Abstract
The G protein α subunits Gpa1, Gpa2, and Gpa3 mediate signal transduction and are important in the growth and virulence of Cryptococcus neoformans. To understand how Gpa1 functions without a conventional Gβ subunit, we characterized a resistance to inhibitors of cholinesterase 8 (Ric8) homolog from C. neoformans, which shares amino acid sequence homology with other Ric8 proteins that exhibit guanine nucleotide exchange factor (GEF) activity toward Gα. We found that the ric8 mutant was reduced in capsule size and melanin formation, which could be suppressed by cyclic AMP (cAMP) supplementation or by introducing the activated GPA1Q284L allele. Consistent with the fact that Ric8 participates in cAMP signaling to regulate virulence, the ric8 mutant was attenuated in virulence toward mice. Interestingly, disruption of RIC8 also resulted in opposing effects on pheromone signaling, as the ric8 mutant showed reduced mating but an enhanced ability to induce the pheromone response in the mating partner. To identify Ric8 functional mechanisms, we examined the interactions between Ric8 and the three Gα proteins. Ric8 interacted with Gpa1 and Gpa2, but not Gpa3. The presence of Gpa1Q284L negatively affected its interaction with Ric8, whereas the activated Gpa2Q203L allele abolished the interaction. Collectively, these findings suggest that Ric8 functions as a GEF to facilitate the activation of Gpa1-cAMP signaling and to promote Gpa2, affecting mating efficiency. Our study highlights the distinct and conserved characteristics associated with G protein signaling and contributes to our overall understanding of how G protein α subunits function with or without a canonical Gβ partner in C. neoformans.
INTRODUCTION
Cryptococcus neoformans is an opportunistic fungal pathogen of global importance, because cryptococcosis annually afflicts nearly 1 million people, resulting in approximately 600,000 deaths worldwide (1). C. neoformans is normally soilborne, and desiccated yeast cells or spores produced by sexual reproduction are thought to be the main propagules of infection through pulmonary inhalation (2, 3). Although many of the disease mechanisms remain obscure, the abilities of C. neoformans to resist harsh environmental conditions, grow at body temperature, and produce the polysaccharide capsule and the melanin pigment are thought to be the main contributors to virulence (4, 5).
Signal transduction pathways play critical roles in the adaptation of fungi such as C. neoformans to different living conditions, including those in a mammalian host, and proliferation. Canonical heterotrimeric GTP-binding proteins, consisting of Gα, Gβ, and Gγ, regulate a myriad of cellular functions, such as growth, sexual and asexual reproduction, development, and virulence, in pathogenic fungi (6–8). In C. neoformans, there are at least two distinct G protein signal transduction pathways. The cyclic AMP (cAMP)-dependent signaling pathway consists of the Gα protein Gpa1, the adenylyl cyclase Cac1, the protein kinase A catalytic subunit Pka1 and regulatory subunit Pkr1, the adenylyl cyclase-associated protein AcaI, and the phosphodiesterases Pde1 and Pde2 (9–12). The second pathway consists of the Gβ protein Gpb1, which couples with either Gpg1 or Gpg2 as a heterodimer to complex with the Gα protein Gpa2 as the typical heterotrimeric complex to govern the pheromone-responsive mating pathway (13, 14). In C. neoformans, cAMP signaling is also closely linked to the production of virulence factors, such as the melanin pigment and the capsule. On the other hand, mating is generally not considered to be closely tied to virulence despite the observation that the basidiospores from sexual reproduction might be more infectious (3).
The G protein signaling pathway is thought to be triggered by the binding of a ligand to the specific G protein-coupled receptor (GPCR) that results in conformational changes of Gα and the exchange of GDP for GTP. The GTP-bound Gα then dissociates from the Gβγ heterodimeric complex. The activated Gα will then resume the inactive GDP-bound state upon GTP hydrolysis, and further signaling is blocked by Gα reassociation with Gβγ (15). There are varieties of accessory proteins that can regulate the activities of heterotrimeric G proteins. Regulators of G protein signaling (RGS) can stimulate the GTPase activity of Gα to inactivate Gα. Guanine nucleotide dissociation inhibitors (GDI) can stabilize GDP-Gα to inhibit the nucleotide exchange for GTP binding, whereas guanine nucleotide exchange factors (GEF) can facilitate the GDP-to-GTP nucleotide exchange to promote Gα activation.
The Ric8 (resistance to inhibitors of cholinesterase 8) protein, also known as synembryn, was first identified in the nematode Caenorhabditis elegans as a noncanonical GEF that exhibits GEF activity toward Gα in either a receptor-dependent or -independent manner (16). Ric8 is also involved in asymmetric cell division during early embryogenesis of C. elegans and in the neural progenitor of the fruit fly Drosophila melanogaster (17, 18). Two Ric8 isoforms are found in humans: Ric8A and Ric8B. Ric8A, identified using the Gα protein as bait to look for interactive partners through a yeast two-hybrid (Y2H) screen (19), preferentially interacts with GDP-bound Gα to facilitate GDP release and forms a complex with Gα until Gα binds to GTP (20). Ric8A also functions as a chaperone protein to fold nascent Gα subunits (21), and it prevents ubiquitination and degradation of Gαi2 and Gαq (22). Ric8B was found to be involved in receptor-dependent signaling processes, because it was able to stimulate cAMP production only after GPCR-Gαs/olf was stimulated first (23), and translocation of Ric8B to the plasma membrane is observed only after GPCR stimulation (19). Ric8 was also identified in Dictyostelium discoideum, where it functions as a receptor-independent GEF for Gα to amplify signaling amplitude (24). Ric8 homologs have also been found to perform various regulatory functions in several fungal organisms. In Neurospora crassa, disruption of the RIC8 gene results in a phenotype similar to that of Gα gna1 and Gα gna3 mutants (25), and expression of a dominant-active GNA3 in the ric8 deletion mutant leads to a significant increase in conidial germination (26). Deletion of the ricA gene (a RIC8 homolog) leads to impaired colony growth and total or partial loss of asexual sporulation in Aspergillus nidulans and Aspergillus fumigatus, respectively (27). In the rice blast fungus Magnaporthe oryzae, MoRic8, a Ric8 homolog, interacts with the Gα protein MoMagB to regulate appressorial differentiation (28). Interestingly, no Ric8 homologs have been found in any plant species or in the budding yeast Saccharomyces cerevisiae.
Previous studies identified three Gα proteins (Gpa1, Gpa2, and Gpa3), one Gβ protein (Gpb1), and two Gγ proteins (Gpg1 and Gpg2) in C. neoformans (13, 14, 29, 30). Several RGS proteins, including Crg1 and Crg2, have also been described (31, 32). While Gpb1 couples only with Gpa2, no conventional Gβ protein that couples with Gpa1 has been found. The identification of Gib2 as a noncanonical Gβ coupling with Gpa1 reveals one of the activation mechanisms for Gpa1. In a search through the Y2H screen for additional proteins that interact with Gpa1 and function in Gpa1 signaling, we examined four candidates, including a Ric8 homolog and three members of the Rho family GTPases (Cdc42, Cdc420, and a Ran1 homolog). In this study, we characterized the Ric8 protein as a potential GEF of Gα proteins in either cAMP signaling that controls virulence or pheromone signaling affecting mating in C. neoformans.
MATERIALS AND METHODS
Strains and media.
The C. neoformans var. grubii archetype H99 (MATα) and the near-congenic KN99a (MATa) strains were used as parental strains in this study (Table 1) (33, 34).
TABLE 1.
