Abstract
In the filamentous cyanobacterium Anabaena sp. strain PCC 7120, heterocysts are formed in the absence of combined nitrogen, following a specific distribution pattern along the filament. The PatS and HetN factors contribute to the heterocyst pattern by inhibiting the formation of consecutive heterocysts. Thus, inactivation of any of these factors produces the multiple contiguous heterocyst (Mch) phenotype. Upon N stepdown, a HetN protein with its C terminus fused to a superfolder version of green fluorescent protein (sf-GFP) or to GFP-mut2 was observed, localized first throughout the whole area of differentiating cells and later specifically on the peripheries and in the polar regions of mature heterocysts, coinciding with the location of the thylakoids. Polar localization required an N-terminal stretch comprising residues 2 to 27 that may represent an unconventional signal peptide. Anabaena strains expressing a version of HetN lacking this fragment from a mutant gene placed at the native hetN locus exhibited a mild Mch phenotype. In agreement with previous results, deletion of an internal ERGSGR sequence, which is identical to the C-terminal sequence of PatS, also led to the Mch phenotype. The subcellular localization in heterocysts of fluorescence resulting from the fusion of GFP to the C terminus of HetN suggests that a full HetN protein is present in these cells. Furthermore, the full HetN protein is more conserved among cyanobacteria than the internal ERGSGR sequence. These observations suggest that HetN anchored to thylakoid membranes in heterocysts may serve a function besides that of generating a regulatory (ERGSGR) peptide.
INTRODUCTION
When confronted with a condition of nitrogen deficiency, some filamentous cyanobacteria produce cells called heterocysts that specialize in the fixation of atmospheric nitrogen and are distributed in a defined pattern along the filament. In strains of the genera Anabaena and Nostoc, heterocysts are found separated by intervals of ca. 10 vegetative cells on average (1, 2). The differentiation of a vegetative cell into a heterocyst results from a program of gene activation events orchestrated by two transcription factors, the global regulator NtcA and the heterocyst-specific regulator HetR (3). In addition, a number of other regulators participate to determine the spatial pattern of heterocyst distribution along the filament. Specifically, inactivation of patS (4) or patU3 (5) or overexpression of hetF (6, 7) produces a multiple contiguous heterocyst (Mch) phenotype in which heterocysts are found in groups, frequently separated by vegetative-cell intervals shorter than those in the wild-type strain. On the other hand, inactivation of patA produces heterocysts mostly at the filament ends (8).
HetN also influences the pattern of heterocyst distribution along the filament. The hetN gene product exhibits similarity to oxidoreductases involved in the synthesis of fatty acids and polyketides and includes an RGSGR sequence that is identical to the five C-terminal residues of the PatS morphogen (9–11). In the genome of Anabaena sp. strain PCC 7120, the hetN gene is preceded by hetM (renamed hglB; the protein product is similar to polyketide synthases and is involved in the formation of heterocyst glycolipids [Hgl]), alr5356 (encoding a hypothetical protein), and other hgl genes and is followed by hetI, which is transcribed from the opposite DNA strand and is annotated as encoding a protein similar to phosphopantetheinyl transferases (9, 12). hetN is expressed starting ca. 6 to 12 h after N stepdown in a monocistronic transcript (9, 13). Contrasting effects of hetN inactivation and overexpression have been reported, with inactivation of hetN by the insertion of different constructs yielding either no change in phenotype, suppression of heterocyst differentiation, or overdifferentiation of heterocysts (9). hetN expression from a heterologous gene promoter (the Cu-inducible promoter PpetE, which is active mainly in vegetative cells) yielded suppression of differentiation, whereas in the absence of Cu, an Mch phenotype was manifest 48 h after N stepdown, but no apparent phenotype was evident at earlier times (10). On the other hand, the presence of hetN on a multicopy plasmid produced suppression of differentiation, but not always (9, 13). To what extent these different effects were due directly to inactivation of hetN, to interference with the effects of hetN transcription on neighboring genes, or to variable protein levels resulting from ectopic gene expression is difficult to discern. Recently, a ΔhetN strain has been reported to yield increased heterocyst frequency and an Mch phenotype 48 h after N stepdown (14). Regarding a possible enzymatic activity of HetN, this protein has been reported to catalyze ATP or GTP hydrolysis in vitro, and conservation of residues integrating a putative catalytic triad necessary for oxidoreductase activity (S142, Y155, and K159) has been noted (15). First it was reported that a K159E substitution, but not an S142D or Y155V substitution, blocked the inhibitory activity of HetN when it was overexpressed from a replicative plasmid, though ATP hydrolysis was only weakly affected (15). Later, conservative replacement of any of the three residues (S142A, Y155F, K159R) was reported to have no effect on the patterning of heterocysts (14).
It has been proposed that an RGSGR peptide resulting from the processing of HetN in the cytoplasm diffuses between cells via direct cytoplasmic exchange (14). However, neither the identity nor the mechanism of action of the actual HetN-derived signaling molecule for the regulation of the heterocyst distribution pattern is known. In this work, we have reconsidered the six C-terminal residues of PatS that are reproduced in the HetN sequence and have confirmed their role in heterocyst patterning. We have also found that HetN-green fluorescent protein (GFP) fusion proteins localized specifically to the heterocyst periphery and cellular poles at thylakoids and that this localization required the presence of an N-terminal sequence including residues 2 to 27, indicating the occurrence of a form of HetN larger than the ERGSGR peptide in these differentiated cells.
MATERIALS AND METHODS
Strains and growth conditions.
Anabaena sp. strain PCC 7120 (also known as Nostoc sp. strain PCC 7120) was grown photoautotrophically in a BG110-based medium supplemented with NH4Cl as described previously (16). For bubbled cultures, the medium was supplemented with NaHCO3 and was sparged with a mixture of air and CO2 (99:1). When required, antibiotics were added to the medium: neomycin (Nm) at a final concentration of 10 μg·ml−1, streptomycin (Sm) and spectinomycin (Sp) at final concentrations of 2 μg·ml−1 in liquid cultures and 5 μg·ml−1 in solid cultures. For spot growth tests on solid medium, filaments grown in liquid BG110 medium supplemented with NH4Cl were washed 3 times with BG110. Filaments were spotted onto petri dishes containing BG110 medium alone or supplemented with NH4Cl, without antibiotics and were incubated under standard conditions for 14 days.
