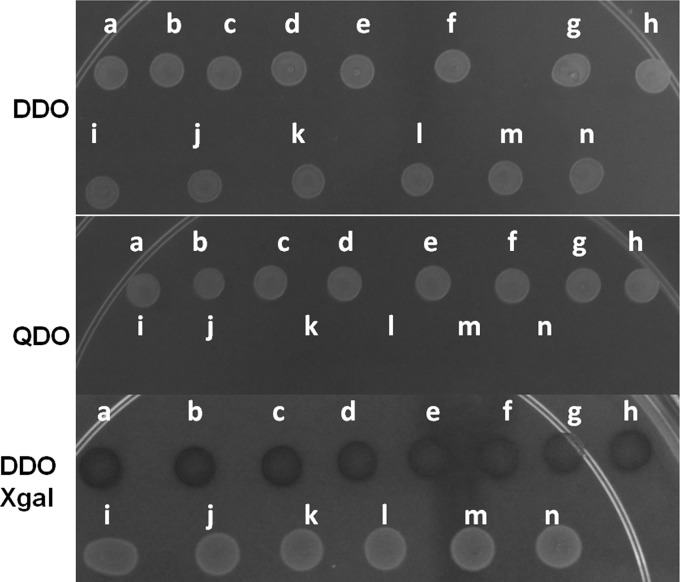
For the directed two-hybrid assay, bait and prey plasmids were introduced into S. cerevisiae strain NMY32 (Dualsystems Biotech) by cotransformation onto double-dropout (DDO) medium (SD without Trp and Leu). Growth was assessed for as long as 5 days on triple-dropout (TDO) medium (SD without Trp, Leu, and His) and quadruple-dropout (QDO) medium (SD without Trp, Leu, His, and Ade; supplemented with 25 mM 3-amino-1,2,4-triazole [3-AT]) to determine the strength of the interactions. Strains bearing each vector alone, both vectors without inserts (not shown), or, for example, the bait plasmid together with an empty prey plasmid, and vice versa, failed to yield growth on QDO plates (see Fig. 1, QDO, plates i to n). Additionally, strains bearing each gene in the bait vector and a “mock” prey, AI-Alg5, also failed to grow on QDO plates (not shown). The strength of the interaction was further assessed by spotting cells onto plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; 20 μg/ml) and disrupting cells on the plate in order to test for the expression of the β-galactosidase reporter that is dependent on interactions in this system. Yeast cells expressing β-galactosidase turned blue within hours, depending on the strength of the interaction.
FIG 1.
Interactions of U. maydis AMTs as determined by a split-ubiquitin assay. Cells (10 μl) of each strain were spotted onto a medium (DDO, QDO, or DDO with X-gal), and the plates were incubated for 24 h at 30°C. (a) Ump1 bait–Ump1 prey; (b) Ump2 bait–Ump2 prey; (c) Ump1 bait–Ump2 prey; (d) Ump2 bait–Ump1 prey; (e) Ump1 prey–Rho1 bait; (f) Ump2 prey–Rho1 bait; (g) Ump1 bait–Rho1 prey; (h) Ump2 bait–Rho1 prey; (i) Ump1 prey–bait vector alone; (j) Ump1 bait–prey vector alone; (k) Ump2 prey–bait vector alone; (l) Ump2 bait–prey vector alone; (m) Rho1 prey–bait vector alone; (n) Rho1 bait–prey vector alone. In each panel, the images are from samples grown on the same plate for the same amount of time. Interactions were measured by growth on QDO medium supplemented with 25 mM 3-AT. (DDO) Strains spotted onto DDO medium, which allows strains bearing both plasmids to grow. (QDO) Growth only of strains in which both plasmids express interacting proteins. (DDO Xgal) Interactions among ammonium transporters and between ammonium transporters and Rho1, observed by a color change indicating β-galactosidase expression (a through h). Yeast strain NMY32 containing a prey vector with an “empty” bait vector (or a bait vector with an “empty” prey vector) was able to grow on DDO medium, showing the presence of plasmids; in contrast, QDO medium did not allow the growth of any mock combinations.
Vector construction for the bimolecular fluorescence complementation (BiFC) assay.
Genetic manipulations involving switching the endogenous copy of the gene with the copy of the gene fused to yellow fluorescent protein (YFP), or the gene fused to the N terminus or C terminus of YFP in U. maydis, were accomplished through homologous recombination as described previously (27). The complete or partial YFP sequence was fused, following fusion PCR and cloning into the pCR 2.1 TOPO vector (Invitrogen), to one of the three genes of interest (ump1, ump2, and rho1). Another fragment, of approximately 1 kb, from downstream (down flank) of the gene of interest was amplified and cloned into pCR 2.1 TOPO. This down flank was digested and was cloned into the vector containing the gene of interest with the YFP fusion (up flank). The 3′ end of the up flank and the 5′ end of the down flank are complementary to the 5′ and 3′ regions of the carboxin and hygromycin resistance cassettes, respectively. The replacement constructs were amplified using high-fidelity Phusion DNA polymerase (Finnzymes, Lafayette, CO), purified, and used to transform U. maydis protoplasts (27). Transformants with the desired replacements were identified and confirmed by PCR and were verified by DNA sequencing in all cases.
The YFP plasmid obtained from the Fungal Genetics Stock Center (Kansas City, MO) was used in an earlier study conducted with Neurospora crassa (28). This version of YFP had not been codon optimized for U. maydis. Thus, to ensure expression of the YFP in U. maydis, the intact YFP was fused to the Ump1 or Ump2 protein, and its expression was detected in cells carrying the respective fusion protein when they were grown under low-ammonium conditions (see Fig. S1D and H in the supplemental material). No fluorescence was detected for these strains on rich medium (see Fig. S1C and G in the supplemental material). Additionally, the cells carrying only one-half of the YFP were scanned in order to detect any background florescence. In cases where we could detect background florescence, it was set at zero, and florescence above this was considered positive for interaction (see Fig. S2 in the supplemental material).