Major strains (C. neoformans var. grubii) used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| H99 | Wild-type MATα | 33 |
| KN99a | Congenic to H99 MATa | 34 |
| CDX6 | gpr4::NEO MATα | 32 |
| JG3 | gpa1::URA5 PGPD1-GPA1Q284L ric8::NAT MATα | This study |
| JG31 | ric8::NEO MATα | This study |
| JG29 | ric8::NAT MATα | This study |
| JG33 | gpa1::ADE2 ric8::NEO MATα | This study |
| JG41 | ric8::NAT RIC8-NEO MATα | This study |
| JG51 | gpr4::NEO ric8::NAT MATα | This study |
| JG258 | gpa1::ADE2 MATα | This study |
| JG259 | gpa2::URA5 MATα | This study |
| JG260 | gpa3::URA5 MATα | This study |
| JG261 | ric8::NAT GPA1Q284L-NEO MATα | This study |
Yeast extract-peptone-dextrose (YPD), synthetic medium (SD), 10% V8 agar (pH 5.0) for mating, synthetic low ammonium dextrose (SLAD) medium for the confrontation assay, Nigerseed agar for melanin production, and Dulbecco's modified Eagle's medium (DMEM) with calcium bicarbonate for capsule formation were prepared following the standard protocols previously described (30, 35).
cDNA synthesis and plasmid construction.
Total RNA was isolated with the TRIzol reagent (Invitrogen, CA), and cDNA was synthesized using an oligo(dT) primer and reverse transcriptase as previously described (35). Rapid amplification of cDNA ends (RACE) was performed to verify the RIC8 open reading frame (ORF), and full-length cDNA was generated by reverse transcription (RT)-PCR using primers PW1624 and PW1625. The cDNA was inserted into the pGADT7 and pGBKT7 plasmids (BD Biosciences) at EcoRI and BamHI sites following the standard protocols (36). cDNAs for GPA1, GPA2, and GPA3 were synthesized using primers PW1654/PW1680, PW1631/PW1632, and PW1633/PW1634, respectively, and the fragments were inserted into pGBKT7. GPA1 was inserted at the NcoI and PstI sites, whereas GPA2 and GPA3 were inserted at the BamHI and PstI sites.
To generate the GPA1Q284L dominant-active allele, primer pairs PW1654/PW1659 and PW1658/PW1680 were used to create two overlapping PCR fragments. These fragments were mixed as the template for a new round of PCR amplification to create the GPA1Q284L allele using primers PW1654 and PW1680. Using a similar method, primer pairs PW1631/PW1657 and PW1656/PW1632 were used to generate the GPA2Q203L dominant-active allele. Similarly, primer pairs PW1633/PW1663 and PW1662/PW1634 were used to generate the GPA3Q206L dominant-active allele. These constructs were inserted into pGBKT7 using the same restriction sites as their wild-type alleles.
For constructs in pGADT7, the primer pair PW1684/PW1685 was used to generate the GPA1 and GPA1Q284L alleles, and the DNA fragments were inserted into pGADT7 at the XmaI and XhoI sites. For GPA3 and GPA3Q206L, primers PW1682 and PW1686 were used for PCR amplification, and DNA was digested by BamHI and XhoI and inserted into pGADT7. The oligonucleotide primers for PCR amplification are listed in Table 2. All plasmid constructs were verified by DNA sequencing.
TABLE 2.
DNA primers used in this study

Semiquantitative RT-PCR.
Cells grown overnight in liquid YPD medium were harvested, washed with sterile distilled water, and divided into three portions. One portion was kept in ice, the second portion was suspended in yeast nitrogen base (YNB), and the third portion was suspended in liquid V8 medium in the presence of the mating partner (KN99a; 1 × 105 cells). Cells in liquid YNB and V8 media were returned to the 30°C shaker (2,500 rpm) for incubation for four additional hours. All the cells were then lyophilized, and RNA was isolated with TRIzol reagent (Invitrogen, CA). Following digestion with RNase-free DNase (RQ1; Promega), RNA was quantified using a NanoVue Plus spectrophotometer (GE Life Sciences). One microgram RNA was used for reverse transcription using random hexamers (SuperScript First Strand; Invitrogen). An equal amount of cDNA (1 μl) was used in the PCR with gene-specific primers. Primer pairs JH7278/JH7279, PW116/PW315, PW54/PW140, PW986/PW1086, and PW1614/PW1615 were used to amplify the partial fragments of GPA1, GPA2, GPA3, RIC8, and ACT transcripts, respectively.
Images of the RT-PCR ethidium bromide-stained agarose gels were acquired with a Kodak Gel Logic 2200 imaging system, and bands were quantified using Carestream MI software (Carestream). The band intensity was expressed as relative absorbance units using the constitutively expressed actin gene (ACT) as a control for normalization of initial variations in the sample concentration and for reaction efficiency. Means and standard deviations were calculated after normalization to actin and were plotted.
Generation and complementation of ric8 mutants.
The RIC8 gene was identified from the C. neoformans var. grubii H99 database (www.broadinstitute.org) by BLAST (Basic Local Alignment Search Tool) search and obtained by PCR amplification. The full-length cDNA sequence was also obtained by RT-PCR amplification. Both sequences were verified by DNA sequencing. To generate the ric8 mutants, overlap PCR was used. Primers PW638 and PW1603 were used to obtain the 3′ partial fragment of the NEO gene cassette for neomycin resistance, and primers PW812 and PW1602 were used to obtain the 5′ partial fragment of the NEO gene. The RIC8 upstream partial fragment was obtained with primers PW1609 and PW1610, and the downstream fragment was obtained by PCR amplification with primers PW1611 and PW1612. The two overlap PCR products amplified by primer pairs PW1609/PW638 and PW812/PW1612 were transformed into H99 or related strains through biolistic transformation. Neomycin-resistant colonies were screened by PCR amplification using primers PW983 and PW638. A ric8::NAT mutant was also generated using a similar strategy that allows verification of phenotype changes associated with RIC8 gene disruption.
The ric8::NAT mutant strain was complemented by reintroduction of a 3,652-bp PCR fragment containing the full-length RIC8 ORF, a 702-bp region preceding the start codon, and a 300-bp fragment downstream of the termination codon. A 4,320-bp fragment was amplified using primers PW983 and PW984 and digested with SacII and NotI, and the cutoff band of 3,652 bp was inserted into pGS200, which contains a NEO selection marker (37). The insert was verified by DNA sequencing before transformation into the ric8::NAT mutant strain. G418-resistant colonies were selected, screened by PCR amplification, and verified by observation for restoration of phenotypes such as capsule and melanin formation.
Heterologous protein expression.
The GPA1 and GPA1Q284L gene alleles were PCR amplified using primers PW1688 and PW1680, digested with KpnI and PstI, and ligated to pET41a(+) (Novagen). Primer pairs PW1668/PW1669 and PW1670/PW1671 were used to obtain GPA2-GPA2Q203L and GPA3-GPA3Q206L, respectively. DNA was digested with BamHI and PstI before ligation to pET41a(+). The primer pair PW1720/PW1721 was used to obtain RIC8 cDNA by PCR amplification, followed by restriction with XhoI and NcoI and ligation into pRSET-B. All insert sequences were verified by DNA sequencing.
