Abstract
The zinc importer ZupT is required for the efficient allocation of zinc to zinc-dependent proteins in the metal-resistant bacterium Cupriavidus metallidurans but not for zinc import per se. The expression of zupT is upregulated under conditions of zinc starvation. C. metallidurans contains three members of the Fur family of regulators that qualify as candidates for the zupT regulator. The expression of a zupT-lacZ reporter gene fusion was strongly upregulated in a ΔfurC mutant but not in a ΔfurA or ΔfurB mutant. Expression of the genes for transition-metal importers (pitA, corA1, corA2, and corA3) was not changed in this pattern in all three Δfur mutants, but they were still downregulated under conditions of elevated zinc concentrations, indicating the presence of another zinc-dependent regulator. FurA was a central regulator of the iron metabolism in C. metallidurans, and furA was constitutively expressed under the conditions tested. Expression of furB was upregulated under conditions of iron starvation, and FurB could be an iron starvation Fur connecting general metal and iron homeostasis, as indicated by the phenotype of a ΔfurB ΔfurC double mutant. FurC was purified as a Strep-tagged protein and retarded the electrophoretic mobility of a DNA fragment upstream of zupT. Binding of FurC to this operator region was influenced by the presence of zinc ions and EDTA. Thus, FurC is the main zinc uptake regulator (Zur) of C. metallidurans and represses synthesis of the central zinc importer ZupT when sufficient zinc is present.
INTRODUCTION
Cupriavidus metallidurans is able to sustain its cellular transition-metal homeostasis even in the presence of mM metal ion concentrations and even in the presence of a mixture of them. This is achieved by highly redundant metal uptake systems, which have only minimal cation selectivity. These uptake systems are in combination with metal efflux systems, which remove surplus cations (1). The most sophisticated efflux systems and batteries of helper proteins are encoded by metal resistance determinants on the two native plasmids of wild-type strain CH34, pMOL28 and pMOL30, but a variety of resistance determinants also are located on the two chromosomes of this betaproteobacterium of the order Burkholderiales (2, 3).
The representatives of secondary uptake systems for zinc ions in C. metallidurans are CorA1, CorA2, and CorA3 of the MIT protein family (TC 1.A.35; TC, transporter classification family [4, 5]), the ZIP protein ZupT (TC 2.A.5), HoxN of the NiCoT family (TC 2.A.52), PitA for metal-phosphate complexes (PiT; TC 2.A.20), and, as uptake or efflux system, the MIT protein ZntB. C. metallidurans does not contain a primary ZnuABC-type ABC uptake system for this important metal (TC 3.A.3) but has two P-type ATPases that may import Mg2+, Ca2+, and/or other divalent cations (1). None of the secondary transport systems alone was essential for zinc import, but ZupT was required under conditions of zinc starvation. A mutant lacking ZupT contained only 20,000 zinc ions per cell when grown in mineral salts medium without additions (MM), while its parent strain, AE104, possessed 70,000 zinc ions per cell. Mutant and parent cells accumulated up to 120,000 zinc ions per cell when 100 μM Zn2+ was added to the growth medium, indicating again that ZupT was not essential for zinc import. Without efflux systems, the cellular zinc content increased to 250,000 per cell, and the cells were growth impaired, marking the upper tolerance level for cellular zinc. The lowest tolerance level might be 20,000 zinc ions per cell (6).
On the other hand, ZupT was needed for efficient allocation of imported zinc ions to important zinc-dependent proteins, such as the RpoC subunit of the RNA polymerase. These data (6) and older results (7) indicated that two zinc pools exist in C. metallidurans, a general pool that needed unspecific uptake and efflux systems for its homeostasis and a ZupT-dependent zinc allocation pipeline. Moreover, C. metallidurans ΔzupT mutant cells were not able to tolerate the presence of the most sophisticated efflux system for zinc, the plasmid pMOL30-encoded cobalt-zinc-cadmium resistance system Czc (6). Thus, ZupT is of central importance for zinc homeostasis of C. metallidurans.
All genes for secondary zinc uptake systems were downregulated at high external zinc concentrations (1), in parallel with upregulation of the zntA and cadA genes for two zinc- and cadmium-exporting PIB2-type ATPases (8). Additionally, zupT expression was upregulated by zinc starvation, e.g., in the presence of metal chelators, such as phosphate or EDTA. This effect could be antagonized best by addition of zinc ions (1), indicating that a zinc-dependent regulator should be in control of zupT expression.
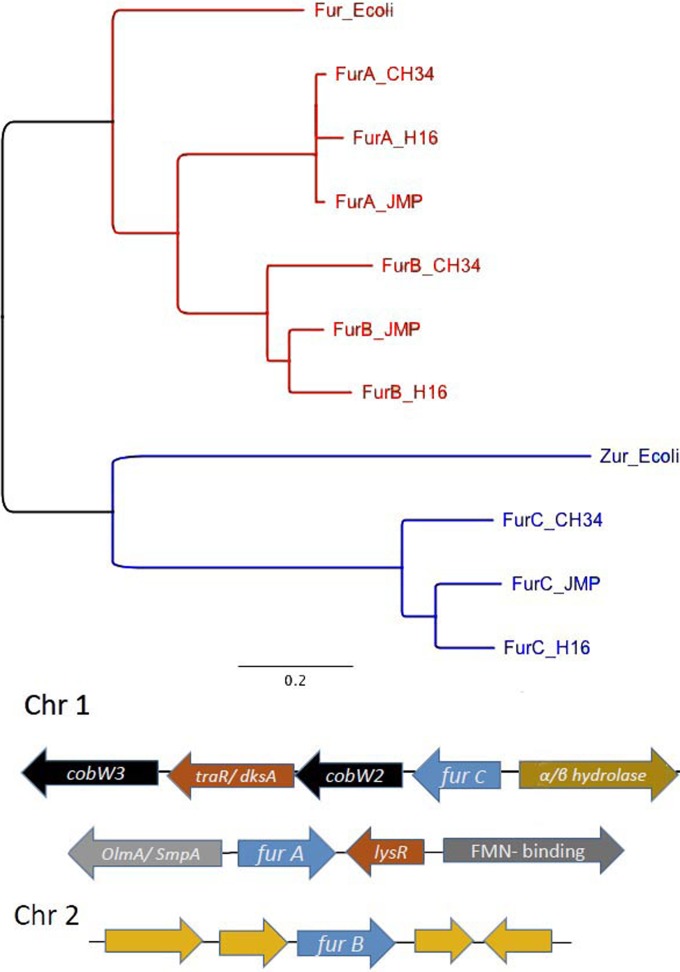
In the gammaproteobacterium Escherichia coli, the Zur protein of the Fur family of regulators controls expression of the operon for the ZnuABC import system for zinc, which has no ortholog in C. metallidurans (9, 10). This family of metalloregulators includes sensors for iron (Fur), zinc (Zur), manganese (Mur), nickel (Nur), and oxidative stress (PerR) (11, 12). Bacterial genomes may contain multiple genes for members of this protein family, which are usually annotated as Fur (12). This is also true for C. metallidurans (3), which possesses the three genes furA and furC on chromosome 1 and furB on chromosome 2 (Fig. 1). This paper demonstrates that FurA and FurB are involved in iron homeostasis and seem to be the main iron regulators, as C. metallidurans does not contain a DtxR-type regulator. FurC is actually the zinc uptake regulator (Zur) of C. metallidurans and controls zupT expression in response to zinc starvation.
FIG 1.
Alignment of members of the Fur protein family. Predicted protein sequences for proteins of the Fur family from C. metallidurans CH34, Cupriavidus eutrophus H16 (synonym Ralstonia eutropha), Cupriavidus pinatubonensis JMP134 (synonym Ralstonia eutropha), and E. coli were globally aligned using the Blosum62 cost matrix and Geneious 6.1.6. The sequences used were YP_585118 (Rmet_2976), YP_587874 (Rmet_5746), and YP_582283 (Rmet_0128 from CH34, YP_727586, YP_728287, and YP_724716 from H16, and YP_297042, YP_299716, YP_294390 from JMP134 for FurA, FurB, and FurC, respectively. The bar represents 0.2 exchanges per site. The Fur cluster is in red, and the Zur cluster is in blue. Shown below the alignment are the genomic environments of the three fur genes from C. metallidurans. Chr 1, chromosome 1; Chr 2, chromosome 2. Genes for Fur are in blue, genes for other functions are in different colors.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. metallidurans strains (see Table S1 in the supplemental material) used in this study all were derivatives of the plasmid-free strain AE104 that lacks pMOL28 and pMOL30 (2). Mutant strains contained deletions in the three fur genes. Plasmids pECD986 to pECD990, pECD1332, and pECD1333 (see Table S1) were used to insert the lacZ reporter gene just downstream of the genes pitA, zupT, corA1, corA2, corA3, zntA, and cadA, respectively. Tris-buffered mineral salts medium (2) containing 2 g/liter sodium gluconate (TMM) was used to cultivate these strains aerobically with shaking at 30°C. Analytical-grade salts of heavy-metal chlorides were used to prepare 1 M stock solutions, which were sterilized by filtration. Solid Tris-buffered media contained 20 g/liter agar. For the MIC determination, the cells were cultivated in TMM for 48 h at 30°C with shaking, diluted 20-fold into fresh TMM, cultivated for another 24 h, diluted 100-fold into fresh TMM, streaked onto TMM plates containing increasing concentrations of metal chlorides, and incubated for 5 days at 30°C.