Microscopy.
Images of cells carrying the complete and partial YFP fusions were acquired using an Olympus FluoView FV1000 confocal scanner coupled to an Olympus IX81 inverted microscope, a PlanApo N 60× objective, and FV10-ASW software, version 2.1.
Imaging protocol.
A single-channel scanning configuration with line scanning was set up for the acquisition of complete or partial YFP-tagged proteins of interest, using the 488-nm line of an argon laser. A transmitted light image was acquired during scanning for visualization of the cell outline in the plane of scanning, by grouping of the transmitted detector with the argon laser. The optimal brightness setting for each channel was configured by determining the high-voltage (HV) setting that yielded maximal intensity without saturation. Each of the settings was tested against samples with cells that did not express tagged proteins in order to ensure the exclusion of nonspecific emission from cells.
RESULTS
Ump1 and Ump2 form homo- and heteromers.
The interaction between the two U. maydis ammonium transporters was first tested using a directed split-ubiquitin two-hybrid screen in the heterologous system in which the screen was developed, S. cerevisiae. Unlike the traditional yeast two-hybrid assay, this modified assay tests for interactions among membrane proteins. The split-ubiquitin assay showed that Ump1 interacts with itself, as does Ump2. This assay also confirmed the hypothesis that the two ammonium transporters in U. maydis would form hetero-oligomers. All three interactions tested were strong, as indicated by growth on the most stringent medium, QDO medium, and also by the activation of transcription of the lacZ reporter gene (Fig. 1, bottom). The interaction between Ump1 and Ump2 was also confirmed by switching the bait and prey for the respective protein (Fig. 1). Control experiments using vectors alone, or using one of the genes of interest expressed from the bait or prey vector and cotransformation with the corresponding empty prey or bait vector, respectively, all gave negative results under these conditions (Fig. 1, QDO and DDO Xgal).
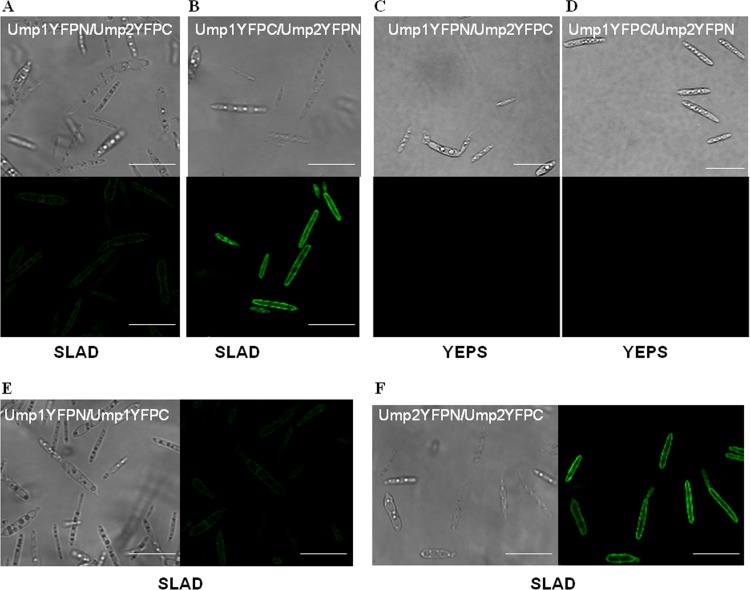
To examine the interactions of these proteins in vivo in U. maydis, we used the bimolecular fluorescence complementation (BiFC) assay. In this assay, the Ump1 and Ump2 proteins were fused to the N and C termini of YFP, respectively, and cells carrying the fusion protein were examined for fluorescence under both nutrient-rich and low-ammonium conditions. The N and C termini of YFP were swapped between Ump1 and Ump2 to ensure that the fluorescence observed was consistent with a true interaction (Fig. 2A and B). Fluorescence from the reconstituted YFP could be detected only when cells carrying the fusion protein were grown under low-ammonium conditions, not under nutrient-rich conditions (Fig. 2C and D). The fluorescence confirmed the localization of the ammonium transporters to the membrane, also suggesting that fusion of the Ump proteins with YFP did not interfere with their localization. Finally, the homomerization by Ump1 and by Ump2 that was observed by the split-ubiquitin assay was confirmed by the BiFC assay (Fig. 2E and F). The cells carrying the fusion proteins also grew like wild-type cells, filamentously, on SLAD agar (i.e., on agar under low-ammonium conditions), signifying no loss of function as a result of the fusion.
FIG 2.
Interaction between Ump1 and Ump2 under low-ammonium conditions. (A and B) Cells expressing Ump1 fused to the N terminus of YFP and Ump2 fused to the C terminus of YFP (A) or Ump1 fused to the C terminus of YFP and Ump2 fused to the N terminus of YFP (B) were grown for 24 h in liquid SLAD medium. (Top) Differential interference contrast images; (bottom) fluorescent images. (C and D) No fluorescence was detected for growth of the cells shown in panels A and B, respectively, on rich YEPS medium. (E and F) Differential interference contrast (left) and fluorescent (right) images of cells in which both the bait and the prey were either Ump1 (E) or Ump2 (F), grown on SLAD medium. Bars, 20 μm. For fluorescence results (and differential interference contrast images) for U. maydis strains in the ½ WT genetic background (40) expressing Ump1 or Ump2 fused to the N or C terminus of YFP, grown in SLAD medium, see Fig. S2 in the supplemental material (note the absence of fluorescence in each case).