Fusion protein induction and purification were performed following the standard protocols or as previously described (14, 38). BL21-CodonPlus (DE3)-RIPL cells were used to express pET41a(+)-based protein constructs, while BL21(DE3)pLysS cells were used to express Ric8 from pRSET B. The glutathione S-transferase (GST)-Gα fusion proteins were purified using Glutathione Sepharose 4B medium (GE Healthcare), while the 6His-Xpress Ric8 fusion protein was purified using a HIS-Select Nickel affinity gel (Sigma-Aldrich).
Coimmunoprecipitation (co-IP) and Western blotting.
Purified GST-Gα and 6His-Xpress-Ric8 fusion proteins were buffer exchanged using PD-10 columns (GE Healthcare) with phosphate-buffered saline (PBS) (10 mM phosphate buffer, pH 7.4, 150 mM NaCl containing 5 mM MgCl2) and 100 μM guanosine 5′-O-(2-thiodiphosphate) trilithium salt. Equimolar amounts of the Gα and Ric8 proteins were mixed and loaded onto glutathione-agarose columns (Sigma-Aldrich). After incubation for 3 h at 4°C, the columns were washed with the same buffer 10 times, and the bound proteins were eluted by 10 mM glutathione in 50 mM Tris-HCl (pH 9.2). The proteins were separated by gel electrophoresis (NuPAGE; Life Technologies), transferred to an Immun-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad), and incubated with the mouse anti-Xpress antibody (1:5,000; Invitrogen) or the mouse anti-GST antibody (1:5,000; Santa Cruz Biotechnology). An anti-mouse IgG horseradish peroxidase (HRP) conjugate (1:7,500; Promega) was used as the secondary antibody.
Phenotypic characterization and virulence assessment.
Cells from each strain were inoculated into 5 ml YPD medium for overnight growth in a 30°C shaker (225 rpm), centrifuged, and washed with sterile distilled water. Cells were counted under a microscope following dilution, and the cell density was adjusted to 1 × 107 per ml. Ten microliters of cell suspension containing 105 cells (1 × 107 cells/ml) was pipetted on l-3,4-dihydroxyphenylalanine (L-DOPA) and Nigerseed agar plates for melanin induction at 30°C and 37°C and into 2 ml liquid DMEM for capsule induction at 30°C with shaking (225 rpm). Cells for capsule formation were mixed with India ink and observed under a Zeiss Axioskop microscope.
Virulence was assessed using the intranasal inoculation (inhalation) route in 4- to 6-week-old female AJC/r mice (Jackson Laboratory). Ten mice per group were each inoculated with 105 cells, and the animals were monitored twice daily. Animals that appeared sick or in pain were sacrificed via CO2 asphyxiation. Mouse survival was analyzed by the Kaplan-Meier method using Prism 4.0 software (Graphpad Software, Inc.) as previously described (35). Animal use was approved by the Louisiana State University Health Sciences Center Institutional Animal Care and Use Committee (IACUC).
RESULTS
Identification and expression of C. neoformans RIC8.
C. neoformans is a haploid basidiomycetous fungus with a bipolar mating system composed of two mating types, MATα and MATa. The pheromone response and mating can be observed in the laboratory environment, from which early knowledge regarding signal transduction in the organism was generated. Studies by Tolkacheva et al. and Alspaugh et al. identified the first G protein subunit (Gpa1), which mediates a cAMP-dependent signaling pathway important in fungal virulence (29, 39). Curiously, a classic G protein β subunit capable of coupling with Gpa1 was not found, leading to propositions that Gpa1 may function as a monomeric G protein whose activation depends on GEF or that certain proteins may function as a bona fide Gβ. We have since characterized a Gpa1 binding protein (Gib2) as a noncanonical Gβ protein forming a heterotrimeric G protein complex with Gpa1 (38, 40). To identify additional proteins capable of binding and potentially activating Gpa1, we used the Y2H screen to examine candidate proteins that included a Ric8 homolog and three members of the Rho family GTPases (Cdc42, Cdc420, and Ran1) (unpublished observations). We sought to characterize Ric8 functions first, as they have yet to be studied in this fungus.
A BLAST search of the C. neoformans H99 (var. grubii) database revealed that the RIC8 gene has a 2,397-bp open reading frame encoding a 692-amino-acid protein. The C. neoformans Ric8 protein shares relatively low amino acid sequence homology (identity and similarity) with the Ric8 proteins of nematodes and humans (23% and 26%, respectively). It shares moderate sequence homology with those of N. crassa (27%) and M. oryzae (30%). In contrast, C. neoformans Ric8 shares a higher amino acid homology with the Ric8 protein of the basidiomycetous jelly fungus Tremella mesenterica (59%). However, the function of the protein is unknown. A phylogenic tree depicting evolutionary relationships among these proteins is shown in Fig. 1A.
FIG 1.
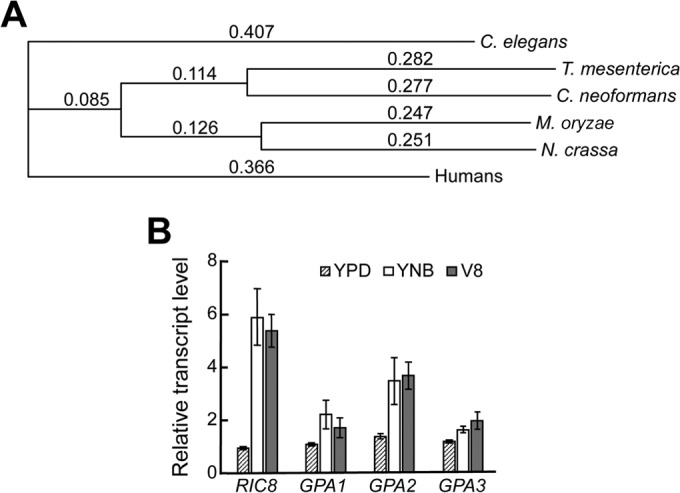
Amino acid sequence alignment of fungal Ric8 proteins and expression of the RIC8 gene of C. neoformans. (A) Phylogenetic diagram showing that C. neoformans Ric8 is within the same node as the Ric8 protein of the basidiomycetous jelly fungus T. mesenterica and is most distant from the Ric8 of C. elegans. The phylogenetic tree was drawn using ClustalW (v1.83) multiple-sequence alignment with the flowing parameters: Open Gap Penalty, 10.0; Extend Gap Penalty, 0.2; Delay Divergent, 30%; Gap Distance, 4; Similarity Matrix, gonnet. The accession numbers for the Ric8 proteins are NP_001023561 (C. elegans), EAA34708 (N. crassa), XP_007006336 (T. mesenterica), XP_003716129 (M. oryzae), and AFR98639 (C. neoformans). (B) The expression of the RIC8 gene is highly inducible by nutrient-limiting medium and by conditions favoring mating. The induction time for cells in YNB and V8 media was 4 h. RT-PCR was repeated twice, and the mean values were used to calculate the expression rate relative to that of the ACT gene encoding actin (averages ± standard deviations are shown). The numbers of PCR cycles were 30 for GPA1 and GPA2, 35 for GPA3 and RIC8, and 25 for ACT.
We thought that examination of RIC8 gene expression would provide invaluable information useful for predicting its function. We opted to use the semiquantitative-RT-PCR approach to examine the expression of RIC8 and three Gα gene transcripts under conditions including growth in nutrient-rich YPD, nutrient-limited YNB, and V8 media in the presence of a mating partner. Gene expression levels were expressed as relative units using the constitutively expressed actin gene as a control. The expression of RIC8 was relatively low in nutrient-rich YPD but increased significantly after 4 h in YNB and V8 media (Fig. 1B). Since the nutrient deprivation conditions permit cAMP signaling and the presence of a mating partner induces mating, this expression profile is consistent with a prediction that Ric8 functions in both signaling pathways. The expression profile is more similar to that of GPA1 and GPA2 than to that of GPA3 (Fig. 1B). The expression of GPA1 is inducible by low-nitrogen conditions, such as SLAD medium (29), while incubation in liquid V8 medium alone permits the expression of GPA3 (13).