Dose-response growth curves were conducted in TMM in cultivation tubes. A preculture was incubated at 30°C with shaking at 250 rpm up to early stationary phase, diluted 1:20 in fresh medium, and incubated for 24 h at 30°C and 250 rpm. This culture was used to inoculate parallel cultures with increasing metal concentrations. Cells were cultivated for 20 h at 30°C and 250 rpm in a shaker (KS501; IKA Labortechnik), and the optical density was determined at 600 nm in a 96-well plate with the Tecan infinite 200 PRO reader (Tecan, Männersdorf, Switzerland).
Genetic techniques.
Standard molecular genetic techniques were used (13, 14). For conjugative gene transfer, overnight cultures of donor strain E. coli S17/1 (15) and of the C. metallidurans recipient strains grown at 30°C in Tris-buffered medium were mixed (1:1) and plated onto nutrient broth agar. After 2 days, the bacteria were suspended in TMM, diluted, and plated onto selective media as previously described (13).
All primer pairs used are listed in Table S1 in the supplemental material. Plasmid pECD1002 was used to construct deletion mutants. It is a derivate of plasmid pCM184 (16). These plasmids harbor a kanamycin resistance cassette flanked by loxP recognition sites. Plasmid pECD1002 additionally carries alterations of 5 bp at each loxP site. Using these mutant lox sequences, multiple gene deletions within the same genome are possible without interference by secondary recombination events (17, 18). Fragments of 300 bp upstream and downstream of the target gene were amplified by PCR, cloned into the vector pGEM T-Easy (Promega), sequenced, and further cloned into plasmid pECD1002. The resulting plasmids were used in a double-crossover recombination in C. metallidurans strains to replace the respective target gene by the kanamycin resistance cassette, which subsequently was deleted by transient introduction of the cre expression plasmid pCM157 (16). Cre recombinase is a site-specific recombinase from the phage P1 that catalyzes the in vivo excision of the kanamycin resistance cassette at the loxP recognition sites. The correct deletions of the respective transporter genes were verified by Southern DNA-DNA hybridization. For construction of multiple deletion strains, these steps were repeated. The resulting mutants carried a small open reading frame instead of the wild-type gene to prevent polar effects.
Gene insertions.
For reporter operon fusions, lacZ was inserted downstream of several targets. This was done without interrupting any open reading frame downstream of the target genes to prevent polar effects. The 300- to 400-bp 3′ ends of the respective target genes were amplified by PCR from total DNA of strain AE104 and the resulting fragments cloned into plasmid pECD794 (pLO2-lacZ) (19). The respective operon fusion cassettes were inserted into the open reading frame of the target gene by single-crossover recombination.
Induction experiments.
C. metallidurans cells with a lacZ reporter gene fusion were cultivated in TMM with shaking at 30°C. A 300-Klett-unit preculture was used to inoculate the main culture to 30 Klett units. At a cell density of 60 to 70 Klett units, heavy-metal salts were added to various final concentrations, and cells were incubated with shaking for an additional 3 h. The specific β-galactosidase activity was determined in permeabilized cells as published previously, with 1 U defined as the activity forming 1 nmol of o-nitrophenol per min at 30°C (20).
CAS agar assay.
The chrome azurol S (CAS) agar assay was done as described previously (21). For 100 ml of agar, 60.5 mg CAS was dissolved in 50 ml H2O (all liquid chromatography-mass spectrometry [LC-MS] grade) and mixed with 10 ml 1 mM FeCl3 × 6H2O (in 10 mM HCl). This solution was added under stirring to 72.9 mg hexadecyltrimethylammonium (HDTMA; in 40 ml H2O) and autoclaved. A mixture of 75 ml H2O, 0.6 g NaOH, 3.02 g piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 10 ml 10× MM9 salts (3 g KH2PO4, 5 g NaCl, 10 g NH4Cl at 1 liter), and 1.5 g Bacto agar was autoclaved separately. After cooling to 50°C, 3 ml Casamino Acids (10%, deferrated), 2 ml sodium gluconate (20%), 0.1 ml MgCl2 (1 M), 0.1 ml CaCl2 (100 mM), and the CAS solution were added slowly, and plates were poured. The petri dishes were inoculated with liquid cultures, which were produced by inoculation of a fresh culture with a 48-h preculture and subsequent incubation with shaking at 30°C for 24 h. The plates were incubated at 30°C until formation of an orange halo around the colonies indicated the production and secretion of siderophores. Three independent assays were performed.
RNA isolation and RT-PCR.
C. metallidurans CH34 cells were cultivated as described above. At a cell turbidity of 150 Klett units, 300 μM metal chlorides was added. After 10 min of incubation at 30°C, the cells were harvested. Total RNA was isolated as described previously (22). After isolation, DNase treatment was performed, followed by purification with phenol-chloroform and precipitation with ethanol. The RNA concentration was determined photometrically, and RNA quality was checked on formamide gels (14). To exclude experimental artifacts resulting from DNA contaminations, only RNA that did not generate products in a PCR with chromosomal primers was used. For the reverse transcriptase (RT) reaction, 1 μg of total RNA and 0.1 μg hexamer primers were incubated at 65°C for 5 min and snap-cooled on ice. After addition of 0.5 mM (each) dATP, dGTP, dTTP, and dCTP, as well as 10 mM dithiothreitol (DTT) and 100 U of reverse transcriptase (Superscript II), in reaction buffer (Invitrogen, Karlsruhe, Germany), reverse transcription proceeded for 10 min at room temperature and then for 1 h at 50°C. After finishing the RT reaction, the enzyme was inactivated at 70°C for 10 min. The resulting cDNA was amplified by PCR as published previously (22).
Purification of Strep-tagged FurC.
The furC gene was amplified by PCR from strain AE104 genomic DNA as the template using primer pair CS14 rmet0128 EcoRI and CS15 rmet0128 SalI and cloned into vector pASK-IBA7. E. coli strain BL21-pLysS (Promega) with pECD1338 (pASK-IBA7::furC) was cultivated in LB medium with shaking at 37°C until an optical density at 600 nm of 0.8. Expression of furC was induced with 200 μg anhydrotetracycline (AHT) per liter, and incubation was continued for 3 h at 30°C or 37°C. Cells were harvested by centrifugation and ultrasonicated. After low-touring centrifugation to remove cell debris, FurC was purified using a Strep-tactin affinity chromatography column according to the manufacturer's protocol (IBA GmbH). FurC was concentrated by using Vivaspin 20 columns (10,000-molecular-weight cutoff [MWCO]; polyethersulfone [PES] membrane; Sartorius Stedim Biotech, Göttingen, Germany). Buffer exchanges were performed by dialysis (ZellusTrans V Serie; 10,000 MWCO; Carl Roth, Karlsruhe, Germany) overnight against the 100-fold volume of protein solution with new buffer or using a PD 10 column (Bio-Rad, Munich, Germany). The protein concentration was determined by a bicinchoninic acid (BCA) kit (Sigma-Aldrich, Steinheim, Germany) or by using a NanoDrop ND-1000 spectrophotometer (the extinction cofactor for FurC-Nstrep, ε280, was 12,865 M−1 cm−1, which was calculated using the ProtParam tool of the ExPASy server [http://web.expasy.org/protparam/]). Protein quality was analyzed on a 12.5% SDS gel stained with Coomassie brilliant blue (23) or by silver staining (24). Fixation was performed in 50% (vol/vol) C2H5OH, 12% (vol/vol) methanol for at least 1 h. Gels were washed for 1 min in 50% (vol/vol) ethanol and sensitized with 1.6 mM Na2S2O3 for 2 min, followed by impregnation with 11.77 mM AgNO3 and 0.185% (vol/vol) formaldehyde for 20 min. Developing solution contained 0.026% (vol/vol) formaldehyde, 0.566 M Na2CO3, and 63.25 μM Na2S2O3. Reactions were stopped with 50% (vol/vol) methanol and 12% (vol/vol) acetic acid.
Western blotting and analysis.
The proteins were transferred with semidry Trans-blot SD (Bio-Rad, Munich, Germany) on a nitrocellulose membrane (porablot NCP; pore size, 0.45 μm; Macherey-Nagel, Duren, Germany) at 20 V and 15 mA for 30 min. The membrane and 12.5% SDS gel were prepared in Towbin buffer (25 mM Tris, 192 mM glycine, 20% methanol) for 10 min. The detection of the N-terminal Strep tag was performed by the Strep tag II detection system (IBA, Göttingen, Germany). The membrane was blocked in 1× phosphate-buffered saline (PBS; 0.4 mM KH2PO4, 1.6 mM Na2HPO4, and 11.5 mM NaCl) with 3% (wt/vol) bovine serum albumin (BSA) and 0.5% Tween 20 overnight at 4°C. Three washing steps then were performed with 1× PBS, 0.1% Tween 20 and incubation with 1:100, 000 Strep-tagged horseradish peroxidase (HRP) conjugate (Bio-Rad, Munich, Germany) in 1× PBS, 0.5% Tween 20, and 5 μg/ml BSA for 1 h at room temperature. To prevent unspecific binding, two washing steps with washing buffer and one with 1× PBS were performed. The Strep tag was detected by an ECL (enhanced chemiluminescence) reaction.