Ump1 and Ump2 interact with Rho1 GTPase protein.
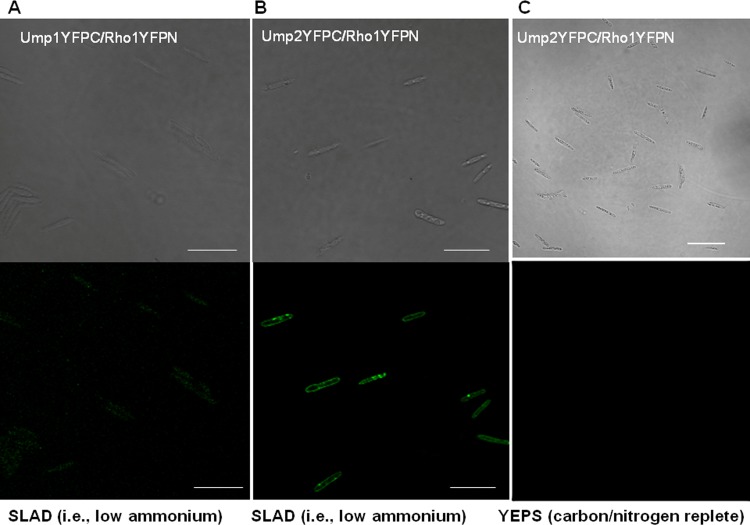
The putative interaction between Ump2 and Rho1 was first identified by yeast two-hybrid screens for interacting partners of Rho1 (26). A directed split-ubiquitin assay was used to confirm this interaction and also to test the interaction of the low-affinity ammonium transporter, Ump1, with Rho1. The interaction of Ump1 or Ump2 with Rho1 was indicated by the growth of yeast cells expressing these proteins on stringent QDO medium, and the strength of the interaction was also confirmed by β-galactosidase activity (Fig. 1, bottom). To assess this interaction in the more biologically relevant context of U. maydis, we used BiFC. When U. maydis was tested in vivo for interaction between Rho1 and Ump1 or Ump2, fluorescence due to reconstitution of the YFP was detected only for cells grown under low-ammonium conditions (Fig. 3A and B), likely due to the very low levels of transcription for either transporter under these conditions (29) (see Fig. S1C and G in the supplemental material). Fluorescence resulting from an interaction between Ump2 and Rho1 was detected at the membrane, whereas fluorescence was mostly dispersed throughout the cell when Ump1 and Rho1 were tested for interaction (Fig. 3A). This suggests that although Ump1 and Rho1 appear to interact under these conditions, the resulting complex is rapidly moved to the cytoplasm for degradation in vacuoles or by the proteosome. No interaction was observable in rich medium (Fig. 3C), again likely because the low levels of expression of Ump1 and Ump2 under such conditions prevented any physical interactions, if present, from being visualized (29). On the other hand, the lack of fluorescence in negative-control cells bearing fusions of the respective proteins with only half-portions of YFP (see Fig. S2 in the supplemental material) reduces the likelihood that the interactions we observed are false-positive results.
FIG 3.
Interaction between Ump1 or Ump2 and Rho1. (A and B) Cells expressing Rho1 fused to the N terminus of YFP and either Ump1 (A) or Ump2 (B) fused to the C terminus of YFP were grown for 24 h in liquid SLAD medium. (Top) Differential interference contrast; (bottom) fluorescence. (C) Differential interference contrast (top) and fluorescence (bottom) images for growth of cells expressing Ump2 and Rho1 on rich YEPS medium. No fluorescence was detected on YEPS medium. Testing of Ump1 and Rho1 on YEPS medium yielded similar results (not shown). Bars, 20 μm. For fluorescence results for U. maydis strains in the ½ WT genetic background (40) expressing Rho1 fused to the N or C terminus of YFP, grown in SLAD medium, see Fig. S2 in the supplemental material (note the absence of fluorescence in each case).
Evidence of genetic interaction between rho1 and ump2.
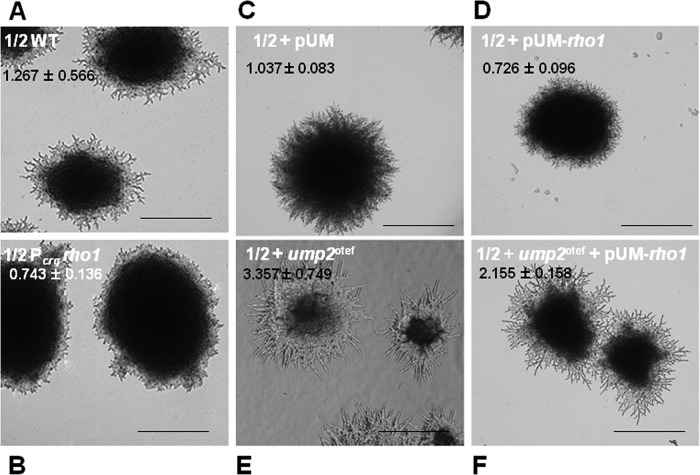
Since BiFC established that Ump2 and Rho1 interact physically under low-ammonium conditions, we wanted to explore further the implications of such interactions at the genetic level. On solid low-ammonium media (e.g., SLAD), wild-type U. maydis cells exhibit extensive filamentation after 3 to 4 days at room temperature (Fig. 4A). We next examined the effects of Rho1 overexpression on filamentation. In one strain, rho1 was placed under the control of the crg1 promoter. In this strain, the expression level of rho1 depends on the growth condition, specifically the carbon source used for growing the cells (i.e., rho1 is induced and overexpressed with arabinose and is repressed with glucose [19, 30]). Additionally, rho1 was placed under the control of the strong constitutive gapd promoter in a plasmid-borne construct (26). In either case, rho1 overexpression led to levels of filamentation on SLAD medium lower than those for the wild type (compare Fig. 4A with 4B and Fig. 4C with Fig. 4D).