Construction of mutants.
To delete different regions in the hetN gene (see Fig. 1A), overlapping PCR primer pairs alr5358-1/alr5358-2 and alr5358-3/alr5358-4 (for complete gene deletion), alr5358-1/alr5358-9 and alr5358-10/alr5358-4 (for deletion of DNA sequences encoding the ERGSGR amino acid sequence), and alr5358-20/alr5358-13 and alr5358-14/alr5358-21 (for deletion of DNA sequences corresponding to amino acids 2 to 27) were used for amplification with Anabaena genomic DNA as the template. All oligodeoxynucleotide primers are described in Table 1. The resulting DNA fragments were sequenced and were cloned first into pMBL-T and then into pRL278 (17) or directly into pRL278. The resulting plasmids were named pCSL23, pCSL42, and pCSL73, respectively.
FIG 1.
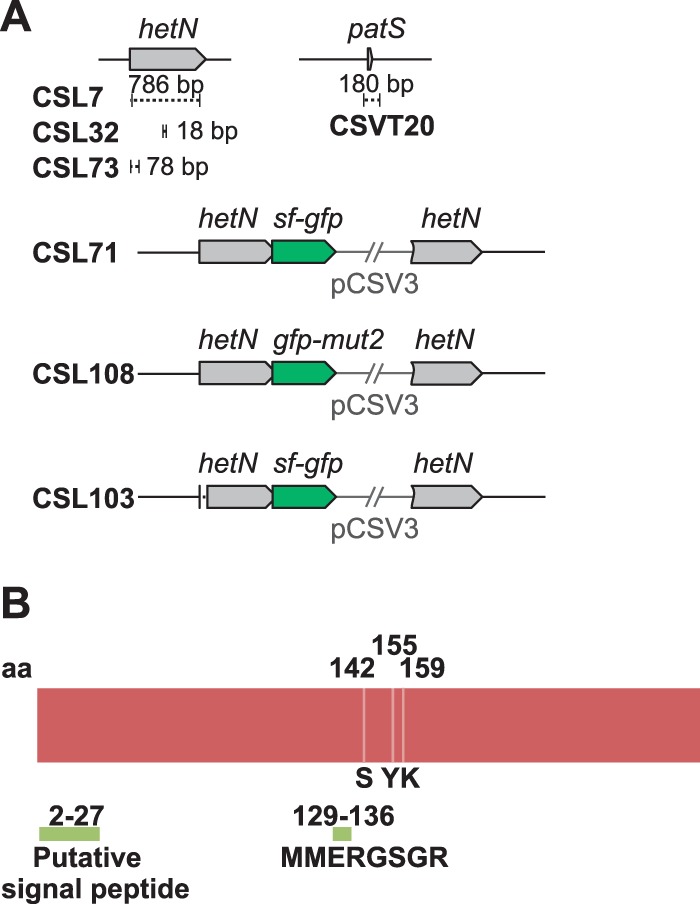
(A) Schematic of the hetN genomic region in Anabaena sp. strain PCC 7120 and mutant derivatives. Genes from the wild-type strain PCC 7120 are represented by gray arrows, and portions (of the specified sizes) deleted in the indicated strains are represented by dashed segments. The two types of gfp genes fused to intact or partially deleted hetN genes are represented by green arrows. (B) Schematic of the HetN protein with the following relevant protein segments indicated: a putative N-terminal signal peptide, the MMERGSGR sequence, and the putative catalytic triad of S142, Y155, and K159 residues.
TABLE 1.
Oligodeoxynucleotide primers used in this work
| Name | Sequence (5′–3′) |
|---|---|
| alr5358-1 | TTATACACCTTGCGTCCCTTCCTC |
| alr5358-2 | CTCAACAGCTACATAGCGTGAAGCGCCGGT |
| alr5358-3 | ACCGGCGCTTCACGCTATGTAGCTGTTGAG |
| alr5358-4 | TGAAGTTCATCTCTGGCGCATTCC |
| alr5358-9 | AGCAATATTGACAATCATCATGCTGGGTAG |
| alr5358-10 | CTACCCAGCATGATGATTGTCAATATTGCT |
| alr5358-13 | TACCGTTGCCTGTTCTTTCATTGTAACCTGCTAGTC |
| alr5358-14 | GACTAGCAGGTTACAATGAAAGAACAGGCAACGGTA |
| alr5358-18 | AACCGGAAGCTTACGCGGTCTTGGTGTCTACATTG |
| alr5358-19 | AAGCAAGGTCTCACGCCTGAGCGATGAGACTC |
| alr5358-20 | GTAGATGGATCCTTATACACCTTGCGTCCCTTCCTC |
| alr5358-21 | TTTTGGCTCGAGTCCACTACACCGAACCAACGAT |
| alr5358-26 | AACCGGGGTACCACGCGGTCTTGGTGTCTACATTG |
| GFP-11 | TCTGGTACCTTATTTGTATAGTTC |
| GFP-13 | TTCTCCTTTGCTAGCACCTCCA |
| GFP-14 | TGGAGGTGCTAGCAAAGGAGAA |
For the construction of a hetN-sf-gfp fusion, primer pair alr5358-18/alr5358-19 was used to amplify the 3′ end of hetN, and the resulting PCR product was cloned into plasmid pCSAL39 (which includes a sequence encoding a tetraglycine linker preceded by a BsaI site just before the sf-gfp sequence). The resulting construct was cloned into pCSV3 (18), producing plasmid pCSL72. For the construction of a hetN-gfp-mut2 fusion, overlapping PCR was performed either using pCSL72 as the template with primer pair alr5358-26/GFP-13 or using pCSL68 (which includes the gfp-mut2 sequence) as the template with primer pair GFP-14/GFP-11. The resulting DNA fragment was cloned using KpnI into pCSV3, producing plasmid pCSL124.
Plasmids pCSL23, pCSL42, pCSL73, pCSL72, and pCSL124 were transferred to strain PCC 7120 by conjugation, performed as described previously (19). Additionally, plasmid pCSL23 was transferred to strain CSVT20 (11) and plasmid pCSL72 to strains CSVT20 and CSL73 (expressing HetN Δ2–27). Exconjugants were selected by their resistance to Sm and Sp, or to Nm; when appropriate, double recombinants were selected by their resistance to sucrose (20) (sucrose sensitivity is conferred by the sacB gene, present in the vector portions of the transferred plasmids); and the chromosome structure in the altered region was tested by PCR and, in the case of pCSL73, by sequencing as well.