Mass spectrometry.
For protein molecular mass determination, the spectra were acquired with an electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) mass spectrometer (Waters, Manchester, United Kingdom) equipped with a nanospray ion source. The samples were injected via a PicoTip (New Objective, Cambridge, MA) with a syringe pump (Harvard Apparatus, MA) with a flow rate of 300 nl/min. The MaxEnt1 algorithm was used for deconvoluting the data to single-charge state.
For fingerprinting, the protein bands were cut out of the gel and the slices incubated for 45 min at 50°C in the presence of 10 mM dithiothreitol (Sigma) in 100 mM ammonium bicarbonate. The solution was discarded, and the slices subsequently were treated with 55 mM iodoacetamide (Sigma) in 100 mM ammonium bicarbonate for 45 min in the dark to modify and block cysteine residues. The solution was removed again; the gel slices were washed three times with water and twice with 10 mM ammonium bicarbonate with 10 mM ammonium bicarbonate in 50% acetonitrile; dried under a gentle stream of nitrogen; reswollen in 20 μl 10 mM ammonium bicarbonate (pH 8.0); and finally digested with trypsin (Promega, Madison, WI) overnight at 37°C. The peptide mass fingerprint spectra were recorded on an Ultraflex-II tandem TOF (TOF/TOF) mass spectrometer (Bruker Daltonic, Bremen, Germany) equipped with a matrix-assisted laser desorption ionization (MALDI) source, nitrogen laser, LIFT cell for fragment ion postacceleration, and gridless ion reflector. The software programs Flex Control 2.4, Flex Analysis 2.4, and Biotools 3.0 were used to operate the instrument and analyze the data. For external calibration, a peptide calibration mixture (Bruker Daltonics, Bremen, Germany) was used. For MALDI sample preparation, 1 μl of a DHB matrix (7 mg 2.5-dihydroxybenzoic acid in 100 μl methanol) was mixed with 1 μl digest and deposited onto the target.
For inductively coupled plasma mass spectrometry (ICP-MS) analysis, HNO3 (trace metal grade; Normatom/PROLABO) was added to the FurC samples to a final concentration of 67% (wt/vol), and the mixture was mineralized at 70°C for 2 h. Samples were diluted to a final concentration of 2% (wt/vol) nitric acid. Indium was added as an internal standard at a final concentration of 10 ppb. Elemental analysis was performed via ICP-MS using ESI sampler SC-2 (Elemental Scientific, Inc., Omaha, NE) and an X-Series II ICP-MS instrument (Thermo Fisher Scientific, Bremen, Germany) operating with a collision cell and flow rates of 5 ml/min of He/H2 (93% He, 7% H2 [25]), with an Ar carrier flow rate of 0.76 liter/min and an Ar make-up flow rate of 15 liter/min. An external calibration curve was recorded with ICP multielement standard solution XVI (Merck) in 2% nitric acid. The sample was introduced via a peristaltic pump and analyzed for its metal content. For blank measurement and quality/quantity thresholds, calculations based on DIN32645 TMM were used.
Electrophoretic mobility shift assay (EMSA).
Gel retardation assays were performed as published previously (22), with modifications. PCR products together with purified Strep-tagged FurC were used. Approximately 0.2 pmol of the zupTp promoter fragment and various concentrations of FurC were used for each assay. The DNA-protein complex was formed at 30°C for 40 min in 30 μl binding buffer (50 mM KCl, 20 mM Tris-HCl [pH 7.8], 5% glycerol, 1 mM DTT). The reactions were cooled on ice and applied to a 10% phosphonoacetic acid (PAA) gel with a running buffer containing 1× native buffer (100 mM Tris, 100 mM glycine). The gel was run at 4°C and 25 mA for 1 to 2 h, stained with ethidium bromide or silver, dried, and scanned.
RESULTS
FurC was required for repression of zupT.
furA, furB, or furC was deleted from the chromosome of the plasmid-free C. metallidurans strain AE104. In each of the resulting single-gene deletion strains, transcriptional lacZ fusions were generated with the most important secondary uptake systems for transition-metal cations, zupT, pitA, corA1, corA2, and corA3. Using these reporter constructs, metal- and EDTA-dependent expression of these genes was characterized (Table 1).
TABLE 1.
Activity of reporter gene fusions with the genes for import systems in Δfur background under various conditionsa
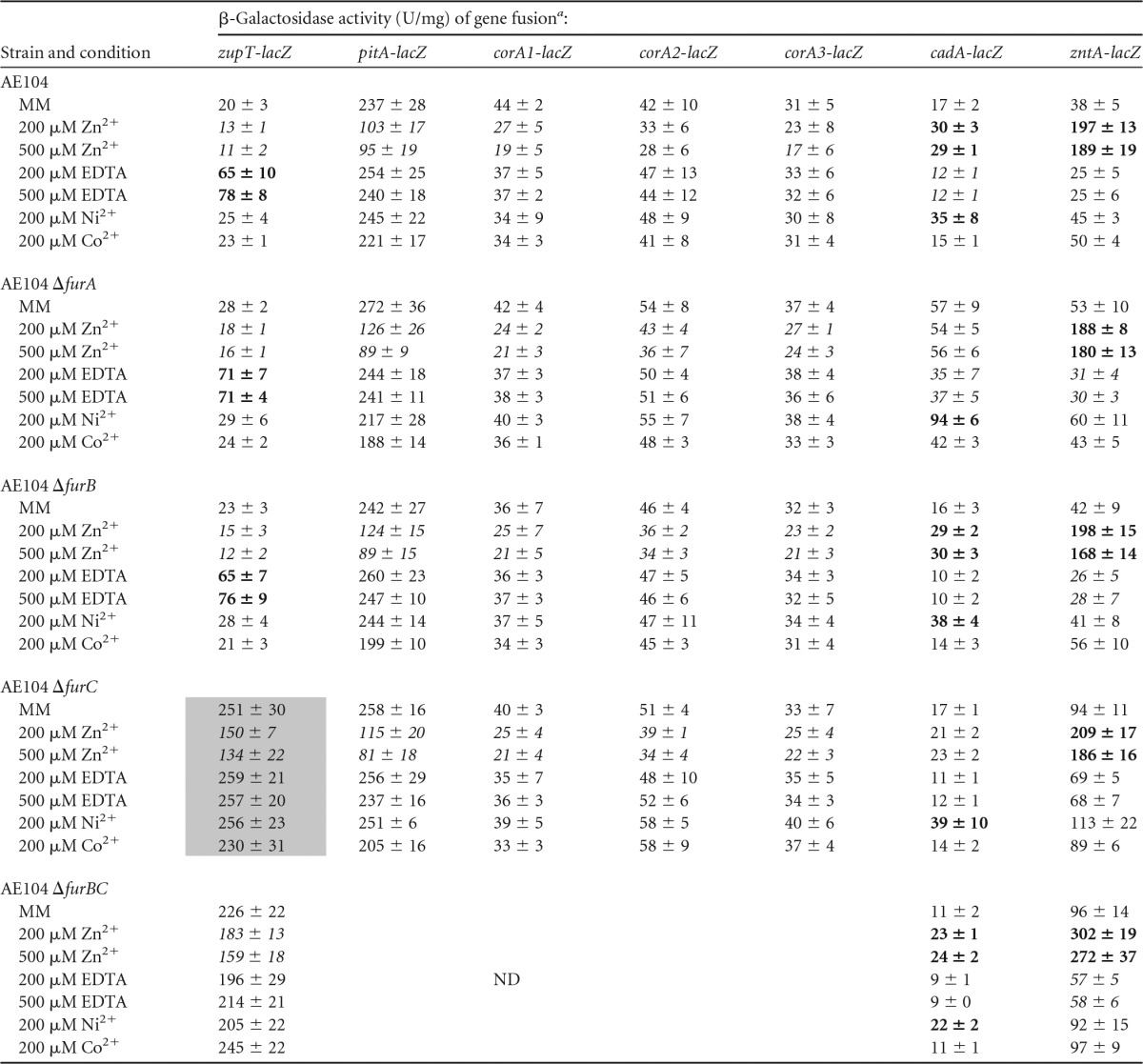
Values in boldface indicate significant upregulation compared to the same strain in mineral salts medium (MM). Values in italics indicate significant downregulation. ND, not done. Eight experiments were performed, and deviations are shown.
Deletion of furC had a strong effect on expression of the zupT gene for the central zinc importer of C. metallidurans (Table 1, shaded values). In the parent strain AE104 and as published previously (1), β-galactosidase activity was in the range of 20 U/mg protein. The presence of sufficient zinc (200 μM) led to downregulation of zupT, while the same concentration of cobalt and nickel cations did not change zupT expression. Addition of 200 μM EDTA, however, led to a 3-fold upregulation of zupT-lacZ expression. Deletion of furA or furB did not change this pattern (Table 1).