FIG 4.
Overexpression of rho1 reduces filamentation due to low-ammonium conditions. (A and B) Haploid cells were grown on SLAA medium, and colony morphology was observed after 4 days. (A) Wild-type strain; (B) strain overexpressing rho1 under the control of the crg1 promoter. (C through E) Additional strains were grown on SLAD medium, and colony morphology was observed after 4 days. (C) Wild type with empty vector (pUM); (D) rho1 overexpression vector (pUM-rho1) with rho1 under the control of the constitutive gapd promoter; (E) ump2 overexpression vector; (F) rho1 overexpression vector (pUM-rho1) in an ump2 overexpression background. Magnification, ×4. Colony edges for the strains depicted in panels C through F are shown at ×10 magnification in Fig. S4 in the supplemental material. For each strain, the ratio of the filamentation area, extended from the edge of a colony, to the colony area is given as the mean ± standard deviation for a minimum of 12 independent measurements.
Overexpression of Ump2 leads to a hyperfilamentous response on low-ammonium medium (Fig. 4E and 5) but leads to filamentation even on rich medium (Fig. 5). As was seen in wild-type cells, overexpression of Rho1 in cells in which Ump2 was concomitantly overexpressed reduced filamentation (Fig. 4F). An alternative way of looking at these results is to compare cells overexpressing rho1 (expressed from a plasmid) with cells overexpressing both rho1 and ump2: overexpression of ump2 improved the filamentation ability of the rho1-overexpressing cells under low-ammonium conditions.
FIG 5.
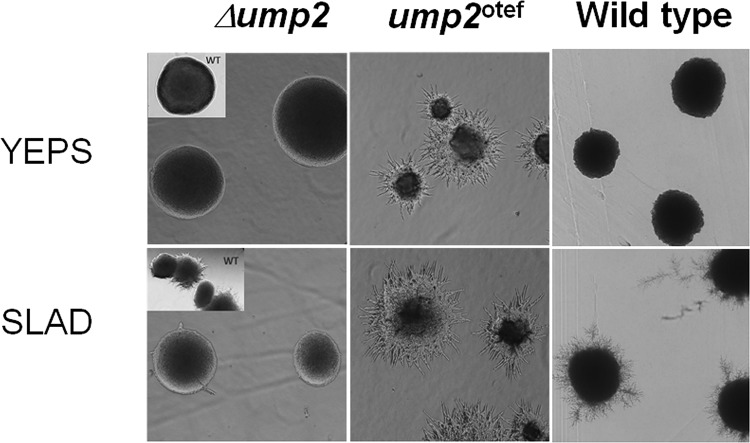
Wild-type haploid cells or haploid cells with ump2 disruption (Δump2) or ump2 overexpression (ump2otef) were grown either on YEPS medium (yeast extract-peptone-dextrose; a carbon/nitrogen-replete medium) or on SLAD (synthetic low-ammonium dextrose) medium for 4 h. Haploid cells overexpressing ump2 showed filamentous growth, even in nitrogen- and carbon-replete medium. Wild-type cells (insets and right panels) were grown under the same conditions as the mutants.
Interestingly, it has been shown that overexpression of another small G protein, Rac1, a “master controller” of polarized cell growth (31), leads to constitutive filamentation on rich medium (see Fig. S3A in the supplemental material) or to hyperfilamentation on SLAD medium (Fig. 6C). Furthermore, since Rho1 acts upstream of Rac1 (GTPase) (26), we tested a model (see Fig. 7) whereby the interaction between Ump2 and Rho1 could sequester the latter protein, interfering with its role in downregulating Rac1. We did this by examining the effect of changes in either rho1 or ump2 expression on the rac1 overexpression phenotype. As we saw above with both wild-type cells and cells overexpressing Ump2, overexpression of Rho1 reduced filamentation associated with Rac1 overexpression on rich medium (see Fig. S3B in the supplemental material) or on a low-ammonium agar (SLAA) (compare Fig. S3C and D in the supplemental material).
FIG 6.
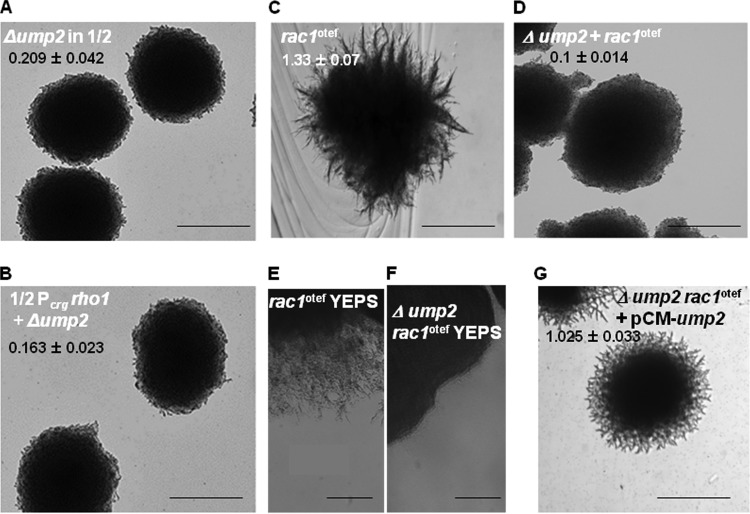
Deletion of ump2 eliminates filamentation when either rho1 or rac1 is overexpressed. (A and B) Haploid cells with ump2 deleted in the ½ WT (A) or Pcrg rho1 (B) background were grown on SLAA, and colony morphology was observed after 4 days. While rho1 overexpression strains exhibit reduced filamentation, ump2 deletion completely eliminates filamentation in all backgrounds examined. (C through F) Strains with rac1 overexpression (C and E) or ump2 deletion in the rac1 overexpression background (D and F) were grown on SLAD (C and D) or YEPS (E and F) medium, and colony morphology was observed after 4 days. ump2 deletion abolishes the filamentation of the rac1 overexpression strains. (G) The rac1 overexpression strain with the ump2 deletion, complemented with a plasmid-borne ump2 gene, displayed an intermediate level of filamentation on SLAD medium, comparable to that seen in the ½ WT strain alone (Fig. 4A) or with an empty vector (Fig. 4C). Magnification, ×4. Colony edges for the strains depicted in panels C, D, and G are shown at ×10 magnification in Fig. S4 in the supplemental material. For each strain, the ratio of the filamentation area, extended from the edge of a colony, to the colony area is given as the mean ± standard deviation for a minimum of 12 independent measurements.
