Abstract
Bacterial uptake of phosphate is usually accomplished via high-affinity transporters that are commonly regulated by two-component systems, which are activated when the concentration of phosphate is low. Mycobacterium smegmatis possesses two such transporters, the widely distributed PstSCAB system and PhnDCE, a transporter that in other bacteria mediates the uptake of alternative phosphorus sources. We previously reported that the transcriptional regulator PhnF controls the production of the Phn system, acting as a repressor under high-phosphate conditions. Here we show that the phnDCE genes are common among environmental mycobacteria, where they are often associated with phnF-like genes. In contrast, pathogenic mycobacteria were not found to encode Phn-like systems but instead were found to possess multiple copies of the pst genes. A detailed biochemical analysis of PhnF binding to its identified binding sites in the phnD-phnF intergenic region of M. smegmatis has allowed us to propose a quantitative model for repressor binding, which shows that a PhnF dimer binds independently to each site. We present the crystal structure of M. smegmatis PhnF at 1.8-Å resolution, showing a homodimer with a helix-turn-helix N-terminal domain and a C-terminal domain with a UbiC transcription regulator-associated fold. The C-terminal domain crystallized with a bound sulfate ion instead of the so far unidentified physiological ligand, allowing the identification of residues involved in effector binding. Comparison of the positioning of the DNA binding domains in PhnF with that in homologous proteins suggests that its DNA binding activity is regulated via a conformational change in the linker region, triggering a movement of the N-terminal domains.
INTRODUCTION
As inorganic phosphorus is an essential and frequently limiting nutrient, the mechanisms of its uptake are important systems for most bacteria. Bacteria possess both high- and low-affinity phosphate transport systems, the former of which are induced under inorganic phosphate (Pi)-limited conditions (1, 2). The high-affinity, ATP-binding cassette (ABC)-type transport system Pst (phosphate-specific transport) consists of four components, PstSCAB, and recognizes free phosphate as its substrate. Pst systems have been well characterized in a number of bacteria, including Escherichia coli (1), Sinorhizobium meliloti (3, 4), and Bacillus subtilis (5, 6). In Mycobacterium tuberculosis, pstS1 and pstS2 have been identified as virulence factors (6, 7). Expression of the pstSCAB operons is generally induced under Pi-limited conditions and is mediated by the two-component regulatory systems PhoBR in Gram-negative microorganisms (8–10), PhoPR in most Gram-positive organisms (11, 12), and SenX3-RegX3 in Mycobacterium smegmatis (13). In E. coli and M. smegmatis, repression of the phosphate starvation response requires the Pst transporter, and mutations in pstS result in constitutive target gene expression (1, 14).
We have previously identified a three-gene operon (phnDCE) in M. smegmatis which encodes a second high-affinity phosphate transport system. Most Phn systems studied to date are responsible for the uptake of alternative phosphorus sources, such as phosphonates (compounds containing a direct carbon-phosphorus bond) or phosphite (15–17). In contrast, the M. smegmatis PhnDCE transporter does not recognize these substrates and is instead specific for Pi (18). Further, we have shown that the transcriptional regulator PhnF controls phnDCE expression by acting as a repressor under phosphate-replete conditions. Induction of phnDCE upon phosphate starvation is further enhanced by the two-component system SenX3-RegX3 (19). The phnF gene is found upstream of and in the opposite orientation to phnDCE, and its expression is autorepressed by PhnF and additionally activated by SenX3-RegX3.
While M. tuberculosis possesses multiple copies of genes encoding Pst systems (20, 21), M. smegmatis contains only a single copy of the pstSCAB genes (4, 22). It therefore appears that the PhnDCE transporter, regulated primarily by PhnF, offers M. smegmatis a mechanism that is an alternative to that of multiple Pst systems for the efficient uptake of Pi under growth-limiting conditions. The work presented here aims to improve our understanding of the role of PhnF by structural and biochemical characterization and to establish a framework for its interaction with both its target DNA and its activating ligand.
MATERIALS AND METHODS
Sequence analysis.
To identify orthologues of the phnDCE and phnF genes, all actinobacterial genomes available in October 2010 at the NCBI website (http://www.ncbi.nlm.nih.gov) were searched with the BLASTP program (23) using the sequences of M. smegmatis PhnD (MSMEG_0649), PhnC (MSMEG_0647), and PhnE (MSMEG_0646) as queries. All genomes containing hits for all three queries encoded by adjacent genes were then subjected to a further BLASTP search using M. smegmatis PhnF (MSMEG_0650) as the query. The results from this analysis were updated in August 2013 using the 154 actinobacterial genomes available in the MicrobesOnline database (24).
Cloning and expression.
The region encoding the C-terminal ligand binding domain of PhnF (C-PhnF; residues 76 to 244) from Mycobacterium smegmatis and the full-length phnF gene were cloned from genomic DNA. The boundary between the N-terminal DNA binding domain and the C-terminal ligand binding domain (C-PhnF) was located by alignment with the gene sequence of PhnF from Escherichia coli (25). C-phnF and full-length phnF were cloned into the Gateway system using the nested two-stage PCR protocol described previously (26, 27) with the following primers: full-length phnF forward primer 5′-GGCAGCGGCGCGGTGACAGCGGGCGCG-3′, C-phnF forward primer 5′-GGCAGCGGCGCGATCAGACAACCCCTCGGCATG-3, and full-length and C-phnF reverse primer 5′-GAAAGCTGGGTGTCACGAAACGATTGCGG-3′. Clones were verified by sequencing and transferred into the pDEST17 expression vector to produce a His-tagged protein (C-PhnF) or pDEST566 to produce a His-tagged maltose-binding protein (MBP) fusion protein (full-length PhnF). Expression was carried out in E. coli BL21(DE3) cells grown in 500 ml of autoinduction medium ZYM-5052 (28) in baffled 2-liter flasks with shaking at 160 rpm. Cultures were grown at 310 K for 4 h and then at 291 K overnight.
Purification.
Cells from the expression culture were harvested by centrifugation at 5,000 × g for 20 min at 277 K. These were resuspended in lysis buffer (20 mM HEPES [pH 7.5], 150 mM NaCl) with 5 or 10 mM imidazole (for full-length PhnF and C-PhnF, respectively) and the addition of lysis mix (final concentrations, 200 μg/ml lysozyme, 50 μg/ml RNase A, 10 μg/ml DNase I, 2 mM MgCl2) and Roche Complete mini EDTA-free protease inhibitor tablets. Cells were lysed using a cell disruptor at 18 kPa. The lysate was clarified by centrifugation at 26,000 × g for 20 min at 277 K, and the soluble fraction was filtered through a 0.45-μm-pore-size filter.