Microscopy.
For light microscopy, filaments grown in BG110 medium plus NH4Cl (in the presence of antibiotics when appropriate) were harvested, washed with nitrogen-free (BG110) medium, and incubated in bubbled cultures at 30°C in the light. At least 300 cells or 100 intervals were counted for each strain in each of three to four independent experiments. Dividing cells were counted as two cells. For the staining of (pro)heterocysts, cell suspensions were mixed (1:10) with a 1% alcian blue (Sigma) solution (11).
Microscopy was also performed with a Leica SP5 confocal microscope equipped with an HCX PL APO lambda blue 63.0× oil UV objective (numerical aperture, 1.40) and with a Leica DM6000 B fluorescence microscope and an ORCA-ER camera (Hamamatsu) using an FITC (fluorescein isothiocyanate) L5 filter (for excitation, a band-pass [BP] 480/40 filter; for emission, a BP 527/30 filter) for GFP fluorescence and a Texas Red TX2 filter (for excitation, a BP 560/40 filter; for emission, a BP 645/75 filter) for cyanobacterial autofluorescence (chlorophyll fluorescence). Treatment of the images, including blind deconvolution for deblurring of 3-dimensional (3D) images, was carried out with Leica Application Suite Advanced Fluorescence (LAS AF) software. For time lapse experiments, Anabaena filaments were placed on BG110 medium solidified with 0.5% agar and were incubated for at least 48 h, growing at 30°C. Sequential pictures were taken for another 27 h.
Sequence analysis.
Genomes were accessed and analyzed from within BioBIKE (21), an online platform that combines a programming language geared to genome analysis with a built-in genomic database. A list of genomes considered is given in Table S1 in the supplemental material. Sequences were compared using BLAST (22). Multiple sequence alignments were made using ClustalW (23) and were visualized using Jalview (24). Graphical alignments of genetic regions were made with a program written by Arnaud Taton in BioBIKE. Phylogenetic trees were made using Phylip (25). Protein domains were predicted through the SMART database (26). All the tools listed above were accessed through built-in BioBIKE functions, which handled any required format transformations.
RESULTS
hetN mutants.
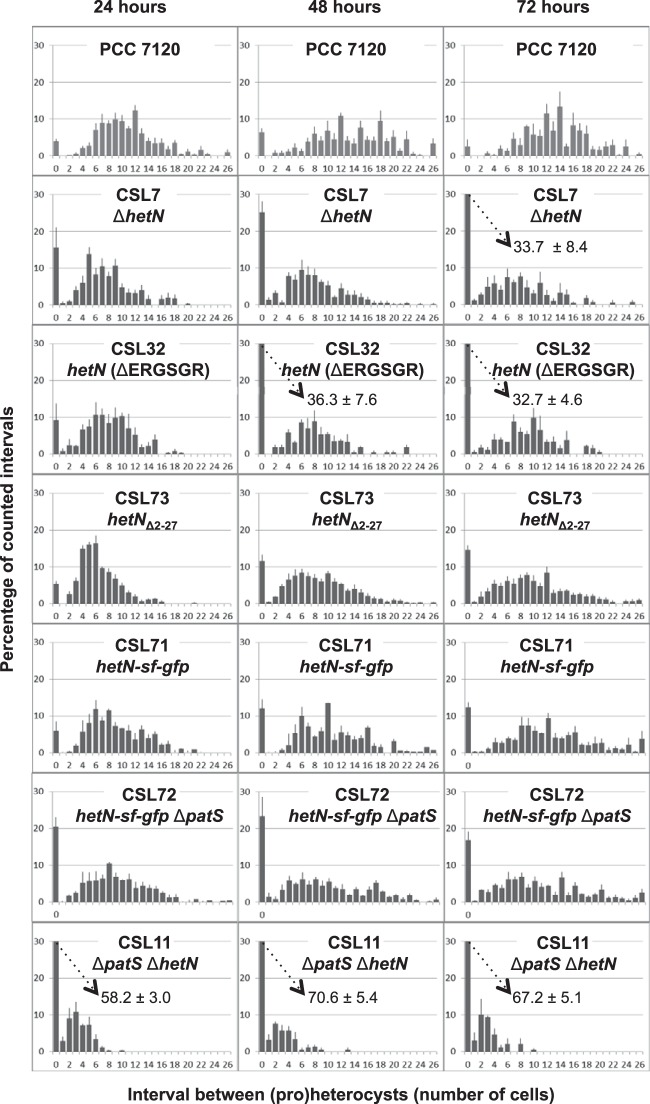
In order to investigate the functional role of HetN in heterocyst spacing, we focused on two regions of the protein (Fig. 1B): (i) an internal ERGSGR sequence (residues 131 to 136), which is the same as the six C-terminal residues of PatS, and (ii) the N terminus of HetN, suggested by Higa et al. (14) to be a signal peptide. Those investigators found that a strain expressing HetN with a deletion of residues 2 to 46 formed a pattern of heterocysts similar to that of the wild type, while different strains with conservative substitutions of the R132, G133, S134, G135, and R136 residues exhibited increased heterocyst frequency and a tendency to form multiple contiguous heterocyts (14). We have constructed strains CSL73 and CSL32, expressing HetN versions lacking residues 2 to 27 and 131 to 136, respectively (Fig. 1A). In these strains we have determined the heterocyst frequency, the frequency of contiguous heterocysts or proheterocysts (for definitions, see reference 11), and the mean size of the vegetative-cell interval between heterocysts (Fig. 2; Table 2). In agreement with previous results, the percentage of total heterocysts, the percentage of contiguous heterocysts, and the mean interval size in strain CSL32 (producing HetN ΔERGSGR) were similar to those of a hetN deletion mutant (strain CSL7, constructed in this work [Fig. 1A]), thus confirming the necessity of the (E)RGSGR stretch for HetN activity influencing heterocyst patterning.
FIG 2.
Heterocyst distribution in Anabaena hetN mutant strains. Filaments from bubbled, ammonium-supplemented cultures of the indicated strains were washed three times with BG110 medium, resuspended in BG110 medium, and incubated under the same culture conditions for 24, 48, or 72 h (as indicated). Cells were counted after staining with alcian blue. Values are means (and standard deviations of the means) of results from 3 to 4 independent experiments (see Materials and Methods). In cases where the frequency is higher than 30%, the actual value is given.