Ric8 regulates capsule and melanin formation.
To characterize Ric8 functional roles, the RIC8 gene was disrupted in the archetypical C. neoformans H99 strain (var. grubii) using the ric8::NEO allele. A ric8 mutation linked to the NAT marker was also obtained using the ric8::NAT allele. The complemented strain was obtained in the ric8::NAT strain (ric8 RIC8).
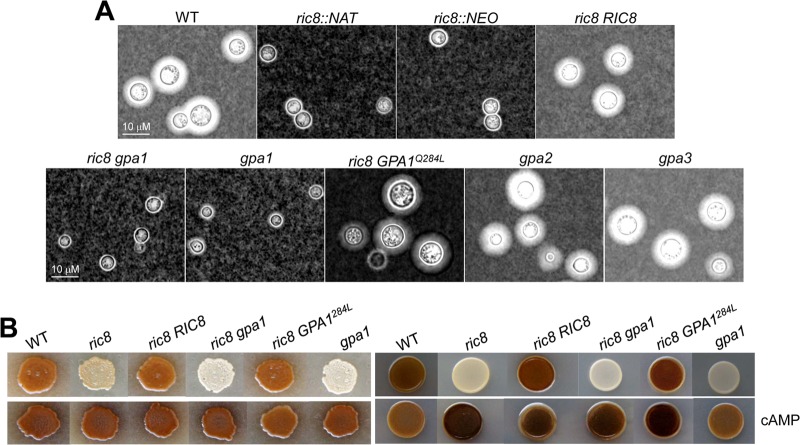
Given that C. neoformans Gpa1 functioning in the cAMP signaling pathway lacks a canonical Gβ protein, Ric8 could exert a conserved GEF function toward Gpa1 by promoting the activation of Gpa1. We sought to test whether this is the case. We first assessed capsule production and melanin formation, traits that are regulated by Gpa1-cAMP signaling. The polysaccharide capsule is regarded as one of the most important virulence factors for C. neoformans, as it protects cells from phagocytosis and killing by macrophages. The ric8 mutant lacked the capsule, indicating that Ric8 is required for capsule formation. This is similar to the gpa1 and ric8 gpa1 mutants that also lacked the capsule (Fig. 2A). Capsule formation was restored in the complemented ric8 RIC8 strain, which was similar to the wild type; the ric8 mutant expressing the constitutively activated Gpa1 allele (ric8 GPA1Q284L); and the gpa2 and gpa3 mutants, all producing normal-size capsules (Fig. 2A). This observation is consistent with a model in which Ric8 and Gpa1 could function in a linear pathway to regulate capsule formation.
FIG 2.
C. neoformans Ric8 regulates capsule production and melanin formation. (A) Ric8 is required for the production of the polysaccharide capsule in vitro. Cells were inoculated in liquid DMEM with calcium carbonate and incubated at 30°C for 2 days. The cells were then washed, stained with India ink, and examined under a microscope (Zeiss Axioskop) equipped with a digital camera. WT, wild type. (B) Ric8 is also required for the formation of melanin in vitro. Cells (105) were spotted on standard Nigerseed agar at 30°C (left) or on L-DOPA medium at 37°C (right) for 2 days. Dark pigments indicate normal melanin production, whereas opaque colonies indicate melanin deficiency. cAMP (10 mM) was added to the melanin induction media shown in the bottom row.
The melanin pigment also protects cells from external damage imposed by reactive oxygen species (ROS) and allows C. neoformans to survive in harsh environments. Melanin formation is regulated by Gpa1-cAMP signaling, as externally supplementing cAMP suppressed the defect in melanin formation that occurred in the gpa1 and cac1 mutants (9, 29). Consistent with this observation, the ric8 mutant strain lacked the melanin pigment when induced. On Nigerseed agar at 30°C (Fig. 2B, left) and on L-DOPA medium at 37°C (Fig. 2B, right), the ric8 mutant produced an opaque colony, in contrast to the wild-type, ric8 RIC8 complemented, and ric8 GPA1Q284L strains. However, similar to capsule induction assays, the defect shown by the ric8 mutant was not as severe as that of the gpa1 and ric8 gpa1 mutants, which showed the most drastic defects (the smallest cell sizes in capsule induction medium and nearly white colonies on melanin induction plates), reinforcing the notion that Ric8 may not be the only “activator” of Gpa1.
Ric8 functions in the Gpa1-cAMP signaling pathway.
Since the activation of the Gpa1-cAMP pathway results in increased intracellular cAMP levels and supplementation of the growth medium with cAMP can suppress pathway defects by temporarily raising cAMP levels, we added cAMP (to 10 mM) to the Nigerseed agar medium and L-DOPA agar medium to examine its effect on melanin formation. Melanin was restored in the ric8 mutant to a level similar to that of the wild-type cells (Fig. 2B), suggesting that Ric8 functions within the Gpa1-cAMP pathway.
To test if Ric8 is directly involved in the regulation of the intracellular cAMP levels, we assayed for the transient levels of cAMP following starvation and glucose triggering (37). The ric8 mutant strain posted 2.9 × 103, 2.5 × 103, 2.6 × 103, and 2.7 × 103 fmol cAMP at 0, 0.5, 1, and 3 min post-glucose stimulation, respectively, which was similar to those posted by the gpa1 mutant strain (2.4 × 103, 2.4 × 103, 2.4 × 103, and 2.6 × 103 fmol) (Fig. 3A). The wild-type strain showed a typical transient spike in cAMP levels following glucose triggering (3.0 × 103, 3.9 × 103, 4.3 × 103, and 3.6 × 103 fmol at the respective time intervals post-glucose stimulation) (Fig. 3A). This assay indicates that Ric8 has a role similar to that of Gpa1 in the positive regulation of transient cAMP levels and the cAMP signaling pathway.
FIG 3.
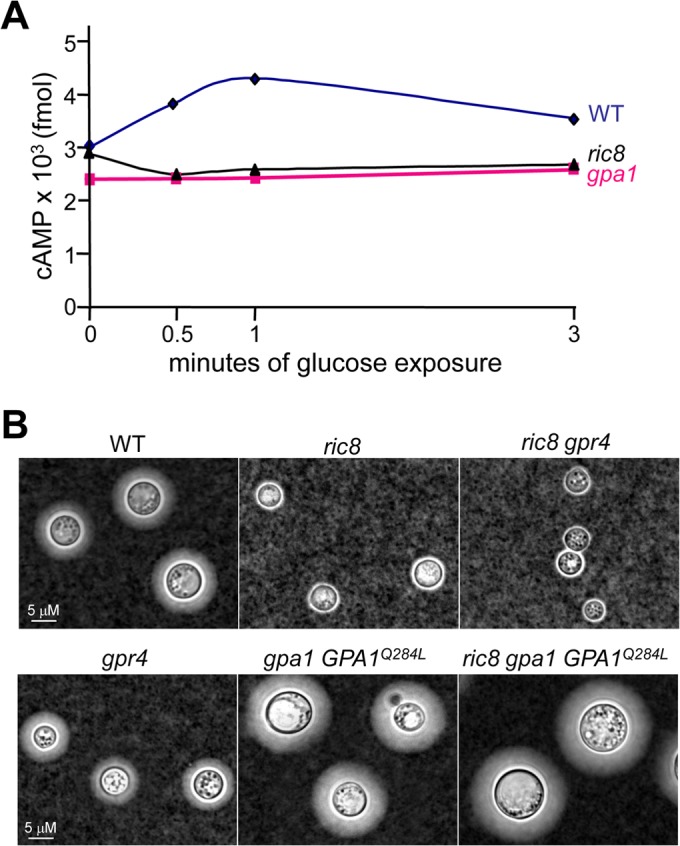
C. neoformans Ric8 positively regulates the intracellular cAMP level and functions independently of Gpr4. (A) Cells of the ric8, gpa1, and wild-type strains were grown in YPD medium overnight and washed twice with sterile distilled water before being subjected to glucose starvation. Cell extraction and cAMP measurement were carried out as previously described (38). The assay was performed in triplicate, and the mean values are plotted. The experiment was repeated twice with similar results. (B) The ric8, ric8 gpr4, ric8 gpa1 GPA1Q284L, gpr4, gpa1 GPA1Q284L, and wild-type strains were examined for capsule formation following induction in DMEM.