C-PhnF was purified by immobilized metal affinity chromatography (IMAC) using a gravity-flow TALON column (Clontech). The clarified supernatant was passed through the column, and the column was washed with lysis buffer containing 20 mM imidazole. Protein was then eluted with lysis buffer containing 150 mM imidazole. This protein was concentrated to 5 ml in a 10-kDa-cutoff centrifuge concentrator and further purified by Superdex 75 size exclusion chromatography using 20 mM HEPES (pH 7.5), 150 mM NaCl. This protein was then used for crystallization. Full-length PhnF was purified by IMAC as described above, the His6-MBP fusion was removed by proteolysis with recombinant tobacco etch virus protease, and the sample was reapplied to the TALON column. The flowthrough containing PhnF was collected and further purified by gel filtration as described above. Protein purity was monitored throughout by SDS-PAGE.
Crystallization and data collection.
Initial crystallization conditions were obtained using a Honeybee (Cartesian Dispensing Systems) nanoliter robotic system in a sitting-drop format with 100 nl of protein solution mixed with 100 nl well solution in Intelliplates (Art Robbins Instruments) and a set of in-house screens covering 480 conditions (27).
(i) C-PhnF.
Diffraction-quality crystals were produced by hanging-drop vapor diffusion, in which 1 or 2 μl of protein (at 2 to 5 mg/ml) was mixed with an equal amount of well solution. Initial crystals were produced using well solution containing 1.0 M ammonium sulfate, 0.1 M lithium sulfate, and 0.1 M Tris HCl (pH 8.5). These crystals were used for microseeding of further droplets in 0.65 M ammonium sulfate, 0.1 M lithium sulfate, and 0.1 M Tris HCl (pH 8.5).
(ii) PhnF.
The best crystals were produced by hanging-drop vapor diffusion by mixing 1 μl of protein solution (at 5 mg/ml) with 1 μl of well solution. The well solution contained 0.1 M sodium cacodylate (pH 8.0) and 0.8 M sodium acetate. All crystals were incubated in a solution of the appropriate mother liquor with 25% (vol/vol) glycerol and flash cooled in liquid nitrogen. Crystals were maintained at 100 K during X-ray exposure in a stream of cooled nitrogen gas.
Data collection, structure solution, and refinement.
X-ray diffraction data were collected to a resolution of 1.8 Å for both PhnF and C-PhnF using an in-house rotating anode X-ray generator (Rigaku Micromax-007 HF) with an image plate detector (Mar Scanner 345). All other computing was carried out in the CCP4 suite unless otherwise stated (29).
For C-PhnF, the structure was solved by molecular replacement using the BALBES system (30) and the structure with PDB accession number 2P19 as the model. Automated model building was performed with the ARP/wARP package (31), and structure refinement was performed with the REFMAC program (32) interspersed with manual rebuilding using the Coot program (33). The structure of PhnF was solved by molecular replacement using PHASER software (34) and C-PhnF as the search model and was refined as described above. Data collection and refinement statistics can be found in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics of M. smegmatis C-PhnF and PhnFa
| Parameterb | Value(s) forc: |
|
|---|---|---|
| C-PhnF | PhnF | |
| Space group | C2 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 114.6, 78.2, 150.5 | 64.5, 77.0, 66.9 |
| α, β, γ (°) | 90.0, 125.4, 90.0 | 90.0, 113.5, 90.0 |
| Data collection statistics | ||
| Wavelength (Å) | 1.54179 | 1.54179 |
| Resolution (Å) | 25.28–1.90 (2.00–1.90) | 23.24–1.80 (1.90–1.80) |
| Rmerge | 0.069 (0.491) | 0.085 (0.466) |
| I/σ〈I〉 | 8.5 (1.6) | 3.7 (1.6) |
| No. of observed reflections | 461,392 (36,151) | 250,529 (35,481) |
| No. of unique reflections | 68,334 (9,909) | 55,650 (8,126) |
| Completeness (%) | 100 (100) | 100 (100) |
| Multiplicity | 6.8 (3.6) | 4.5 (4.4) |
| Refinement statistics | ||
| Resolution (Å) | 98.06–1.90 (1.95–1.90) | 61.31–1.80 (1.85–1.80) |
| No. of reflections | 64,877 (4,751) | 52,806 |
| Completeness (%) | 99.95 | 99.9 |
| Rwork/Rfree | 0.181/0.196 (0.255/0.277) | 0.213/0.248 (0.377/0.371) |
| RMSD | ||
| Bond length (Å) | 0.015 | 0.015 |
| Bond angle (°) | 1.60 | 1.55 |
| Mean B factor (Å2) | 30.1 | 28.8 |
| PDB accession no. | 3F8L | 3F8M |
Of the modeled residues for C-PhnF, 98.6% lie in the preferred region, 1.4% lie in the allowed region, and no residues lie in the disallowed areas. For PhnF, 98.0% of the modeled residues lie in the preferred region, 1.9% lie in the allowed region, and one residue (Gln 92 on chain C) lies just within the disallowed area; on inspection, this residue lies on a loop region where the electron density for the side chain is weak, aside from that for the terminal amide group, which is clear.
Rmerge = Σ|I – 〈I〉|/ΣI × 100, where I is the intensity of a reflection and 〈I〉 is the average intensity; RMSD, root mean square deviation.
Values for the highest-resolution shell are shown in parentheses.
Electrophoretic mobility shift assays (EMSAs).
A 377-bp fragment containing the intergenic region between phnF and phnD was PCR amplified using primers PphnFR and PphnDR (19) and end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Roche) according to the manufacturer's instructions. Target DNA carrying mutations in the PhnF binding sites was created as described previously (19). Binding reactions were carried out in binding buffer (20 mM Tris HCl [pH 8], 50 mM KCl, 1 mM dithiothreitol, 4% [wt/vol] glycerol, 5 μg ml−1 salmon sperm DNA), and the reaction mixtures contained ca. 2,000 cpm radiolabeled DNA and various concentrations of PhnF, as indicated below. For competition experiments, 100 ng of unlabeled DNA containing either the wild-type sequence (TGGTATAGACCA for both binding sites) or a sequence with mutations in both PhnF binding sites (TGTGATAGACAC] in site F1, TGGTATAGCACA in site F2 [the mutated sites are underlined]) (19) was added to the binding reaction mixture. PhnF was diluted in 20 mM Tris HCl (pH 8), 150 mM NaCl, 50 μg ml−1 bovine serum albumin. The reaction mixtures were incubated at room temperature for 20 min, followed by electrophoresis on a 6% (wt/vol) acrylamide (19:1 acrylamide/bisacrylamide) gel in 1× TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA) at 300 V for 20 min. Bands were detected using a phosphorimager.