FIG 7.
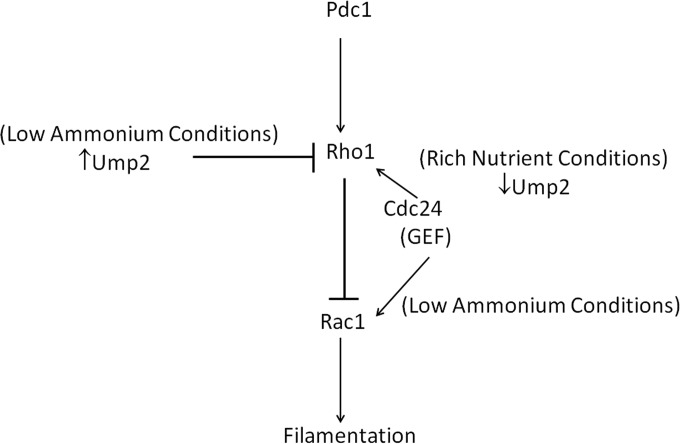
Model for Ump2 interaction with Rho1 signaling. Previous work provided evidence for the role of Rho1 in negatively affecting Rac1 and thereby blocking filamentation (26). Conditions of ammonium limitation are sensed by Ump2, which relays the signal to downstream effectors to help decide the fate of the cells under such nutrient limiting conditions. Under such conditions, when Ump2 is expressed, it interacts with Rho1 and sequesters it away from its competition with Rac1, thereby allowing/causing filamentation. Specifically, since Cdc24 is the GEF for both Rho1 and Rac1, the model posits that Ump2 sequestration of Rho1 allows Cdc24 to activate Rac1 preferentially under conditions of ammonium limitation.
Ump2 is required for filamentation on low-ammonium medium, independently of Rho1 or Rac1 expression levels.
Since all three proteins (Ump2, Rho1, and Rac1) appear to affect filamentation as a response to ammonium availability, we wanted to see whether Ump2 expression would be a prerequisite for the filamentation associated with Rac1 expression. Otherwise wild-type cells in which the ump2 gene is deleted are unable to form filaments on SLAD medium (20). In the current studies, the ump2 gene was also deleted from haploid U. maydis strains where rho1 was overexpressed. Like otherwise wild-type cells, cells in which ump2 was deleted and rho1 was overexpressed failed to display filamentation on low-ammonium medium. This suggests either epistatic or pleiotropic effects of the ump2 deletion on filamentation on solid low-ammonium medium. Similarly, as with the wild-type and Rho1 overexpression strains in which ump2 was deleted (Fig. 6A and B), deletion of ump2 abolished the filamentation associated with rac1 overexpression (compare Fig. 6C and D). This effect was also apparent on rich medium, where deletion of ump2 abolished the filamentation associated with rac1 overexpression (compare Fig. 6E and F) (also, see reference 31 for rac1 overexpression on rich media). As a control, the strain complemented with ump2 on a plasmid displayed filamentation (Fig. 6G) similar to that of the original Rac1 overexpression strain. These results, along with previous observations (26), indicate that the ability of Rac1 to affect filamentation may be controlled by the interaction of Rho1 with Ump2.
DISCUSSION
Two lines of evidence suggest interaction between and among the two ammonium transporters identified in U. maydis. First, the split-ubiquitin assay suggested the interaction between the two ammonium transporters: it demonstrated that Ump1 and Ump2 each interact with themselves to form homo-oligomers and also that they interact with each other to form hetero-oligomers. These findings are consistent with a large number of studies suggesting physical and genetic interactions among AMTs and between AMTs and highly conserved signaling pathways (4–6, 9, 14, 17, 18, 25, 32). For example, support for interactions between S. cerevisiae Mep proteins comes from trans-dominant point mutants of Mep1 or Mep3 that interfere with the function of the wild-type versions of the respective putative partners (4, 6). Similarly, homo-oligomerization of AMTs is suggested by the fact that coexpression of a mutant and a wild-type form of LeAMT1;1 from tomato in Xenopus oocytes inhibits ammonium transport in a dominant negative fashion; this interaction was further confirmed by the split-ubiquitin system (17). Moreover, several AMTs have now been crystallized (33–35), and these studies have provided convincing evidence for the trimeric organization of these members of the AMT family (33–35). Similarly, the role of heteromerization of AMTs in Arabidopsis (36) was recently shown, in that oligomerization of plant AMTs in the heterologous yeast system is critical for allosteric regulation of transport activity, in which the conserved cytosolic C terminus functions as a trans-activator. Further, the Arabidopsis thaliana transporters examined, AMT1;1 and AMT1;3, form functional homo- and heterotrimers in yeast and plant roots; additionally, AMT1;3 carrying a phosphomimetic residue in its C terminus regulated both homo- and heterotrimers in a dominant negative fashion in vivo. In the current study, the physical interaction between Ump1 and Ump2 was validated in vivo in U. maydis by using the bimolecular fluorescence assay. Expression data obtained via transcriptome sequencing (RNA-Seq) (M. Perlin, unpublished data) suggest that ump2 is approximately 10-fold more highly transcribed under low-ammonium conditions than is ump1. This suggests that the most of the Ump2 protein may be more involved in homomerization than in heteromer formation with Ump1. Although the functional importance of the interaction between Ump1 and Ump2 is not evident at present, the interaction is not essential for the transport of ammonium across the membrane in U. maydis, since mutants missing either protein grow on low-ammonium media (20); in addition, there is no obvious phenotype for loss of the Ump1 protein alone. No evidence of allosteric regulation between the two ammonium transporters has yet been identified in U. maydis, in contrast to what has been observed in other fungi, such as S. cerevisiae and A. nidulans (14, 18) and, more recently, Arabidopsis thaliana (36). However, this does not rule out the possibility that such interactions provide a mechanism for regulating the responses of these proteins and the cell to the changing availability of external ammonium.