In C. neoformans, Gpr4 was identified as a GPCR that senses environmental cues, such as the amino acid methionine, and functions upstream of Gpa1 (41). Disruption of GPR4 resulted in attenuated capsule formation to a modest degree in comparison to that of the gpa1 or ric8 mutant (41) (Fig. 3B). The ric8 gpr4 mutant displayed an acapsular phenotype similar to that of the ric8 and ric8 gpa1 mutants, suggesting that Ric8 function might be independent of Gpr4. In addition, expression of the constitutively active GPA1Q284L allele in the gpa1 mutant resulted in enhanced capsule formation, indicative of upregulated cAMP signaling. The enhanced capsule formation is not affected by disruption of the RIC8 gene (Fig. 3B). Collectively, these findings are consistent with the hypothesis that Ric8 likely functions in Gpa1-cAMP signaling by providing a GEF function toward Gpa1 in the absence of stimulation by Gpr4.
Ric8 has opposing roles in pheromone response and mating.
Previous studies showed that mutants such as the gpa1 mutant were also attenuated in mating, although the mechanism is unclear (29). The ric8 mutant showed similar attenuation in mating. To examine whether Ric8 functions in the pheromone-responsive mating pathway, we carried out confrontation and mating assays.
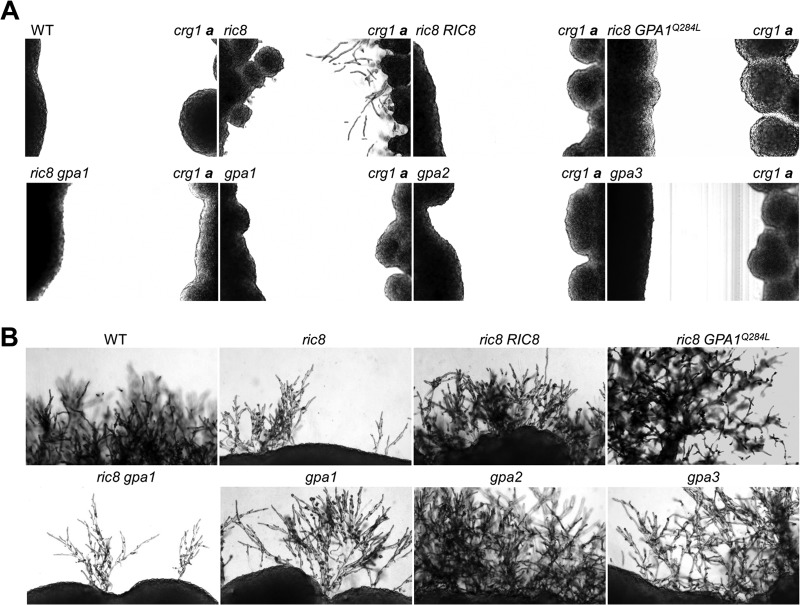
C. neoformans secretes diffusible pheromones, and the placement of compatible mating type strains in proximity results in the formation of conjugation tubes whose fusion precedes meiosis and basidiospore formation. Assessment of this type of pheromone response is referred to as a confrontation assay (42). In contrast to C. neoformans var. neoformans, where conjugation tubes are readily seen, C. neoformans var. grubii strains rarely form conjugation tubes even when one of the partners carries a crg1 mutation with an enhanced pheromone response (35). When streaked next to the MATa crg1 mutant in filament agar, a small number of “stubby” conjugation tubes were seen in the ric8 mutant, while abundant and long conjugation tubes were seen in the crg1 mutant strain (Fig. 4A). This enhanced pheromone response cannot be caused by the wild-type, complemented ric8 RIC8, ric8 gpa1, or ric8 GPA1Q284L strain expressing the dominant-active GPA1 allele, suggesting that Ric8 may negatively influence pheromone production or responses.
FIG 4.
C. neoformans Ric8 protein regulates the pheromone response and mating. (A) The ric8, ric8 RIC8 complemented, ric8 gpa1, ric8 GPA1Q284L, gpa1, gpa2, gpa3, and wild-type strains were streaked in proximity to the MATa crg1 strain on filament agar, and the plates were incubated in the dark at room temperature for 3 days. Only the ric8 mutant showed stubby conjugation tube-like structures, and the paired crg1 strain showed abundant conjugation tubes. (B) The same strains were also mixed with KN99a by patching onto V8 agar and incubated for 7 days. The cross with the ric8 mutant resulted in attenuated formation of dikaryotic filaments and basidiospores, similar to that of gpa1. The attenuation was most severe in the ric8 gpa1 double-mutant strain. The assays were repeated multiple times, and representative images are shown.
In a cross by mixing cells with the MATa strain KN99a, the wild-type strain H99 formed abundant branched and elongated dikaryotic filaments followed by basidium and basidiospore production (Fig. 4B). However, this type of dikaryotic filament was much less apparent in crosses with the ric8 mutant. This degree of attenuation appeared to be similar to that of the gpa1 mutant, and interestingly, further attenuation was seen in the ric8 gpa1 double-knockout mutant strain (Fig. 4B), which is consistent with the reasoning that Ric8 and Gpa1 have certain functional distinctions. The cross with the ric8 RIC8 complemented strain resulted in dikaryotic-filament and basidiospore formation near the wild-type level, while crosses with the ric8 GPA1Q284L, gpa2, and gpa3 strains yielded elongated and profusely branched dikaryotic-filament formation that was consistent with previous studies (13, 14). These assays, repeated multiple times with similar results, indicate that Ric8 has a positive role in mating and a negative role in pheromone production and in the response. We inferred that these opposite roles are likely to be mediated by distinct mechanisms or targets, and this hypothesis is supported by our further findings described below.
Ric8 associates with Gpa1 and Gpa2 in vivo and in vitro.