Mathematical model for PhnF-DNA binding.
On the basis of the palindromic recognition sequence of PhnF in the intergenic region of phnF-phnD, it seemed likely that each of the F1 and F2 sites can be bound by a PhnF2 dimer. Assuming that proteins dimerize prior to DNA binding with dimerization dissociation constant Kd, the free dimer concentration, [PhnF2], depends on the total PhnF concentration, [PhnFtot], as follows:
| (1) |
To model the DNA binding probabilities shown in Fig. 1D and E, a thermodynamic framework for protein-DNA interactions was applied (35). In DNA fragments containing only site F1 (Fig. 1E), the probability that site F1 bound by PhnF2 would be found (p1) reads
| (2) |
where is the PhnF2-F1 dissociation constant. Accordingly, the probability that site F1 will be found to be unbound () is given by 1 − p1. Expansion of the square root in equation 1 shows that only for small ratios of [PhnFtot]/Kd does the dimer concentration depend quadratically on the total PhnF level, such that in this limit the binding probability of site F1 approximates a Hill function with a Hill coefficient of n equal to 2:
| (3) |
where is the effective DNA binding constant of PhnFtot. Analogous expressions were derived for site F2 with the PhnF2-F2 dissociation constant . In the DNA fragment containing both the F1 and the F2 sites (Fig. 1D), the probability that site F1 or site F2 exclusively would be found to be bound (p1/2; corresponding to a partial band shift in Fig. 1B) is given by
| (4) |
and the probability that both sites would simultaneously be bound (p1+2; corresponding to a supershift in Fig. 1B) is given by
| (5) |
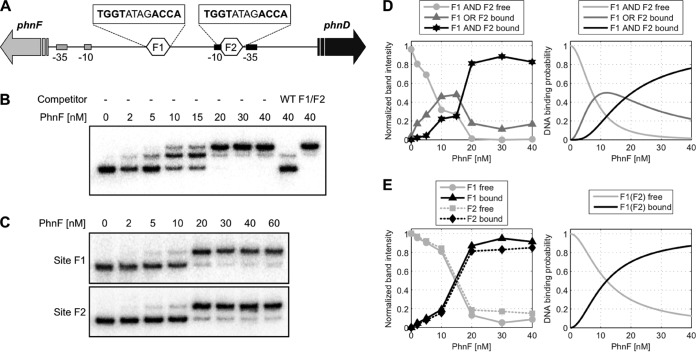
FIG 1.
Electrophoretic mobility shift assays with PhnF. Radiolabeled DNA containing the phnF-phnD intergenic region was incubated with various concentrations of PhnF, as indicated, followed by electrophoresis on a 6% acrylamide gel. (A) Schematic of target DNA region. The intergenic region between phnF and phnD is shown. The two PhnF target sites are indicated as hexagons labeled F1 and F2, and their sequences are given above. The −10/−35 promoter elements for phnF and phnD are indicated by gray and black rectangles, respectively. (B) Target DNA of wild-type sequence. Competitor DNA was 100 ng the unlabeled PCR product of the wild-type (WT) sequence (TGGTATAGACCA for both binding sites) or the same fragment carrying mutations in both PhnF binding sites (F1/F2; TGTGATAGACAC in site F1, TGGTATAGCACA in site F2 [the mutated sites are underlined]). (C) Target DNA of the same fragment used in the assay whose results are presented in panel B, but with only the phnF-proximal (site F1) or phnDCE-proximal (site F2) PhnF binding site being intact. (D) Quantification of experimental band intensities from the bands in panel B (left) and model prediction (right). To correct for variations in the total amount of DNA loaded per lane, the graph shows the experimental band intensities relative to the total intensity in each lane. The theoretical curves correspond to the probability of finding both F1 and F1 free (light gray), either F1 or F2 occupied (dark gray), and both sites occupied by a PhnF2 dimer (black). (E) Quantification of experimental band intensities from the bands in panel C (left) and model prediction (right). Normalization of the experimental band intensities was done as described in the legend to panel D. The theoretical curves show the DNA binding probability for binding of PhnF2 to F1, which is the same as that for binding to F2. The model parameters for panels D and E are Kd equal to 100 nM for the PhnF2 dimerization constant and equal to equal to 1 nM for the PhnF2-DNA binding constant.
Protein structure accession numbers.
The structure factors and final structural models both for the C-terminal domain of PhnF and for full-length PhnF have been deposited in the Protein Data Bank (PDB) with the accession numbers 3F8L and 3F8M, respectively.
RESULTS
Distribution of Phn systems in actinobacteria.
Previous studies of phosphate transport in mycobacteria have shown that M. tuberculosis contains several copies of genes encoding a PstSCAB transporter (21), whereas M. smegmatis contains one operon each for a PstSCAB transporter and a PhnDCE transporter (18). To gain a better understanding of the distribution of these transport systems among the actinobacteria, we searched the currently available genomes of these bacteria for orthologues of M. smegmatis PhnDCE. Our results showed that none of the pathogenic mycobacteria for which genomes are available encode a PhnDCE-like phosphate transport system but instead have several copies of the Pst phosphate transport system (Table 2). In contrast, 15 of the analyzed genomes of environmental mycobacteria and related actinobacteria possessed a complete putative phnDCE operon, with Mycobacterium vanbaalenii and Clavibacter michiganensis containing two such operons. None of these bacteria contained multiple copies of pst genes (Table 2).
TABLE 2.
Comparative genome analysis of phosphate transport systems and PhnF-like regulators in actinobacteria
| Species | Presence of multiple Pst systemsa | PhnDCE systemb | PhnF orthologuec |
|---|---|---|---|
| Mycobacterium tuberculosis H37Rv | Yes (3) | ||
| Mycobacterium bovis AF2122/97 | Yes (3) | ||
| Mycobacterium leprae Br4923 | Yes (2+) | ||
| Mycobacterium avium 104 | Yes (3+) | ||
| Mycobacterium avium subsp. paratuberculosis K10 | Yes (4) | ||
| Mycobacterium smegmatis mc2155 | No | MSMEG_0649 | MSMEG_0650 |
| Mycobacterium vanbaalenii PYR-1 | No | Mvan_2904 | Mvan_0056d |
| Mvan_5877 | Mvan_5874 | ||
| Mycobacterium sp. strain JLS | No | Mjls_1320 | Mjls_1316 |
| Corynebacterium glutamicum R | No | cgR_2192 | cgR_2194 |
| Corynebacterium amycolatum SK46 | No | CORAM0001_1913 | CORAM0001_1911 |
| Corynebacterium ammoniagenes DSM 20306 | No | HMPREF0281_02511 | HMPREF0281_02513 |
| Nocardia farcinica IFM 10152 | No | nfa35300 | nfa35320 |
| Rhodococcus opacus B4 | No | ROP_24270 | ROP_24260 |
| Streptomyces violaceusniger Tu 4113 | No | Strvi_5929 | Strvi_5948d |
| Frankia sp. strain CcI | No | Francci3_2155 | Francci3_4270d |
| Brevibacterium linens BL2 | No | BlinB01000793 | BlinB01000795 |
| Arthrobacter aurescens TC1 | No | AAur_1205 | AAur_2055d |
| Saccharopolyspora erythraea NRRL 2338 | No | SACE_6645 | SACE_6649 |
| Clavibacter michiganensis NCPPB 382 | No | CMM_0367e | CMM_0381e |
| CMM_2193 | |||
| Stackebrandtia nassauensis DSM 44728 | No | Snas_4961 | Snas_5635e |
The number of pstS genes present in the genome is given in parentheses; +, the presence of an additional pseudogene.