Additional genetic evidence in S. cerevisiae and Candida albicans points to interaction between the putative transceptor proteins (i.e., the respective Mep2 proteins) and specific signaling pathways (3, 4, 32). Overexpression of Mep2 in S. cerevisiae induces a transcriptional profile consistent with activation of the MAPK pathway, and epistasis analysis further supports the linkage of Mep2 to the MAPK pathway in pseudohyphal growth (4). In C. albicans, Mep2 activates the MAPK pathway and, in a Ras1-dependent fashion, the cAMP-dependent PKA pathway (3). The split-ubiquitin assay has also been a valuable tool in assessing membrane protein interactions. A recent study (25) supported the effectiveness of this technique in identifying proteins interacting with membrane proteins. In particular, the interacting partners for yeast Mep2 and Gap1, transceptors involved in both transport and signaling functions, were identified (25).
Our study confirms and provides the first evidence of a physical interaction in vivo between an ammonium transporter and the signaling protein Rho1. In U. maydis, Rho1 is the only member of its family of proteins required for cell viability. Rho1 is involved in controlling cytokinesis and cell polarity. Although overexpression of Rho1 led to reduced filament production in U. maydis cells, cells depleted of Rho1 showed nonpolarized growth followed by cell death, suggesting a role for Rho1 in the dimorphic transition (26). Pham et al. (26) have shown that the lethality of the rho1 deletion strain can be rescued when another gene, rac1, is deleted in this background. Rac1 is another GTPase protein and has been shown to be the master regulator controlling filamentation in U. maydis. A constitutively active allele of Rac1 causes isotropic delocalized cell extension and, eventually, cell death (31). Epistasis analyses suggest that Rho1 acts upstream and negatively regulates Rac1 to control cytokinesis and cell polarity. According to the model suggested by Pham et al. (26), Rho1 could be interfering with the localization of Rac1 at the polar tips, thereby preventing polar growth. Alternatively, it could be sequestering the effector of Rac1, Cla4 (a p21-activated kinase), or utilizing the common guanine-nucleotide exchange factor (GEF) of Rho1 and Rac1 (Cdc24), to prevent polar growth.
We demonstrate here the interaction of Rho1 with the ammonium transporters Ump1 and Ump2. The interaction of Rho1 with Ump2 is clear and appears to be localized to the cytoplasmic membrane (Fig. 3B). On the other hand, unlike wild-type Ump1-YFP, the interaction of Ump1 with Rho1 does not localize to the membrane, and what we see (Fig. 3A) may reflect an interaction that is rapidly removed and sent to the proteasome. It is known that an interaction between N-terminal green fluorescent protein (GFP) and C-terminal GFP in the BiFC assay can stabilize otherwise transient protein interactions (37). This might explain the different localizations of Ump1-YFP in the wild type (see Fig. S1 in the supplemental material) and Ump1 fused to the C terminus of YFP in the strain expressing Rho1 fused to the N terminus of YFP and may indicate that only a small proportion of Ump1 molecules interact with Rho1 and then move into the cytoplasm. In any case, the Ump1-Rho1 interaction is unexpected, and its role remains unresolved.