TABLE 2.
Pattern of heterocysts in Anabaena strains expressing altered hetN versions
| Genotype | Straina | Time (h) | % heterocystsb | % contiguous heterocystsc | Mean intervald |
|---|---|---|---|---|---|
| Wild type | PCC 7120 | 24 | 10.4 ± 0.4 | 3.9 ± 0.6 | 10.5 ± 0.7 |
| 48 | 8.6 ± 0.4 | 6.4 ± 1.0 | 13.1 ± 0.8 | ||
| 72 | 7.5 ± 0.7 | 2.5 ± 2.0 | 13.3 ± 0.8 | ||
| ΔpatS | CSVT20 | 24 | 18.6 ± 2.2 | 27.8 ± 8.0 | 5.5 ± 0.7 |
| 48 | 17.2 ± 1.0 | 23.4 ± 6.8 | 6.8 ± 0.4 | ||
| 72 | 17.6 ± 1.0 | 33.7 ± 2.5 | 6.0 ± 0.6 | ||
| ΔhetN | CSL7 | 24 | 14.3 ± 1.0 | 15.7 ± 5.5 | 6.8 ± 0.1 |
| 48 | 13.9 ± 0.6 | 25.0 ± 3.1 | 6.2 ± 0.2 | ||
| 72 | 17.7 ± 1.0 | 33.7 ± 8.4 | 5.6 ± 1.2 | ||
| hetN ΔERGSGR | CSL32 | 24 | 13.8 ± 2.4 | 9.2 ± 4.6 | 7.5 ± 1.0 |
| 48 | 14.2 ± 3.2 | 36.3 ± 7.6 | 5.5 ± 0.5 | ||
| 72 | 14.6 ± 2.8 | 32.7 ± 4.6 | 6.3 ± 0.7 | ||
| hetNΔ2–27 | CSL73 | 24 | 15.9 ± 0.9 | 5.3 ± 0.9 | 6.2 ± 0.1 |
| 48 | 15.2 ± 0.8 | 11.6 ± 1.6 | 8.1 ± 0.2 | ||
| 72 | 13.9 ± 0.3 | 14.6 ± 1.2 | 8.8 ± 0.5 | ||
| hetN-sf-gfp | CSL71 | 24 | 10.0 ± 0.1 | 5.9 ± 0.8 | 8.7 ± 0.3 |
| 48 | 12.2 ± 2.0 | 12.0 ± 4.0 | 9.8 ± 0.6 | ||
| 72 | 11.2 ± 1.4 | 12.4 ± 1.4 | 11.1 ± 1.2 | ||
| hetN-sf-gfp ΔpatS | CSL72 | 24 | 16.3 ± 0.9 | 20.5 ± 1.6 | 7.5 ± 0.2 |
| 48 | 15.7 ± 0.1 | 23.3 ± 5.2 | 8.1 ± 0.6 | ||
| 72 | 14.2 ± 0.6 | 16.8 ± 2.4 | 9.4 ± 0.4 | ||
| ΔpatS ΔhetN | CSL11 | 24 | 42.7 ± 4.8 | 58.2 ± 3.0 | 1.5 ± 0.2 |
| 48 | 62.5 ± 10.0 | 70.6 ± 5.4 | 1.1 ± 0.3 | ||
| 72 | 65.0 ± 6.7 | 67.2 ± 5.1 | 1.1 ± 0.02 |
The CSVT20 (11) and CSL11 mutants are included as controls.
Heterocyst frequency is expressed as a percentage of total cells.
Vegetative cell interval size = 0, expressed as the percentage of total intervals.
Number of vegetative cells between heterocysts.
In strain CSL73 (producing HetN Δ2–27), the percentage of heterocysts was higher (1.5% at 24 h and 1.8% at 48 or 72 h), and the mean interval size shorter (0.6 at 24 and 48 h and 0.7 at 72 h), than in the wild type. The percentage of contiguous heterocysts was considerably greater in CSL73 than in the wild type, and the difference increased with time after N stepdown (1.4-fold at 24 h, 1.8-fold at 48 h, and 5.9-fold at 72 h) (Fig. 2; Table 2). With regard to all three parameters measured, the phenotype of strain CSL73 appeared intermediate between those of the wild type and the hetN deletion mutant, strain CSL7. Thus, deletion of the N-terminal region of HetN up to residue 27 leads to impairment of its activity.
The capability for diazotrophic growth was tested in strains expressing altered hetN versions. The two strains with partial deletions in hetN, CSL73 and CSL32, were capable of growth in the absence of combined nitrogen, as observed also for strain CSL7, in which hetN is fully deleted (data not shown).
HetN localization.
A hetN-gfp translational fusion placed on a plasmid has been reported to give faint fluorescence primarily in proheterocysts 17 h after N stepdown (10), and HetN has been detected in association with cytoplasmic and thylakoid membrane fractions and in heterocyst preparations from strain PCC 7120 (27). The localization of HetN in the Anabaena filament was studied by means of C-terminal fusions to GFP. Derivative strains producing a HetN-sf-GFP construct as the only HetN version in a wild-type (strain CSL71) or patS (strain CSL72) background were generated (Fig. 1A; Table 2). The superfolder version of GFP (sf-GFP) can fold properly even in the periplasm (28). The frequency and distribution of heterocysts in strains CSL71 and CSL72 were more similar to those in their parental strains (PCC 7120 and CSVT20, respectively) than to those in the strains with hetN deleted in the same backgrounds (CSL7 and CSL11, respectively) (Table 2). Thus, the HetN-sf-GFP fusion protein retains appreciable HetN activity.