Only locus tags for the first gene (phnD orthologue) are shown for each operon.
Gene identifiers are given as locus tags.
PhnF orthologue not directly associated with Phn transporter genes.
Separated from the phnDCE operon by 10 other phn genes; probably involved in phosphonate metabolism.
The phnDCE operon of M. smegmatis is associated with an additional gene, phnF, which encodes a transcriptional regulator that acts as a repressor of phnDCE expression (19). We therefore searched the actinobacterial genomes encoding a PhnDCE transporter for the presence of orthologues of M. smegmatis PhnF. Eleven of the transporter operons possessed a phnF-like gene in their genomic neighborhood. In the remaining four genomes, a phnF-like gene was encoded elsewhere on the chromosome, and in the two species containing two phnDCE operons, only one operon was associated with such a gene (Table 2). PhnDCE transporters were originally identified to be uptake systems for phosphonates, an alternative source of phosphorus that can be utilized by many bacteria, especially inhabitants of the soil (36). Interestingly, only two of the transport operons identified by our analysis were associated with the remaining phn genes required for the degradation of phosphonates (Table 2). The PhnDCE system of M. smegmatis does not recognize phosphonates as a substrate, consistent with the absence of the degradative genes (18), and this may also be the case for the remaining actinobacterial systems identified here.
Our findings indicate that possession of a PhnDCE phosphate uptake system is a common trait in environmental actinobacteria, where such systems may have originated as phosphonate transporters. The pathogenic mycobacteria appear to have lost these systems and have possibly compensated for this loss through duplication of their pst genes. On the basis of the conserved genomic arrangement and co-occurrence of phnDCE and phnF genes, it appears likely that production of PhnDCE transporters is generally regulated by PhnF-like repressors in the actinobacteria.
PhnF binds to two sites in the phnF-phnD intergenic region.
We have previously proposed that PhnF binds to two sites in the phnF-phnD intergenic region in M. smegmatis, based on genetic evidence (19). To determine whether purified PhnF is indeed able to bind to these putative binding sites, we carried out DNA binding studies using electrophoretic mobility shift assays (EMSAs) with a DNA fragment encompassing the intergenic region between phnF and phnD (Fig. 1A). A band shift of this fragment was detected in the presence of PhnF at a concentration of 2 nM or higher, and a supershift occurred at in the presence of PhnF at a concentration of 10 nM or higher (Fig. 1B). A complete shift of all DNA was observed from 20 nM PhnF. This is consistent with PhnF binding to two separate sites on the target DNA, where only one site is occupied at low protein concentrations (giving a shift), followed by occupation of both sites simultaneously at higher protein concentrations (resulting in a supershift). Binding of PhnF was specific, as the addition of excess unlabeled target DNA abolished the shift, whereas the addition of unlabeled DNA carrying mutations in both proposed binding sites had no effect (Fig. 1B, last two lanes). This confirmed that the two inverted repeats in the phnF-phnD intergenic region indeed constitute the PhnF binding sites.
The presence of two binding sites in the intergenic region raised the question of whether the binding displayed cooperative behavior. To address this, additional binding assays were carried out with fragments containing only the phnF-proximal binding site (site F1) or only the phnD-proximal binding site (site F2). As shown in Fig. 1C, PhnF binds to both of these fragments in a very similar pattern, with a partial shift observed from 5 nM protein and a complete shift observed at 20 nM protein. No supershift occurred, consistent with the presence of only one binding site in the target DNA. In contrast to the more gradual shifting of DNA containing both binding sites, the single-site fragments both showed a steep response between 10 nM and 20 nM protein concentrations, which was also reflected in the sigmoidal curve after quantification of band intensities (Fig. 1E, left).
These results showed that the two sites displayed almost identical binding behavior and, due to the sigmoidal dependency, suggested that binding of PhnF to each individual site is cooperative. In fact, on the basis of the palindromic recognition sequence (TGGT-N4-ACCA) (19), it seemed likely that each site is bound by a dimer of PhnF. In order to examine this, we set up a theoretical model that included protein dimerization prior to binding of DNA. This model indeed produced the same dose-response behavior observed experimentally (Fig. 1E, right). In order to reproduce the sigmoidal shape of the experimental data, the protein dimerization affinity constant was required to be high (Kd ≈ 100 nM) compared to the total PhnF concentrations used in the band-shift assays. Under these conditions, the PhnF monomer-dimer equilibrium shifted toward the dimer with increasing total PhnF concentration, and together with a high protein dimer-DNA binding affinity ( nM), this led to an effective Hill coefficient of n nearly equal to 2. Importantly, both the dimerization and the DNA binding affinity constants were chosen from a typical range of in vivo parameters for bacterial transcription factors (37–40), suggesting that the observed sigmoidal binding curves can be explained by dimerization of PhnF alone and do not require further, unknown mechanisms.
Adapting the same model with identical parameters for a DNA fragment containing two binding sites for PhnF could also accurately reproduce the more complex binding behavior observed for the wild-type DNA fragment (Fig. 1D). Here the model predicted that for increasing PhnF concentrations the probability of finding either site F1 or site F2 occupied first increases and then decreases again (Fig. 1D, right, dark gray curve). Within the model this decrease is due to occupation of the second site at higher PhnF concentrations, which, accordingly, increases the probability of PhnF binding to F1 and F2 simultaneously (Fig. 1D, right, black curve). Together, these results show that binding of PhnF to each individual binding site is cooperative and that this cooperativity likely arises from dimerization prior to DNA binding. Moreover, the binding of both sites in the intergenic region is consistent with a model without cooperativity between the individual sites, suggesting that PhnF binds sites F1 and F2 independently.