In contrast, deletion of furC resulted in a strongly increased expression level of zupT-lacZ, which reached values of 250 U/mg (Table 1) in cells cultivated in mineral salts medium without additions. This was a 50% higher level of activity than the maximum expression value obtained in all experiments in the past in the parent strain (1). The addition of EDTA, nickel, or cobalt salts did not change this expression level, but 200 μM or 500 μM Zn(II) still was able to downregulate the zupT-lacZ expression level by half (Table 1, shaded, italicized values).
This indicated that FurC acts as a repressor for the zupT gene. FurA or FurB, however, had no influence on zupT. Interestingly, even in the ΔfurC strain, zinc-dependent downregulation of zupT-lacZ still was possible, indicating that at least one more regulator should exist for zupT.
No influence of the three Fur proteins on the genes for other metal transport systems.
PitA is an important importer for metal phosphate complexes in C. metallidurans. Very similar to zupT and as published previously (1), the expression of pitA-lacZ was 60% downregulated in the presence of ample zinc salts (Table 1), but EDTA, nickel, or cobalt salts had no effect. Deletion of furA, furB, or furC did not change this pattern. This indicated that neither FurC nor one of the two other Fur proteins was involved in the control of pitA expression and that another zinc-dependent regulator exists in C. metallidurans that regulated pitA.
The corA1 gene, as the most important secondary importer for magnesium and other divalent metal cations, exhibited the same effect as pitA, namely, downregulation by zinc but not by EDTA, nickel, or cobalt and no influence of either fur deletion (Table 1). Compared to that of pitA, however, the expression level of corA1 was 5-fold lower, as has been published previously (1). The same pattern was visible with corA2 and corA3. Thus, FurC was in control of zupT expression, but neither FurC, FurA, nor FurB was in control of the zinc-dependent regulation of pitA, corA1, corA2, or corA3. This function and the residual zinc-dependent regulation of zupT was performed by an unknown regulator.
The influence of the fur genes on expression of the genes for the two most important chromosomally encoded efflux systems for zinc and cadmium, the P-type ATPases ZntA and CadA, also was investigated. As published previously (8), zntA-lacZ was 5-fold upregulated by 200 μM or 500 μM Zn(II) in the growth medium (Table 1). EDTA decreased the expression level significantly, but nickel or cobalt ions had no effect. Deletion of furA or furB had no effect. Deletion of furC, in contrast, did not change the regulatory pattern of zntA-lacZ and also did not change the maximum expression level obtained by zinc treatment. However, the expression levels of zntA-lacZ in untreated cells in the presence of nickel or cobalt ions and in the presence of EDTA were higher level than those of zntA-lacZ in the parent strain (Table 1). This indicated that FurC did not control zntA expression but that another regulator should be involved and that upregulation of zupT in the ΔfurC mutant obviously led to higher cellular zinc contents, which consequently resulted in the elevated expression of zntA.
As judged by the cadA-lacZ fusion, expression of the gene for the second PIB2-type ATPase in strain AE104, cadA, was only slightly upregulated by zinc and nickel ions. This was expected, because CadA primarily removes cadmium ions. Deletion of furB had no effect on cadA-lacZ. Deletion of furC mollified the slight upregulation of cadA-lacZ by zinc ions even more and showed a small but significant upregulation by nickel ions, all probably resulting from the high expression level of zupT. Surprisingly, deletion of furA had a significant effect on the expression level of this cadmium efflux system. The overall expression level of the reporter gene was twice as high as that in the parent strain AE104, zinc had no effect, EDTA or cobalt ions decreased the expression level, and nickel ions increased it (Table 1).
Thus, deletion of furC led to increased expression of zupT, probably resulting in increased import of Zn2+ and other divalent metal cations, which subsequently led to the upregulation of ZntA. Similarly, deletion of furA could mediate the upregulation of genes for import routes for other transition-metal cations. Such a hampered regulation of the expression of transport systems should lead to decreased metal resistance.
Metal resistance of the Δfur mutant strains.
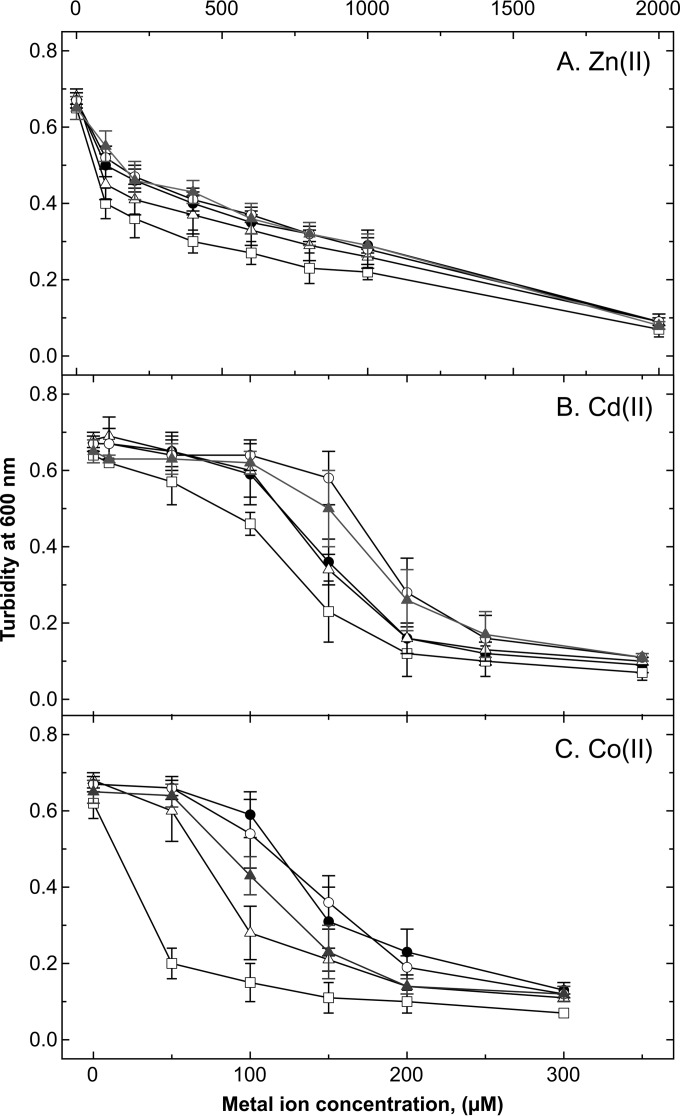
The ΔfurA mutant displayed decreased metal resistance in dose-response curves (Fig. 2) and on solid mineral salts medium (Table 2), indicating the upregulation of import routes for transition-metal cations. The ΔfurC deletion increased cadmium resistance to a small extent due to the known mollifying effect of zinc on toxicity of the chemically related cadmium ion (Fig. 2). The effect of the ΔfurB deletion was between the resistance levels of the ΔfurA mutant and those of the parent strain AE104. Since the ΔfurA deletion and, to some degree, the ΔfurB deletion influenced cobalt resistance but the expression levels of the genes for the three magnesium and cobalt importers CorA1, CorA2, and CorA3 were not changed in these mutants, FurA and FurB may be involved in the regulation of the import of iron, the neighbor of cobalt in the periodic system of the elements.
FIG 2.
Resistance of the Δfur mutant strains to transition metals. Dose-response experiments were performed with parent strain AE104 (closed circles; n = 6 or 7) and its ΔfurA (open squares; n = 6), ΔfurB (open triangles; n = 6), ΔfurC (open circles; n = 8), and ΔfurBC (gray triangles; n = 5 or 6) mutant strains in the presence of zinc (A), cadmium (B), or cobalt (C). Deviation bars are shown.
TABLE 2.
Influence of furA and furB gene deletions on siderophore biosynthesis and metal resistance
| Parent and mutant strain | Diam of the haloa (mm) | MIC (mM) ofb: |
|||
|---|---|---|---|---|---|
| Cd2+ | Co2+ | Ni2+ | Zn2+ | ||
| CH34 wild type | 22 ± 2 | 2.5 | 11.5 | 7.5 | 7.0 |
| ΔfurA | 24 ± 3 | 2.0 | 6.0 | 5.0 | 4.5 |
| ΔfurB | 19 ± 2 | 2.0 | 6.0 | 7.5 | 5.0 |
| ΔRmet_1113-Rmet_1114 (iucA-iucC) | 0 ± 0*** | 2.5 | 6.0 | 6.5 | 4.75 |
| ΔRmet_1118 (aleB) | 17 ± 2* | 1.5 | 6.0 | 5.5 | 4.75 |
| ΔRmet_5806-Rmet_5807 (fecA1-fecA2) | 22 ± 4 | 2.0 | 7.0 | 6.0 | 5.0 |
| AE104 wild type | 19 ± 2 | 0.2 | 0.35 | 0.7 | 0.15 |
| ΔfurA | 27 ± 3** | 0.2 | 0.3 | 0.5 | 0.15 |
| ΔfurB | 17 ± 3 | 0.2 | 0.35 | 0.5 | 0.15 |
| ΔRmet_1113-Rmet_1114 (iucA-iucC) | 0 ± 0*** | 0.2 | 0.3 | 0.6 | 0.15 |
| ΔRmet_1118 (aleB) | 16 ± 2 | 0.2 | 0.3 | 0.5 | 0.15 |
| ΔRmet_5806-Rmet_5807 (fecA1-fecA2) | 17 ± 3 | 0.2 | 0.35 | 0.5 | 0.25 |
A CAS assay was performed with C. metallidurans wild-type strain CH34, its plasmid-free derivative strain AE104, and single-deletion mutants of these strains. The diameter of the yellow halo around the wells filled with the cell suspensions was determined. Three experiments were performed, and mean values and deviations are indicated. Significance was determined with Student's t test. ***, >99.9%; **, >99%; *, >95%.