While we do not have direct evidence, we suspect that, at least for Ump2, the interaction with Rho1 is likely relevant for signaling and may be occurring at the C terminus. Mutations in the C-terminal portions of putative transceptors have been able to separate the transporter from the signaling portions (3, 4, 38). The Ras1-dependent activation of the PKA pathway in C. albicans depends on the cytoplasmic C-terminal tail of Mep2 (3); moreover, a G349C mutation in the protein, also in its C-terminal portion, renders it independent of Npr1 kinase for ammonium transport (38). The interactions we observed (Fig. 3) occur under low-ammonium conditions, as evidenced by the detection of fluorescence only under ammonium-limiting conditions (compare Fig. 3B and C), although we recognize the possibility that the expression of Ump1 and Ump2 under nutrient-rich conditions is so low (29) as to limit our ability to detect such interactions under these conditions. In our model (Fig. 7), we hypothesize that this interaction would be functionally relevant because the interaction of Rho1 with the ammonium transporters under low-ammonium conditions could inhibit Rho1 from negatively regulating Rac1, thereby causing filamentation under low-nutrient conditions. When ump2 was deleted, the filamentation associated with low-ammonium conditions was completely lacking (Fig. 6A, B, and D), regardless of the background examined. This is likely because, absent Ump2, Rho1 is completely free to block Rac1, probably by competing for its GEF, Cdc24 (39). Of course, this could also be due to some other pleiotropic effect of deletion of the ump2 gene. The reduced filamentation observed when Rho1 is overexpressed (Fig. 4B and D) could be due to the altered stoichiometry of the Rho1 protein; in this case, interaction with Ump2 is not adequate to avoid inhibition of Rac1, thereby reducing filamentation. Overexpression of Ump2 at least partially alleviated this phenotype (Fig. 4F). This is consistent with a better balance of stoichiometry of these two proteins, relieving the negative effect of Rho1 on Rac1. Furthermore, although the expression levels of Rho1 under nutrient-rich and nutrient-limiting conditions are similar (29), the expression of ammonium transporters is much reduced under nutrient-rich conditions (29) (see also Fig. S1 in the supplemental material), and thus, we cannot detect interaction of Umps with Rho1. Under such conditions, Rho1 is also likely free and inhibits the activity of Rac1, thereby blocking filamentation. Interestingly, this could also explain the filamentation phenotype observed under nutrient-rich conditions when Ump2 is overexpressed (Fig. 5). Under these conditions, Ump2 would scavenge all available Rho1, leaving the Cdc24 pool high, thus relieving the normal repression on Rac1, resulting in filamentation. In fact, the filamentation associated with Rac1 overexpression is abolished in the absence of Ump2 (Fig. 6D). Consistent with all these results is a model where interaction of Rho1 with Ump2 sequesters Rho1, allowing Cdc24 to activate Rac1, thereby positively affecting filamentation. In situations where ump2 is not expressed, i.e., growth under nutrient-rich conditions or deletion of ump2, Rho1 would be free to interact with Cdc24, thereby blocking the activation of Rac1 and suppressing filamentous growth. To summarize, we propose that Rho1 interacts with the ammonium transporter protein Ump2 and coordinates with Rac1 to block filaments under conditions of abundant ammonium, suggesting that ammonium transporter proteins add a fundamental layer to this signaling pathway to affect downstream effectors and control of the filamentous growth response.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joe Heitman and Yen-Ping Hsueh for help with the split-ubiquitin assay and especially the former for insightful suggestions regarding the manuscript. We also thank the reviewers for the journal, who provided constructive criticisms that improved the quality of the final version. The technical assistance of Seth Adams, Eleanor Castro, Swathi Kuppireddy, Margaret Wallen, Victoria Mosely, and Madison Furnish is also appreciated.
This work was supported, in part, by IRIG-RIG grants to M.H.P. from the Office of the Vice President for Research at the University of Louisville.
J.A.P. and M.H.P. conceived and designed the experiments. J.A.P., M.B., M.C., and M.H.P. performed the experiments. J.A.P. and M.H.P. analyzed the data and wrote the paper.
Footnotes
Published ahead of print 15 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00150-14.
REFERENCES
- 1.Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746–785. 10.1128/MMBR.64.4.746-785.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan X, Heitman J. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas K, Morschhäuser J. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56:649–669. 10.1111/j.1365-2958.2005.04576.x [DOI] [PubMed] [Google Scholar]
- 4.Rutherford JC, Chua G, Hughes T, Cardenas ME, Heitman J. 2008. A Mep2-dependent transcriptional profile links permease function to gene expression during pseudohyphal growth in Saccharomyces cerevisiae. Mol. Biol. Cell 19:3028–3039. 10.1091/mbc.E08-01-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorenz MC, Heitman J. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236–1247. 10.1093/emboj/17.5.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorenz MC, Heitman J. 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150:1443–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau G, Hamer JE. 1996. Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea. Plant Cell 8:771–781. 10.1105/tpc.8.5.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellier A-L, Laugé R, Veneault-Fourrey C, Langin T. 2003. CLNR1, the AREA/NIT2-like global nitrogen regulator of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Mol. Microbiol. 48:639–655. 10.1046/j.1365-2958.2003.03451.x [DOI] [PubMed] [Google Scholar]
- 9.Wirén N, Merrick M. 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants, p 95–120 In Boles E, Kramer R. (ed), Molecular mechanisms controlling transmembrane transport. Springer, Berlin, Germany [Google Scholar]
- 10.Marini AM, Boeckstaens M, André B. 2006. From yeast ammonium transporters to Rhesus proteins, isolation and functional characterization. Transfus. Clin. Biol. 13:95–96. 10.1016/j.tracli.2006.03.002 [DOI] [PubMed] [Google Scholar]