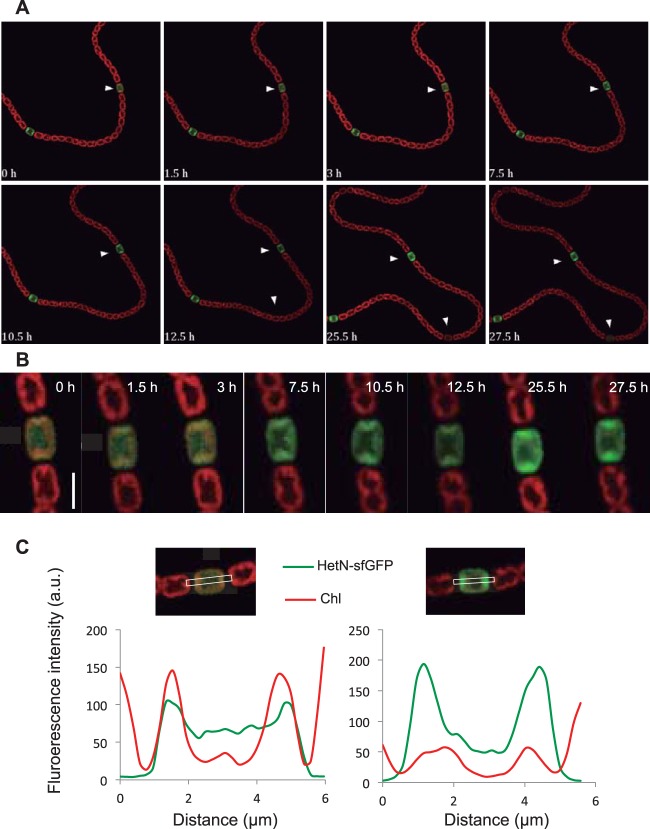
Time lapse microscopy with strain CSL71 growing on N2 showed that upon initiation of heterocyst differentiation, the GFP fluorescence was first distributed throughout all the area of a differentiating cell and thereafter localized progressively to the periphery and the polar region of the cell but apparently did not enter the heterocyst neck (Fig. 3A and B and 4A). Quantification of the GFP fluorescence and the chlorophyll fluorescence along the heterocyst showed a relatively uniform distribution of GFP fluorescence within early differentiating heterocysts. In mature heterocysts, however, GFP fluorescence was markedly concentrated at the cell poles, at places coincident with chlorophyll, whose fluorescence decreased during heterocyst differentiation (Fig. 3C).
FIG 3.
Localization of HetN-GFP during heterocyst differentiation. (A) Time lapse sequence of confocal microscopic images (merged GFP [green] and chlorophyll [red] fluorescence) of strain CSL71 (expressing a HetN-sf-GFP fusion in a wild-type background) grown in solid (0.5% agar) BG110 medium. Each image was taken at the specified time after the arbitrarily chosen start of observations (see Materials and Methods). For GFP fluorescence, the background value of strain PCC 7120 was set at zero. Arrowheads point to differentiating heterocysts. (B) Magnification of the cell marked with an arrowhead in panel A, 0 h. Bar, 4 μm. (C) Quantification of chlorophyll (Chl) and GFP fluorescence across the longitudinal axis of the differentiating heterocysts shown in panel B, 0 h (left) and 27.5 h (right). The areas monitored are outlined in white in the images above the graphs.
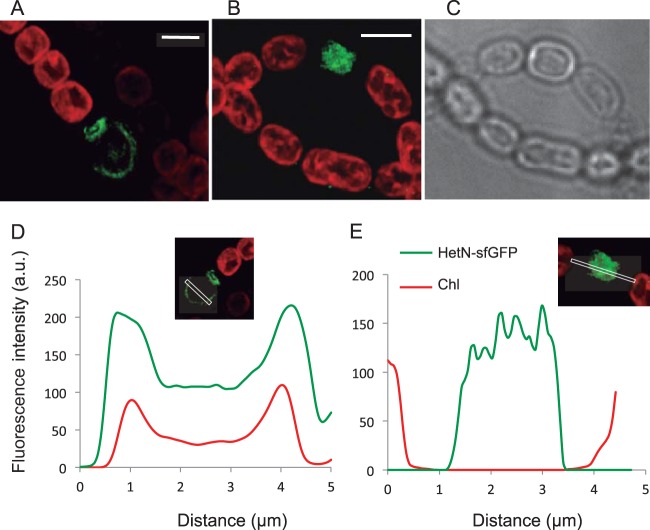
FIG 4.
Localization of HetN-GFP in strains CSL71 and CSL103. (A and B) Deconvolved fluorescence microscopy images (merged GFP [green] and chlorophyll [red] fluorescence) of filaments of strains CSL71 (expressing HetN-sf-GFP) (A) and CSL103 (expressing HetN Δ2–27-sf-GFP) (B) grown in solid BG110 medium. Bars, 4 μm. (C) The bright-field image of the field shown in panel B shows a heterocyst with two polar plugs, indicating its maturity. A polar plug was seen also in the heterocyst shown in panel A (bright-field image not shown). (D and E) Quantification of chlorophyll fluorescence and GFP fluorescence across the transverse axis of the differentiating heterocyst shown in panel A (CSL71) (D) and across the longitudinal axis of the differentiating heterocyst shown in panel B (CSL103) (E). The areas monitored are outlined in white in the images above the graphs. The arbitrary units (a.u.) in panels D and E are different.
The localization of HetN-sf-GFP in strain CSL72 (Fig. 5A) was similar to that in CSL71. Thus, the absence of PatS did not influence the localization of HetN. A similar sf-GFP fusion was generated with the HetN protein lacking residues 2 to 27, which was the product of the only hetN gene version present in strain CSL103 (Fig. 1). In this strain, regardless of the differentiation stage, fluorescence from GFP remained homogeneously distributed throughout the cell area with no apparent preference for the cell periphery or the cell poles (a mature heterocyst, as indicated by morphology, is shown in Fig. 4B and C), a pattern also evidenced by fluorescence quantification (Fig. 4E). Thus, the deleted stretch of the protein (residues 2 to 27) is important for HetN localization during heterocyst differentiation.
FIG 5.