PhnF represses transcription of the phnDCE operon and of its own structural gene under phosphate-replete conditions, and repression is relieved when the cells enter phosphate limitation (19). To test whether phosphate had an effect on DNA binding, the EMSAs were repeated in the presence of different phosphate concentrations. Only very high concentrations (above 10 mM) had any adverse effects on binding (data not shown), but as the internal free phosphate concentration of M. smegmatis is below 0.5 mM (our unpublished data), this is not likely to be of physiological relevance. Because most GntR-family transcriptional regulators repress their target genes in the absence of their ligand (41–46), it appears likely that PhnF responds to a molecule present in phosphate-starved but not phosphate-replete cells. This is discussed in more detail below.
Structure of PhnF.
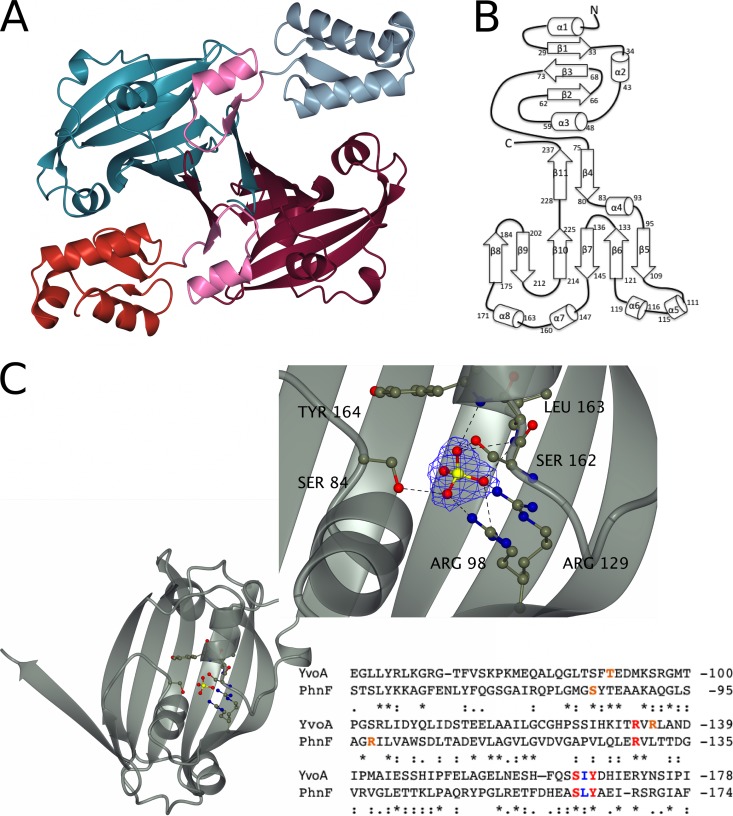
In order to better understand the nature of phosphate regulation by PhnF, we set out to determine its structure. PhnF crystallizes as a homodimer with one dimer in the asymmetric unit (Fig. 2A). PhnF is composed of two domains. The N-terminal domain, comprising residues 10 to 73, consists of a canonical winged helix-turn-helix (wHtH) DNA binding domain with an α1-β1-α2-α3-β2-β3 topology (Fig. 2B) and is connected to the ligand binding domain by a linker region of 21 amino acids (Fig. 2A, pink residues), 5 of which form the β4 strand. The C-terminal domain has a chorismate lyase fold (47), comprising a six-stranded β-sheet at its core, which is extended by strands β11 and β4 from the second monomer. Dimerization is mediated by two equivalent sets of interactions to give a dimer containing two twisted eight-stranded β-sheets. This mode of dimerization is identical to that observed in the crystal structures of other members of the GntR/HutC family of transcriptional regulators, including YvoA, YurK, and YydK from Bacillus subtilis and PhnF from E. coli (PDB accession numbers 2WVO, 2IKK, 3BWG, and 2FA1) (48, 49). We also independently crystallized the C-terminal domain of PhnF (C-PhnF). The structure of this domain alone has a fold and a dimerization interface identical to those seen in the full-length protein.
FIG 2.
Key structural features of PhnF from Mycobacterium smegmatis. (A) Crystal structure of the PhnF homodimer. One monomer is shown in gray (N-terminal domain) and cyan (C-terminal domain), and the other monomer is shown in red (N-terminal domain) and purple (C-terminal domain). The linker region of both monomers is shown in pink. (B) Topology plot of the PhnF monomer. (C) Ribbon representation of the effector domain showing the residues involved in SO42− binding in ball-and-stick representation. (Inset) A detailed view of the binding site. Electron density shown is a 2Fo − Fc synthesis contoured at approximately 1σ (0.55 electron/Å2). An alignment of the protein sequences for the C-terminal domains of YvoA and PhnF is shown for comparison. Red, amino acids that form the same contacts with SO42− through their side chains in both proteins; blue, amino acids that form equivalent contacts through their main chains; orange, amino acids that form similar contacts with SO42− but do not occupy similar positions in the binding site. Alignment was performed with the Clustal Omega program (56).
The program DaliLite (50) reports (as of 20 May 2014) 22 different proteins with Z-scores greater than 10 to be near structural neighbors. The nearest structural neighbor was identified as being the GntR/HutC-family transcriptional regulator YvoA from B. subtilis, which is involved in the regulation of the N-acetylglucosamine-degrading pathway (PDB accession number 2WV0, Z-score = 21.5, sequence identity = 31%) (Fig. 3A, green strand) (48). The second-nearest neighbor was identified to be an uncharacterized gene product from Staphylococcus aureus (PDB accession number 2OOI) with a Z-score of 20.3 and a sequence identity of 12%. The vast majority of matches are results from structural genomics programs and consist of structures encoding putative GntR/HutC-family regulators. This demonstrates that the fold of PhnF is confined to enzymes of the GntR/HutC family of regulators, of which PhnF is a member; Fig. 3A shows a structural comparison between the C-terminal domains of PhnF and three of its related proteins.
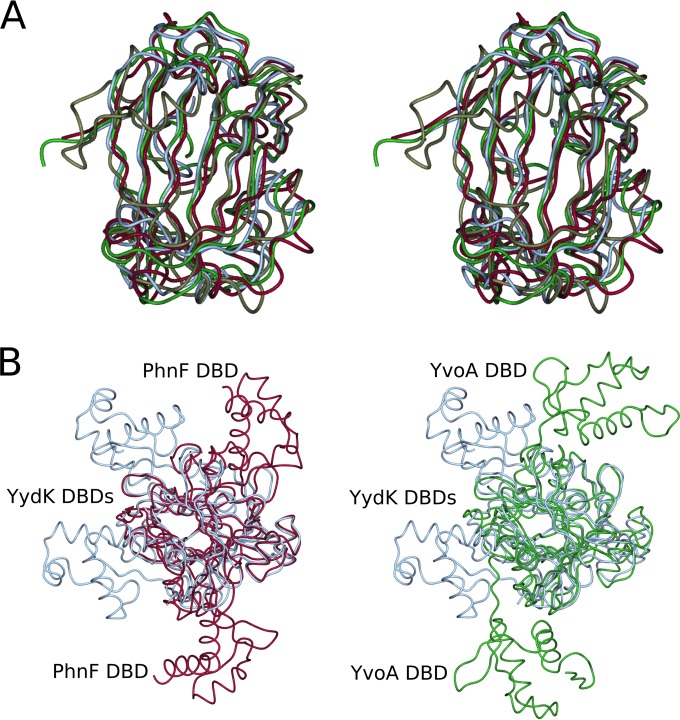
FIG 3.