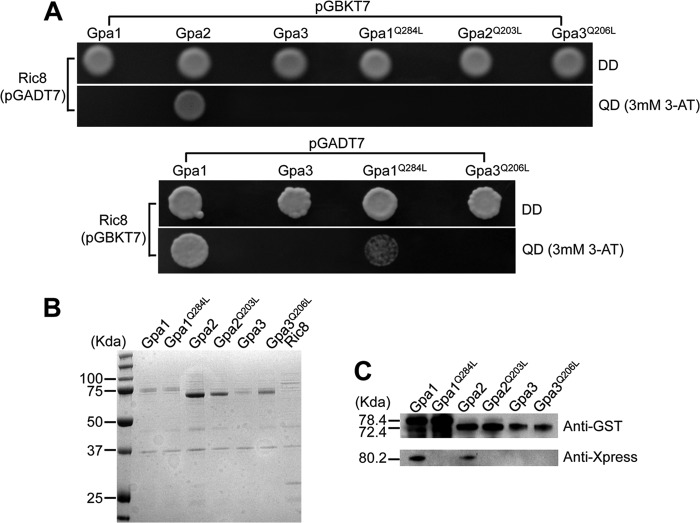
Phenotype characterization leads to the proposition that Ric8 functions in both cAMP signaling and pheromone-responsive mating pathways. To examine whether Ric8 indeed functions by modulating the activation of Gα proteins as a GEF, protein-protein interaction assays by means of Y2H screening and coimmunoprecipitation were performed. In the Y2H screen, pGADT7-RIC8 was cotransformed into host yeast AH109 cells with pGBKT7-GPA1, -GPA1Q284L, -GPA2, -GPA2Q203L, -GPA3, and -GPA3Q206L, and the same amount of cells (105) were plated on selective medium. Gpa1Q284L, Gpa2Q203L, and Gpa3Q206L are the GTPase-deficient and constitutively activated forms of Gpa1, Gpa2, and Gpa3, respectively. Only cells cotransformed with pGADT7-RIC8 and pGBKT7-GPA2 showed growth on selective medium, indicating that Ric8 interacts with Gpa2 (Fig. 5A, top). In addition, this interaction was abolished when Gpa2 was replaced by Gpa2Q203L (Fig. 5A, top). Interestingly, when the vectors were switched by inserting RIC8 in pGBKT7 and GPA1, GPA1Q284L, GPA3, and GPA3Q206L in pGADT7, AH109 cells transformed with pGBKT7-RIC8/pGADT7-GPA1 and pGBKT7-RIC8/pGADT7-GPA1Q284L showed growth, suggesting Ric8 also interacts with Gpa1 (Fig. 5A, bottom). In comparison to cells with pGBKT7-RIC8/pGADT7-GPA1, the growth of cells expressing Ric8 and Gpa1Q284L was very limited, indicating that the introduction of the dominant-active Gpa1Q284L allele weakened its binding to Ric8. This assay did not find any interactions between the Ric8 and Gpa3 proteins (Fig. 5A).
FIG 5.
Ric8 interacts with Gpa1 and Gpa2 in vivo and in vitro. (A) Yeast two-hybrid screen showing host cell growth in SD-Leu-Trp medium (DD) and SD-Leu-Trp-His-Ade medium plus 3 mM 3-amino-1,2,4-triazole (3-AT) [QD (3mM 3-AT)], suggesting that Ric8 interacts with Gpa2 (top) and Gpa1 and Gpa1Q284L (bottom). (B) Heterologous expression of Gpa1, Gpa1Q284L, Gpa2, Gpa2Q203L, Gpa3, Gpa3Q206L, and Ric8 proteins revealed by Coomassie blue staining following SDS-PAGE. The expected molecular masses of these proteins are 78.4, 78.3, 72.5, 72.5, 73.1, 73.1, and 80.2 kDa, respectively. (C) Protein pulldown assay indicating that Ric8 physically interacts with Gpa1 and Gpa2, but not Gpa3. The GST-tagged Gpa1, Gpa1Q284L, Gpa2, Gpa2Q203L, Gpa3, and Gpa3Q206L and Xpress-tagged Ric8 proteins were prepared as previously described (14). The GST fusion Gα proteins were adsorbed to glutathione Sepharose beads and washed, and Ric8 was then added. Bound proteins were separated and analyzed by blotting with either the anti-GST antibody for Gαs (top) or the anti-Xpress antibody for Ric8 (bottom).
A co-IP experiment was performed to validate the interactions established through the Y2H screen. Ric8 and all six Gα proteins were expressed heterologously, and the protein masses were verified (Fig. 5B). Indeed, interactions were established between Ric8 and Gpa1 and between Ric8 and Gpa2 (Fig. 5C). The weak interaction between Ric8 and Gpa1Q284L through the Y2H screen could not be substantiated through co-IP, in which the stable GDP analog guanosine 5′-O-(2-thiodiphosphate) trilithium salt was present (Fig. 5C). Collectively, these findings suggest that Ric8 can function as a GEF toward both Gpa1 and Gpa2, but not Gpa3. Ric8 can facilitate GTP binding to Gpa1 and allows Gpa1 activation to regulate cAMP signaling. Likewise, Ric8 may also promote GDP-to-GTP exchange of Gpa2 to modulate mating efficiency. Disruption of the RIC8 gene negatively impacts the Gpa1-cAMP signaling amplitude and delays dissociation of Gpa2 from Gpb1, resulting in attenuation in the formation of dikaryotic filaments and basidiospores. How Ric8 might influence the pheromone response and whether it interacts with Gpa3 in vivo remain unknown and await further studies.
Mutation of ric8 results in attenuated virulence toward animals.
Gpa1 and the Gpa1-mediated cAMP pathway are known to have major regulatory roles in the expression of virulence traits, including melanin formation and capsule production (10, 29). Disruption of GPA1 or PKA1 resulted in significant reductions in melanin and capsule formation, and the gpa1 and pka1 mutant strains were severely attenuated in virulence toward animals. To examine whether Ric8 plays a role in virulence, we infected 10 AJC/r mice with the ric8 mutant, the H99 strain, and two independent ric8 RIC8 complemented strains with 105 cells through intranasal inhalation (35). Mouse survival was monitored twice daily, and the results were plotted against time (in days). The mice infected with the ric8 mutant survived 78 days, with a median survival of 55 days, in contrast to mice infected with H99, which survived 23 days with a median survival of 20 days (P < 0.001), and two complemented strains survived 24 and 29 days (median survival, 20 and 24 days, respectively; P < 0.001) (Fig. 6A). Brain smears of mice infected with the ric8 mutant showed the presence of yeast cells maintaining the acapsular form in comparison to the capsular wild-type strain (Fig. 6B). The observation that the ric8 mutant had a significant delay in causing cryptococcosis of mice demonstrates that Ric8 positively regulates fungal virulence.
FIG 6.

Disruption of the C. neoformans RIC8 gene attenuates virulence in a murine model of cryptococcosis. (A) The wild-type strain H99, the ric8 mutant, and two independent ric8 RIC8 complemented strains (105 cells) were each inoculated via the nasal cavities into 10 AJC/r mice. Mouse survival was monitored twice daily and plotted against time. The difference in survival between the ric8 mutant and the control strains is statistically significant (P < 0.001). (B) Fungal cells of the ric8 mutant and the wild-type H99 strain were visible in the brain homogenates of moribund mice following India ink staining.