The MICs were determined for the CH34 and AE104 derivatives after 5 days of growth at 30°C on TMM. Three experiments were performed with identical results.
FurA seems to be the main Fur of C. metallidurans.
C. metallidurans is able to produce a siderophore, staphyloferrin B, previously designated alcaligin E (26, 27). The gene locus responsible for its production contains an rpoI gene for an extracytoplasmic-function sigma factor, its anti-sigma factor, FecR, a TonB-dependent receptor, AleB, an importer of the major facilitator protein superfamily, and biosynthesis genes (Rmet_1120 to Rmet_1112). Production of staphyloferrin B strictly depends on the sigma factor RpoI (28).
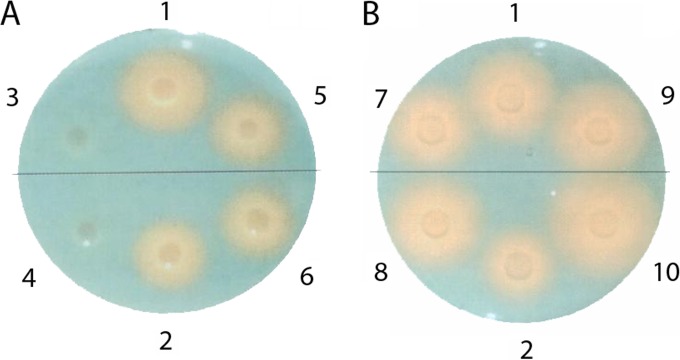
A CAS assay that visualizes production of siderophores was performed with the ΔfurA or ΔfurB mutant strain and its parent. For this experiment, ΔfurA and ΔfurB mutants also were constructed in the wild-type strain CH34. This was not done with the ΔfurC mutant, because ΔzupT mutant cells were not able to tolerate the presence of the most sophisticated efflux system for zinc, the plasmid pMOL30-encoded cobalt-zinc-cadmium resistance system Czc (6). Control experiments were performed with deletion strains in two of the biosynthesis genes (ΔiucA and iucCI-CII; Rmet_1112 and Rmet_1113, respectively) and ΔaleB (Rmet_1118). The halo produced by strain AE104 was smaller than that of strain CH34 (Fig. 3), but the difference was not significant (>90%).
FIG 3.
CAS assay for siderophore excretion. Five μl of a cell suspension of C. metallidurans cells was transferred onto a CAS agar plate and incubated for 24 h at 30°C. A yellow halo around the droplet of cell suspension indicates excretion of siderophores that sequester Fe(III) away from the chromazurol indicator (21). Wild-type strain CH34 and its plasmid-free derivative AE104 are shown at positions 1 and 2 on each plate, respectively. CH34 mutants are above the line, AE104 mutants are below. Mutants from CH34 or AE104 are ΔiucA-iucC on positions 3 and 4, ΔaleB on 5 and 6, ΔfurB on 7 and 8, and ΔfurA on 9 and 10, respectively.
Strains with the ΔiucA–iucCI-CII deletion were not able to produce the siderophore at all, as expected (Fig. 3A and Table 2). Deletion of the aleB gene led to a somewhat decreased halo diameter in strain CH34 (significance of only >90% in strain AE104). Deletion of furA increased the halo diameter in strain AE104 (Table 2) on a plate in which strain CH34 also was present (Fig. 3B). Other deletions did not change the halo diameters. This experiment also indicated that the overall transition-metal homeostasis, which is different in strain CH34, with its multiplicity of plasmid-encoded metal efflux pumps, of strain AE104 has an influence on iron homeostasis as well. The deletion of furB had no effect.
FurA was involved in the control of the production of the sole siderophore of C. metallidurans; thus, it may be an important iron uptake regulator of this bacterium.
FurB could be a second Fur needed under iron starvation.
One or more unknown regulators were able to downregulate zupT expression and upregulate zntA and cadA expression under conditions of surplus external zinc, even in the ΔfurC mutant strain. FurB could be this unknown regulator. To investigate this possibility, a ΔfurB ΔfurC double deletion strain was constructed. Expression of zupT-lacZ or cadA-lacZ was not changed in the ΔfurB ΔfurC double mutant strain compared to the ΔfurC (Table 1), but the zntA-lacZ expression level was 50% higher in the double mutant in the presence of elevated zinc concentrations, indicating some influence of FurB on transition-metal uptake. Metal resistance was not changed in the ΔfurB ΔfurC double mutant compared to that of the ΔfurB single mutant (Fig. 2).
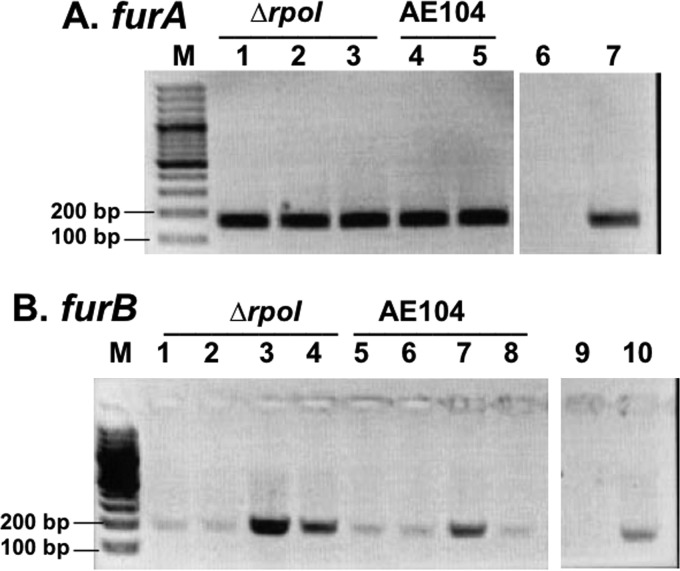
RT-PCR experiments were done to investigate the expression of the three fur genes. While furC was slightly upregulated in cells after EDTA treatment (data not shown), the expression of furA was constitutive in strain AE104 and a ΔrpoI mutant of this strain (Fig. 4A). The sigma factor RpoI was not required for expression of the furA gene.
FIG 4.
Expression of the genes furA and furB. C. metallidurans strain AE104 and its ΔrpoI deletion mutant that is unable to produce staphyloferrin B were treated for 10 min with 500 μM Fe(III) (B, lanes 1 and 5), 100 μM Fe(III) (A, lanes 1 and 4; B, lanes 2 and 6), 80 μM DIP (A, lanes 2 and 5; B, lanes 3 and 7), or without additions (A, lane 3; B, lanes 4 and 8). RNA was isolated, treated with DNase I, and reverse transcribed with random hexameric primers, and the cDNA subsequently was amplified by PCR using furA (A)- or furB (B)-specific primers. M, marker lane with two bands labeled. Lanes 6 and 9 in panels A and B, respectively, show the negative control (no template control). Lanes 7 and 10 in panels A and B, respectively, show the positive control (genomic DNA instead of RNA). As a loading control, the constitutively expressed gene rpoZ was amplified by PCR showing the same amounts of product for all RNA samples.
In contrast to furA, expression of furB was upregulated when AE104 cells were incubated in the presence of the iron chelator 2,2′-dipyridyl (DIP; 80 μM) (Fig. 4B), but furB expression was not influenced by iron addition, even in the presence of high concentrations (Fig. 4B, lanes 1 and 5). In the ΔrpoI mutant strain that was not able to synthesize staphyloferrin B, furB was upregulated compared to the parent strain, AE104, in mineral salts medium without additions (Fig. 4B, lanes 4 and 8), and it was still more upregulated in the presence of DIP (Fig. 4B, lanes 3 and 7). This indicated that FurB was not a FurC-substituting regulator but is needed under circumstances of iron starvation, generated in this experiment by removing the ability to produce a siderophore in combination with DIP treatment. FurB could be involved in the control of the production of iron import systems with low substrate specificity, which would generate an increased cellular content of other divalent metal cations, such as Zn(II), subsequently resulting in the increased expression level of zntA in the ΔfurB ΔfurC mutant strain compared to the single-gene deletion mutants (Table 1).
Purification of FurC.
FurC could be involved in direct or indirect control of zupT expression. If in direct control, FurC should bind to the zupT upstream region. To purify FurC, the furC gene was cloned in vector plasmid pASK-IBA 7 in E. coli strain BL21. This added the amino-terminal sequence MASWSHPQFEKIEGRRDRGPEF to FurC instead of the N-terminal methionine residue. This amino acid sequence contained a Strep tag II (underlined). Moreover, a C-terminal VDLQGDHGL sequence was fused to the resulting Strep-tagged FurC. After purification, the sodium dodecyl sulfate polyacrylamide gels always showed two bands (see Fig. S1 in the supplemental material) for Strep-tagged FurC, which also were visible in the anti-Strep tag Western blot (data not shown). Various chemical treatments, e.g., with zinc chloride, EDTA, and reducing agents, did not change this pattern (data not shown).