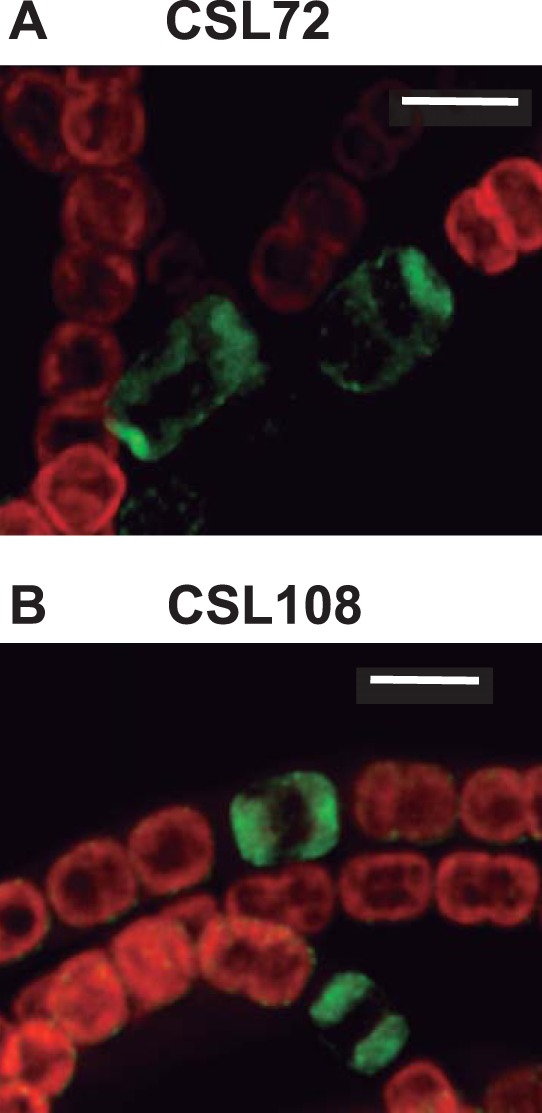
Localization of HetN-GFP in strains CSL72 and CSL108. Shown are deconvolved fluorescence microscopy images (merged GFP [green] and chlorophyll [red] fluorescence) of ammonium-grown filaments of strains CSL72 (expressing a HetN-sf-GFP fusion in a patS background) (A) and CSL108 (expressing HetN-GFP-mut2 in a wild-type background) (B) incubated for 20 h in bubbled BG110 medium. Bars, 4 μm.
Finally, an Anabaena derivative producing a HetN-GFP-mut2 construct as the only HetN version in a wild-type background (strain CSL108) was generated. The localization of GFP fluorescence in this strain (Fig. 5B) appears similar to that in strain CSL71 (Fig. 3). Because the GFP encoded by the gfp-mut2 gene folds properly only in the cytoplasm (29), which also excludes the thylakoid lumen, these results suggest a cytoplasmic localization of the C terminus of HetN.
HetN in other cyanobacteria.
We examined the sequences of the 113 proteins most similar to HetN, which exhibited broad similarity over the N-terminal 70% of the protein. That region in many, if not most, of the proteins showed a significant match (E value, <10−25) with Pfam family adh_short (short-chain dehydrogenases) and a less significant match with Pfam family KR (polyketide synthases), in accordance with the biochemical activity measured by Liu and Chen (15). A cluster of eight proteins also exhibited similarity over the C-terminal 30% of HetN amino acids, and these formed group a, one of the seven groups of HetN-like proteins defined by sequence similarity (see Fig. S1 to S3 in the supplemental material).
The ERGSGR peptide of HetN lies in the large N-terminal region, within a poorly conserved part of an otherwise well conserved protein (Fig. 6). It is found in only three related cyanobacteria (Anabaena sp. strain PCC 7120, Anabaena variabilis, and Nostoc sp. strain PCC 7524) (see Fig. S3B in the supplemental material) and two distantly related single cellular strains (Synechococcus sp. strains JA-3-3Ab and JA-2-3B [data not shown]). One can see from the alignment (Fig. 6) two potential start codons internal to HetN, coding for methionine residues immediately upstream of ERGSGR. The more upstream methionine is conserved in 95 of the 113 aligned sequences, but the other methionine is found only in the HetN protein of Anabaena sp. strain PCC 7120, the other 2 most closely related proteins, and 3 proteins in group g. The two methionines are not preceded by any sequence resembling a ribosome-binding site (data not shown), but if either served to initiate translation, the resulting peptide would be similar to those shown by Corrales-Guerrero et al. (11) to partially complement the absence of PatS.
FIG 6.
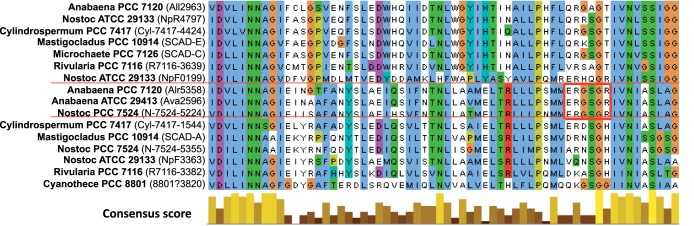
Alignment of proteins most similar to Anabaena sp. strain PCC 7120 HetN (Alr5358). Shown is a portion of the alignment of the HetN-like proteins described in Fig. S3 in the supplemental material, in the region corresponding to Anabaena sp. strain PCC 7120 HetN coordinates 84 to 145. The red horizontal lines demarcate the three proteins from heterocyst-forming cyanobacteria that contain ERGSGR, and thick red lines surround the ERGSGR region. The consensus score histogram indicates the degree to which each residue is conserved (23), calculated over all 111 sequences (35). The entire alignment is given in Fig. S1 in the supplemental material.
The hetN gene was originally discovered within a cluster dense with genes important in heterocyst differentiation (9, 30). Its position in that cluster is shared by the other two genes encoding RGSGR-containing HetN-like proteins of group a, but in other heterocyst-forming cyanobacteria, that position is occupied by a gene encoding a distantly related oxidoreductase or by nothing at all (see Fig. S4 in the supplemental material). The genes encoding other HetN-like proteins of group a are in different regions of the chromosome.
DISCUSSION
HetN is a factor influencing the heterocyst pattern in Anabaena sp. strain PCC 7120 through a negative effect on differentiation. Our results indicate that a 26-residue N-terminal stretch of the protein (residues 2 to 27) is required for the normal activity of HetN in modulating heterocyst patterning and that this region is required for the correct localization of HetN in mature heterocysts.
The nature of the N terminus is not clear. The region was identified as a possible signal peptide by one prediction program (SignalP, version 3.0 [D value, 0.547, with a nominal cutoff of 0.44 for significance]) but not by others (e.g., SignalP, version 4.1 [D value, 0.269, with a nominal cutoff of 0.57 for significance], or TatP, version 1.0). SignalP, version 3.0, predicts a signal sequence at the positions of residues 1 to 27, with the most likely cleavage site between residues 27 and 28. It should be noted that Higa et al. (14) reported that deletion of HetN residues 1 to 46 had no effect on patterning, but in support they provided only the heterocyst percentages at an unspecified time after N stepdown. Because the largest difference between our strain CSL73 (HetN Δ2–27) and the wild type was the difference in the percentage of contiguous heterocysts, which is indeed the most definitive feature of the Mch phenotype, especially at 72 h after N stepdown, differences in strain characterization could be at the basis of the contrasting findings of the effects of N-terminal deletions in HetN. The two experiments also differed in the choice of promoters to drive the expression of the constructs: PhetN in the current work versus the much stronger promoter PpetE in the work of Higa et al. (14). The lesser ability of a HetN mutant to affect differentiation may be masked by higher expression.