Structural comparison of PhnF with related proteins. (A) Stereo image of the C-terminal domain of PhnF (purple) superposed with chorismate lyase (gray, PDB accession number 1G1B), B. subtilis YydK (blue, PDB accession number 3BWG), and B. subtilis YvoA (green, PDB accession number 2WVO). (B) Superpositions of YydK (blue, PDB accession number 3BWG) with YvoA (green, PDB accession number 2WVO) and PhnF (purple) looking down the β-sheet and showing the different positions of the N-terminal DNA binding domains (DBDs).
The C-terminal domain of PhnF was crystallized in a high concentration of sulfate ions, as was YvoA from B. subtilis (48), and a sulfate ion is found in the structures of both these proteins. This ion is bound in the same position in both C-PhnF and YvoA, being contacted by residues from strands β5 and β6, the loop region between helices α7 and α8, and helix α4 (Fig. 2C). This sulfate ion forms hydrogen bonds with the side chains of R129, S162, R98, and S84 and with the main chain peptide NH groups of L163 and Y164. The sulfate-binding site of PhnF shows a great degree of similarity with that of YvoA, with several of the residues being conserved. R129, S162, and Y164 form contacts equivalent to those made by R133, S165, and Y167 in the B. subtilis protein, and L163 forms the same contact through the backbone as I166 of YvoA. There are, however, some differences in binding that likely allow the two proteins to have different substrate specificities. Whereas in the B. subtilis protein R135 and T90 make side chain hydrogen bonds with the sulfate ion, these interactions are replaced in C-PhnF by hydrogen bonds with R98 and S84. The interaction between the sulfate ion and R135 is also observed in the related trehalose repressor from B. subtilis TreR (R128; PDB accession number 2OGG) (51). Although anions commonly bind to the N termini of two or more α-helices, mutations in the residues involved in binding the sulfate ion effectively obliterated effector binding in YvoA (48), suggesting that this is indeed the region of the protein involved in effector binding.
DISCUSSION
Regulation of PhnF activity.
PhnF is a regulator of both the phnDCE operon and of its own transcription (19). When M. smegmatis is grown under phosphate-limiting conditions, the repression of the phnDCE operon is relaxed. As mentioned above, other members of the HutC regulator family bind to their target DNA only in the absence of their respective ligands (41–46). Accordingly, our hypothesis for the mechanism of repression by PhnF is that the protein recognizes a molecule present only under phosphate-starved conditions, which would trigger the derepression of the PhnF target genes. In the absence of this ligand, i.e., under phosphate-replete conditions, PhnF remains bound to DNA and the target genes are repressed.
However, the identity of the ligand to which PhnF responds is unknown. Unlike E. coli, M. smegmatis is unable to utilize phosphonates or phosphite as a phosphorus source (18). Furthermore, repression of phnF and phnDCE expression by PhnF in M. smegmatis is relieved by phosphate starvation alone, regardless of the presence of any alternative phosphorus compounds. It is therefore unlikely that M. smegmatis PhnF responds to a phosphonate ligand. A simpler hypothesis—that PhnF binds phosphate directly and that this allows it to bind DNA—is also unlikely to be correct, because DNA binding was not affected by the absence or presence of physiological concentrations of phosphate. It therefore appears likely that M. smegmatis PhnF responds to a small molecule that is produced or accumulates in the cell as it enters phosphate starvation. Due to the large-scale metabolic rearrangements of cellular metabolism upon phosphate limitation, including changes in energy metabolism and adaptations that reduce the cell's phosphorus demand (52, 53), it is difficult to predict candidate molecules. The presence of sulfate in the ligand binding pocket of C-PhnF may suggest that the true ligand is a phosphate-containing species, based on the structural similarity between sulfate and phosphate groups, but we have no experimental evidence to date.
Structural mechanism of DNA binding by PhnF.
PhnF crystallized as a homodimer, with the N-terminal DNA binding domains being positioned on opposite sides of the effector domains (Fig. 3B, purple strand). This position means that in their current orientation the two domains would be unable to simultaneously contact the same DNA molecule. As described above, we have shown that each dimer of PhnF binds to one DNA binding site and that PhnF dimerization occurs before binding to the DNA. Thus, the structure observed in the crystal must undergo a rearrangement that brings the two N-terminal DNA binding domains closer together in order to allow PhnF to functionally bind DNA. The structurally related GntR/HutC transcription factor YvoA from B. subtilis has a similar arrangement of its DNA binding domains (Fig. 3B, green strand) (48). In this case, the authors postulated that the structure that they determined is that of the non-DNA binding form of the protein, which then undergoes a shift in N-terminal domain orientation in order to allow the formation of a 1:1 DNA-protein complex. In addition, they observed a sulfate ion bound to the effector domain, which they suggest mimics the true ligand and so triggers the protein to adopt a non-DNA binding conformation, thus explaining the relative positioning of the N-terminal domains.
The existence of an alternative orientation of the N-terminal DNA binding domains is supported by the report of the structure of YydK, which is structurally closely related to PhnF (Z-score = 19.7) (PDB accession number 3BWG). The N-terminal domains in this structure are positioned closely together in such a way that they could make contact with the same DNA binding site (Fig. 3B, blue strand). That the structure of YydK shows the authentic DNA binding orientation of the domains is lent weight by the observation of how the GntR/HutC-family regulator FadR from E. coli binds to its DNA binding site (54). The DNA binding domain dimer of FadR can be overlaid with that of YydK, with a reported C-α root mean square deviation of 5.3 Å, while differences between the single DNA binding domains are much smaller. This confirms that the orientation of the domains is very similar in both instances. The structure of YydK does not contain an anion in the effector domain, unlike the structures of YvoA and the related proteins YurK and TreR (described above), which all contain sulfate ions bound in equivalent positions and show DNA binding domains in the non-DNA binding orientation on opposing faces of their effector domains. Comparison of these structures with the structure of PhnF shows that the N-terminal domains in the latter occupy a position between the positions seen in YvoA and YydK (PDB accession number 3BWG) (48).