Both bands were isolated from the SDS-PAGE gels, the cysteine residues were modified using iodoacetamide, the resulting protein was digested with trypsin, and a mass spectrum of the resulting peptides was acquired (see Fig. S2 in the supplemental material). Both spectra were identical within 74.9% of the protein sequence initially covered. Only small predicted tryptic peptides were not found, such as IEGR (molecular mass, 473.5 Da), LR (287.4 Da), QLGAR (543.6 Da), VTQPR (599.7 Da), AGNDR (531.5 Da), VFR (420.5 Da), AEVHR (610.7 Da), CDR (392.4 Da), SVAPR (528.6 Da), and two single-arginine residues (data not shown). The underlined peptides subsequently could be identified manually (data not shown). The only difference between the samples concerned a peptide with a molecular mass of 2,542 Da, which was lacking in the lower band and could not be accounted for. This could be a modification of the 2,528-Da band (see Fig. S2). The peptide pattern showed only traces of oxidation, e.g., of methionine residues.
The peak with the third highest intensity (molecular mass, 1,216 Da) was not identified by the analysis program. This peak corresponded to the N-terminal tryptic fragment ASWSHPQFEK of Strep-tagged FurC with the N-terminal M1 removed. Thus, FurC was produced in E. coli as a protein with N-terminal methionine residues removed (198 amino acyl residues; predicted molecular mass of 21,796 Da), and the protein sequence now was recovered to 87.9%. Both protein bands were identical within this >87.9% of the sequence. They could represent different conformations or contain different cofactors or modifications.
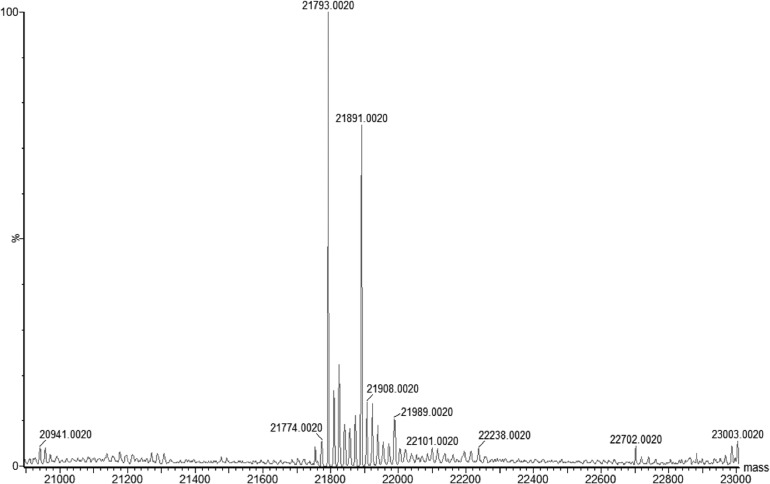
A mass spectrogram of both protein bands was obtained without digestion (Fig. 5), giving two bands with molecular masses of 21,793 and 21,891 Da. The 21,793-Da signal or peak could be the predicted 21,796-Da Strep-tagged FurC with the N-terminal fMet1 removed, with three protons additionally being removed and/or an error of 0.014%. The second signal, identical to the first band in the mass spectrum after tryptic digestion, was 98 Da larger. The ratio of the peak areas was 0.57:0.43 (Fig. 5).
FIG 5.
Mass spectrum of the two Strep tag FurC bands without trypsin digestion. Concentrated Strep tag-FurC was desalted by using a ZipTip C4 column (Millipore, Billerica, MA) and analyzed by an ESI-Q-TOF 2 mass spectrometer.
A size difference of 98 Da could correspond to a phosphate group (95 Da; unprotonated water released by the phosphorylation step was not counted). However, extensive treatment of Strep-tagged FurC with alkaline phosphatase did not change the double-band appearance of this protein (see Fig. S3 in the supplemental material).
The size difference of 98 Da would fit perfectly to 1.5 zinc ions per monomer (3 zinc ions per dimer are 98.055 Da, with the six protons occupying the zinc ligands in the zinc-free form not counted) or, alternatively, one zinc ion (65.37 Da) plus another component, such as a chloride (35.4 Da) or two hydroxide groups (34 Da). Indeed, when the metal content of Strep-tagged FurC was analyzed using ICP-MS, the protein contained 0.71 ± 0.01 mol zinc per mol protein (data not shown). No other elements, such as Fe, Cu, Se, Mo, W, Mg, Ni, and Cd, could be found. If one form of FurC contained 3 zinc ions per dimer and the second no zinc, the zinc-containing form would represent 47% ± 1% (71% divided by 1.5) of the total mixture, agreeing sufficiently with the ratio of the peak areas of 0.57:0.43. An asymmetric, partially active dimer with three zinc ions also has been described for the Bacillus subtilis Zur (29). Thus, FurC probably was produced in E. coli as a mixture of 45% ± 2% of a form that contained 3 zinc ions per dimer and of 55% ± 2% of a form without zinc.
Binding of FurC to the zupTp promoter.
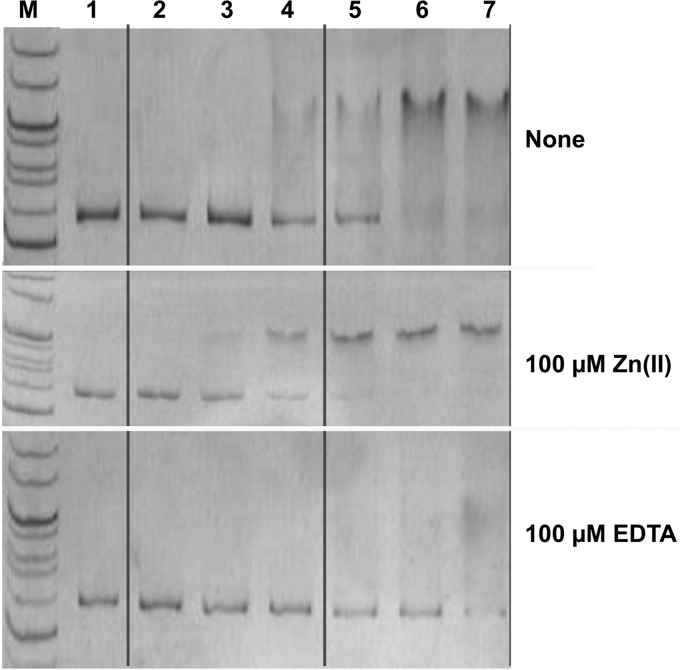
To verify the direct regulation of zupT expression by FurC, a 563-bp NotI/NcoI DNA fragment covering the upstream region of zupT or parts of it was amplified by PCR and used in mobility shift assays. FurC clearly retarded migration of the 563-bp fragment, while this could not be observed in the controls (heat-inactivated FurC, bovine serum albumin, and the DNA-binding protein Sco0212 from Streptomyces coelicolor). The presence of 50 μM EDTA, a mixture of 100 μM Zn(II) and 100 μM EDTA, of 100 μM Zn(II), or of 500 μM EDTA did not change the pattern (see Fig. S4 in the supplemental material).
When 0.2 pmol of the zupT promoter region was titrated with FurC (Fig. 6), the promoter fragment occurred with a 50:50 ratio of retarded to nonretarded DNA at about 12 pmol of ZupT (see Fig. S5 in the supplemental material) in this individual experiment, representing a concentration of 800 nM in the reaction mixture. In reproductions, the 50:50 ratio occurred between 4 and 8 pmol FurC and at about 6 pmol (data not shown), but scanning all these gels and calculating the ratios and the mean values from them did not yield significant results (see Fig. S5). Thus, FurC retarded half of the zupTp promoter fragments between 6 pmol and 12 pmol, representing a concentration of 400 nM to 800 nM protein and a FurC:zupTp ratio between 30 and 60. This fluctuation of the data indicates that FurC produced in E. coli was indeed a mixture of an inactive zinc-free form and a partially active, asymmetric form with 3 zinc ions per dimer and that the ratio of both forms depends on the individual purification batch and storage time of the purified protein.
FIG 6.
Mobility shift of the zupT upstream region by Strep tag FurC. The zupT upstream region was amplified as a 563-bp NotI-NcoI fragment. In a reaction volume of 15 μl, 0.2 pmol of this DNA fragment was incubated for 30 min at 30°C without Strep tag FurC (lane 1) or with 1 pmol (lane 2), 4 pmol (lane 3), 8 pmol (lane 4), 12 pmol (lane 5), 16 pmol (lane 6) or 20 pmol Strep-tagged FurC (lane 7). Moreover, the reaction mixture contained no addition (upper), 100 μM Zn2+ (middle), or 100 μM EDTA (lower). M, size marker lane. A silver-stained 10% (wt/vol) native polyacrylamide gel was used. Vertical lines were drawn to allow better orientation.
When 100 μM Zn(II) was added to FurC and the promoter fragment, the 50:50 ratio appeared at a smaller amount of FurC in three independent experiments at 4 pmol (Fig. 6 and data not shown; also see Fig. S5 in the supplemental material), representing a concentration of 270 nM FurC and a FurC:zupTp ratio of 20. Zinc ions seem to stabilize the zinc-containing form. In contrast, more FurC was needed for half-maximum retardation in the presence of 100 μM EDTA, 20 pmol (Fig. 6; also see Fig. S5) in one experiment, while the other two experiments did not yield a difference between addition of EDTA and no addition (data not shown). Thus, FurC bound to the zupTp promoter region, and more FurC was needed at low zinc concentrations (600 ± 200 nM) than at high zinc concentrations (270 nM FurC) for half-maximum retardation of this promoter region.