HetN-GFP is first seen localized throughout the proheterocyst area; later in heterocyst differentiation, it is seen localized at the cell periphery, appearing more concentrated at the heterocyst polar regions (Fig. 3). Previously, a HetN-C-YFP translational fusion expressed from the PhetN promoter on a plasmid in a hetR mutant background insensitive to HetN was reported to give fluorescence uniformly at the periphery of heterocyst (14). The lack of polar specificity in this strain, in contrast to our results with CSL71 and CSL108, could result from higher hetN expression or from structural differences in heterocysts arising from the lack of a fully functional HetR protein (employed to allow heterocyst differentiation despite the high level of HetN).
Under our conditions, normal localization of HetN to the poles of heterocysts requires the N-terminal stretch of the protein comprising residues 2 to 27 (Fig. 4). This stretch of HetN may include an unconventional signal sequence for sorting the protein. Moreover, the requirement of this stretch for regulation of the heterocyst pattern as well indicates that the correct localization of HetN may be needed for proper function.
HetN-GFP at the heterocyst poles appears not to enter the heterocyst neck (Fig. 3B and 4A), but the arrangement resembles dispositions of internal membranes such as the “honeycomb” membrane system that exhibits oxidase activity (31), and HetN-GFP may coincide with the thylakoid membranes that are concentrated at the poles in these cells (32). Indeed, the coincidence of HetN-GFP fluorescence with chlorophyll fluorescence (Fig. 3C) supports a localization of HetN in the thylakoids. This would be consistent with the previous detection by immunoblotting of HetN in photosynthetic membrane fractions of whole filaments (27). The localization of HetN in heterocyst thylakoids could involve anchoring to the thylakoid membranes. In principle, this could rely on an internal transmembrane segment. However, analyses for transmembrane segment predictions in HetN were inconclusive (a transmembrane domain was predicted at residues 173 to 193 by Toppred, at residues 112 to 127 by MEMSAT-SVM, and at residues 61 to 79 by MEMSAT; no segment was predicted by SOSUI, PSORT, or TMHMM). Thus, rather than involving a transmembrane segment, HetN could be localized to the heterocyst thylakoid membrane by interactions that do not imply membrane spanning. HetN could represent a good marker for following the dynamics of thylakoid reorganization during heterocyst differentiation, from a peripheral location throughout the vegetative cells to a peripheral polar location in the heterocysts.
Both conservative amino acid substitutions within the chromosomal hetN gene (14) and deletion of residues 131 to 136 (strain CSL32 [Table 2]) point to the importance in heterocyst spacing of the (E)RGSGR region of HetN, which is identical in sequence to the 6 C-terminal residues of the PatS morphogen. On the other hand, Li et al. (27) reported that similar site-specific mutations in this region had no effect on the ability of a second copy of hetN, placed on a multicopy plasmid, to suppress heterocyst differentiation, and Higa et al. (14) reported only a modest reduction in suppression ability under these circumstances when G133, S134, or G135 had been mutated. Again, differences in expression levels may explain the discrepancies between these two types of experiments.
Studies of the molecular mechanism governing heterocyst differentiation have focused to a large degree on Anabaena sp. strain PCC 7120. If the presence of the (E)RGSGR peptide within HetN is an indispensable part of the machinery governing heterocyst spacing, then one would expect these residues to occur in HetN orthologs in all heterocyst-forming cyanobacteria, but this is not the case. They are found only in cyanobacteria very closely related to Anabaena sp. strain PCC 7120. It seems likely that the ability of HetN to provide the (E)RGSGR peptide was gained relatively recently. If such a function served by HetN is present in other cyanobacteria with spaced heterocysts, it must be provided by a thus far unreported protein.
In summary, regarding the role of the HetN (E)RGSGR peptide, the pattern of fluorescence from HetN-GFP fusions argues against the idea that HetN is transported intact outside the heterocyst. However, it remains possible that the (E)RGSGR peptide is excised from a superficial region of HetN within the cytoplasm and is then transported out of the heterocyst. Also, one of the internal ATG codons 5′ to the ERGSGR region in the hetN gene (Fig. 6) may initiate the translation of a short peptide directly by a Shine-Dalgarno sequence-independent mechanism (33).
In addition to the role of the (E)RGSGR peptide, the full HetN protein could serve a different function, which could be related to photosynthesis/respiration in the heterocysts, consistent with its colocalization with internal membranes, its similarity to oxidoreductases, and its reported enzymatic activity (see reference 15). Indeed, the occurrence in heterocysts of a form of the protein larger than the ERGSGR peptide is evident from the localized fluorescence observed from GFP fused to the C terminus of HetN (compare Fig. 4A in the present work with Fig. 1 in reference 34, showing the homogeneous distribution of free GFP), confirming the immunological findings of Li et al. (27).
Supplementary Material
ACKNOWLEDGMENTS
We thank Ivan Khudyakov for helpful discussions.
Research in Seville was supported by grant BFU2010-17980 from the Spanish Government, cofinanced by FEDER. L.C.-G. was the recipient of a JAE-predoc fellowship from the CSIC. D.J.N. was supported by a Queen Mary College studentship. Sequence analysis was supported in part by grant DBI-0850146 from the U.S. National Science Foundation.
Footnotes
Published ahead of print 21 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01922-14.