Although the structure of full-length PhnF described in this work does not contain a bound anion and would be postulated to be in the DNA binding form, this does not seem to be the case. Nevertheless, a small movement in the linker region between the N- and C-terminal domains would be sufficient to bring the N-terminal domains into the position occupied by those of YydK and thus render them capable of binding DNA. Comparison of this region of the two structures shows that there is a turn in the backbone of YydK at residues 71 to 73, which orients the N-terminal domain to lie above the dimerization interface. In contrast, this turn is absent in PhnF, but the equivalent residues, 73 to 75, form only one hydrogen bond with the rest of the protein, between the backbone of Lys75 and the side chain of Gln92, and this might easily be broken to allow movement of the N-terminal domain about this hinge point. Use of the web server HINGEPROT (55) identified a hinge region for the PhnF dimer at residue 74, providing further support for the possibility of movement in this region of the protein.
Effector binding site of PhnF.
The identity of the effector molecule for PhnF is currently unknown, but it seems likely that the endogenous ligand is a phosphate-containing molecule, as is postulated to be the case for YvoA. While there are large similarities in the binding sites of M. smegmatis PhnF and B. subtilis YvoA, the differences in side chain orientation are likely to be responsible for the different specificities of the proteins (51). It is possible that any allosteric change that would take place on ligand binding is not observed in PhnF in the case of binding sulfate, because the anion is too small in comparison to the size of the physiological effector molecule to make the contacts necessary to drive a conformational change.
Conclusions.
The crystal structure of PhnF shows that the protein consists of an N-terminal winged helix-turn-helix DNA binding domain and a C-terminal ligand-binding domain of the UbiC transcription regulator-associated (UTRA) fold, as predicted by sequence alignment (19). The protein forms a homodimer, mainly through interactions between the C-terminal domains. Comparison with structurally related proteins suggests that a conformational change in the linker region upon ligand release is likely to facilitate DNA binding in the absence of the ligand by triggering movement of the N-terminal domains. We have demonstrated that phnDCE genes in environmental actinobacteria are generally associated with a phnF gene and are therefore likely to be regulated by PhnF-mediated repression. In most cases there are no associated phosphonate degradation genes, which implies that phosphate specificity in these actinobacteria is likely to be similar to that in M. smegmatis. Moreover, we showed that PhnF binds to two independent binding sites in the phnD-phnF intergenic region and that binding to each site is cooperative and compatible with a thermodynamic model assuming PhnF dimerization prior to DNA binding. The hypothesis that dimerization occurs in the absence of the effector ligand is further supported by the observation of a dimer in the crystal structure where no ligand is present. While the identity of the physiological effector ligand for PhnF remains elusive, it is likely to be a small, phosphate-containing molecule produced in phosphate-starved cells.
ACKNOWLEDGMENTS
V.A.M. thanks EMBO and the Royal Society for funding. J.N.B. and N.J.M. were supported by contract UOAX0301 from the New Zealand Foundation for Research, Science and Technology, awarded to E.N.B. and J.S.L. G.M.C. thanks the Health Research Council New Zealand for funding. G.F. was supported through the priority program SPP1617 (Phenotypic Heterogeneity and Sociobiology of Bacterial Populations) of the Deutsche Forschungsgemeinschaft. Work in the group of S.G. is supported by grants from the Deutsche Forschungsgemeinschaft (GE2164/3-1) and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print 21 July 2014
REFERENCES
- 1.Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon, p 1357–1381 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 2.van Veen HW. 1997. Phosphate transport in prokaryotes: molecules, mediators and mechanisms. Antonie Van Leeuwenhoek 72:299–315. 10.1023/A:1000530927928 [DOI] [PubMed] [Google Scholar]
- 3.Yuan ZC, Zaheer R, Finan TM. 2006. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 188:1089–1102. 10.1128/JB.188.3.1089-1102.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qi Y, Kobayashi Y, Hulett FM. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the Pho regulon. J. Bacteriol. 179:2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allenby NE, O'Connor N, Pragai Z, Carter NM, Miethke M, Engelmann S, Wipat A, Ward AC, Harwood CR. 2004. Post-transcriptional regulation of the Bacillus subtilis pst operon encoding a phosphate-specific ABC transporter. Microbiology 150:2619–2628. 10.1099/mic.0.27126-0 [DOI] [PubMed] [Google Scholar]
- 6.Collins DM, Kawakami RP, Buddle BM, Wards BJ, de Lisle GW. 2003. Different susceptibility of two animal species infected with isogenic mutants of Mycobacterium bovis identifies phoT as having roles in tuberculosis virulence and phosphate transport. Microbiology 149:3203–3212. 10.1099/mic.0.26469-0 [DOI] [PubMed] [Google Scholar]
- 7.Peirs P, Lefèvre P, Boarbi S, Wang XM, Denis O, Braibant M, Pethe K, Locht C, Huygen K, Content J. 2005. Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect. Immun. 73:1898–1902. 10.1128/IAI.73.3.1898-1902.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardin SD, Finan TM. 1998. Regulation of phosphate assimilation in Rhizobium (Sinorhizobium) meliloti. Genetics 148:1689–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Kruger WM, Humphreys S, Ketley JM. 1999. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 145:2463–2475 [DOI] [PubMed] [Google Scholar]
- 10.Wanner BL, Chang BD. 1987. The phoBR operon in Escherichia coli K-12. J. Bacteriol. 169:5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sola-Landa A, Moura RS, Martin JF. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U. S. A. 100:6133–6138. 10.1073/pnas.0931429100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulett FM, Lee J, Shi L, Sun G, Chesnut R, Sharkova E, Duggan MF, Kapp N. 1994. Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J. Bacteriol. 176:1348–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR., Jr 2007. The two-component regulatory system SenX3-RegX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189:5495–5503. 10.1128/JB.00190-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriakov J, Lee S, Jacobs WR., Jr 2003. Identification of a regulated alkaline phosphatase, a cell surface-associated lipoprotein, in Mycobacterium smegmatis. J. Bacteriol. 185:4983–4991. 10.1128/JB.185.16.4983-4991.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metcalf WW, Wanner BL. 1993. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J. Bacteriol. 175:3430–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metcalf WW, Wanner BL. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173:587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imazu K, Tanaka S, Kuroda A, Anbe Y, Kato J, Ohtake O. 1998. Enhanced utilization of phosphonate and phosphite by Klebsiella aerogenes. Appl. Environ. Microbiol. 10:3754–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebhard S, Tran SL, Cook GM. 2006. The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate. Microbiology 152:3453–3465. 10.1099/mic.0.29201-0 [DOI] [PubMed] [Google Scholar]