Complementation of the ΔfurC mutant strain.
The furC gene with the N-terminal Strep tag was cloned into plasmids pBBR1-MCS2 and pBBR1-MCS3 (30) and transferred into C. metallidurans strain AE104 and the ΔzupT mutant. No repression of zupT-lacZ could be observed (Table 3). Nevertheless, the Strep-tagged FurC protein could be isolated from cells of C. metallidurans strain AE104 (see Fig. S6 in the supplemental material). After production in C. metallidurans and isolation of the protein by affinity chromatography (see Fig. S6), a band corresponding to the 46-kDa dimer of FurC was clearly visible, but this band did not yield a signal in the Western blot, as the Strep tag was masked. Moreover, the protein seemed to be rapidly degraded in C. metallidurans to an N-terminal Strep-tagged FurC of 14 kDa (see Fig. S6C).
TABLE 3.
Activity of a zupT-lacZ reporter gene fusion in a ΔfurC background complemented in trans under various conditionsa
| Condition | Activity of fusion in AE104 strain: |
|||||
|---|---|---|---|---|---|---|
| Wild type | ΔfurC | ΔfurCb | ΔfurCc | ΔfurCd | ΔfurCe | |
| MM | 20 ± 3 | 251 ± 30 | 229 ± 14 | 220 ± 37 | 11 ± 1 | 24 ± 5 |
| 200 μM Zn2+ | 13 ± 1 | 150 ± 7 | 140 ± 18 | 120 ± 35 | 8 ± 2 | 11 ± 3 |
| 500 μM Zn2+ | 11 ± 2 | 134 ± 22 | 111 ± 20 | 111 ± 27 | 7 ± 3 | 12 ± 2 |
| 200 μM EDTA | 65 ± 10 | 259 ± 21 | 256 ± 20 | 261 ± 25 | 59 ± 15 | 94 ± 6 |
| 500 μM EDTA | 78 ± 8 | 257 ± 20 | 243 ± 38 | 269 ± 29 | 61 ± 16 | 98 ± 7 |
Three experiments were performed, and deviations are shown.
With plasmid pBBR1.
With plasmid pBBR::furC with an N-terminal FurC tag.
With plasmid pBBR::furC with a C-terminal FurC tag.
With plasmid pBBR::furC.
Cloning from plasmid pASK7 to the broad-host-range vector pBBR1 added the amino acid sequence MTMITPSAQLTLTKGNKSWVPGP upstream of the sequence containing the Strep tag (in boldface), MASWSHPQFEKIEGRRDRGPEF, and these additional amino acids might be responsible for instability. Thus, FurC also was produced as a C-terminal tagged and untagged protein in C. metallidurans. In both cases, the regulatory pattern of the parent strain AE104 was restored (Table 3).
DISCUSSION
The zinc homeostasis system of C. metallidurans.
Transition-metal cations may form complex compounds, which are needed for many sophisticated reactions in the cellular biochemistry. On the other hand, transition-metal cations produce oxidative stress due to their interaction with thiol compounds, some may catalyze production of dangerous hydroxyl radicals in Fenton-type reactions, and, since these cations have very similar ionic diameters, they may interfere with each other's homeostasis (31).
Thus, how transition-metal cations can be efficiently allocated to the corresponding proteins without causing collateral damage is an interesting question (32). One solution seems to be mélange control: not only the cellular quota of individual transition-metal cations but also the composition of the overall transition-metal mélange are being strictly controlled. Interestingly, the transition-metal composition of the bacterial cell resembles that of seawater, leading to the idea that it is sufficient in most environments to take up the mixture of transition-metal cation as they come along and later handle surplus or nonsufficient individuals (33).
The metal-resistant betaproteobacterium C. metallidurans of the order Burkholderiales (3) is a suitable subject to study transition-metal homeostasis, since it is able to maintain its multiple-metal homeostasis even in a mixture of highly concentrated transition-metal salts (1). Zinc is the second most important transition-metal cation in C. metallidurans after iron, but it is required in larger amounts than the cations of the other transition metals. Surplus zinc is removed from the C. metallidurans cell by (i) the PIB2-type ATPase ZntA, providing a basal resistance level; (ii) the back-up PIB2-type ATPase CadA that also provides basal-level resistance to Cd2+; and (iii) the plasmid-encoded high resistance-mediating Czc system composed of the CDF exporter CzcD and the high-rate PIB4-type ATPase CzcP for cytoplasmic Zn2+ in combination with the CzcCBA transenvelope efflux complex for periplasmic Zn2+ and many other Czc components. A variety of secondary import systems may interact with these exporters to form a kinetic flow equilibrium that adjusts the concentration of individual cations as well as the overall mélange. These importers, ZupT, PitA, CorA1, CorA2, and CorA3, do not form shunts of grouped exporters and importers, with flow and gene expression controlled by individual cations, but compose a highly redundant battery of metal uptake systems with only minimal cation selectivity. Indeed, as suggested by the seawater idea, transition-metal cations may be imported as they come along, with subsequent mélange control by the efflux systems (1).
This seawater model, however, is too simple. Although ZupT was not essential for zinc import into C. metallidurans and may have a broad substrate specificity as its ortholog from E. coli, ΔzupT mutants were not able to tolerate the presence of a CzcCBA efflux system and did not allocate zinc as efficiently as the parent strains to zinc-dependent proteins, such as the RpoC subunit of the RNA polymerase (6). On the basis of the fact that the PIB4-type high-zinc resistance ATPase CzcP alone is not able to mediate a basic level of resistance to zinc (7) despite its high transport rate of this metal, at least two zinc pools might exist in C. metallidurans. A pool of loosely bound Zn(II) also can be filled up with other transition-metal cations, zinc is provided to this pool by the low-specificity secondary importers, and surplus zinc is removed by high-rate exporters, such as CzcP and CzcD. Another pool of more firmly bound zinc needs ZupT for replenishment and ZntA or CadA for zinc removal (6). Thus, a zinc allocation pipeline may start at ZupT and lead to important zinc-dependent proteins. This pipeline may complement the other, more unspecific metal allocation system and may be involved in metal discrimination.
Members of the Fur protein family in C. metallidurans.
Unraveling the molecular identity of the zinc pools and zinc pipeline in C. metallidurans starts best at ZupT due to its importance as a central zinc importer. The regulator of zupT expression also might be involved in the control of the synthesis of other components of this pipeline. Here, FurC of C. metallidurans was identified as the Zur in this bacterium. FurC is closely related to orthologs in other Cupriavidus/Ralstonia species and to the Zur from E. coli, the patriarch of the Zur tribe (Fig. 1). The other two members of the Fur protein family in C. metallidurans, FurA and FurB, form a cluster of proteins related to Fur from E. coli (Fig. 1). A more detailed BLAST analysis of the 500 proteins most related to FurB from C. metallidurans (data not shown) identified 37 sequences coming, with one exception, exclusively from bacteria of the families Burkholderiaceae, Comamonadaceae, and Oxalobacteraceae of the order Burkholderiales. The exception came from the genus Thauera from the Rhodocyclales, also an order of the class Betaproteobacteria. This cluster of Burkholderiales FurB protein was related to 224 sequences from gammaproteobacteria, which contain Fur orthologs from Enterobacteriaceae, and 213 FurA-like proteins, again from betaproteobacteria. Twenty-four sequences from Burkholderiales were outside these clusters. In contrast, the 500 FurC/Zur proteins formed clusters of sequences coming from all kinds of bacterial phyla. Together with the location of the furB gene on chromosome 2 from C. metallidurans, this indicates that during the evolution of Burkholderiales, the fur gene was duplicated into furA and furB, with their gene products also differentiating in their function, with FurA being the main iron uptake regulator while FurB supports FurA under conditions of iron limitation. Burkholderiales might have adapted to environments where iron is scarce or other transition metals interfere with iron acquisition.
Zur/FurC-type regulators.
The Zur regulator, as a member of the Fur family of regulators, was found by Klaus Hantke (9). Zur in Escherichia coli controls the expression of the znuA<>znuCB divergon for an ABC-type zinc importer (10). Zur is a dimer and binds to a defined Zur box in the znu operator (34), and Zn(II) binds with subfemtomolar affinity to at least two sites, one in an S-3-N(O) environment and a second in a different environment (35). In other proteobacteria, the Zur regulons were only roughly characterized, mostly in (plant) pathogens.
FurC and the zinc homeostasis system in C. metallidurans.
FurC with an N-terminal Strep tag needed for purification was produced in two different conformations in E. coli and C. metallidurans. In E. coli, nearly no dimer was visible, but there were two conformations of the monomer (see Fig. S1 in the supplemental material). A mass spectrogram of the tryptic digest indicated that the amino acid sequence of both bands was identical and that both started with Ala2 after the removal of the N-terminal fMet1 (see Fig. S2). A mass spectrogram of the nondigested protein displayed this 21.8-kDa form plus a second form that was 98 Da larger in size. Since FurC contained 0.71 zinc ions per monomer, this second form could represent a FurC dimer in solution that was loaded with 3 zinc ions per dimer. Such a form has been described for Zur in B. subtilis (29) and would represent 45% of the FurC preparations.