- 11.Veenhoff LM, Heuberger EHML, Poolman B. 2002. Quaternary structure and function of transport proteins. Trends Biochem. Sci. 27:242–249. 10.1016/S0968-0004(02)02077-7 [DOI] [PubMed] [Google Scholar]
- 12.Grasberger B, Minton AP, DeLisi C, Metzger H. 1986. Interaction between proteins localized in membranes. Proc. Natl. Acad. Sci. U. S. A. 83:6258–6262. 10.1073/pnas.83.17.6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marini AM, Soussi-Boudekou S, Vissers S, André B. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marini AM, Springael JY, Frommer WB, André B. 2000. Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol. Microbiol. 35:378–385. 10.1046/j.1365-2958.2000.01704.x [DOI] [PubMed] [Google Scholar]
- 15.Blakey D, Leech A, Thomas GH, Coutts G, Findlay K, Merrick M. 2002. Purification of the Escherichia coli ammonium transporter AmtB reveals a trimeric stoichiometry. Biochem. J. 364(Part 2):527–535. 10.1042/BJ20011761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loqué D, Lalonde S, Looger LL, von Wirén N, Frommer WB. 2007. A cytosolic trans-activation domain essential for ammonium uptake. Nature 446:195–198. 10.1038/nature05579 [DOI] [PubMed] [Google Scholar]
- 17.Ludewig U, Wilken S, Wu B, Jost W, Obrdlik P, El Bakkoury M, Marini AM, André B, Hamacher T, Boles E, von Wirén N, Frommer WB. 2003. Homo- and hetero-oligomerization of ammonium transporter-1 uniporters. J. Biol. Chem. 278:45603–45610. 10.1074/jbc.M307424200 [DOI] [PubMed] [Google Scholar]
- 18.Monahan BJ, Unkles SE, Tsing IT, Kinghorn JR, Hynes MJ, Davis MA. 2002. Mutation and functional analysis of the Aspergillus nidulans ammonium permease MeaA and evidence for interaction with itself and MepA. Fungal Genet. Biol. 36:35–46. 10.1016/S1087-1845(02)00004-X [DOI] [PubMed] [Google Scholar]
- 19.Brachmann A, Weinzierl G, Kämper J, Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047–1063. 10.1046/j.1365-2958.2001.02699.x [DOI] [PubMed] [Google Scholar]
- 20.Smith DG, Garcia-Pedrajas MD, Gold SE, Perlin MH. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50:259–275. 10.1046/j.1365-2958.2003.03680.x [DOI] [PubMed] [Google Scholar]
- 21.Lovely CB, Perlin MH. 2011. Cla4, but not Rac1, regulates the filamentous response of Ustilago maydis to low ammonium conditions. Commun. Integr. Biol. 4:670–673. 10.4161/cib.17063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho E, Cahill M, Saville B. 2007. Gene discovery and transcript analyses in the corn smut pathogen Ustilago maydis: expressed sequence tag and genome sequence comparison. BMC Genomics 8:334. 10.1186/1471-2164-8-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horst RJ, Doehlemann G, Wahl R, Hofmann J, Schmiedl A, Kahmann R, Kämper J, Voll LM. 2010. Ustilago maydis infection strongly alters organic nitrogen allocation in maize and stimulates productivity of systemic source leaves. Plant Physiol. 152:293–308. 10.1104/pp.109.147702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aydogdu M, Boyraz N. 2011. Effects of nitrogen and organic fertilization on corn smut (Ustilago maydis (DC) Corda.). Afr. J. Agric. Res. 6:4539–4543 [Google Scholar]
- 25.Van Zeebroeck G, Kimpe M, Vandormael P, Thevelein JM. 2011. A split-ubiquitin two-hybrid screen for proteins physically interacting with the yeast amino acid transceptor Gap1 and ammonium transceptor Mep2. PLoS One 6:e24275. 10.1371/journal.pone.0024275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham CD, Yu Z, Sandrock B, Bölker M, Gold SE, Perlin MH. 2009. Ustilago maydis Rho1 and 14-3-3 homologues participate in pathways controlling cell separation and cell polarity. Eukaryot. Cell 8:977–989. 10.1128/EC.00009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brachmann A, König J, Julius C, Feldbrügge M. 2004. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272:216–226. 10.1007/s00438-004-1047-z [DOI] [PubMed] [Google Scholar]
- 28.Bardiya N, Alexander WG, Perdue TD, Barry EG, Metzenberg RL, Pukkila PJ, Shiu PK. 2008. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178:593–596. 10.1534/genetics.107.079384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul JA. 2011. Regulation of morphogenesis by ammonium transporters in Ustilago maydis. Ph.D. dissertation University of Louisville, Louisville, KY [Google Scholar]
- 30.Bottin A, Kämper J, Kahmann R. 1996. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 253:342–352. 10.1007/PL00008601 [DOI] [PubMed] [Google Scholar]
- 31.Mahlert M, Leveleki L, Hlubek A, Sandrock B, Bölker M. 2006. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 59:567–578. 10.1111/j.1365-2958.2005.04952.x [DOI] [PubMed] [Google Scholar]
- 32.Morschhäuser J. 2011. Nitrogen regulation of morphogenesis and protease secretion in Candida albicans. Int. J. Med. Microbiol. 301:390–394. 10.1016/j.ijmm.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 33.Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li X-D. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 101:17090–17095. 10.1073/pnas.0406475101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khademi S, O'Connell J, III, Remis J, Robles-Colmenares Y, Miercke LJW, Stroud RM. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305:1587–1594. 10.1126/science.1101952 [DOI] [PubMed] [Google Scholar]
- 35.Andrade SLA, Dickmanns A, Ficner R, Einsle O. 2005. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U. S. A. 102:14994–14999. 10.1073/pnas.0506254102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan L, Gu R, Xuan Y, Smith-Valle E, Loqué D, Frommer WB, von Wirén N. 2013. Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell 25:974–984. 10.1105/tpc.112.108027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. 2005. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127:146–157. 10.1021/ja046699g [DOI] [PubMed] [Google Scholar]
- 38.Neuhäuser B, Dunkel N, Satheesh SV, Morschhäuser J. 2011. Role of the Npr1 kinase in ammonium transport and signaling by the ammonium permease Mep2 in Candida albicans. Eukaryot. Cell 10:332–342. 10.1128/EC.00293-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frieser SH, Hlubek A, Sandrock B, Bölker M. 2011. Cla4 kinase triggers destruction of the Rac1-GEF Cdc24 during polarized growth in Ustilago maydis. Mol. Biol. Cell 22:3253–3262. 10.1091/mbc.E11-04-0314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gold SE, Brogdon SM, Mayorga ME, Kronstad JW. 1997. The Ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell 9:1585–1594. 10.1105/tpc.9.9.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.