DISCUSSION
In eukaryotic cells, heterotrimeric G protein-mediated signaling transduction pathways play critical roles in sensing and relaying external cues so that cells can adapt. In the pathogenic fungus C. neoformans, G protein signaling also becomes an integral part of the virulence repertoire. C. neoformans encodes three Gα proteins (Gpa1, Gpa2, and Gpa3), one Gβ protein (Gpb1), one Gβ-like protein (Gib2), and two Gγ proteins (Gpg1 and Gpg2). Functionally, these proteins exhibit remarkable conservation with homologous proteins of many other eukaryotic organisms and even higher eukaryotes, such as mammals. Gpa1, Cac1, and Pka1 constitute a major cAMP-dependent signal transduction pathway that controls growth, differentiation, and the production of virulence characteristics (reviewed in references 6, 42, and 43). Mutations occurring among the components of this pathway lead to attenuated cAMP signaling and altered capsule size and melanin formation. Meanwhile, there is growing evidence indicating that G protein signaling has evolved to be quite distinct in this fungus. A GPCR homolog, Gpr4, was found to stimulate Gpa1 upon the sensing of methionine, but a GPCR capable of sensing glucose and activating the cAMP pathway identified in other fungi, such as S. cerevisiae, was not found (41). Also, a canonical Gβ protein capable of binding to Gpa1 was not found, but the noncanonical Gβ protein Gib2 proved to form a heterotrimeric complex with Gpa1 and Gpg1 or Gpg2 (38). In addition, Gib2 functions as a scaffolding protein that positively regulates cAMP levels through novel functions involving the Ras1 and Cac1 proteins (40). The current identification of the Ric8 protein provides another mechanistic explanation of how Gpa1 functions without a classic Gβ subunit.
In contrast to the Gpa1-cAMP signaling pathway, the mechanism of pheromone response and mating regulated by Gpb1 through a conventional Gαβγ heterotrimeric complex appears to be conserved. However, the specific functions of Gpa2 and, in particular, Gpa3 in pheromone responses and mating are still unresolved. We found that a mutation of either Gpa2 or Gpa3 results in enhanced pheromone response and mating, while a companion study showed that Gpa2 promotes mating while Gpa3 inhibits mating (13, 14). We showed that Gpa2, but not Gpa3, interacted with Gpb1 and Crg1, a regulator of G protein signaling that negatively regulates pheromone responses and mating through Y2H screening (35). Consistent with a role in pheromone responses and mating, Gpa2 interacted with the pheromone receptor homolog Ste3α. In contrast, Gpa3 failed to interact with Gpb1 but interacted with Crg1. While we were unable to detect an interaction between Gpa3 and Ste3α, Hsueh et al. were able to establish an interaction using a different version of Y2H screening (13). Finally, Gpa3 is able to interact with Crg2, which is the main negative regulator of Gpa1 in cAMP signaling (31, 32).
The present study shows that Ric8 has a role in promoting mating (dikaryotic-filament and basidiospore formation), and it is conceivable that Ric8 may perform this function as a GEF toward Gpa2, dependent on or independently of Ste3α. Disruption of RIC8 delays dissociation of Gpa2 from Gpb1, resulting in attenuated formation of dikaryotic filaments and basidiospores. However, Ric8 also appears to negatively regulate the secretion of pheromones and conjugation tube formation, but an interaction cannot be found between Ric8 and Gpa3, which also lacks a Gβ partner. This calls for additional studies to address whether the pheromone response and mating pathways are separate events regulated by different Gα proteins and to further distinguish functional differences between Gpa2 and Gpa3 (Fig. 7).
FIG 7.
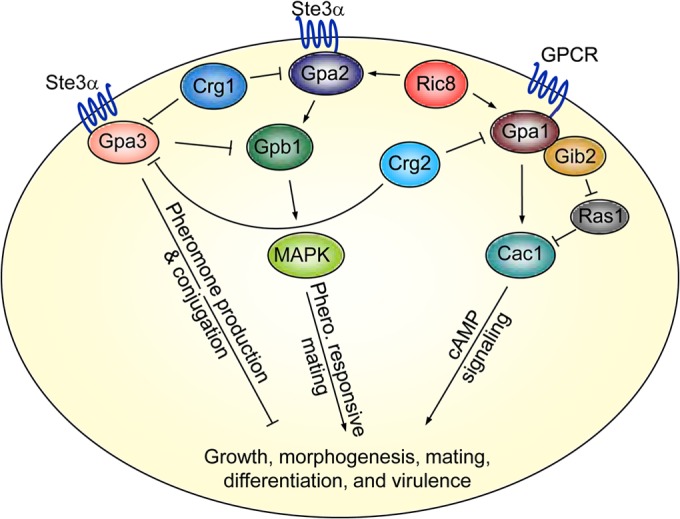
The Ric8 protein regulates the mating and cAMP signal transduction pathways in C. neoformans. The pheromone-responsive mating pathway consists of the upstream components, including the pheromone receptor Ste3α, the RGS protein Crg1, the Gα protein Gpa2, the Gβ protein Gpb1, and either the Gγ protein Gpg1 or the Gγ protein Gpg2. The cAMP signaling pathway consists of the G protein-coupled receptor homolog Gpr4, the Gα protein Gpa1, the Gβ-like protein Gib2, the Gγ proteins Gpg1/Gpg2, and the RGS protein Crg2. Ric8 performs a GEF function toward Gpa1 and Gpa2. It facilitates GDP-to-GTP exchange of Gpa1 to promote cAMP signaling that regulates growth, differentiation, and the production of virulence factors. It also promotes Gpa2 activation to subsequently prolong the positive regulatory function of Gpb1 in mating. Gpa3 likely functions as a distinctive Gα protein in a pathway that senses pheromones but counters the activity of Gpa2. No physical interactions have been established between Gpa3 and Gpb1 or between Gpa3 and Ric8.
Ever since being originally identified in nematodes and fruit flies, Ric8 proteins have been found in many other eukaryotes, including animals and even certain fungi, to possess a wide variety of functions. In addition to being a GEF of Gα proteins to amplify and prolong G protein-mediated intracellular signal transduction, Ric8 proteins were also found to exhibit various other functions. Ric8A was found to prevent ubiquitination and degradation of Gαi2 and Gαq (22) and to be a chaperone protein for folding nascent Gα subunits (21). Our studies of C. neoformans Ric8 revealed that it has many functions, such as affecting polysaccharide capsule and melanin production, attenuating pheromone responses, and decreasing mating efficiency. Such effects could contribute to the function of Ric8 as a GEF for Gpa1 and Gpa2. Nevertheless, additional functions are likely to be found through further research efforts.
We conclude that the C. neoformans Ric8 homolog protein functions as a GEF to facilitate the binding of GTP to Gpa1. This amplifies the cAMP-dependent signaling pathway that regulates capsule size, melanin formation, and virulence. The Ric8 protein also provides a GEF function for Gpa2, affecting mating efficiency. Our study highlights the distinct, as well as conserved, G protein signaling mechanisms and contributes to research efforts leading to the discovery of novel antifungal targets.
ACKNOWLEDGMENTS
This research was supported in part by a fund from the Research Institute for Children, Children's Hospital of New Orleans, New Orleans, LA (CHNOLA), and grants from the National Institute of Allergy and Infectious Diseases (NIAID) (R01AI054958 and R01AI074001) to P.W. J.D.G. is the recipient of a Center for Engaged Learning and Teaching (CELT) Fund grant from Tulane University. Research in the Z.Z. laboratory was supported in part by a special professorship from Jiangsu, China.
The content is solely our responsibility and does not necessarily represent the official views of CHNOLA, NIAID, or other sponsors.