This indicates that E. coli cells were not able to allocate sufficient zinc to the FurC protein. Nevertheless, at least one of the two forms, probably the dimer with 3 zinc ions, was able to bind to the zupTp promoter. The protein/promoter ratios of at least 20:1 in this experiment also agree with the hypothesis that FurC produced in E. coli was not fully folded and active.
In C. metallidurans, N-terminally tagged FurC was unstable, probably due to 23 additional amino acids upstream of the tag, which resulted from the cloning of the gene to the broad-host-range vector. The C-terminal and untagged versions of FurC were able to restore the ΔfurC mutant to the phenotype of the parent, as indicated by the regulatory pattern of the zupT-lacZ reporter gene fusion (Table 3).
The additional zinc-dependent regulator.
Expression of reporter gene fusions for other secondary metal uptake systems, such as corA1-lacZ, were zinc dependent in the ΔfurC mutant, and even the zupT-lacZ fusion showed some zinc dependence in its regulation of expression in this strain (Table 1). This indicated the presence of another zinc-dependent regulator. FurB could contribute to this regulation but is not the only additional zinc-dependent regulator, as demonstrated by the phenotype of the ΔfurB ΔfurC double mutant (Table 1). The furC gene is the first in an operon region, followed by genes for CobW proteins of the COG0523 family that might represent zinc chaperones (36) and a gene for a DksA-type regulator (Fig. 1).
DksA-type regulators bind to the RNA polymerase and not to operators on the DNA, and they link the transcriptional activity to the alarmone guanosine pentaphosphate (37). E. coli possesses one DksA that is a typical zinc finger protein, and C. metallidurans has four putative DksA proteins. Three of these also are typical zinc finger proteins and are encoded by genes close to genes for UspA universal stress proteins (38) plus for a GntR-type regulator of these genes. All are in the vicinity of an old chromosomal czc-like metal resistance determinant that was inactivated in C. metallidurans during evolution by native transposon insertion and chromosome reorganization (39). The three zinc finger DksA proteins could not be found in the proteome of C. metallidurans AE104 (M. Herzberg and D. H. Nies, unpublished data). In contrast, in the product of the dksA gene downstream of furC, the typical zinc finger has been removed and there are sufficient copies of this DksA protein in the cell to saturate all RNA polymerase complexes. Such a zinc-free DksA could have the function of decreasing the number of zinc ions in the functional RNA polymerase. Since DksA is linked to the global stress response of C. metallidurans at a high hierarchical level, drastic effects could result, such as the degradation of all proteins preventing import of zinc ions, e.g., CzcA (6). Thus, DksA is a candidate for a global zinc regulator in C. metallidurans.
It seems clear that FurC is the central zinc uptake regulator Zur in C. metallidurans and is responsible for control of zupT expression and subsequently for full function of the zinc allocation pipeline starting at ZupT (6). This is similar to the situation described for many other bacteria. On the other hand, FurC may have features additional to those of the Zur proteins coming from non-metal-resistant bacteria: it may be closely linked to the filling level of the zinc pools by providing a feedback mechanism between ZupT-mediated filling up of this pool and zupT regulation. Moreover, FurC also could be involved in global zinc-dependent metabolic control in C. metallidurans by DksA- and UspA-type proteins. Thus, FurC might be more than just the Zur in C. metallidurans.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this work was provided by the Deutsche Forschungsgemeinschaft (Ni262/10-2).
We thank Grit Schleuder and Karola Otto for skillful technical assistance, Martin Herzberg for ICP-MS, and Angelika Schierhorn for MS analysis of FurC.
Footnotes
Published ahead of print 21 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01713-14.
REFERENCES
- 1.Kirsten A, Herzberg M, Voigt A, Seravalli J, Grass G, Scherer J, Nies DH. 2011. Contributions of five secondary metal uptake systems to metal homeostasis of Cupriavidus metallidurans CH34. J. Bacteriol. 193:4652–4663. 10.1128/JB.05293-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, van Gijsegem F. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Medigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M. 2010. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433. 10.1371/journal.pone.0010433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busch W, Saier MHJ. 2002. The transporter classification (TC) system. Crit. Rev. Biochem. Mol. Biol. 37:287–337. 10.1080/10409230290771528 [DOI] [PubMed] [Google Scholar]
- 5.Saier MHJ, Tran CV, Barabote RD. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acid Res. 34:D181–D186. 10.1093/nar/gkj001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herzberg M, Bauer L, Nies DH. 2014. Deletion of the zupT gene for a zinc importer influences zinc pools in Cupriavidus metallidurans CH34. Metallomics 6:421–436. 10.1039/c3mt00267e [DOI] [PubMed] [Google Scholar]
- 7.Scherer J, Nies DH. 2009. CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol. Microbiol. 73:601–621. 10.1111/j.1365-2958.2009.06792.x [DOI] [PubMed] [Google Scholar]
- 8.Legatzki A, Anton A, Grass G, Rensing C, Nies DH. 2003. Interplay of the Czc-system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J. Bacteriol. 185:4354–4361. 10.1128/JB.185.15.4354-4361.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hantke K. 2005. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8:196–202. 10.1016/j.mib.2005.02.001 [DOI] [PubMed] [Google Scholar]
- 10.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199–1210. 10.1046/j.1365-2958.1998.00883.x [DOI] [PubMed] [Google Scholar]
- 11.Stojiljkovic I, Bäumler AJ, Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531–545 [DOI] [PubMed] [Google Scholar]
- 12.Helmann JD, Soonsange S, Gabriel S. 2007. Metallalloregulators: arbiters of metal sufficiency, p 37–71 In Nies DH, Silver S. (ed), Molecular microbiology of heavy metals, vol 6 Springer-Verlag, Berlin, Germany [Google Scholar]
- 13.Nies D, Mergeay M, Friedrich B, Schlegel HG. 1987. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J. Bacteriol. 169:4865–4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 15.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 16.Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062–1067 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H. 2005. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl. Environ. Microbiol. 71:8472–8480. 10.1128/AEM.71.12.8472-8480.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albert H, Dale EC, Lee E, Ow DW. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7:649–659. 10.1046/j.1365-313X.1995.7040649.x [DOI] [PubMed] [Google Scholar]
- 19.Lenz O, Schwartz E, Dernedde J, Eitinger T, Friedrich B. 1994. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J. Bacteriol. 176:4385–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nies DH. 1992. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 174:8102–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- 22.Große C, Grass G, Anton A, Franke S, Navarrete Santos A, Lawley B, Brown NL, Nies DH. 1999. Transcriptional organization of the czc heavy metal homoeostasis determinant from Alcaligenes eutrophus. J. Bacteriol. 181:2385–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber K, Osborn M. 1969. Reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406–4412 [PubMed] [Google Scholar]
- 24.Blum H, Beier H, Gross HJ. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99. 10.1002/elps.1150080203 [DOI] [Google Scholar]
- 25.Wagegg W, Braun V. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J. Bacteriol. 145:156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilis A, Khan AM, Cornelis P, Meyer JM, Mergeay M, van der Lelie D. 1996. Siderophore-mediated iron uptake in Alcaligenes eutrophus CH34 and identification of aleB encoding the ferric-alcaligin E receptor. J. Bacteriol. 178:5499–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munzinger M, Taraz K, Budzikiewicz H. 1999. Staphyloferrin B, a citrate siderophore of Ralstonia eutropha. Z. Naturforsch. 54:867–875 [Google Scholar]
- 28.Große C, Friedrich S, Nies DH. 2007. Contribution of extracytoplasmic function sigma factors to transition metal homeostasis in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 12:227–240. 10.1159/000099644 [DOI] [PubMed] [Google Scholar]
- 29.Ma Z, Gabriel SE, Helmann JD. 2011. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 39:9130–9138. 10.1093/nar/gkr625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- 31.Nies DH. 2007. Bacterial transition metal homeostasis, p 118–142 In Nies DH, Silver S. (ed), Molecular microbiology of heavy metals, vol 6 Springer-Verlag, Berlin, Germany [Google Scholar]
- 32.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460:823–830. 10.1038/nature08300 [DOI] [PubMed] [Google Scholar]
- 33.Nies DH. 2012. Zinc starvation response in a cyanobacterium revealed. J. Bacteriol. 194:2407–2412. 10.1128/JB.00257-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321–24332. 10.1074/jbc.M001775200 [DOI] [PubMed] [Google Scholar]
- 35.Outten CE, Tobin DA, Penner-Hahn JE, O'Halloran TV. 2001. Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry 40:10417–10423. 10.1021/bi0155448 [DOI] [PubMed] [Google Scholar]
- 36.Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crecy-Lagard V. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. 10.1186/1471-2164-10-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. 2004. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell 118:297–309. 10.1016/j.cell.2004.06.030 [DOI] [PubMed] [Google Scholar]
- 38.Nachin L, Nannmark U, Nystrøm T. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187:6265–6272. 10.1128/JB.187.18.6265-6272.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Rozycki T, Nies DH. 2009. Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 96:115–139. 10.1007/s10482-008-9284-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.