REFERENCES
- 1.Flores E, Herrero A. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8:39–50. 10.1038/nrmicro2242 [DOI] [PubMed] [Google Scholar]
- 2.Kumar K, Mella-Herrera RA, Golden JW. 2010. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2:a000315. 10.1101/cshperspect.a000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrero A, Picossi S, Flores E. 2013. Gene expression during heterocyst differentiation. Adv. Bot. Res. 65:281–329. 10.1016/B978-0-12-394313-2.00008-1 [DOI] [Google Scholar]
- 4.Yoon HS, Golden JW. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935–938. 10.1126/science.282.5390.935 [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Du Y, Khudyakov I, Fan Q, Gao H, Ning D, Wolk P, Xu X. 2007. A cluster that regulates both heterocyst differentiation and pattern formation in Anabaena sp. strain PCC 7120. Mol. Microbiol. 66:1429–1443. 10.1111/j.1365-2958.2007.05997.x [DOI] [PubMed] [Google Scholar]
- 6.Wong FC, Meeks JC. 2001. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J. Bacteriol. 183:2654–2661. 10.1128/JB.183.8.2654-2661.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risser DD, Callahan SM. 2008. HetF and PatA control levels of HetR in Anabaena sp. strain PCC 7120. J. Bacteriol. 190:7645–7654. 10.1128/JB.01110-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang J, Scarpino L, Haselkorn R. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. U. S. A. 89:5655–5659. 10.1073/pnas.89.12.5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black TA, Wolk CP. 1994. Analysis of a Het− mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol. 176:2282–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callahan SM, Buikema WJ. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40:941–950. 10.1046/j.1365-2958.2001.02437.x [DOI] [PubMed] [Google Scholar]
- 11.Corrales-Guerrero L, Mariscal V, Flores E, Herrero A. 2013. Functional dissection and evidence for intercellular transfer of the heterocyst-differentiation PatS morphogen. Mol. Microbiol. 88:1093–1105. 10.1111/mmi.12244 [DOI] [PubMed] [Google Scholar]
- 12.Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakazaki N, Shimpo S, Sugimoto M, Takazawa M, Yamada M, Yasuda M, Tabata S. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205–213. 10.1093/dnares/8.5.205 [DOI] [PubMed] [Google Scholar]
- 13.Bauer CC, Ramaswamy KS, Endley S, Scappino LA, Golden JW, Haselkorn R. 1997. Suppression of heterocyst differentiation in Anabaena PCC 7120 by a cosmid carrying wild-type genes encoding enzymes for fatty acid biosynthesis. FEMS Microbiol. Lett. 151:23–30. 10.1111/j.1574-6968.1997.tb10390.x [DOI] [PubMed] [Google Scholar]
- 14.Higa KC, Rajagopalan R, Risser DD, Rivers OS, Tom SK, Videau P, Callahan SM. 2012. The RGSGR amino acid motif of the intercellular signaling protein, HetN, is required for patterning of heterocysts in Anabaena sp. strain PCC 7120. Mol. Microbiol. 83:682–693. 10.1111/j.1365-2958.2011.07949.x [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Chen W-L. 2009. Characterization of HetN, a protein involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 297:17–23. 10.1111/j.1574-6968.2009.01644.x [DOI] [PubMed] [Google Scholar]
- 16.López-Igual R, Picossi S, López-Garrido J, Flores E, Herrero A. 2012. N and C control of ABC-type bicarbonate transporter Cmp and its LysR-type transcriptional regulator CmpR in a heterocyst-forming cyanobacterium, Anabaena sp. Environ. Microbiol. 14:1035–1048. 10.1111/j.1462-2920.2011.02683.x [DOI] [PubMed] [Google Scholar]
- 17.Black TA, Cai Y, Wolk CP. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77–84. 10.1111/j.1365-2958.1993.tb01670.x [DOI] [PubMed] [Google Scholar]
- 18.Valladares A, Rodríguez V, Camargo S, Martínez-Nöel GMA, Herrero A, Luque I. 2011. Specific role of the cyanobacterial PipX factor in heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 193:1172–1182. 10.1128/JB.01202-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elhai J, Taton A, Massar J, Myers JK, Travers M, Casey J, Slupesky M, Shrager J. 2009. BioBIKE: a Web-based, programmable, integrated biological knowledge base. Nucleic Acids Res. 37:W28–W32. 10.1093/nar/gkp354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clamp M, Cuff J, Searle SM, Barton GJ. 2004. The Jalview Java alignment editor. Bioinformatics 20:426–427. 10.1093/bioinformatics/btg430 [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. 2005. PHYLIP (phylogeny inference package), version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 26.Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40:D302–D305. 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Huang X, Zhao J. 2002. Expression of hetN during heterocyst differentiation and its inhibition of hetR up-regulation in the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 517:87–91. 10.1016/S0014-5793(02)02582-6 [DOI] [PubMed] [Google Scholar]
- 28.Dinh T, Bernhardt TG. 2011. Using superfolder green fluorescent protein for periplasmic protein localization studies. J. Bacteriol. 193:4984–4987. 10.1128/JB.00315-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammeyer T, Tinnefeld P. 2012. Engineered fluorescent proteins illuminate the bacterial periplasm. Comput. Struct. Biotechnol. J. 3:e201210013. 10.5936/csbj.201210013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Q, Huang G, Lechno-Yossef S, Wolk CP, Kaneko T, Tabata S. 2005. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 58:227–243. 10.1111/j.1365-2958.2005.04818.x [DOI] [PubMed] [Google Scholar]
- 31.Murry MA, Olafsen AG, Benemann JR. 1981. Oxidation of diaminobenzidine in the heterocysts of Anabaena cylindrica. Curr. Microbiol. 6:201–206. 10.1007/BF01566972 [DOI] [Google Scholar]
- 32.Kumazaki S, Akari M, Hasegawa M. 2013. Transformation of thylakoid membranes during differentiation from vegetative cell into heterocyst visualized by microscopic spectral imaging. Plant Physiol. 161:1321–1333. 10.1104/pp.112.206680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang B, Halgamuge S, Tang S-L. 2006. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene 373:90–99. 10.1016/j.gene.2006.01.033 [DOI] [PubMed] [Google Scholar]
- 34.Mariscal V, Herrero A, Flores E. 2007. Continuous periplasm in a filamentous, heterocyst-forming cyanobacterium. Mol. Microbiol. 65:1139–1145. 10.1111/j.1365-2958.2007.05856.x [DOI] [PubMed] [Google Scholar]
- 35.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, Calteau A, Cai F, Tandeau de Marsac N, Rippka R, Herdman M, Sivonen K, Coursin T, Laurent T, Goodwin L, Nolan M, Davenport KW, Han CS, Rubin EM, Eisen JA, Woyke T, Gugger M, Kerfeld CA. 2013. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 110:1053–1058. 10.1073/pnas.1217107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.