Footnotes
Published ahead of print 1 August 2014
REFERENCES
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 2.Casadevall A, Perfect JR. 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 3.Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77:4345–4355. 10.1128/IAI.00542-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozel TR. 1995. Virulence factors of Cryptococcus neoformans. Trends Microbiol. 3:295–299. 10.1016/S0966-842X(00)88957-X [DOI] [PubMed] [Google Scholar]
- 5.Buchanan KL, Murphy JW. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4:71–83. 10.3201/eid0401.980109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen W-C, Wang P, Pan X, Waugh M, Heitman J. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746–785. 10.1128/MMBR.64.4.746-785.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kays AM, Borkovich KA. 2004. Signal transduction pathways mediated by heterotrimeric G proteins. Springer-Verlag, Berlin, Germany [Google Scholar]
- 8.Hoffman CS. 2005. Except in every detail: comparing and contrasting G-protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryot. Cell 4:495–503. 10.1128/EC.4.3.495-503.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, Perfect JR, Kronstad JW, Heitman J. 2002. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75–84. 10.1128/EC.1.1.75-84.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179–3191. 10.1128/MCB.21.9.3179-3191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J. 2004. Adenylyl cyclase-associated protein AcaI regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot. Cell 3:1476–1491. 10.1128/EC.3.6.1476-1491.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks JK, Bahn YS, Heitman J. 2005. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell 4:1971–1981. 10.1128/EC.4.12.1971-1981.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsueh YP, Xue C, Heitman J. 2007. G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans. Mol. Biol. Cell 18:3237–3249. 10.1091/mbc.E07-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Shen G, Zhang ZG, Wang YL, Thompson JK, Wang P. 2007. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol. Biol. Cell 18:4201–4209. 10.1091/mbc.E07-02-0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilman AG. 1987. G-proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56:615–649. 10.1146/annurev.bi.56.070187.003151 [DOI] [PubMed] [Google Scholar]
- 16.Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. 1996. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. U. S. A. 93:12593–12598. 10.1073/pnas.93.22.12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afshar K, Willard FS, Colombo K, Siderovski DP, Gonczy P. 2005. Cortical localization of the Galpha protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development 132:4449–4459. 10.1242/dev.02039 [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Ng KH, Qian H, Siderovski DP, Chia W, Yu F. 2005. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat. Cell Biol. 7:1091–1098. 10.1038/ncb1317 [DOI] [PubMed] [Google Scholar]
- 19.Klattenhoff C, Montecino M, Soto X, Guzman L, Romo X, Garcia MA, Mellstrom B, Naranjo JR, Hinrichs MV, Olate J. 2003. Human brain synembryn interacts with Gsalpha and Gqalpha and is translocated to the plasma membrane in response to isoproterenol and carbachol. J. Cell Physiol. 195:151–157. 10.1002/jcp.10300 [DOI] [PubMed] [Google Scholar]
- 20.Tall GG, Krumins AM, Gilman AG. 2003. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J. Biol. Chem. 278:8356–8362. 10.1074/jbc.M211862200 [DOI] [PubMed] [Google Scholar]
- 21.Chan P, Thomas CJ, Sprang SR, Tall GG. 2013. Molecular chaperoning function of Ric-8 is to fold nascent heterotrimeric G protein alpha subunits. Proc. Natl. Acad. Sci. U. S. A. 110:3794–3799. 10.1073/pnas.1220943110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chishiki K, Kamakura S, Yuzawa S, Hayase J, Sumimoto H. 2013. Ubiquitination of the heterotrimeric G protein alpha subunits Galphai2 and Galphaq is prevented by the guanine nucleotide exchange factor Ric-8A. Biochem. Biophys. Res. Commun. 435:414–419. 10.1016/j.bbrc.2013.04.103 [DOI] [PubMed] [Google Scholar]
- 23.Von Dannecker LE, Mercadante AF, Malnic B. 2005. Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Galphaolf. J. Neurosci. 25:3793–3800. 10.1523/JNEUROSCI.4595-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kataria R, Xu X, Fusetti F, Keizer-Gunnink I, Jin T, van Haastert PJ, Kortholt A. 2013. Dictyostelium Ric8 is a nonreceptor guanine exchange factor for heterotrimeric G proteins and is important for development and chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 110:6424–6429. 10.1073/pnas.1301851110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright SJ, Inchausti R, Eaton CJ, Krystofova S, Borkovich KA. 2011. RIC8 is a guanine-nucleotide exchange factor for Galpha subunits that regulates growth and development in Neurospora crassa. Genetics 189:165–176. 10.1534/genetics.111.129270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eaton CJ, Cabrera IE, Servin JA, Wright SJ, Cox MP, Borkovich KA. 2012. The guanine nucleotide exchange factor RIC8 regulates conidial germination through Galpha proteins in Neurospora crassa. PLoS One 7:e48026. 10.1371/journal.pone.0048026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon NJ, Park HS, Jung S, Kim SC, Yu JH. 2012. The putative guanine nucleotide exchange factor RicA mediates upstream signaling for growth and development in Aspergillus. Eukaryot. Cell 11:1399–1412. 10.1128/EC.00255-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Yan X, Wang H, Liang S, Ma WB, Fang MY, Talbot NJ, Wang ZY. 2010. MoRic8 is a novel component of G-protein signaling during plant infection by the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 23:317–331. 10.1094/MPMI-23-3-0317 [DOI] [PubMed] [Google Scholar]
- 29.Alspaugh JA, Perfect JR, Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206–3217. 10.1101/gad.11.23.3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P, Perfect JR, Heitman J. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20:352–362. 10.1128/MCB.20.1.352-362.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen G, Wang YL, Whittington A, Li L, Wang P. 2008. The RGS protein Crg2 regulates pheromone and cyclic AMP signaling in Cryptococcus neoformans. Eukaryot. Cell 7:1540–1548. 10.1128/EC.00154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue C, Hsueh YP, Chen L, Heitman J. 2008. The RGS protein Crg2 regulates both pheromone and cAMP signalling in Cryptococcus neoformans. Mol. Microbiol. 70:379–395. 10.1111/j.1365-2958.2008.06417.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perfect JR, Toffaletti DL, Rude TH. 1993. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 61:4446–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 71:4831–4841. 10.1128/IAI.71.9.4831-4841.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, Cutler JE, King J, Palmer D. 2004. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot. Cell 3:1028–1035. 10.1128/EC.3.4.1028-1035.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Russell DG. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Shen G, Whittington A, Song K, Wang P. 2010. Pleiotropic function of intersectin homologue Cin1 in Cryptococcus neoformans. Mol. Microbiol. 76:662–676. 10.1111/j.1365-2958.2010.07121.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer DA, Thompson JK, Li L, Prat A, Wang P. 2006. Gib2, a novel Gbeta-like/RACK1 homolog, functions as a Gbeta subunit in cAMP signaling and is essential in Cryptococcus neoformans. J. Biol. Chem. 281:32596–32605. 10.1074/jbc.M602768200 [DOI] [PubMed] [Google Scholar]
- 39.Tolkacheva T, McNamara P, Piekarz E, Courchesne W. 1994. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein α-subunit homolog. Infect. Immun. 62:2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Shen G, Gong J, Shen D, Whittington A, Qing J, Treloar J, Boisvert S, Zhang Z, Yang C, Wang P. 2014. Noncanonical Gbeta Gib2 is a scaffolding protein promoting cAMP signaling through functions of Ras1 and Cac1 in Cryptococcus neoformans. J. Biol. Chem. 289:12202–12216. 10.1074/jbc.M113.537183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue C, Bahn YS, Cox GM, Heitman J. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667–679. 10.1091/mbc.E05-07-0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P, Heitman J. 1999. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2:358–362. 10.1016/S1369-5274(99)80063-0 [DOI] [PubMed] [Google Scholar]
- 43.Alspaugh JA, Perfect JR, Heitman J. 1998. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet. Biol. 25:1–14. 10.1006/fgbi.1998.1079 [DOI] [PubMed] [Google Scholar]