- 19.Gebhard S, Cook GM. 2008. Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon. J. Bacteriol. 190:1335–1343. 10.1128/JB.01764-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braibant M, Lefèvre P, de Wit L, Ooms J, Peirs P, Huygen K, Wattiez R, Content J. 1996. Identification of a second Mycobacterium tuberculosis gene cluster encoding proteins of an ABC phosphate transporter. FEBS Lett. 394:206–212. 10.1016/0014-5793(96)00953-2 [DOI] [PubMed] [Google Scholar]
- 21.Lefèvre P, Braibant M, de Wit L, Kalai M, Roeper D, Grotzinger J, Delville JP, Peirs P, Ooms J, Huygen K, Content J. 1997. Three different putative phosphate transport receptors are encoded by the Mycobacterium tuberculosis genome and are present at the surface of Mycobacterium bovis BCG. J. Bacteriol. 179:2900–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatt K, Banerjee SK, Chakraborti PK. 2000. Evidence that phosphate specific transporter is amplified in a fluoroquinolone resistant Mycobacterium smegmatis. Eur. J. Biochem. 267:4028–4032. 10.1046/j.1432-1327.2000.01437.x [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 24.Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. 2005. The MicrobesOnline website for comparative genomics. Genome Res. 15:1015–1017. 10.1101/gr.3844805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelik M, Lunin VV, Skarina T, Savchenko A. 2006. Structural characterization of GntR/HutC family signaling domain. Protein Sci. 15:1506–1511. 10.1110/ps.062146906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Listwan P, Cowieson N, Kurz M, Hume DA, Martin JL, Kobe B. 2005. Modification of recombinatorial cloning for small affinity tag fusion protein construct generation. Anal. Biochem. 346:327–329. 10.1016/j.ab.2005.06.016 [DOI] [PubMed] [Google Scholar]
- 27.Moreland N, Ashton R, Baker HM, Ivanovic I, Patterson S, Arcus VL, Baker EN, Lott JS. 2005. A flexible and economical medium-throughput strategy for protein production and crystallization. Acta Crystallogr. D Biol. Crystallogr. 61:1378–1385. 10.1107/S0907444905023590 [DOI] [PubMed] [Google Scholar]
- 28.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 30.Long F, Vagin AA, Young P, Murshudov GN. 2008. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64:125–132. 10.1107/S0907444907050172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. 2001. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57:1445–1450. 10.1107/S0907444901014007 [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–253. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. 2004. Coot: model building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R. 2005. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 15:116–124. 10.1016/j.gde.2005.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kononova SV, Nesmeyanova MA. 2002. Phosphonates and their degradation by microorganisms. Biochemistry (Mosc.) 67:184–195. 10.1023/A:1014409929875 [DOI] [PubMed] [Google Scholar]
- 37.Donner AL, Paa K, Koudelka GB. 1998. Carboxyl-terminal domain dimer interface mutant 434 repressors have altered dimerization and DNA binding specificities. J. Mol. Biol. 283:931–946. 10.1006/jmbi.1998.2136 [DOI] [PubMed] [Google Scholar]
- 38.Hobart SA, Ilin S, Moriarty DF, Osuna R, Colón W. 2002. Equilibrium denaturation studies of the Escherichia coli factor for inversion stimulation: implications for in vivo function. Protein Sci. 11:1671–1680. 10.1110/ps.5050102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swint-Kruse L, Zhan H, Matthews KS. 2005. Integrated insights from simulation, experiment, and mutational analysis yield new details of LacI function. Biochemistry 44:11201–11213. 10.1021/bi050404+ [DOI] [PubMed] [Google Scholar]
- 40.Stormo GD, Fields DS. 1998. Specificity, free energy and information content in protein-DNA interactions. Trends Biochem. Sci. 23:109–113. 10.1016/S0968-0004(98)01187-6 [DOI] [PubMed] [Google Scholar]
- 41.Rigali S, Derouaux A, Giannotta F, Dusart J. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507–12515. 10.1074/jbc.M110968200 [DOI] [PubMed] [Google Scholar]
- 42.Kalivoda KA, Steenbergen SM, Vimr ER, Plumbridge J. 2003. Regulation of sialic acid catabolism by the DNA binding protein NanR in Escherichia coli. J. Bacteriol. 185:4806–4815. 10.1128/JB.185.16.4806-4815.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peekhaus N, Conway T. 1998. Positive and negative transcriptional regulation of the Escherichia coli gluconate regulon gene gntT by GntR and the cyclic AMP (cAMP)-cAMP receptor protein complex. J. Bacteriol. 180:1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quail MA, Dempsey CE, Guest JR. 1994. Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEBS Lett. 356:183–187. 10.1016/0014-5793(94)01264-4 [DOI] [PubMed] [Google Scholar]
- 45.Hu L, Allison SL, Phillips AT. 1989. Identification of multiple repressor recognition sites in the hut system of Pseudomonas putida. J. Bacteriol. 171:4189–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miwa Y, Fujita Y. 1988. Purification and characterization of a repressor for the Bacillus subtilis gnt operon. J. Biol. Chem. 263:13252–13257 [PubMed] [Google Scholar]
- 47.Gallagher DT, Mayhew M, Holden MJ, Howard A, Kim KJ, Vilker VL. 2001. The crystal structure of chorismate lyase shows a new fold and tightly retained product. Proteins 44:304–311. 10.1002/prot.1095 [DOI] [PubMed] [Google Scholar]
- 48.Resch M, Schiltz E, Titgemeyer F, Muller YA. 2010. Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA. Nucleic Acids Res. 38:2485–2497. 10.1093/nar/gkp1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorelik M, Lunin VV, Skarina T, Savchenko A. 2006. Structural characterization of GntR/HutC family signaling domain. Protein Sci. 15:1505–1511. 10.1110/ps.062146906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:545–549. 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rezacova P, Krejcirikova V, Borek D, Moy SF, Joachimiak Otwinowski AZ. 2007. The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacillus subtilis 168 reveals a unique quarternary assembly. Proteins 69:679–682. 10.1002/prot.21516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allenby NEE, O'Connor N, Prágai Z, Ward AC, Wipat A, Harwood CR. 2005. Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis. J. Bacteriol. 187:8063–8080. 10.1128/JB.187.23.8063-8080.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antelmann H, Scharf C, Hecker M. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478–4490. 10.1128/JB.182.16.4478-4490.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Aalten DM, DiRusso CC, Knudsen J. 2001. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 20:2041–2050. 10.1093/emboj/20.8.2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. 2008. HingeProt: automated prediction of hinges in protein structures. Proteins 70:1219–1227. 10.1002/prot.21613 [DOI] [PubMed] [Google Scholar]
- 56.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539–544. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]